Abstract
Pseudomonas putida KT2440, a paradigm organism in biodegradation and a good competitive colonizer of the maize rhizosphere, was the subject of studies undertaken to establish the genetic determinants important for its rhizospheric lifestyle. By using in vivo expression technology (IVET) to positively select single cell survival, we identified 28 rap genes (root-activated promoters) preferentially expressed in the maize rhizosphere. The IVET system had two components: a mutant affected in aspartate-β-semialdehyde dehydrogenase (asd), which was unable to survive in the rhizosphere, and plasmid pOR1, which carries a promoterless asd gene. pOR1-borne transcriptional fusions of the rap promoters to the essential gene asd, which were integrated into the chromosome at the original position of the corresponding rap gene, were active and allowed growth of the asd strain in the rhizosphere. The fact that five of the rap genes identified in the course of this work had been formerly characterized as being related to root colonization reinforced the IVET approach. Up to nine rap genes encoded proteins either of unknown function or that had been assigned an unspecific role based on conservation of the protein family domains. Rhizosphere-induced fusions included genes with probable functions in the cell envelope, chemotaxis and motility, transport, secretion, DNA metabolism and defense mechanism, regulation, energy metabolism, stress, detoxification, and protein synthesis.
The rhizosphere is a densely populated area in which plant roots interact with soilborne microorganisms, including bacteria, fungi, and invertebrates, feeding on an abundant source of organic material (43). Many colonization traits and genes have been identified by random mutagenesis of good competitive root-colonizing bacteria (mainly Pseudomonas fluorescens and Pseudomonas chlororaphis) and through screening for gain or loss of competitive root tip colonization ability (see reference 28 for a recent review). Often these genes have been found to be unimportant for growth in the laboratory, and their role in bacterial fitness has required competition studies involving wild-type and mutant strains.
Information on gene expression in the rhizosphere is, however, limited and partial. Previous results obtained by our group have shown that utilization of the imino acid proline by Pseudomonas putida KT2440, which involves uptake and catabolism, was induced by maize root exudates (50). In addition, an aminotransferase involved in the catabolism of lysine was also identified in a screen to select for maize root exudate-induced genes by using a promoter probe transposon (15).
The most extensive study so far based on positive selection of bacterial traits as a consequence of their contribution to root colonization is that of Rainey (38). In that work, the in vitro expression technology (IVET) approach was used to select P. fluorescens genes activated during sugar cane root colonization. The study was partial, since only 10% of the genome was analyzed.
Our work represents an attempt to expand our knowledge of bacterial gene expression in a complex environment, such as the rhizosphere, using another model system. We faced the identification of P. putida genes (rap, for root-activated promoters) induced during maize root colonization by this bacterium by taking advantage of the fact that the complete genome sequence is available for P. putida KT2440.
P. putida KT2440 is being used by our research group as a model organism in studies of plant-microbe interaction to establish the molecular bases of the initial seed adhesion and subsequent root colonization. KT2440 is a TOL plasmid-cured derivative of the natural isolate P. putida mt-2, which was isolated in 1960 from a planted field in Japan and whose historical itinerary has been reviewed by Nakazawa (35). Not surprisingly, P. putida KT2440, besides its potential for removing xenobiotics, exhibits high fitness in the colonization of the rhizosphere of a large number of plants (32).
Our approach was to use a positive selection system based on IVET. This strategy has been previously used to identify Pseudomonas in vivo-induced genes under different conditions (17), including a recent study of P. fluorescens genes induced in soil (47). The system was originally developed to select Salmonella enterica serovar Typhimurium virulence genes that were induced during host infection (29). For IVET implementation in P. putida, we used a derivative of the pIVPRO plasmid containing a promoterless gene expression reporter cassette (asd) (20), which is useful in generating transcriptional fusions, and as a host to select appropriate fusions, we used an asd null mutant unable to survive in the rhizosphere (42), which allowed selection on the basis of their in vivo induction. The asd gene product is involved in the biosynthesis of aspartate-β-semialdehyde dehydrogenase, a key intermediate in the biosynthesis of diaminopimelic acid (DAP), which is required for integrity of the cell wall, and of amino acids such as lysine, methionine, and threonine (21).
MATERIALS AND METHODS
Bacterial strains, culture conditions, and solutions.
P. putida KT2440, a derivative of the P. putida soil isolate mt-2, has been described previously (35). In the capture of rhizosphere-activated promoters, an asd::xylE null mutant of KT2440 was used, as it is unable to survive in the rhizosphere because of its dependence on DAP, Lys, Thr, and Met (42). Plasmids with an oriR6K replication origin were maintained in Escherichia coli DH5αλpir (25), and E. coli S17-1λpir (48) was used for conjugation. E. coli strains were grown at 37°C in Luria-Bertani (LB) medium (44). P. putida strains were grown at 30°C in either LB or minimal medium, which was basal M9 medium (44) supplemented with Fe-citrate (6 μg/liter), MgSO4 (1 mM), and trace metals as described before (1) and with glucose (25 mM) or sodium citrate (10 mM) as the carbon source, unless otherwise specified. When appropriate, the following antibiotics were added to the media at the given concentrations (in μg/ml): ampicillin, 100; chloramphenicol, 30; kanamycin, 25 and 50 for E. coli and Pseudomonas strains, respectively; and rifampin, 10. Diaminopimelic acid was added at a final concentration of 0.5 mg/ml. The l amino acids Lys, Met, and Thr were supplied at 40 μg/ml.
DNA techniques.
Plasmid DNA was isolated with a QIAGEN miniprep kit. Preparation of chromosomal DNA, digestion with restriction enzymes, dephosphorylation, ligation, and electrophoresis were carried out using standard methods (4, 44). DNA fragments were recovered from agarose gels with a QIAGEN gel extraction kit.
Construction of pOR1.
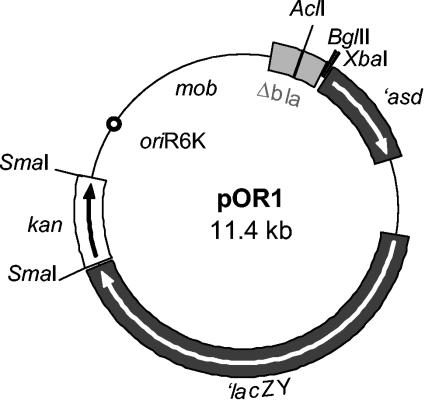
pIVPRO promoter probe vector (20) was optimized for in vivo expression technology in P. putida as follows: (i) a kanamycin cassette contained in the SmaI fragment of plasmid p34S-Km3 (12) was cloned at the unique SmaI site of pIVPRO and (ii) in the resulting plasmid, a partial deletion of the bla gene was performed by removing the 0.4-kb AclI fragment internal to this gene. This deletion was carried out to avoid recombination between plasmid and chromosome through the ampC homologous gene present in the chromosome of P. putida KT2440, which encodes PP2876. As a result, plasmid pOR1 (11.4 kb) was obtained. Figure 1 shows that this plasmid carries a promoterless asd gene followed by the promoterless lacZY as reporter genes. Plasmid pOR1 is the IVET vector used to construct a promoter library of KT2440.
FIG. 1.
Map of pOR1 plasmid (IVET vector) used to identify P. putida root-activated promoters. Modifications to the original plasmid pIVPRO (20) are described in the text. This plasmid has the replication origin oriR6K and thus requires protein Π for stable maintenance. The plasmid is therefore a suicide in Pseudomonas and is mobilizable as tra functions are supplied in trans.
Construction of libraries.
P. putida KT2440 genomic DNA was partially digested with Sau3AI, size selected by agarose gel electrophoresis (1 to 4 kb), and cloned into the BglII site 5′ of the promoterless asd-lacZY encoded by pOR1 (Fig. 1). The pool of fusions was electrotransformed in E. coli S17-1λpir, and about 12,000 clones were obtained. Eighty percent of the GenBank clones exhibited inserts in the plasmids as analyzed by double digestion with XbaI and AclI. The pOR1-KT2440 GenBank was then transferred en masse by bacterial conjugation to P. putida Δasd using as selective medium for the transconjugants LB supplied with DAP, kanamycin, and chloramphenicol, which counterselected E. coli. The frequency of exconjugants containing integrated fusions was 10−4 to 10−5 per recipient cell.
Identification of the sequences captured by the rap fusions.
DNA sequences from the rap isolates were determined by arbitrary PCR (8). A first round of amplification was done by using the chromosomal DNA of the mutants as a template, with an arbitrary primer (ARB1; 5′-GGCACGCGTCGACTAGTACNNNNNNNNNNGATAT-3′) and an internal primer of the ′asd gene borne by pOR1 (Asdext; 5′-TCGCTGTATGAGTACGGAACCC-3′). The first round was as follows: 3 min at 95°C; six cycles of 1 min at 95°C, 1 min at 30°C, and 1 min at 72°C; 30 cycles of 30 s at 95°C, 30 s at 50°C, and 1 min at 72°C; and an extension period of 7 min at 72°C. A second round of amplification was done using as the template 5 μl of the first-round reaction as follows: 3 min at 95°C; 30 cycles of 30 s at 95°C, 30 s at 60°C, and 1 min at 72°C; and 7 min at 72°C. Primers used for the second round were those corresponding to the conserved region of ARB1 (ARB2; 5′-GGCACGCGTCGACTAGTAC-3′) and a second internal primer of the ′asd gene closer to the 5′ end (Asdint; 5′-CGATCAGACCTACACGCTTCATC-3′). Reaction mixtures were electrophoresed, and the most intense bands were isolated and sequenced. Sequencing was done on an ABI PRISM 310 automated sequencer using oligonucleotide Asdint as a primer. The ∼50-bp distance between this primer and the BglII site used for cloning provided an internal control to ensure that the sequence obtained corresponded to the junction between the promoterless asd gene and the flanking chromosome in the cointegrate.
Sequences were analyzed and compared with the GenBank database by using BLAST programs (3). Sequence data for the P. putida, Pseudomonas syringae pv. tomato strain DC3000, and Pseudomonas aeruginosa genomes were obtained from The Institute for Genomic Research (www.tigr.org) and the Pseudomonas Genome Project (www.pseudomonas.com).
Surface sterilization, germination of seeds, and root colonization assays.
Corn seeds were surface sterilized by rinsing with sterile deionized water and washing twice for 10 min with 70% (vol/vol) ethanol and once with 20% (vol/vol) bleach, followed by thorough rinsing with sterile deionized water. Surface-sterilized seeds were germinated on a petri dish in the presence of penicillin G (500 μg/ml) (37) at 30°C for 2 days for root colonization assays or for 3 days if seedlings were to be transferred to a tube and grown hydroponically in the specified solution. For root colonization assays, overnight cultures grown in LB were diluted in M9 salts to a turbidity at 660 nm of about 1, and seeds were inoculated with an appropriate dilution of bacterial suspensions (5 μl of the suspension per ml of M9). After incubation for 1 h without shaking at 30°C, the seeds were washed and planted in pots containing vermiculite or used to determine the number of bacteria attached to the seed. To analyze gene expression in the maize rhizosphere, Sterilin tubes (50 ml) containing sterile sea sand (40 g) were used, and plant nutrient solution (PNS) was added (10% [vol/wt]). PNS was Ca(NO3)2 (5 mM), KNO3 (5 mM), MgSO4 (5 mM), KH2PO4 (1 mM) (pH 5) supplemented with Fe-EDTA (100 μM) and micronutrients of MS medium as described previously (34). Controls without plant seeds were run similarly as required. Plants were maintained in a controlled chamber at 24°C and 55 to 65% humidity with a daily light period of 16 h for 1 to 2 weeks. To recover bacteria from the rhizosphere, plants were removed and roots were cut, weighed, and placed in sterile 50-ml screw-cap tubes containing 20 ml M9 and 4 g glass beads (diameter, 3 mm). The tubes were vortexed for 2 min, and the number of CFU per gram of root was determined for each plant by plating serial dilutions on selective media consisting of either LB medium supplied with DAP and antibiotics or M9 minimal medium supplied with citrate, DAP, Lys, Met, Thr, and antibiotics. The same process was used with inoculated seeds to determine the number of attached bacteria, i.e., the initial inoculum on the seeds, except that 2 ml of M9 and 10 to 12 glass beads were used. To recover bacterial cells for β-galactosidase assays, glass beads were not used.
β-Galactosidase activity assay.
Specific β-galactosidase activity from bacterial suspensions growing on either liquid cultures or seedling hydroponic cultures was determined spectrophotometrically (31). Given that samples showed color and contained low numbers of cells, activities of the rap fusion strains as recovered from the maize rhizosphere were determined with 4-methylumbelliferyl-β-d-galactoside as the substrate and detection of the fluorescent product methyl-umbellipherone with a spectrofluorometer (excitation wavelength, 365 nm; emission wavelength, 450 nm).
RESULTS
Development of IVET strategy for P. putida.
For the selection of root-activated promoters with IVET, we took advantage of the asd null mutant developed previously in our laboratory. This mutant did not survive in the maize rhizosphere because of its inability to synthesize three amino acids (Lys, Met, and Thr) and DAP; however, when the asd gene was supplied in trans in a plasmid, colonization ability was restored to the wild-type level (42). Plasmid pOR1 was constructed as described in Materials and Methods to generate transcriptional fusions of chromosomal DNA to the promoterless cassette asd-lacZ. Relevant characteristics of this plasmid are shown in Fig. 1. There is a unique BglII site for cloning upstream from the 5′ end of ′asd. The mob functions confer mobilization properties to the plasmid so that it can be transferred by bacterial conjugation to P. putida Δasd. The pOR1 plasmid is unstable in Pseudomonas (due to the replication origin oriR6K), so that selection for kanamycin resistance forces integration of its chimeric derivatives in the chromosome by homologous recombination through the DNA inserts cloned in BglII at their corresponding loci. Thus, the so-generated IVET fusions strains, as merodiploids, mostly present intact copies of the targeted loci, while their promoters, in their native chromosomal context, are responsible for the expression of the essential ′asd gene.
Screening of the P. putida cointegrate library in planta to select rap genes.
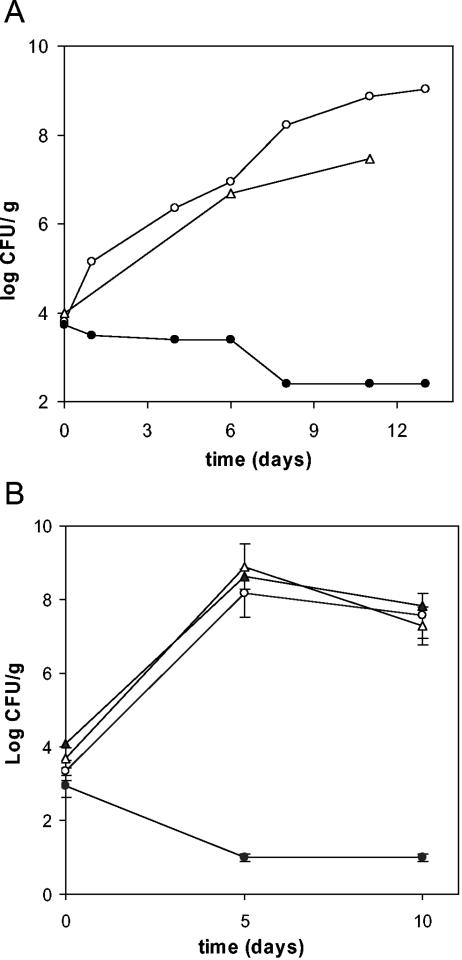
We developed the IVET strategy for P. putida, as reported above, to identify specific bacterial functions expressed during root colonization. The screen was based on that used for P. fluorescens (38). Cointegrate strains containing fusions were screened for survival in the rhizosphere in pools of about 500 clones. Initially, four pools were screened by inoculating 12 seeds with 106 bacteria. Because bacterial adhesion was 0.5% of the inoculum on average, the number of cells attached to the seed was ∼5 × 103. Bacterial pools were recovered from the rhizosphere at different times, and survival was analyzed. Maximal CFU/g of root were obtained after 1 week (107 to 108), and therefore subsequent screening runs were carried out for 1 to 2 weeks. P. putida Δasd was kept as a negative control for survival in the rhizosphere (Fig. 2A). Up to 6,000 clones were analyzed. Bacteria from the rhizosphere were removed as described in Materials and Methods. Up to 10,000 clones containing fusions of putative rhizosphere-activated promoters were selected on M9-citrate agar supplied with DAP, Lys, Met, Thr, and kanamycin. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was also added to monitor inactivity of the rap promoters in vitro. Fusion strains carrying constitutive promoters, detected for their ability to grow in M9-citrate agar without the addition of DAP, Lys, Met, and Thr, were discarded. Seventy-eight rap fusions were selected for their auxotrophy or diminished growth on citrate-supplied M9 minimal medium. Because active promoters upstream to ′asd were required for the survival of these fusions in the rhizosphere, lack of growth (or residual growth) of the fusions in vitro suggested that expression from the promoters was preferential in the rhizosphere.
FIG. 2.
Effect of rap promoter activity on colonization ability. (A) Survival of chromosomal IVET cointegrates pool in the rhizosphere. ○, KT2440R; •, Δasd; ▵, a pool of approximately 500 clones of P. putida IVET cointegrates. (B) Survival of rap fusion strains. ○, KT2440R; •, Δasd; ▵, rap1-2 fusion; and ▴, rap1-4 fusion. Maize seeds were inoculated and plants grown as described in Materials and Methods. Bacterial cells were recovered on plates at the indicated times. At time zero, CFU was per g of seed; afterwards, CFU is plotted per g of rhizosphere including root. Means and standard deviations for three independent experiments are shown.
Sequence analysis of rap fusions.
To identify the genes whose promoters were responsible for the survival of the asd mutant in the rhizosphere, arbitrary PCR was performed with each fusion as described in Materials and Methods. This technique allowed us to amplify fragments containing 300 to 750 bp of DNA flanking the promoterless asd gene, which was responsible for the expression of this gene in the fusion strains. Twenty-eight independent fusions were identified after sequencing the DNA adjacent to the ′asd gene with the primer Asdint. To confirm the legitimacy of these fusions, they were recovered from the genome by conjugative cloning as described by Rainey and coworkers (39) and sequenced with the primer pOR1fw (CATGAGCGGATACATATTTGAATG), which allowed us to determine the flanking DNA at the 3′ end of the fusion.
The rap genes are listed and organized by function in Table 1. Genes whose translation products play a role in energy generation, DNA metabolism, transport and secretion, stress and detoxification, motility, and synthesis of components of the cell envelope were identified. Nine genes (32%) of unknown function were identified, including genes predicted to have a regulatory function.
TABLE 1.
Function of the P. putida rap genes identified during maize root colonization
| Function and/or role | Gene and/or rap fusion | Locus |
|---|---|---|
| Cell envelope (GDP-mannose 6-dehydrogenase) | algD (rap2-45) | PP1288 |
| Chemotaxis and motility (flagellar assembly protein) | fliO (rap1-8) | PP4356 |
| Transport | ||
| ABC-type sulfate transport system, periplasmic component | rap2-16 | PP4305 |
| Gamma-aminobutyrate (GABA) transporter permease | rap2-21 | PP2543 |
| Sodium/proline symporter | putP (rap2-40) | PP4946 |
| Secretion | ||
| Protein chaperone | secB (rap1-2) | PP5053 |
| Preprotein translocase subunit | yidO (rap2-26) | PP0006 |
| DNA metabolism and defense mechanism | ||
| Type 1 restriction-modification system, M subunit | hsdM (rap1-19) | PP4741 |
| Single-stranded DNA-specific exonuclease | recJ (rap2-28) | PP1477 |
| DNA topoisomerase IV, A subunit | parC (rap2-37) | PP4912 |
| Energy metabolism | ||
| Pyruvate dehydrogenase, decarboxylase component | aceE (rap2-2) | PP0339 |
| Isocitrate lyase putative (glyoxylate bypass) | aceA (rap2-14) | PP4116 |
| 6-Phosphogluconate dehydrogenase | gnd (rap2-15) | PP4043 |
| 2,3-Biphosphoglycerate-independent phosphoglycerate mutase | pgm (rap2-77) | PP5056 |
| Regulatory functions | ||
| SAM-dependent methyltransferase transcriptional regulator | rap1-4 | PP4966 |
| Transcriptional regulator, AsnC family | rap1-12 | PP4424 |
| Transcriptional regulator, AraC family | rap2-44 | PP2070 |
| Sensor histidine kinase | colS (rap2-63) | PP0902 |
| Stress (general stress protein Ctc [ribosomal 5S rRNA E-loop binding protein Ctc/L25/TL5]) | rap1-9 | PP0721 |
| Detoxification (lactoylglutathione lyase [methylglyoxal metabolism]) | gloA (rap2-53) | PP3766 |
| Protein synthesis (glutamyl-tRNA synthetase tRNA-Ala, tRNA-Glu) | gltX (rap2-1) | PP1977 |
| Unknown | ||
| Seed adhesion, antisense | mus-5 (rap2-74) | PP4615 |
| Hypothetical protein | rap2-7 | PP5390 |
| Hypothetical putative lipoprotein of unknown function | rap2-18 | Between PP3854 and PP3855a |
| Conserved hypothetical protein alpha/beta hydrolase fold superfamily, antisense | rap2-19 | PP4634 |
| Amino oxidase, putative | rap2-23 | PP0383 |
| Conserved hypothetical protein (predicted SAM-dependent methyltransferases) | rap2-39 | PP4306 |
| Hypothetical protein | rap2-73 | PP2298 |
The junction was at a position between the two indicated proteins. A putative 348-bp open reading frame was detected (4.378.820-4.379.167). See the text for details.
Activity of rap promoters versus root colonization.
Because the DAP auxotrophic mutant P. putida Δasd was unable to colonize the rhizosphere (Fig. 2), rap genes must be induced in the rhizosphere to promote asd transcription and thus growth of the strains bearing the corresponding fusions on their chromosome. The survival of a representative pool of 500 clones of the IVET cointegrates library is shown in Fig. 2A. All rap fusion strains were analyzed individually for their colonization capacity. Figure 2B shows an example of the colonization ability of two rap fusion strains. After 10 days, the number of bacteria per g of rhizosphere was not notably different from P. putida KT2440R, a rifampin-resistant derivative of KT2440 used as a positive control strain (16). Slightly different behavior was observed in four rap fusions, rap2-44, rap2-45, rap2-73, and rap2-74, which exhibited 1 to 1.5 log lower survival on average (not shown). In the absence of induction, colonization by the Δasd mutant is not possible, so induction must take place from the rap fusion for colonization of the rhizosphere.
Expression from the rap promoters in conditions of hydroponically growing seedlings.
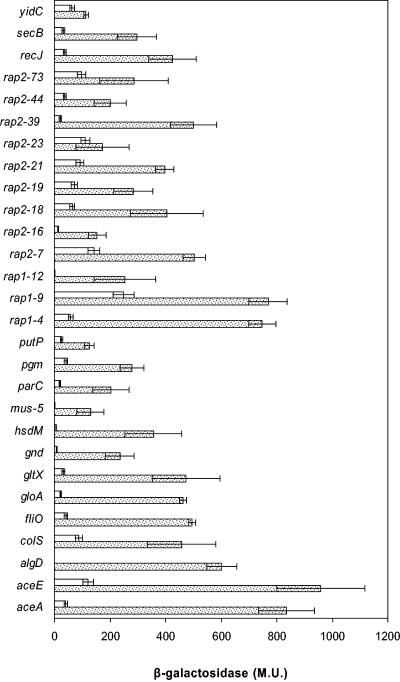
Induction by plant exudates of the rap promoters was initially evaluated by incubating the rap fusion strains in M9 in the presence of 3-day-old seedlings for 72 h. Bacteria were also cultivated in M9-citrate (2.5 mM) supplied with DAP, Lys, Met, and Thr up to an optical density at 600 nm of about 0.3, which was on average the growth supported by the maize seedlings under these conditions. β-Galactosidase activity was determined by spectrophotometric assay (Fig. 3). All of the fusions exhibited induction in the presence of the seedling in comparison to the same cells incubated in citrate-supplied minimal M9 medium. The highest induction rate was normally observed in strains which exhibited lower (or null) basal activity under laboratory conditions in the absence of plant exudates. Two fusions were remarkable: those identified as algD (up to 100-fold) and hsdM (higher than 50-fold). The algD upstream regulatory region was identified as active in the presence of the corn seedling. algD is the first gene in the algD-8-44-KEGXLIJFA operon, a cluster of 12 genes responsible for the biosynthesis of the exopolysaccharide alginate in Pseudomonas strains and other bacteria that synthesize alginate. hsdM encodes a methylase of the type I restriction-modification system. Another interesting fusion was rap2-74 (ddcA), which corresponded to mus-5 (in its antisense strand), reported to have a role in adhesion (16). Two metabolic genes, aceA and aceE, showed high expression levels. The rap1-9 and rap1-4 fusions were also highly expressed, although the former had shown considerable basal expression.
FIG. 3.
β-Galactosidase activity (Miller units) of rap fusions during seedling colonization in hydroponic culture. Dotted bars show values from cells in suspension in the presence of maize seedling. Open bars are values in 2.5 mM citrate-supplied M9 minimal medium with all requirements to support the growth of the rap fusions, as described in Materials and Methods. Inoculum size was 2.5 × 106 to 5 × 106 CFU/ml. Means and standard deviations from three independent experiments are shown.
Analysis of rap gene expression in the rhizosphere.
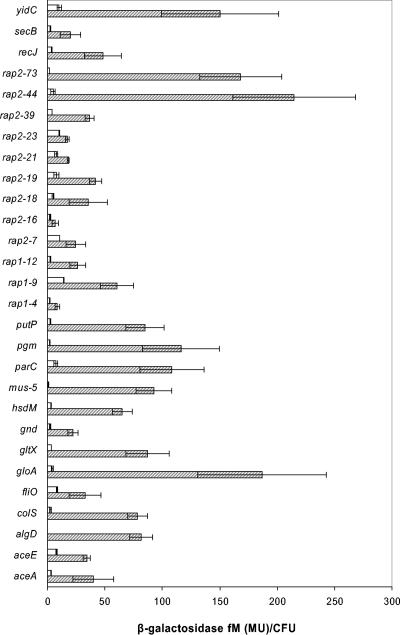
To confirm that rap promoters were active preferentially during root colonization, we evaluated their activity in cells that had been colonizing the maize rhizosphere for 1 week. As a control, cells were incubated in the absence of plants on sea sand supplied with all of the requirements of rap strains and citrate (10 mM) as the sole carbon source. Data for gene expression measured as β-galactosidase activity are shown in Fig. 4. For all of the rap fusions assayed, gene expression measured as β-galactosidase activity per cell was higher in cells recovered from the rhizosphere than in control cells. One of the genes with the highest expression was yidC. This gene encodes a 60-kDa inner membrane protein responsible for the insertion of membrane proteins into the lipid bilayer. The basal level of expression observed for this gene was consistent with YidC essentiality in E. coli (46). Other fusions that were expressed highly during colonization were rap2-73 and rap2-44. The former encodes a hypothetical protein located upstream to the trigger factor, which—like the ribosomal chaperone trigger factor—is also involved in protein fate. The latter is an AraC family transcriptional regulator of unknown function. Rap2-73 homologues are present in P. syringae and P. fluorescens. Intriguingly, neither of these fusions was highly expressed during hydroponic growth. Several fusions exhibited high rates of induction during colonization as a consequence of their low basal levels of gene expression. Among them, the most silent rap promoter was the algD promoter. Also silent were the control cells of rap2-74 and hsdM fusions, which also exhibited high induction during hydroponic growth. Three other fusions were also highly induced during colonization. The rap2-63 fusion, which involves the promoter of the two-component system colRS, was previously reported to play a role in colonization (11), the rap2-40 fusion substantiated the induction of the proline transport system PutP in the rhizosphere (50), and the rap2-77 fusion revealed that the phosphoglycerate mutase encoded by pgm was induced in vivo.
FIG. 4.
β-Galactosidase activity of rap fusions in the rhizosphere. Hatched bars show values from cells recovered from the maize rhizosphere supplied with PNS. Open bars are values from control cells in sand supplied with PNS, citrate, and all requirements to support the growth of the rap fusions as described in Materials and Methods. Inoculum size for controls was 1 × 104 to 2.5 × 104, similar to the number of cells attached to the seeds. Means and standard deviations from three independent experiments are shown. Units are given as the concentration of 4-methyl-umbelliferone (femtomolar) per CFU.
DISCUSSION
In this report we identify P. putida genes that are induced during maize root colonization as a way to explore the biology of this saprophyte in the rhizosphere. In view of its isolation origin, a crop field (35), the rhizosphere must constitute a familiar niche for this bacterium.
The IVET method utilizes a variant of the promoter trap vector pIVPRO, which we optimized and named pOR1. The capture of an active promoter in pOR1 commanded transcription of the asd gene and therefore conferred survival in the rhizosphere to the asd mutant carrying the root-activated fusions. These derivatives of pOR1 were maintained stably in Pseudomonas only as a consequence of their integration in the chromosome through homologous recombination by the promoter region. Although the IVET fusion strains, as merodiploids, mostly present intact copies of the targeted loci, exceptions causing the loss of the host gene(s) cannot be discarded, in particular for those fusions involving operon internal promoters. In the hypothetical case of gene inactivation, there would be a possibility of change in promoter activity of autoregulated genes, which could lead to their initial isolation. Even so, this fact would not affect the conclusions reached for rap genes in this study, since controls in the absence of plants are included. Nevertheless, it is impossible to select inactivation involving loci essential for survival in the rhizosphere with the IVET strategy.
The bla gene was removed from the ancestor pIVPRO because this plasmid itself, before capturing promoters, used to form cointegrates by recombination with a 98% identical ampC gene in the chromosome of KT2440.
With our selection strategy, 28 transcriptionally active units of P. putida that were induced in the rhizosphere of maize seedlings were isolated. About 17% of the genome was represented in the IVET library. A similar system based on the dap gene was used with P. fluorescens to identify genes induced during sugar cane root colonization (38). In that species, about 10% of the genome was analyzed. Interestingly, no gene overlap was observed between P. fluorescens and P. putida. Further analyses will be needed to discern whether the reason for this was the partial nature of the genome analysis in both cases or species-dependent gene expression specificity. It cannot be ruled out that different genes were identified as a consequence of the different host strains used for the IVET fusions, i.e., asd in this work versus dap in P. fluorescens.
Our approach was validated by the fact that our positive selection scheme based on pOR1 isolated five fusions that identified bacterial functions previously highlighted in Pseudomonas-plant interactions. These fusions were rap2-40, which identified putP, encoding a proline permease (50), rap2-74, corresponding to mus-5, which is involved in seed adhesion (16), rap2-63, identifying colS, which encodes the sensor histidine kinase element of the double component system ColR/ColS (11), rap1-2, which identified secB, encoding a protein chaperone (26), and rap1-8, corresponding to fliO, which is involved in the synthesis of the flagella export apparatus (14). The putative roles of these genes in colonization are described in detail below as appropriate in the context of this discussion.
In the system reported here, survival and β-galactosidase activity were both affected by the promoter activity and so contributed together to the level of gene expression. Thus, lower and higher values of β-galactosidase activity, as plotted in Fig. 3 and 4, did not necessarily correlate with lower and higher cell survival.
The expression level of rap genes was measured under two different bacterial lifestyles: planktonic during plant hydroponic growth (Fig. 3) and as a result of rhizosphere colonization (Fig. 4). All rap genes were induced under both conditions in comparison to the control situation. However, differences in gene expression were observed in the two data sets, and further work will be required to unveil the basis for these differences.
The rap2-45 fusion isolated the algD promoter, a possible indication of alginate biosynthesis in the rhizosphere, thus establishing an important parallelism with the biofilm lifestyle (6). It was recently reported that algA, the last gene in the algD-A operon, is controlled by water stress in P. putida (49). Nelson and coworkers (36) suggested that the absence of the regulatory gene mucC from the algT-mucC operon might explain the P. putida nonmucoid morphotype under standard culturing conditions. However, the same divergence in the algT-mucC operon has been described for P. syringae, a bacterium that does synthesize alginate (23). Hence, alginate-defective mutants of P. syringae are compromised in their ability to colonize plant tissue (51). As in P. syringae, alginate biosynthesis regulation in P. putida might respond to environmental signals. Further research is required to document the synthesis of alginate by KT2440 and a role for alginate in colonization.
The IVET strategy identified genes involved in nutrient acquisition. This is the role of putP (which encodes proline permease), a gene known to be induced in the presence of maize exudates (50). This finding was not surprising, since proline can be used by P. putida as the sole carbon and nitrogen source and maize exudates are rich in this imino acid (C. Ramos, personal communication). A permease for the nonprotein amino acid gamma-aminobutyric acid (GABA) was also identified in this screening. GABA is produced from putrescine in E. coli, and putrescine has been found in tomato root exudate (28). Thus, the induction of GABA transporter permease in the rhizosphere is not unexpected, since Pseudomonas strains are able to use GABA as the sole nutrient source. In connection with amino acid utilization, a mutant in the gene identified with the rap2-23 fusion (which codes for a putative amino oxidase) is not able to use l-lysine as the sole nitrogen and carbon source (O. Revelles, personal communication), whereas wild-type KT2440 is able to grow on this amino acid. This gene was erroneously annotated as a putative tryptophan-2-monooxygenase (36). Genes involved in the catabolism of Lys have been shown to be induced by maize exudates, and this is an indication of the presence of Lys itself or any other Lys-related intermediary metabolites in the plant exudates (15).
Specific studies of bacteria of the genus Pseudomonas were recently compiled (40). However, many aspects of the metabolism and the general physiology of these bacteria still remain unknown. Knowledge about the entry of carbon sources, such as sugars and carboxylic acids, into the central metabolic pathways is limited and incomplete, and the same applies to the biosynthesis and catabolism of amino acids. Nevertheless, important aspects of energy metabolism have been revealed in this work. The induction of gnd (which codes for 6-phosphogluconate dehydrogenase) indicates that the pentose-phosphate pathway, an alternative to the Embden-Meyerhoff pathway, is active in the rhizosphere. Ribulose-5-phosphate, an important precursor for the synthesis of purine nucleotides and histidine (27), is generated via the so-called pentose shunt. Evidence that the Embden-Meyerhoff pathway was also induced comes from the isolation of the rap2-77 fusion. Interestingly, the cofactor-independent phosphoglycerate mutase appeared to be essential for growth and pathogenicity of P. syringae in its host tomato plant (33). Pyruvate dehydrogenase, an enzyme with a major role in central metabolism, was also induced in the rhizosphere. The aceE gene, which encodes the pyruvate dehydrogenase E1 component, was identified as being induced in vivo in E. coli during septicemic infection in a murine model (24). In addition, isocitrate lyase, an enzyme of the glyoxylate bypass pathway, was probably induced in response to the presence of acetate and/or fatty acids, which have been identified as major components of the plant exudates (28).
Genes with a known role in protein trafficking have been shown to be induced in vivo. One example is secB, which encodes the component of the general secretory pathway SecB. This gene was previously identified as being essential for competitive root tip colonization in P. fluorescens, but it is not required for competitive growth in laboratory culture media (26). In E. coli, the protein chaperone SecB facilitates the targeting of periplasmic and outer membrane proteins, whereas insertase YidC is used for the insertion of membrane proteins (45). The gene encoding the inner membrane protein YidC was also identified in our screening.
Of special interest is the rap2-74 fusion, which isolated a transcriptionally active region, apparently commanding the expression of putative antisense ddcA (mus-5). The mus-5 gene was first identified by our group as playing a role in corn seed adhesion. It was recently reported that ddcA encodes a putative membrane polypeptide and expression of this gene is directly dependent on cell density and seed exudates (16). However, the detection of antisense expression in vivo during root colonization reported here might be a consequence of read-through transcription from the convergent gene coding for PP4614, since no rho-independent termination sequence determinant was detected. Further studies will be needed to determine whether this messenger is an indication of ddcA silencing after the adhesion stage. Putative genes carried in the strand opposite to that of known genes have been also identified using IVET to recognize soil-induced genes in P. fluorescens (47).
Regulatory genes were also isolated in our IVET screening. colS was identified as highly induced during colonization (Fig. 4). A P. fluorescens mutant with a mutation in the sensor kinase member of the ColR/ColS two-component system was seriously impaired in competitive root colonization in several plants (11). The rap1-4 fusion also identified a regulatory gene. The 3′ end of this probable transcriptional regulator of the ArsR family encoded by rap1-4 is located 18 base pairs upstream from metK. In E. coli, the metK gene, which codes for S-adenosylmethionine synthetase (SAM), has been found to be induced in vivo during the biofilm lifestyle (41) and septicemic infection (24). In this bacterium, SAM is a corepressor of met genes, the donor of most methyl groups (9), and the source of the propylamino group of spermidine (19).
In each situation of the two assayed in vivo, a regulatory gene of unknown function was highly expressed and induced in comparison to the control (rap1-12 in hydroponic culture and rap2-44 in root colonization). This finding suggests a regulatory role for Rap1-12 in exudate nutrient utilization. In support of this hypothesis is the finding that rap1-12 is located convergent to genes that encode an amino acid ABC transporter.
Flagella also seem to be involved in seed adhesion (10). Our work identified a promoter of the operon comprising fliO as induced in vivo. P. fluorescens flagellar mutants are impaired in the ability to colonize developing roots (14), and a role for the flagella in chemotactic motility toward exudate components has been reported (13).
Our results identified the parC gene, which encodes DNA topoisomerase subunit A, as being induced in vivo. Interestingly, parE (which encodes subunit B) was found to be induced in vivo in E. coli during septicemia infection (24).
Strain KT2440 has been reported to be a restriction-minus derivative (hsdR1) of P. putida mt-2 (18). In this work, we identified the hsdM gene as being induced during colonization. hsdM exhibits translational coupling with hsdS, and both genes encode a functional modification methylase. All three hsdRMS genes encode a type IA restriction-modification system commonly found in Enterobacteriaceae. These enzymes are believed to play a role not only as a defense mechanism for bacterial cells against foreign DNA but also in a specialized recombination system thought to control the flow of genes between bacterial strains (5). It does not seem exceptional that the expression of genes that code for methyltransferases and endonucleases is tightly regulated, since it is essential that the methyltransferase completely modifies the cellular genome at all times to protect it from the lethal action of the endonuclease. This regulation must be especially important when restriction-modification genes first enter a cell, on conjugative plasmids, for example, but it might also be important during changes in the physiology of the cell (e.g., entry into the stationary phase, starvation associated with a deficiency of methyl donors, and other stresses).
The gloA gene, with a probable role in detoxification, was identified. Lactoylglutathione lyase (glyoxalase I) encoded by gloA catalyzes the isomerization of hemithioacetal to S-lactoylglutathione. Hemithioacetal is formed by the spontaneous reaction between the cytotoxic methylglyoxal and glutathione. The synthesis of methylglyoxal has been related to glucose flux exceeding the potential for growth (22) and growth on some carbon sources in the presence of cyclic AMP (2).
Fusion rap1-9 identified the ribosomal protein L25, which belongs to the ribosomal L25p family. This family includes Ctc from Bacillus subtilis, which is induced by stress (7).
More than 30% of the rap fusions were to genes of unknown function. Fusion rap2-19 was to a gene that encodes a hypothetical protein: a predicted hydrolase of the alpha/beta-fold family of unknown function. No similar protein was identified in other Pseudomonas species. However, a conserved hypothetical protein showing 55% identity and 65% similarity was found in Burkholderia (BPSL2346). Fusion rap2-7 identified a hypothetical protein that exhibited translational coupling to a putative protein-S-isoprenyl cysteine methyltransferase. Homologues were not present in other species of Pseudomonas, although a homologous protein was found in the cyanobacteria Nostoc sp. (55% identity, 66% similarity). Fusion rap2-18 identified what corresponded to an intergenic region in the P. putida KT2440 annotation by Nelson et al. (36). However, using the blastX program (BLAST 2.2.10) we identified in this region an unknown hypothetical protein conserved in P. aeruginosa (PA3399) (47% identity and 62% similarity), between PP3854 and PP3855. Rap2-39 is a predicted SAM-dependent methyltransferase highly conserved among Pseudomonas species. Fusions rap2-16 and rap2-39 were to convergent genes with no loop structure sequence observed in the intergenic region. The rap2-16 fusion identified the periplasmic component of an ABC-type sulfate transport system, and ABC transport system components have been suggested to act as adhesins (30). Fusion rap2-73 identified a hypothetical protein, which exhibited homology to a hypothetical protein found only in P. syringae and P. fluorescens, two bacteria that also colonize plant roots.
Our work with in vivo expression technology allowed us to identify 28 rap genes, which have the potential to elucidate the colonization process and adaptation of P. putida to the rhizosphere environment. Given the biodegradative potential of KT2440, identifying promoters of preferential induction in the rhizosphere may open new avenues in rhizoremediation. The strictest rap promoters, such as those identified by fusions rap2-45 (algD) and rap1-19 (hsdM), might represent attractive genetic tools for exploiting the potential of this strain for biocontrol or rhizoremediation purposes.
Acknowledgments
M.I.R.-G. is the recipient of a grant from the Junta de Andalucía. Work in our laboratory was supported by a PETRI-NBT grant (2001261-P/, MCyT: BIO2003-00515), other grants from Génomica España (Gen2001-4698-C05-03), and the European Commission (QLRT-2000-00170).
We are grateful to D. Lehoux for kindly providing us with plasmid pIVPRO and to O. Revelles for initial modifications in this plasmid. We thank I. Peralta for technical assistance and A. Hurtado for DNA sequencing. Our sincere appreciation goes to M. Espinosa-Urgel for helpful comments on and revision of the manuscript and to K. Shashok for checking the use of English.
REFERENCES
- 1.Abril, M. A., C. Michán, K. N. Timmis, and J. L. Ramos. 1989. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J. Bacteriol. 171:6782-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackerman, R. S., N. R. Cozzarelli, and W. Epstein. 1974. Accumulation of toxic concentrations of methylglyoxal by wild-type Escherichia coli K-12. J. Bacteriol. 119:357-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 5.Bickle, T. A., and D. H. Kruger. 1993. Biology of DNA restriction. Microbiol. Rev. 57:434-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd, A., and A. M. Chakrabarty. 1995. Pseudomonas aeruginosa biofilms: role of the alginate exopolysaccharide. J. Ind. Microbiol. 15:162-168. [DOI] [PubMed] [Google Scholar]
- 7.Boylan, S. A., A. R. Redfield, M. S. Brody, and C. W. Price. 1993. Stress-induced activation of the sigma B transcription factor of Bacillus subtilis. J. Bacteriol. 175:7931-7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caetano-Annoles, G. 1993. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 3:85-92. [DOI] [PubMed] [Google Scholar]
- 9.Cantoni, G. L. 1953. S-adenosylmethionine: a new intermediate formed enzymatically from L-methionine and adenosinetriphosphate. J. Biol. Chem. 204:403-416. [PubMed] [Google Scholar]
- 10.DeFlaun, M. F., B. M. Marshall, E. Kulle, and S. B. Levy. 1994. Tn5 insertion mutants of Pseudomonas fluorescens defective in adhesion to soil and seeds. Appl. Environ. Microbiol. 60:2637-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dekkers, L. C., C. J. Bloemendaal, L. A. de Weger, C. A. Wijffelman, H. P. Spaink, and B. J. Lugtenberg. 1998. A two-component system plays an important role in the root-colonizing ability of Pseudomonas fluorescens strain WCS365. Mol. Plant-Microbe Interact. 11:45-56. [DOI] [PubMed] [Google Scholar]
- 12.Dennis, J. J., and G. J. Zylstra. 1998. Improved antibiotic-resistance cassettes through restriction site elimination using Pfu DNA polymerase PCR. BioTechniques 25:772-774, 776. [DOI] [PubMed] [Google Scholar]
- 13.De Weert, S., H. Vermeiren, I. H. Mulders, I. Kuiper, N. Hendrickx, G. V. Bloemberg, J. Vanderleyden, R. De Mot, and B. J. Lugtenberg. 2002. Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol. Plant-Microbe Interact. 15:1173-1180. [DOI] [PubMed] [Google Scholar]
- 14.De Weger, L. A., C. I. van der Vlugt, A. H. Wijfjes, P. A. Bakker, B. Schippers, and B. J. Lugtenberg. 1987. Flagella of a plant-growth-stimulating Pseudomonas fluorescens strain are required for colonization of potato roots. J. Bacteriol. 169:2769-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Espinosa-Urgel, M., and J. L. Ramos. 2001. Expression of a Pseudomonas putida aminotransferase involved in lysine catabolism is induced in the rhizosphere. Appl. Environ. Microbiol. 67:5219-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espinosa-Urgel, M., and J. L. Ramos. 2004. Cell density-dependent gene contributes to efficient seed colonization by Pseudomonas putida KT2440. Appl. Environ. Microbiol. 70:5190-5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Espinosa-Urgel, M., and M. I. Ramos-Gonzalez. 2004. In vivo gene expression: the IVET system, p. 351-366. In J. L. Ramos (ed.), Pseudomonas, vol. I. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 18.Franklin, F. C., M. Bagdasarian, M. M. Bagdasarian, and K. N. Timmis. 1981. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc. Natl. Acad. Sci. USA 78:7458-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greene, R. C. 1957. Incorporation of the carbon chain of methionine into spermidine. J. Am. Chem. Soc. 79:3929. [Google Scholar]
- 20.Handfield, M., H. P. Schweizer, M. J. Mahan, F. Sanschagrin, T. Hoang, and R. C. Levesque. 1998. ASD-GFP vectors for in vivo expression technology in Pseudomonas aeruginosa and other gram-negative bacteria. BioTechniques 24:261-264. [DOI] [PubMed] [Google Scholar]
- 21.Hoang, T. T., S. Williams, H. P. Schweizer, and J. S. Lam. 1997. Molecular genetic analysis of the region containing the essential Pseudomonas aeruginosa asd gene encoding aspartate-β-semialdehyde dehydrogenase. Microbiology 143:899-907. [DOI] [PubMed] [Google Scholar]
- 22.Hopper, D. J., and R. A. Cooper. 1971. The regulation of Escherichia coli methylglyoxal synthase; a new control site in glycolysis? FEBS Lett. 13:213-216. [DOI] [PubMed] [Google Scholar]
- 23.Keith, L. M., and C. L. Bender. 2001. Genetic divergence in the algT-muc operon controlling alginate biosynthesis and response to environmental stress in Pseudomonas syringae. DNA Sequence 12:125-129. [DOI] [PubMed] [Google Scholar]
- 24.Khan, M. A., and R. E. Isaacson. 2002. Identification of Escherichia coli genes that are specifically expressed in a murine model of septicemic infection. Infect. Immun. 70:3404-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolter, R., M. Inuzuka, and D. R. Helinski. 1978. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell 15:1199-1208. [DOI] [PubMed] [Google Scholar]
- 26.Kuiper, I. 2001. Ph.D. thesis. Molecular characterization of root colonizing Pseudomonas strains for rhizoremediation. Leiden University, Leiden, The Netherlands.
- 27.Lessie, T. G., and P. V. Phibbs, Jr. 1984. Alternative pathways of carbohydrate utilization in pseudomonads. Annu. Rev. Microbiol. 38:359-388. [DOI] [PubMed] [Google Scholar]
- 28.Lugtenberg, B. J., and G. V. Bloemberg. 2004. Life in the rhizosphere, p. 403-430. In J. L. Ramos (ed.), Pseudomonas, vol. I. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 29.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1993. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259:686-688. [DOI] [PubMed] [Google Scholar]
- 30.Matthysse, A. G., H. A. Yarnall, and N. Young. 1996. Requirement for genes with homology to ABC transport systems for attachment and virulence of Agrobacterium tumefaciens. J. Bacteriol. 178:5302-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 32.Molina, L., C. Ramos, E. Duque, M. C. Ronchel, J. M. García, L. Wyke, and J. L. Ramos. 2000. Survival of Pseudomonas putida KT2440 in soil and in the rhizosphere of plants under greenhouse and environmental conditions. Soil Biol. Biochem. 32:315-321. [Google Scholar]
- 33.Morris, V. L., D. P. Jackson, M. Grattan, T. Ainsworth, and D. A. Cuppels. 1995. Isolation and sequence analysis of the Pseudomonas syringae pv. tomato gene encoding a 2,3-diphosphoglycerate-independent phosphoglyceromutase. J. Bacteriol. 177:1727-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murashige, T., and F. Skoog. 1962. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15:473-497. [Google Scholar]
- 35.Nakazawa, T. 2002. Travels of a Pseudomonas, from Japan around the world. Environ. Microbiol. 4:782-786. [DOI] [PubMed] [Google Scholar]
- 36.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. Chris Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. A. Eisen, K. N. Timmis, A. Dusterhoft, B. Tummler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 37.Oehrle, N. W., D. B. Karr, R. J. Kremer, and D. W. Emerich. 2000. Enhanced attachment of Bradyrhizobium japonicum to soybean through reduced root colonization of internally seedborne microorganisms. Can. J. Microbiol. 46:600-606. [DOI] [PubMed] [Google Scholar]
- 38.Rainey, P. B. 1999. Adaptation of Pseudomonas fluorescens to the plant rhizosphere. Environ. Microbiol. 1:243-257. [DOI] [PubMed] [Google Scholar]
- 39.Rainey, P. B., D. M. Heithoff, and M. J. Mahan. 1997. Single-step conjugative cloning of bacterial gene fusions involved in microbe-host interactions. Mol. Gen. Genet. 256:84-87. [DOI] [PubMed] [Google Scholar]
- 40.Ramos, J. L. (ed.). 2004. Pseudomonas. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 41.Ren, D., L. A. Bedzyk, S. M. Thomas, R. W. Ye, and T. K. Wood. 2004. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 64:515-524. [DOI] [PubMed] [Google Scholar]
- 42.Ronchel, M. C., and J. L. Ramos. 2001. Dual system to reinforce biological containment of recombinant bacteria designed for rhizoremediation. Appl. Environ. Microbiol. 67:2649-2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryan, P. R., E. Delhaize, and D. L. Jones. 2001. Function and mechanism of organic anion exudation from plant roots. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52:527-560. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 45.Samuelson, J. C., F. Jiang, L. Yi, M. Chen, A. Kuhn, and R. E. Dalbey. 2001. Function of YidC for the insertion of M13 procoat protein in E. coli: translocation of mutants that show differences in their membrane potential dependence and Sec-requirement. J. Biol. Chem. 276:34847-34852. [DOI] [PubMed] [Google Scholar]
- 46.Samuelson, J. C., M. Chen, F. Jiang, I. Möller, M. Wiedmann, A. Kuhn, G. J. Phillips, and R. E. Dalbey. 2000. YidC mediates membrane protein insertion in bacteria. Nature 406:637-641. [DOI] [PubMed] [Google Scholar]
- 47.Silby, M. W., and B. L. Stuart. 2004. Use of in vivo expression technology to identify genes important in growth and survival of Pseudomonas fluorescens Pf0-1 in soil: discovery of expressed sequences with novel genetic organization. J. Bacteriol. 186:7411-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 49.van de Mortel, M., and L. J. Halverson. 2004. Cell envelope components contributing to biofilm growth and survival of Pseudomonas putida in low-water-content habitats. Mol. Microbiol. 52:735-750. [DOI] [PubMed] [Google Scholar]
- 50.Vilchez, S., L. Molina, C. Ramos, and J. L. Ramos. 2000. Proline catabolism by Pseudomonas putida: cloning, characterization, and expression of the put genes in the presence of root exudates. J. Bacteriol. 182:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu, J., A. Peñaloza-Vazquez, A. M. Chakrabarty, and C. L. Bender. 1999. Involvement of the exopolysaccharide alginate in the virulence and epiphytic fitness of Pseudomonas syringae pv. syringae. Mol. Microbiol. 33:712-720. [DOI] [PubMed] [Google Scholar]