Abstract
The committed biosynthetic reaction to benzoyl-coenzyme A in the marine bacterium “Streptomyces maritimus” is carried out by the novel prokaryotic phenylalanine ammonia lyase (PAL) EncP, which converts the primary amino acid l-phenylalanine to trans-cinnamic acid. Recombinant EncP is specific for l-phenylalanine and shares many biochemical features with eukaryotic PALs, which are substantially larger proteins by ∼200 amino acid residues.
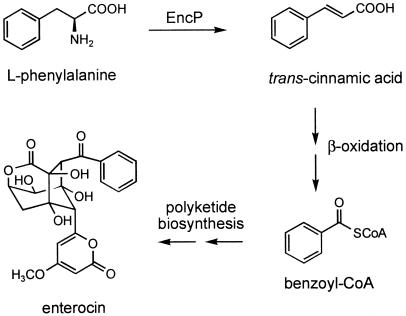
The bacteriostatic agent enterocin is a natural product of the marine bacterium “Streptomyces maritimus” whose biosynthesis involves a number of unusual features (9, 16, 17, 22). Among these is the formation of the rare polyketide synthase starter unit benzoyl-coenzyme A (benzoyl-CoA) (15). The initial biochemical reaction involves the conversion of the amino acid l-phenylalanine to trans-cinnamic acid by the novel bacterial phenylalanine ammonia lyase (PAL; EC 4.3.1.5) EncP (24). Activation of cinnamic acid to its CoA thioester, followed by a single round of β-oxidation, yields benzoyl-CoA (7, 8, 23), which primes the enterocin type II polyketide synthase for chain extension with seven molecules of malonyl-CoA (Fig. 1).
FIG. 1.
EncP-catalyzed conversion of phenylalanine to trans-cinnamic acid and biosynthesis of benzoyl-CoA-derived enterocin in “S. maritimus.”
Although PAL is a ubiquitous higher-plant enzyme that catalyzes the nonoxidative deamination of phenylalanine to cinnamic acid in the committed step to phenylpropanoid metabolites (6), it has only been encountered in a few bacteria, where it is involved in benzoyl-CoA biosynthesis in “S. maritimus” (24) and Sorangium cellulosum (10) and in the biosynthesis of cinnamamide in Streptomyces verticillatus (2). We previously characterized the first prokaryotic PAL-encoding gene (encP) and showed that its inactivation resulted in the abolishment of de novo cinnamic acid and enterocin synthesis in “S. maritimus” (12, 24). Enterocin biosynthesis could be restored in encP-inactivated mutants through supplementation with cinnamic or benzoic acid, as well as complementation with plasmid-borne encP. Furthermore, the heterologous expression of the encP gene under the control of the ermE* promoter in Streptomyces coelicolor led to the production of cinnamic acid in the fermented cultures (24). Here we report the biochemical characterization of this novel bacterial PAL, including substrate specificity, pH dependence, and kinetics, as well as its inhibition with the known plant PAL inhibitor 2-aminoindan-2-phosphonic acid (AIP) (1).
Sequence homology, expression, and purification of EncP.
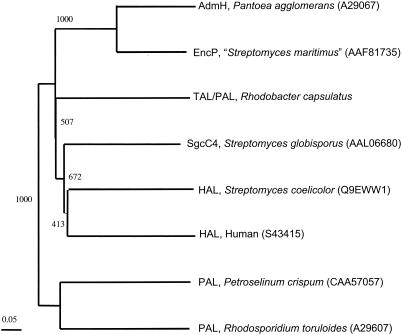
The encP gene encodes a 522-amino-acid protein that is considerably smaller than eukaryotic PALs by nearly 200 amino acid residues. Although sequence homologous to plant PALs such as from Petroselinum crispum (19) (CAA57056; 30% identical and 48% similar), it rather shares greater homology to bacterial histidine ammonia lyases (HALs; EC 4.3.1.3) such as from Pseudomonas putida (21) (A35251; 36% identical and 54% similar) and to tyrosine ammonia lyase from Rhodobacter capsulatus (13) (Fig. 2). The homology includes the conserved active-site serine residue at position 143 of the phenylalanine/histidine/tyrosine family of ammonia lyases that is the probable precursor of the modified dehydroalanine residue in the 4-methylideneimidazole-5-one prosthetic group (14, 18, 21). EncP has the greatest sequence homology with AdmH (AAO39102; 63% identical and 76% similar), a putative phenylalanine aminomutase involved in andrimid biosynthesis in Pantoea agglomerans that is related to the tyrosine aminomutase Sgc4 from Streptomyces globisporus (4, 5).
FIG. 2.
Relatedness tree of aromatic amino acid ammonia lyases from prokaryotes and eukaryotes. Sequences were retrieved from GenBank (accession numbers are given in parentheses) and aligned with ClustalX (1.83) using the neighbor-joining method.
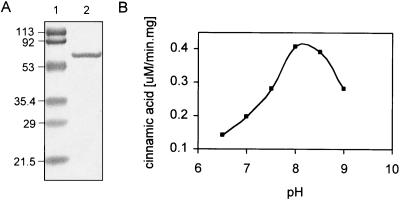
The gene encP was PCR amplified from “S. maritimus” cosmid clone pJP15F11 (17) with forward primer 5′-AAAGGATCCTTCGTCATAGAGCTCGAC-3′ (BamHI in bold) and reverse primer 5′-AAAAAGCTTCCAGGTGCTGCTTCAGTG-3′ (HindIII in bold) and cloned into pCRR-Blunt (Invitrogen). Its sequence was verified, and it was digested with BamHI and HindIII and cloned into expression plasmid pHIS8 (11). Recombinant EncP was overexpressed as an N-terminal octahistidyl-tagged fusion protein in Escherichia coli BL21(DE3)/pLysS (Invitrogen). A colony of the plasmid-transformed E. coli bacteria was grown overnight in 3 ml LB broth containing 50 μg/ml kanamycin and 37 μg/ml chloramphenicol at 37°C. One milliliter of the resultant culture was inoculated into 100 ml TB broth with the same antibiotics in a 500-ml Erlenmeyer flask and grown until the optical density at 600 nm reached 0.7. After induction with 0.2 mM isopropyl-β-d-thiogalactopyranoside, the cells were cultured for another 20 h at 28°C. The recombinant EncP protein was purified by Ni2+ affinity chromatography over a nickel-nitrilotriacetic acid column. Its mobility upon sodium dodecyl sulfate-polyacrylamide gel electrophoresis corresponded to a mass of ∼60 kDa, in close agreement with the value of 58.7 calculated for the recombinant protein (Fig. 3).
FIG. 3.
(A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of purified, octahistidyl-tagged EncP. Lane 1, molecular size markers (kDa); lane 2, His8-EncP (calculated molecular mass is 58.7 kDa). (B) pH dependence on the rate of trans-cinnamic acid formation from l-phenylalanine by EncP.
Characterization of recombinant EncP.
To confirm that the purified ammonia lyase was indeed specific for l-phenylalanine, the recombinant protein was incubated with phenylalanine, histidine, and tyrosine and monitored spectrophotometrically at 30°C by measuring the increase in absorbance at 280 nm accompanying the formation of the conjugated aryl acid. Imidazole was removed from the elution buffer due to significant enzyme inhibition. The standard assay conditions contained 100 mM Tris-HCl buffer (pH 8.0), 0.2 mM substrate, and 200 μg/ml EncP in a 1-ml total volume. Incubation with the substrate l-phenylalanine provided trans-cinnamic acid, which was confirmed by reversed-phase high-performance liquid chromatography (HPLC)-mass spectrometry with an authentic standard. d-Phenylalanine, l-tyrosine, and l-histidine had no detectable activity under similar conditions with up to 2 mM substrate, even when the reaction proceeded overnight and was monitored by HPLC. The pH dependency of the PAL activity was furthermore determined spectrophotometrically and showed a pH optimum of approximately 8.0 (Fig. 3).
Kinetic parameters of cinnamic acid formation.
The steady-state kinetic parameters of the recombinant “S. maritimus” PAL EncP were determined spectrophotometrically. Two hundred micrograms of the enzyme in 1 ml 100 mM Tris-HCl (pH 8.0) was preincubated at 30°C for 5 min before 0.02 to 0.2 mM l-phenylalanine was added to initiate the 20-min reaction. Kinetic constants were calculated from initial velocity measurements at 280 nm (ɛ = 17,423 for trans-cinnamic acid) in which product formation was linear over the time periods monitored. The kinetic experiments showed that recombinant EncP had a slightly larger Km and a smaller kcat than that of PAL from P. crispum, as summarized in Table 1.
TABLE 1.
Kinetic constants of wild-type PALs and EncP mutantsa
| Enzyme | Km (mM) | kcat (s−1) | kcat/Km (mM−1 · s−1) |
|---|---|---|---|
| EncP | 0.023 ± 0.005 | 4.79 ± 0.28 | 208 |
| V83A EncP | 0.120 ± 0.018 | 99.58 ± 1.78 | 829 |
| V83H EncP | —b | — | — |
| P. crispum PAL | 0.12 ± 0.004 | 13.5 ± 0.1 | 112.5 |
| R. capsulatus TAL/PAL | 1.277 | 15.1 | 11.8 |
PAL activity was measured by monitoring the formation of trans-cinnamic acid at 280 nm. The kinetic values for EncP are averaged from two independent measurements. The values for P. crispum PAL are from reference 19 and the values for R. capsulatus TAL/PAL relative to L-phenylalanine are from reference 13.
The activity of V83H was too low to calculate Km and kcat.
The three-dimensional X-ray structures of HAL from P. putida (20, 21) and, more recently, of PAL from Rhodosporidium toruloides (3) revealed the active-site residues of these tetrameric enzymes that are important for substrate binding, catalysis, and 4-methylideneimidazole-5-one formation. All active-site residues in HAL are present in EncP, except for H83 and E414, which are replaced with valine and glutamine residues, respectively (24). H83 in HAL is proposed to bind and orient the imidazole moiety of l-histidine at the active site and to stabilize an enzyme-bound cationic intermediate, whereas the carboxylate group of E414 may act as a base in catalysis. To examine the contribution of V83 to cinnamic acid formation by EncP, we generated the V83A and V83H mutants by site-directed mutagenesis using the QuickChange Multi-Site Directed Mutagenesis method (Stratagene). The V83H mutation was introduced into pHIS8-EncP with primers M13F 5′-CGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAG-3′ and 5′-CCAGGAGAACCTGATCAACGCGCACGCCACCAACGTGGGGGCG-3′ (the underlined bases CA were mutated from GT). The V83A mutation was similarly introduced with primers M13F and 5′-CCAGGAGAACCTGATCAACGCGGCCGCCACCAACGTGGGGGC-3′ (the underlined base C was mutated from T). The mutations were confirmed by DNA sequencing. The mutated encP genes were digested by BamHI-HindIII and cloned separately into pHIS8. In both cases, similar expression levels of the recombinant mutant enzymes showing the same monomeric size as observed with wild-type EncP were measured.
While the V83H mutant lost its PAL activity, the V83A mutant was more active than wild-type EncP (Table 1). The V83A mutant showed a slightly lower affinity to l-phenylalanine with a Km of 120 μM versus 23 μM for the wild-type enzyme. On the other hand, kcat was 20-fold higher, resulting in a mutant that was about six times more active than the wild-type enzyme. The V83H mutant was further tested for HAL activity as the conserved histidine residue in HALs coordinates its imidazole group through a hydrogen bond with that of the bound histidine substrate (20, 21). PALs, on the other hand, carry aliphatic residues such as valine and isoleucine at this position, which is consistent with that of EncP, to provide a hydrophobic environment for the benzene ring of its substrate (19). We unfortunately did not observe any HAL activity in the V83H mutant by HPLC analysis, as similarly reported by Rétey and coworkers for the parsley PAL (19).
Kinetic analysis of the inhibition of EncP by AIP.
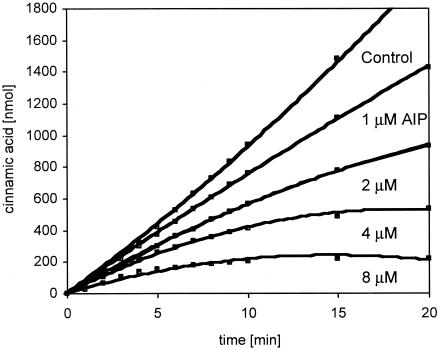
The conformationally restricted phenylalanine analogue AIP has been shown to be an effective inhibitor of plant PAL enzymes both in vivo (25) and in vitro (1). AIP is a competitive inhibitor of P. crispum PAL with a Ki of 25 ± 4 nM and inhibits the enzyme in a time-dependent manner (1). We similarly analyzed the in vitro interaction of AIP with EncP and likewise measured its concentration-dependent inhibition (Fig. 4). The Ki of EncP was calculated from the equation Km′ = (1 + [I]/Ki), where [I] is the concentration of AIP. A Ki of 1.91 ± 0.07 μM was obtained for EncP, which was about 76 times higher than that of P. crispum PAL.
FIG. 4.
Assay progress curves in the presence of AIP. EncP was incubated at 40°C in 100 mM Tris-HCl (pH 8.0) containing 0.4 mM l-phenylalanine and increasing concentrations of AIP. The reaction was started by the addition of the enzyme. Points represent averaged values from duplicate experiments.
Acknowledgments
This work was supported by the NIH (AI47818).
We thank Joseph P. Noel (Salk Institute for Biological Studies, La Jolla, CA) for the vector pHIS8, Jerzy Zon (Wroclaw University, Wroclaw, Poland) for generously providing the inhibitor AIP, and Yoshimitsu Hamano (University of Arizona) for helpful discussions.
REFERENCES
- 1.Appert, C., J. Zon, and N. Amrhein. 2003. Kinetic analysis of the inhibition of phenylalanine ammonia-lyase by 2-aminoindan-2-phosphonic acid and other phenylalanine analogues. Phytochemistry 62:415-422. [DOI] [PubMed] [Google Scholar]
- 2.Bezanson, G. S., D. Desaty, A. V. Emes, and L. C. Vining. 1970. Biosynthesis of cinnamamide and detection of phenylalanine ammonia-lyase in Streptomyces verticillatus. Can. J. Microbiol. 16:147-151. [DOI] [PubMed] [Google Scholar]
- 3.Calabrese, J. C., D. B. Jordan, A. Boodhoo, S. Sariaslani, and T. Vannelli. 2004. Crystal structure of phenylalanine ammonia lyase: multiple helix dipoles implicated in catalysis. Biochemistry 43:11403-11416. [DOI] [PubMed] [Google Scholar]
- 4.Christenson, S. D., W. Liu, M. D. Toney, and B. Shen. 2003. A novel 4-methylideneimidazole-5-one-containing tyrosine aminomutase in enediyne antitumor antibiotic C-1027 biosynthesis. J. Am. Chem. Soc. 125:6062-6063. [DOI] [PubMed] [Google Scholar]
- 5.Christenson, S. D., W. Wu, M. A. Spies, B. Shen, and M. D. Toney. 2003. Kinetic analysis of the 4-methylideneimidazole-5-one-containing tyrosine aminomutase in enediyne antitumor antibiotic C-1027 biosynthesis. Biochemistry 42:12708-12718. [DOI] [PubMed] [Google Scholar]
- 6.Hahlbrock, K., and D. Scheel. 1989. Physiology and molecular biology of phenylpropanoid metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 40:347-369. [Google Scholar]
- 7.Hertweck, C., A. P. Jarvis, L. Xiang, B. S. Moore, and N. J. Oldham. 2001. A mechanism of benzoic acid biosynthesis in plants and bacteria that mirrors fatty acid β-oxidation. ChemBioChem 2:784-786. [DOI] [PubMed] [Google Scholar]
- 8.Hertweck, C., and B. S. Moore. 2000. A plant-like biosynthesis of benzoyl-CoA in the marine bacterium “Streptomyces maritimus.” Tetrahedron 56:9115-9120. [Google Scholar]
- 9.Hertweck, C., L. Xiang, J. A. Kalaitzis, Q. Cheng, M. Palzer, and B. S. Moore. 2004. Context-dependent behavior of the enterocin iterative polyketide synthase: a new model for ketoreduction. Chem. Biol. 11:461-468. [DOI] [PubMed] [Google Scholar]
- 10.Hill, A. M., B. L. Thompson, J. P. Harris, and R. Segret. 2003. Investigation of the early stages in soraphen A biosynthesis. Chem. Commun. 1358-1359. [DOI] [PubMed]
- 11.Jez, J. M., J.-L. Ferrer, M. E. Bowman, R. A. Dixon, and J. P. Noel. 2000. Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry 39:890-902. [DOI] [PubMed] [Google Scholar]
- 12.Kalaitzis, J. A., M. Izumikawa, L. Xiang, C. Hertweck, and B. S. Moore. 2003. Mutasynthesis of enterocin and wailupemycin analogues. J. Am. Chem. Soc. 125:9290-9291. [DOI] [PubMed] [Google Scholar]
- 13.Kyndt, J. A., T. E. Meyer, M. A. Cusanovish, and J. J. Van Beeumen. 2002. Characterization of a bacterial tyrosine ammonia lyase, a biosynthetic enzyme for the photoactive yellow protein. FEBS Lett. 512:240-244. [DOI] [PubMed] [Google Scholar]
- 14.Langer, B., M. Langer, and J. Rétey. 2001. Methylidene-imidazolone (MIO) from histidine and phenylalanine ammonia-lyase, p. 175-214. In J. P. Klinman and J. E. Dove (ed.), Advances in protein chemistry, vol. 58. Academic Press, Inc., New York, N.Y. [DOI] [PubMed] [Google Scholar]
- 15.Moore, B. S., and C. Hertweck. 2002. Biosynthesis and attachment of novel bacterial polyketide synthase starter units. Nat. Prod. Rep. 19:70-99. [DOI] [PubMed] [Google Scholar]
- 16.Piel, J., C. Hertweck, P. R. Shipley, D. M. Hunt, M. S. Newman, and B. S. Moore. 2000. Cloning, sequencing and analysis of the enterocin biosynthesis gene cluster from the marine isolate “Streptomyces maritimus”: evidence for the derailment of an aromatic polyketide synthase. Chem. Biol. 7:943-955. [DOI] [PubMed] [Google Scholar]
- 17.Piel, J., K. Hoang, and B. S. Moore. 2000. Metabolic diversity encoded by the enterocin biosynthesis gene cluster. J. Am. Chem. Soc. 122:5415-5416. [Google Scholar]
- 18.Poppe, L. 2001. Methylidene-imidazolone: a novel electrophile for substrate activation. Curr. Opin. Chem. Biol. 5:512-524. [DOI] [PubMed] [Google Scholar]
- 19.Röther, D., L. Poppe, G. Morlock, S. Viergutz, and J. Rétey. 2002. An active site homology model of phenylalanine ammonia-lyase from Pretroselinum crispum. Eur. J. Biochem. 269:3065-3075. [DOI] [PubMed] [Google Scholar]
- 20.Röther, D., L. Poppe, S. Viergutz, B. Langer, and J. Rétey. 2001. Characterization of the active site of histidine ammonia-lyase from Pseudomonas putida. Eur. J. Biochem. 268:6011-6019. [DOI] [PubMed] [Google Scholar]
- 21.Schwede, T. F., J. Rétey, and G. E. Schulz. 1999. Crystal structure of histidine ammonia-lyase revealing a novel polypeptide modification as the catalytic electrophile. Biochemistry 27:5355-5361. [DOI] [PubMed] [Google Scholar]
- 22.Xiang, L., J. A. Kalaitzis, and B. S. Moore. 2004. EncM, a versatile enterocin biosynthetic enzyme involved in Favorskii oxidative rearrangement, aldol condensation, and heterocycle-forming reactions. Proc. Natl. Acad. Sci. USA 101:15609-15614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiang, L., and B. S. Moore. 2003. Characterization of the benzoyl coenzyme A biosynthesis genes in the enterocin-producing bacterium “Streptomyces maritimus.” J. Bacteriol. 185:399-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiang, L., and B. S. Moore. 2002. Inactivation, complementation and heterologous expression of encP, a novel bacterial phenylalanine ammonia-lyase gene. J. Biol. Chem. 277:32505-32509. [DOI] [PubMed] [Google Scholar]
- 25.Zon, J., and N. Amrhein. 1992. Inhibitors of phenylalanine ammonia-lyase: 2-aminoindan-2-phosphonic acid and related compounds. Liebigs Ann. Chem. 625-628.