Abstract
Thioredoxins are important thiol-reactive proteins. Most knowledge about this class of proteins is derived from proteome studies, and little is known about the global transcriptional response of cells to various thioredoxin levels. In Bacillus subtilis, thioredoxin A is encoded by trxA and is essential for viability. In this study, we report the effects of minimal induction of a strain carrying an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible trxA gene (ItrxA) on transcription levels, as determined by DNA macroarrays. The effective depletion of thioredoxin A leads to the induction of genes involved in the oxidative stress response (but not those dependent on PerR), phage-related functions, and sulfur utilization. Also, several stationary-phase processes, such as sporulation and competence, are affected. The majority of these phenotypes are rescued by a higher induction level of ItrxA, leading to an approximately wild-type level of thioredoxin A protein. A comparison with other studies shows that the effects of thioredoxin depletion are distinct from, but show some similarity to, oxidative stress and disulfide stress. Some of the transcriptional effects may be linked to thioredoxin-interacting proteins. Finally, thioredoxin-linked processes appear to be conserved between prokaryotes and eukaryotes.
Thioredoxins are relatively small (∼13-kDa), heat-stable, ubiquitous proteins, characterized by the presence of a conserved amino acid motif, WCGPC. In the bacterial cell, thioredoxins are involved in a number of important reactions (for reviews, see references 1, 14, and 22). First of all, they are implied in the maintenance of the intracellular redox state. Secondly, thioredoxins are involved in the response of cells to oxidative stress. Thirdly, this class of proteins can serve as efficient oxidoreductases of disulfides in low-molecular-weight compounds and proteins through thiol-disulfide exchange reactions. In this respect, thioredoxins are thought to be key players in the prevention of the formation of disulfide bonds, especially for cytoplasmic proteins. Finally, thioredoxins can exert an effect through functional association with other proteins. Many of these functions depend on the reducing potential of thioredoxin. Recycling of this reducing potential of thioredoxins is mediated by a thioredoxin reductase in a NADPH-dependent manner.
Thioredoxin A (TrxA).
In the gram-positive bacterium Bacillus subtilis, thioredoxin A is encoded by the trxA gene. Its product is essential for cell viability (33) and is induced by multiple kinds of stresses (54). Expression of the gene is driven from a sigma A (housekeeping) or sigma B (general stress) promoter (54). Although thioredoxin in several other organisms has been rather well studied, little is known about the B. subtilis protein and its function. Interestingly, none of the other nine thioredoxin-like genes (ybdE [skfH {23}], ydbP, ydfQ, ykuV, ykvV [spoIVH/stoA {16, 29, 58}], yneN, yosR, ytpP, and yusE) encoded on the genome of this bacterium is essential, indicating a critical role for thioredoxin A in the cell (J.-Y. F. Dubois, unpublished observations).
Aim of this study.
Most of the knowledge about thioredoxin A is inferred from proteome studies with organisms other than B. subtilis. Furthermore, little is known about the possible pleiotropic effects of thioredoxin depletion on global transcription levels. In this study, we assessed the effects of thioredoxin A levels on transcription when B. subtilis is grown in minimal medium, by comparing the gene expression profiles of an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible trxA strain (ItrxA) with those of a wild-type strain in the early stationary growth phase. This analysis was primed by the observation that several stationary-phase processes in the ItrxA mutant seemed affected in a phenotype screen. The minimal medium is frequently used in B. subtilis research and contains a small amount of redox-active compounds, if any. By induction with minimal amounts of IPTG (25 μM), thioredoxin A is effectively depleted, whereas induction with 100 μM of IPTG restores the protein to an approximately wild-type level (this study; J.-Y. F. Dubois, unpublished observations). To our knowledge, this is the first assessment of the transcriptional effects of thioredoxin depletion in bacteria, which contributes to the understanding of the cellular functions of the thioredoxin protein.
MATERIALS AND METHODS
Strains.
The transcriptional effects of thioredoxin depletion were investigated by use of a derivative of B. subtilis 168 (trpC2) (35) containing an IPTG-inducible trxA gene that was essentially constructed as described by Scharf et al. (54) (laboratory strain collection; J.-Y. F. Dubois). In order to compare the in vivo redox state of the cytoplasm of this strain with different induction levels of IPTG, a xylose-inducible redox-sensitive green fluorescent protein (GFP) was introduced in the amyE locus. This strain was constructed as follows. By use of primers rbs-gfp-f (5′-GCTCTAGAAGGAGGGCTGAATTCATGAGTAAAGGAGA-3′) and GFP-R (5′-CTCGGATCCTTATTATTTGTATAG-3′), a PCR was performed on pRSETb encoding a previously described redox-sensitive GFP (15, 27). The resulting fragment was cleaved with XbaI/BamHI (the sites are underlined in the primer sequences) and ligated into SpeI/BamHI-digested pX (32). The resulting plasmid (pXro2GFP) was checked by restriction and introduced by transformation into B. subtilis 168. Transformants were selected on plates containing appropriate antibiotics and screened for an amyE-negative phenotype on starch-containing plates. Subsequently, the ItrxA mutation was introduced by transformation with chromosomal DNA from the ItrxA strain. Transformants were checked for IPTG dependency because of the unstable nature of the ItrxA mutation.
Media and growth conditions.
B. subtilis cells were grown in minimal medium (61) supplemented with IPTG. The ItrxA strain was grown with 25 and 100 μM of IPTG, and the control strain was cultured in medium containing 25 μM IPTG, since, for this condition, the largest effects were expected based on the phenotypic screen. Twenty-five milliliters of medium was inoculated to an optical density at 600 nm of 0.01 from an overnight culture and incubated at 37°C under vigorous shaking. Cultures were continuously monitored for optical density at 600 nm, and cells were harvested 3 h after the transition point to stationary growth phase. For the in vivo comparison of the redox state of the ItrxA strain with a range of IPTG induction levels, cells were grown in minimal medium in which glucose was replaced by fructose in order to alleviate the glucose repression of the xylose-inducible promoter (32) of the pXro2GFP plasmid. This medium was supplemented with 0.5% xylose and 25 to 500 μM of IPTG to induce the expressions of ro2GFP and ItrxA, respectively.
To determine the effects of cysteine on the growth of the ItrxA strain, cells were grown in chemically defined medium (CDM) supplemented with 25 or 100 μM of IPTG. CDM was based on B. subtilis minimal salts (per liter) as follows: 2 g K2SO4, 10.8 g K2HPO4, 6 g KH2PO4, 1 g Na citrate, and 0.02 g MgSO4. After the adjustment of the pH to 7.0 and sterilization, the following components were added to complete the CDM: 0.5% glucose, 0.1% sodium glutamate, 50 μM MnCl2, 20 mg/liter tryptophan. For cysteine-containing medium, 20 mg/liter cysteine was added.
RNA isolation, cDNA preparation, and hybridization.
RNA was isolated by spinning down the cells from 4 ml of culture and resuspending the pellet in 300 μl of ice-cold starvation medium (61). The cell suspension was added to a 1.5-ml screw-cap Eppendorf tube containing 1.5 g of glass beads (75 to 150 μm; Sigma), 0.5 ml phenol-chloroform-isoamyl alcohol (12:12:1), 50 μl 10% sodium dodecyl sulfate (SDS), and 50 μl 3 M sodium acetate (pH 5.2). All solutions were prepared with diethylpyrocarbonate-treated water. After being vortexed, the tube was frozen in liquid nitrogen and stored at −80°C. Cells were broken in a Bio101 FastPrep machine (QBiogene) for 45 seconds at speed 6. Following 5 min of centrifugation (10,000 rpm, 4°C; Eppendorf centrifuge), the water phase (0.4 ml) was transferred to a clean tube containing 0.4 ml chloroform. After a vortexing and centrifugation (2 min, 14,000 rpm, 4°C; Eppendorf centrifuge), the water phase was transferred to a clean tube, and the RNA was isolated with a High Pure RNA isolation kit (Roche). RNA was eluted in 50 μl elution buffer and quantified with GeneQuant (Amersham). RNA integrity was checked with a BioAnalyzer 2100 (Agilent) according to the instructions of the supplier.
The reverse transcription and purification of cDNA were carried out as described previously (26). 32P-labeled cDNA was hybridized to Bacillus subtilis Panorama arrays (Sigma-Genosys) as described by the manufacturer. After hybridization and a washing, Cyclone phosphorimager screens (Packard) were exposed for 46 to 48 h. The Cyclone readouts were analyzed with Array-Pro Analyzer 4.5 (Media Cybernetics). After background subtraction, duplicate spots were averaged, and the signal was normalized against the total signal of all open reading frames on the array.
Data analysis and visualization.
The normalized array data were subjected to a statistical analysis by using Cyber-T, a program based on a t test variant combined with a Bayesian statistical framework as described previously (26), yielding for each gene a P value that indicates the probability of differential expression. The raw and normalized data for the complete gene sets can be downloaded from the MolGen RUG website (http://molgen.biol.rug.nl/publication/trx_data/). Genes with a P of <0.025 and a >2-fold up-regulation or down-regulation were considered to be significantly affected. Spots were associated with gene names by use of the spreadsheet “B. subtilis Array information.xls” provided by Sigma-Genosys. Gene descriptions were derived from this file and checked against the SubtiList database (http://genolist.pasteur.fr/SubtiList/) and PubMed website (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi) for newer annotations. Some affected genes without descriptions in either of these sources and a >3-fold change in expression were used as queries in an online BlastP search (http://www.ncbi.nlm.nih.gov/BLAST/) and checked for conserved domains with the CD-search option. Results from the analysis were visualized on a linear genome map by using Genome2D (2).
Flow cytometric analysis.
Cells were diluted 12.5 × to 50 × in 0.2 μM filtered starvation medium (26) and directly analyzed on a Coulter Epics XL-MCL flow cytometer (Beckman Coulter, Mijdrecht, The Netherlands) operating an argon laser at 488 nm. For each sample, 20,000 cells were analyzed. GFP signals were collected through a fluorescein isothiocyanate filter with the photomultiplier voltage set between 700 and 800 V. Data were captured by using EXPO32 software (Beckman Coulter) and further analyzed by using WinMDI 2.8 (http://facs.scripps.edu/software.html). Figures were prepared for publication by using WinMDI 2.8, Microsoft Excel 2000, and Corel Graphics Suite 11.
SDS-polyacrylamide gel electrophoresis and Western blotting analyses.
The presence of TrxA in cell lysates was assayed by Western blotting. For this purpose, cellular or secreted proteins were separated by SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Roche Molecular Biochemicals). Subsequently, TrxA was detected with specific polyclonal antibodies. The detection of bound antibodies was performed with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G and the Super Signal West Dura extended-duration substrate (Pierce).
RESULTS AND DISCUSSION
Changes in global transcription levels.
In this study, we present the first overview of the global effects of thioredoxin A levels on transcription in the gram-positive bacterium B. subtilis. To this end, we made use of the ItrxA strain harboring an IPTG-inducible variant of trxA, which encodes thioredoxin A. Induction with 25 μM of IPTG permits wild-type growth, yet levels of thioredoxin A are below the detection limits of a Western blot. In contrast, when the strain is induced with more than 100 μM of IPTG, thioredoxin A levels are similar to those of the wild type (Fig. 1). Hence, a comparison of the transcription profiles of the ItrxA strain grown in 25 μM or 100 μM of IPTG to those of a wild-type B. subtilis strain allows the assessment of the transcriptional response to thioredoxin A depletion and indicates if these can be reverted to wild-type profiles by inducing to near-wild-type levels of thioredoxin A.
FIG. 1.
Western blot detection of TrxA protein in wild-type B. subtilis 168 and the ItrxA strain grown in the presence of 25 or 100 μM IPTG.
A total number of 636 genes showed a >2-fold change in transcription level, combined with a P value below 0.025 in at least one of the conditions tested. This list is available in the supplemental material (see Table S1 in the supplemental material). The number of affected genes corresponds to about 15% of all the open reading frames of the B. subtilis genome. Of these, 435 were affected only in the ItrxA cells induced with 25 μM of IPTG (hereafter referred to as ItrxA 25 cells), in which thioredoxin A is effectively depleted, and these are hereafter referred to as class I genes. A small group, class II (consisting of 63 genes), was affected only in the ItrxA cells induced with 100 μM of IPTG (hereafter referred to as ItrxA 100 cells). The third group (class III), of 138 genes, contains genes that have P values below 0.025 for both conditions tested and show a >2-fold change in transcription in at least one of the ItrxA transcription profiles. In fact, the majority of the genes from this class shows a >1.5-fold restoration of transcription levels in the direction of a wild-type strain in the ItrxA 100 cells, suggesting that the phenotype associated with thioredoxin depletion might be rescued by higher induction levels of the ItrxA strain. Remarkably, only a small group of genes shows an increase in the difference (n-fold) in expression in the ItrxA 100 cells compared to that in the ItrxA 25 cells. Together, these results indicate that the thioredoxin A levels, rather than constitutive expression, cause the transcriptional changes in the ItrxA mutant.
Thioredoxin-related proteins.
It can be expected that B. subtilis, in response to the depletion of thioredoxin A, caused by the minimal induction of ItrxA, employs a strategy to counter the effects of the resulting stress. Other thioredoxins or thioredoxin-related proteins are excellent candidates for this response. Indeed, in the ItrxA 25 cells, transcription of the thioredoxin B gene, which encodes thioredoxin reductase, is about eightfold increased. Thioredoxin reductase (TrxB) is required to recycle the reducing potential of thioredoxin, and an increased transcription of trxB could therefore indicate a high ratio of oxidized TrxA compared to its reduced form or a low availability of TrxA in general. In the ItrxA 100 cells, the difference between the transcription of trxB in the wild type and that in the mutant is reduced to 2.3-fold. Another gene, ydbP, encodes a thioredoxin A-like protein. The transcription of this gene is increased moderately in the ItrxA 25 cells (2.3-fold) but shows no significant difference in expression level in the ItrxA 100 cells. Interestingly, a gene encoding a protein with high homology to glutaredoxin (PFAM000462 or COG0695), ytnI, is up-regulated about 450-fold in ItrxA 25 cells, whereas the difference for the wild type in the ItrxA 100 cells is only 5-fold. The ytnI gene is part of the putative ytmI operon, encoding a transporter for l-cystine uptake (11). It has also been reported that ytnI (renamed moxB in that study, for methionine sulfoxide oxidase) is part of a transcript containing ytmO (56). The genes in this region show similar up-regulation (see “Stationary-phase processes” below). As B. subtilis lacks the enzymes to synthesize glutathione (35), an increase of the glutaredoxin-like protein YtnI could serve to reduce oxidized disulfides. Alternatively, it can be envisaged that glutathione from the medium is taken up and used as cellular reductant, as has been suggested for Lactococcus lactis SK11 (39). In this case, the YtnI could serve to regenerate functional glutathione.
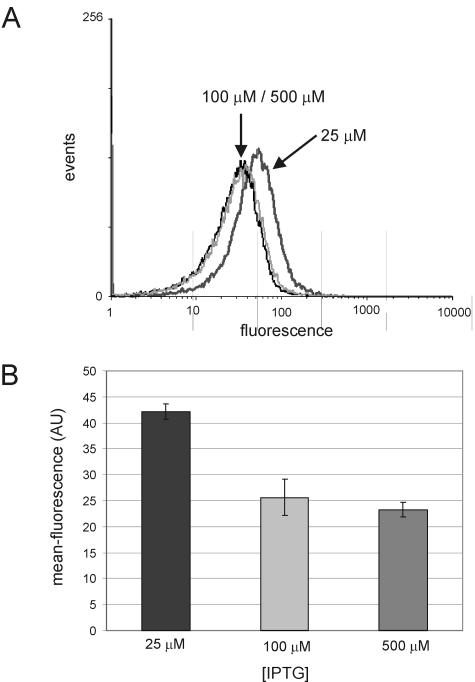
Thioredoxin A is thought to be a key player in maintaining the intracellular redox state. Since no obvious effect of minimal induction of trxA on growth or viability was observed, we wondered whether the observed up-regulation of the thioredoxin-like genes might restore the intracellular redox state to levels that approximate those observed for wild-type B. subtilis. To investigate this, a xylose-inducible redox-sensitive variant of GFP was introduced in the amyE locus of the B. subtilis chromosome. Previous work has demonstrated that the fluorescent properties of this variant of GFP change depending on the bonding state of the cysteines that have been introduced (15, 27). The mean fluorescence of the cells, when analyzed by flow cytometry, increases as the cytoplasm becomes more oxidized. From Fig. 2, it can be seen that fluorescence in the ItrxA 25 cells is significantly higher than that in the same strain induced with 100 or 500 μM IPTG. Thus, despite the up-regulation of thioredoxin-like genes, the cytoplasm in the ItrxA 25 cells is more oxidized than that in the ItrxA 100 cells. There is no appreciable difference between the ItrxA 100 and 500 cells, consistent with biochemical data that show that levels of TrxA are similar to those of the wild type in both cases (data not shown).
FIG. 2.
Flow cytometric analysis of xylose-induced expression of a redox-sensitive GFP in the ItrxA mutant upon induction with 25, 100, or 500 μM of IPTG. (A) Fluorescence distribution of a typical experiment; (B) mean fluorescence of a culture based on flow cytometric data.
The results described above demonstrate that although the depletion of thioredoxin A leads to increased transcription of trxB, ydbP, and ytnI encoding (putative) thiol-reactive proteins, this up-regulation is insufficient to restore the cytoplasmic redox state of TrxA-depleted cells to that observed in cells with wild-type levels of thioredoxin. Thioredoxin A thus plays a critical role in maintaining the intracellular redox state of B. subtilis.
Redox catalytic proteins.
For several enzymes, it has been demonstrated that transiently formed disulfide bridges are part of their catalytic cycle. These proteins include ribonucleotide reductases, methionine sulfoxide reductases, and phosphoadenosyl phosphosulfate (PAPS) reductases. The thiol-dependent oxidoreductase activity of thioredoxin is thought to be of importance for the functionality of these enzymes. Therefore, we examined the effects of thioredoxin depletion on the genes coding for these proteins. In B. subtilis, nrdEF codes for ribonucleoside diphosphate reductase (55). A putative homologue of these genes, yosP, is also found in the ItrxA results. The genes behave like class I genes in the transcriptome experiments with the ItrxA strain, showing a 3.2- to 3.5-fold up-regulation in ItrxA 25 cells. Methionine sulfoxide reductases are encoded by the msrA gene (28) and its putative homologue yppQ. Both genes show a >2-fold up-regulation only in the ItrxA 25 cells. cysH, finally, was identified as the gene encoding the PAPS sulfotransferase or thioredoxin-dependent PAPS reductase (9, 41). Upon depletion of thioredoxin A, this gene is 7.7-fold up-regulated.
Phage-related functions.
A single group of genes stood out clearly, being strongly induced upon thioredoxin A depletion. In the ItrxA 25 cells, operons and genes with annotated phage-related functions (xhlAB, yqaK, and the xkd, xtm, xtr, yqb, yqc, and yqx operons), in general, show an up-regulation of 2- to 72-fold, whereas these genes in the ItrxA 100 cells are not affected. Most of the genes belong to the PBSX prophage that is present in the B. subtilis chromosome (63). Thus, it seems that thioredoxin A depletion stress is a novel trigger for the induction of the PBSX prophage located in the B. subtilis genome. Despite the transcriptional response of the PBSX genes, we observed no lysis of ItrxA 25 cells in the time frame of our transcriptome study or under any other condition of growth tested. At least partially, the induction of the prophage genes can be explained by the fact that PBSX induction is part of the so-called SOS response (44). However, the severity of the transcriptional up-regulation of (pro)phage-related genes in general is much greater than those of the other SOS-related genes (uvrA, dinB, and tagC), indicating that another regulatory mechanism may be involved. A known transcriptional repressor of the PBSX prophage, Xre (43), contains no cysteines. A positive regulator of PBSX, Xpf (44), contains a single cysteine residue, but this residue is predicted to be in a nonbonded state, based on a CysPred analysis (http://gpcr.biocomp.unibo.it/cgi/predictors/cyspred/pred_cyspredcgi.cgi). This indicates that, if thioredoxin should have an interaction with either one of these regulators, it is not likely to be due to thiol reactivity. Together, these results indicate that thioredoxin stress-dependent induction of (pro)phage genes may rely on a yet unknown mechanism in addition to the SOS response.
Stationary-phase processes.
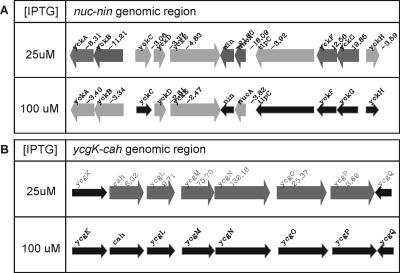
Among the genes that are most strongly repressed in the ItrxA 25 cells, a large group is formed by those involved in competence for genetic transformation. The expression of late competence genes, and therefore of competence development as a whole, is governed by the key regulator ComK (60). The macroarray results from the ItrxA strain clearly reveal that the low expression of the late competence genes in the ItrxA 25 cells is due to low expression levels of the gene encoding the key regulator for competence, ComK. The levels of comK gene expression are >8.5-fold lower in the ItrxA 25 cells than those in the wild-type cells, which is similar to the results obtained with a comK-disrupted strain (26). All but three genes of the so-called core ComK regulon, defined in the same study, were similarly repressed in the ItrxA 25 cells. Moreover, several other genes for which ComK dependency was demonstrated were also represented in the results from this study (Table 1). In the ItrxA 100 cells, transcription levels of most of the late competence genes were comparable to those of the wild-type cells or showed at least a substantial shift towards wild-type levels. Consistent with these findings, transformation frequencies of ItrxA 25 cells as tested in a standard assay (26) were extremely low, whereas ItrxA 100 cells were readily transformed (data not shown). Further characterization of the level at which thioredoxin A acts in the competence signal transduction cascade is currently in progress (C. Eschevins, personal communication). A comparison of the results from the DNA macroarrays carried out with a comK disruption mutant (26) and the ItrxA strain led to some additional interesting observations. For instance, it was reported that the nucA-nin region of the genome of B. subtilis contains several ComK-dependent genes, yet based on the ComK-regulon study, it was difficult to comment on transcriptional organization, possibly because of the presence of antisense RNA detection (2, 26) and indirect effects. In the ItrxA arrays, most of the genes in this region behave like ComK-regulated genes (i.e., severely down-regulated in the ItrxA 25 cells) (Fig. 3 and Table 1). However, two genes, yckG and yckF (hxlA and hxlB [66]), show opposite behavior. This indicates that these genes do not strictly depend on ComK for their regulation or that they are subject to regulation by another factor. A logical candidate (yckH, or hxlR) which has been shown to positively regulate these genes (66) behaves like a ComK-dependent gene. The results thus suggest that the putative ComK-dependent induction of yckGF (hxlAB) may be through yckH (hxlR). Similarly, the genes dinB, tagC, trpC, yorB, and yqeN do not behave like classical ComK-dependent genes. In the cases of the first two, this may be because these are members of the SOS regulon (65), which is induced by oxidative stress. yqeN, whose product shows similarity to DNA polymerase III, is a gene located downstream of comEC. It was suggested that the observed ComK dependency was due to transcriptional read-through from the comE operon (2).
TABLE 1.
ComK-regulated genes affected in the ItrxA mutanta
| Similarity of gene to comK | Classb | Gene namec |
|---|---|---|
| Similar | I | bmrU |
| III | comC | |
| III | comEABC | |
| III | comER | |
| III | comFABC | |
| III | comGABCDEFG | |
| I | comK | |
| I | cwlJ | |
| II | maf | |
| III | med | |
| I | nin | |
| III | nucA | |
| I | rapH | |
| I | sacX | |
| III | sacY | |
| III | spoIIB | |
| I | sucC | |
| I | tlpC | |
| III | ybdK | |
| I | ybyB | |
| III | ycbP | |
| III | ycbR | |
| III | yckBA | |
| III | yckCDE | |
| I | yckH | |
| III | yhxD | |
| III | ykzA | |
| I | yqgJ | |
| III | yqzE | |
| III | yrhL | |
| III | ysxA | |
| I | yvrNM | |
| I | yvrP | |
| III | ywfL | |
| III | ywfM | |
| III | ywpH | |
| I | yxiP | |
| III | yyaF | |
| Not similar | I | dinB |
| I | tagC | |
| I | trpC | |
| I | yckF | |
| I | yckG | |
| I | yorB | |
| III | yqeN |
The study of Hamoen and colleagues (26) was used as a reference because of similarities in experimental procedures.
I, significantly affected in ItrxA 25; II, significantly affected in ItrxA 100; III, significantly affected in both ItrxA 25 and 100.
Genes indicated in bold belong to the core ComK regulon (26).
FIG. 3.
(A) The transcriptional response of the nucA-nin genome region in the ItrxA mutant cells; (B) the transcriptional response of the cah-ycg genome region in the ItrxA mutant cells. Thin black arrows indicate no significant difference (n-fold) from the wild type. Thick gray arrows indicate a transcriptional response upon thioredoxin A depletion (P < 0.025). Differences (n-fold) in expression levels are given next to the gene names.
In our list of genes, we also noted a transcriptional effect on several genes involved in sporulation, of which the regulatory network is closely intertwined with competence development. However, the directionality of the response of the sporulation genes was not conserved. For instance, the gene encoding the stage II protease spoIIGA, which activates sigma E (31), shows a consistent up-regulation of about 12-fold in both the ItrxA 25 and the ItrxA 100 cells. Similarly, spo0F, spoIIP, and spoVD are up-regulated. In contrast, spoIIB, spoIIIAD, and spoIVCB (encoding the N-terminal part of sigma K) are down-regulated. Therefore, a standard sporulation test was performed with sporulation medium, which revealed that in the ItrxA 25 cells, no chloroform-resistant spores are formed (data not shown). Most likely, TrxA influences spore formation at the later stages of sporulation. Consistent with this hypothesis, it was recently reported that the thiol-disulfide oxidoreductase YkvV (SpoIVH or StoA) (16, 29, 58) is essential for cortex formation, and it was suggested that its action depends on thioredoxin A (16). Depletion of TrxA could lead to an altered functionality of this protein and the loss of a chloroform-resistant cortex.
Sulfur uptake and utilization.
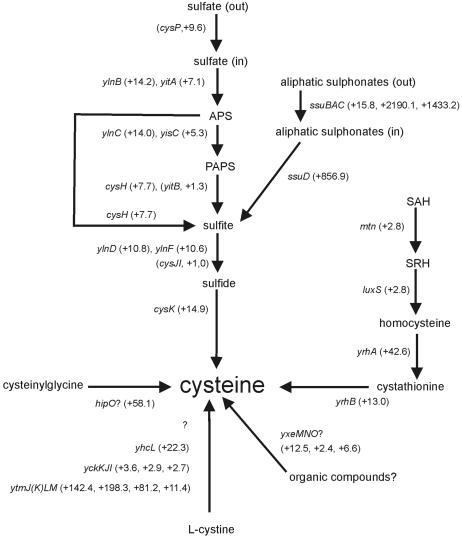
In the list of genes affected in the ItrxA 25 cells, those involved in cysteine metabolism and the closely related sulfur metabolism are clearly up-regulated (for a review, see reference 24) (summarized in Fig. 4). The effects extend to multiple pathways, such as the uptake and utilization of sulfates (ylnBC [now renamed sat and cysC], cysH, ylnDF, and cysK) and sulfonates (the ssu operon), and the synthesis of cysteine from S-adenosyl homocysteine via S-ribosyl homocysteine, homocysteine, and cystathionine (requiring mtn [also known as mntN], luxS, and yrhAB). In addition, recently identified uptake systems of cystine (the oxidized form of cysteine) and, possibly, organic sulfur-containing compounds (ytmJKLM [also known as tcyJKLMN], yhcL [tcyP], yckKJI [tcyABC], and yxeMNO) show a similar pattern (11; I. Martin-Verstraete, personal communication). ytmJKLM has also been implied in methionine sulfoxide degradation by Sekowska and coworkers and was renamed mdgOPQT in that study (56). In fact, only three genes from the sulfur uptake and utilization pathways are not significantly affected: cysP and cysJI. The cysP gene product is the transporter involved in the uptake of sulfate from the environment (42). Upon closer inspection, this gene showed a rather large up-regulation, but it was ignored because of the cutoff settings imposed in the experiment (P < 0.025). The products of cysJI (previously known as yvgRQ) are responsible for the conversion of the sulfite pool in the cell into sulfide. This operon is under the additional control of CysL (YwfK) (25). The differential regulation of cysJI and the other sulfur-related genes suggests that an additional regulator involved in this process may exist and that CysL is probably not a target for thioredoxin A in B. subtilis.
FIG. 4.
The effect of thioredoxin A depletion on the transcription of genes involved in sulfur uptake and utilization. Gene names are indicated in italics. Alternative gene names for some of the genes are given in the text (see “Sulfur uptake and utilization”). Regulation (n-fold) of genes in ItrxA 25 cells compared to that of the wild type is given in parentheses next to each gene name. When gene names are in parentheses too, P values did not meet our criteria. SAH, S-adenosyl homocysteine; SRH, S-ribosyl homocysteine. Question marks indicate the involvement of an unknown factor or a postulated function.
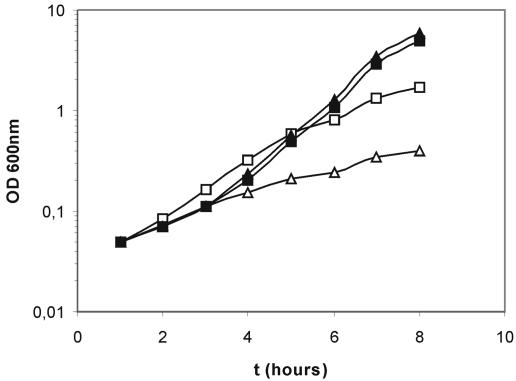
In a number of studies, a link between oxidative stress and auxotrophy for certain amino acids has been reported (6, 8, 12). Most notably, auxotrophy for cysteine in Escherichia coli has been attributed to the leakage of sulfite from the cells (7). Based on proteome studies with other organisms (see the supplemental material), several candidate proteins interacting with thioredoxin and involved in cysteine metabolism were identified (cysteine synthetase, sulfite reductase, and sulfate adenylyl transferase). CysK, cysteine synthetase (59), is suggested to be a central checkpoint for sulfur metabolism (I. Martin-Verstraete, personal communication), and reduced activity of this enzyme is thought to be responsible for the leakage of sulfite from the cells (8). In addition, Berndt and colleagues recently reported that the B. subtilis CysH, a 3′-phospho/adenosine-phosphosulfate-sulfonucleotide reductase complex (Fig. 4), can use thioredoxin as a reductant (9). CysJI is the B. subtilis sulfite reductase (25, 59). Although transcription of the genes encoding this enzyme complex in the ItrxA mutants is not affected, this does not exclude an interaction between the protein(s) and thioredoxin A. Finally, a TRX1/2 double mutant of Saccharomyces cerevisiae has been proven to be unable to grow in a defined medium without methionine or cysteine (46). Considering this information, we wondered whether the ItrxA 25 cells would be auxotrophic for cysteine. To test this, we grew the ItrxA strain in a chemically defined medium containing either 25 or 100 μM of IPTG, with or without cysteine (Fig. 5). The results demonstrate that the ItrxA strains with 100 μM of IPTG show similar growth characteristics in the media with and without cysteine. However, the ItrxA 25 cells are severely impaired in growth in this CDM. Note, however, that in the minimal medium used for the array experiment, no significant difference in growth was observed. The growth defect of the ItrxA 25 cells in CDM is partially restored when the cells are grown in the presence of cysteine, yet it does not reach ItrxA 100 levels. The fact that no absolute auxotrophy is observed can be due to the small amount of thioredoxin that is still present in the ItrxA 25 cells (the thioredoxin A gene is essential [33]).
FIG. 5.
Growth of the ItrxA mutant in chemically defined medium with cysteine (squares) or without cysteine (triangles). Medium was supplemented with 25 μM (open symbols) or 100 μM (closed symbols) of IPTG. Times (t) are given in hours after inoculation.
In conclusion, the results from this study suggest a critical role for thioredoxin A in sulfur utilization of B. subtilis. This characteristic might contribute to the essentiality of the TrxA protein.
Comparison with transcriptome results from related stresses in B. subtilis.
The results from the DNA macroarray analysis of ItrxA show substantial similarity to the results from studies on the transcriptional response of B. subtilis to related stresses. Therefore, a more careful comparison seems to be justified.
Thioredoxin A is involved in maintaining the intracellular redox state, which is greatly influenced by the presence or absence of intracellular reactive oxygen species (ROS). ROS in the cell can be formed as a by-product of natively occurring chemical reactions, such as the respiratory chain reactions, or as a result of the exposure of the cells to ionizing radiation or chemicals, such as hydrogen peroxide. Also, thioredoxin A of B. subtilis has been implied in the oxidative stress response (54). In B. subtilis, Mostertz and colleagues performed transcriptome and proteome analyses of cells in response to superoxide and peroxide stresses (oxidative stress) induced by treatment with paraquat and hydrogen peroxide, respectively (45). When the data from the ItrxA strain are compared with the list of 355 genes identified in the study by Mostertz and colleagues, 94 genes were found to be common between the two data sets. Interestingly, these genes do not include the PerR regulon (13, 19) or genes involved in the stringent response (17). On the other hand, we observed a similar effect with respect to some genes from the SOS regulon (dinB, tagC, and uvrA), Fur regulon (ydbN, yuiI, dhbACE, and ykuNOP), and many genes involved in sulfur metabolism.
Thioredoxin A is thought to keep cytoplasmic proteins in a reduced state (53). Loss of the reducing potential by depletion of thioredoxin A could therefore lead to illegitimate disulfide bond formation. This is also observed when cells are treated with thiol-cross-linking agents, such as diamide. Leichert and colleagues performed transcriptome and proteome analyses of B. subtilis upon treatment with diamide (disulfide stress) (37). A comparison of the data from the disulfide stress experiments with the data obtained from the ItrxA arrays shows that 170 genes were identified in both experiments. Interestingly, more than half of these are genes with no annotated function (so-called “y-genes”). It is striking that also in this case, some of the major effects of disulfide stress, such as induction of the canonical oxidative stress genes (PerR regulon) and induction of class III heat shock and stringent-response genes, are absent from the ItrxA data set.
Recently, it was reported that transcriptional control by the protein Spx is induced by thiol-specific oxidative stress (49). spx is a gene that was initially identified as a bypass mutation for the competence-deficient phenotype of a clpP or clpX mutant strain (47). Furthermore, Nakano and colleagues identified genes that are affected by an interaction between Spx and RNA polymerase, and it was shown that Spx is responsible for the induction of genes involved in thiol homeostasis, including the genes encoding thioredoxins A and B (50). The transcription activation of trxA and trxB by Spx was subsequently shown to depend on the formation of an intramolecular disulfide bond under oxidizing conditions, making Spx a redox-sensitive regulator (48). Considering the facts described above, we also made a comparison of the ItrxA macroarray data with the results from the study of Nakano and colleagues. A total of 81 genes from this study are also found in the thioredoxin A array studies. The most striking similarities are the down-regulation of several so-called rap/phr modules, which are involved in cell-cell signaling, and the up-regulation of trxB, ydbP, and the l-cystine transporter yckIJK (tcyABC) (11). It is reasonable to assume that part of the transcriptional response of cells to thioredoxin A depletion, resulting in more oxidizing conditions, is mediated through the redox-sensitive Spx protein.
In Table S1, available in the supplemental material, it is indicated whether or not the genes showing altered transcription patterns in the ItrxA macroarray study were also identified in the related studies discussed above. The comparison is numerically summarized in Table 2. These data indicate that, although there is generally a good correlation between the behaviors of genes in response to diamide, reactive oxygen species, Spx-overproduction, and thioredoxin depletion, the genes react oppositely in some cases. A total of 402 of the affected genes, upon thioredoxin depletion stress, do not seem to be affected by any of the other aforementioned stress conditions. This may be in part due to differences in the experimental setup and criteria for calling a gene differentially expressed but could also indicate that some of the effects are specific for thioredoxin depletion stress. In this respect, it is interesting that an ItrxA spx double-mutant strain displays a severe growth defect while the corresponding single-mutant strains show no growth defects. Moreover, thioredoxin A is indispensable for growth and viability of B. subtilis, whereas Spx is not (33). These findings underscore the view that the effects of the individual ItrxA and spx mutations are not identical. Accordingly, it is well conceivable that the cellular responses to thioredoxin A depletion and the absence of Spx are at least partially different.
TABLE 2.
Comparison of thioredoxin depletion stress with related stresses in B. subtilis
| Type of stressa (no. of genes; %) | Directionb (no. of genes) | No. of responsive ItrxA genes (by concn)c
|
|||
|---|---|---|---|---|---|
| 25
|
100
|
||||
| Down | Up | Down | Up | ||
| Diamide (170; 18) | Down (64) | 38 | 21 | 13 | 5 |
| Up (106) | 9 | 94 | 4 | 13 | |
| ROS (94; 26) | Down (23) | 8 | 4 | 10 | 3 |
| Up (71) | 7 | 62 | 3 | 7 | |
| Spx (81; 29) | Down (49) | 34 | 12 | 8 | 3 |
| Up (32) | 1 | 30 | 0 | 7 | |
| Unique | 164 | 195 | 83 | 64 | |
Numbers and percentages indicate the overlap in the results of the studies on diamide stress (38), oxidative stress (ROS) (46), and Spx overproduction (50) and this study.
Down, lowered expression; up, elevated expression in response to the specific stresses (38, 46, 50).
The values 25 and 100 μM correspond to the concentration of IPTG in the medium. Down and up indicate the directionality of the response in the ItrxA array experiments.
The largest groups of genes unique to thioredoxin stress are those involved in stationary-phase processes (especially competence and sporulation) and the group of prophage and phage-related genes. About half of the genes uniquely identified in this study belong to the group of genes with no defined function, yet many have putative functions which fall into one of the categories already discussed. In Table S2, available in the supplemental material, y-genes for which a function could be inferred or a “cluster of orthologous groups” class was described are summarized. Interestingly, many genes encode proteins with thioredoxin-related functions, such as a predicted oxidoreductase (yvaA), a putative tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase (yrrA, or trmU, on the basis of homology), a flavodoxin (yhcB), a NADH dehydrogenase (yumB), and a protein with similarity to organic hydroperoxide resistance proteins (ykzA). The fact that the transcription of these genes is altered by thioredoxin A levels supports their putative or inferred functions. Further experiments will be needed to confirm the functions of these proteins biochemically, as was recently done for ykzA (now renamed ohrB) (18).
Indirect effects of thioredoxin A on transcription.
So far, thioredoxin has not been reported to act as a transcription factor. As such, the severe transcriptional responses to thioredoxin A depletion as observed in the DNA macroarray experiments shown here are likely to represent indirect effects. These may arise from transcription factors that show altered transcription patterns in the ItrxA strain (such as abrB, senS, or one of the many putative regulators) (see Table S1 in the supplemental material), but if the effects on certain transcription factors are exerted posttranslationally, their role may remain undetected. The presence of almost the entire Fur regulon (yclP, dhbAC, ydbN, ykuNOP, yuiI, yclN, and fhuB) (3) as class I genes in our data set, for instance, suggests that the activity of Fur is modulated by thioredoxin in B. subtilis, a supposition that is supported by the identification of Fur in a proteome analysis of thioredoxin-interacting proteins in E. coli (34). Alternatively, the activity of redox-sensitive regulators, such as Spx for instance (48), can be modulated by the more oxidized state of the cytoplasm (Fig. 2) (see “Changes in global transcription levels” above). Also, some other regulators, such as the LysR-type regulator YcgK (Fig. 3B) or YtlI, seem to be associated with genes that show a significant change in transcription in the ItrxA array analysis.
In addition to the effect on transcription factors described above, thioredoxin A may interact with or influence proteins other than transcription factors by direct association (30, 57) or thiol reactivity (9, 10). It would be interesting to compare an analysis of thioredoxin-interacting proteins from B. subtilis with the results from our transcriptome data, but currently, these thioredoxin-interacting protein data are not available. In order to evaluate whether some of the results from the transcriptome study could be linked to thioredoxin-interacting proteins, we therefore took a closer look at the available proteome data from other organisms. Although the relationship between a direct protein-protein interaction and a transcriptional response of the gene encoding the protein is unclear, there is a remarkable overlap between the two approaches. Primarily through affinity chromatography, a substantial number of thioredoxin-interacting partners was identified in cereal starchy endosperm (62), plant chloroplasts and mitochondria (4, 5), Synechocystis sp. strain PCC 6803 (40), Chlamydomonas reinhardtii (38), Arabidopsis thaliana cytosol (64), and E. coli cytosol (34, 36). We compared the lists of thioredoxin-interacting proteins from these proteome studies to the list of genes affected by thioredoxin A levels from our study. Interestingly, many of the affected genes from the ItrxA array encode proteins with similarity to the identified thioredoxin-interacting partners. Aside from the well-characterized interactions (such as with ribonucleoside diphosphate reductase), the similarities include dehydrogenases, (Clp) proteases, chaperones, catalases, oxidoreductases, proteins involved in tetrapyrrole synthesis and sulfur, carbon, and purine metabolism, oligopeptide and phosphate uptake systems, and ribosomal proteins. A detailed description of the comparison of the data from this study and the proteome studies is available in the supplemental material (see the supplemental material). The possible links between eukaryotic and prokaryotic proteome data on the one side and our study on the other suggest that many thioredoxin-linked processes may be conserved between prokaryotes and eukaryotes. In Table S1, available in the supplemental material, the final column indicates genes encoding putative thioredoxin-interacting proteins based on the proteome analyses discussed above. The proteins identified may account for at least part of the transcriptional response to thioredoxin A depletion.
Finally, two other mechanisms through which thioredoxin A can modulate transcription can be envisaged. Thioredoxin could alter the availability of cofactors such as NADH or metal ions, which are necessary for proper functioning of an enzyme. In the list of genes affected by thioredoxin depletion (see Table S1 in the supplemental material), for instance, a zinc uptake system (ykvW/zosA) (20) which might influence proteins that require zinc as a cofactor is identified. In this respect, it is interesting that two genes of the Zur regulon (yciA and ytiA), which responds to zinc availability (21, 51), are among the few true class III genes, showing no restoration in the ItrxA 100 cells. It is also conceivable that thioredoxin A affects transcription by altering transcriptional or translational efficiency. It was demonstrated that thioredoxin modulates the processivity of T7 DNA polymerase (30, 57), and thioredoxin has been identified as an RNA polymerase interacting partner in Synechocystis (40). It was reported that disulfide stress (which shows significant similarities to thioredoxin stress) in Streptomyces coelicolor affects translation (52), and multiple proteome studies identified thioredoxin as an interacting partner for elongation factors (4, 5, 34, 38, 40, 64). In our study, we identified rplA, yhzA, and ytiA as class III genes. All the mechanisms described above are likely to contribute to the pleiotropic effect of thioredoxin A on transcription.
Concluding remark.
Thioredoxin A is the product of 1 of the 271 essential genes of B. subtilis (33). This implies that thioredoxin A is much more important for B. subtilis than for other organisms, such as E. coli, in which thioredoxin A is not essential. Nevertheless, very little experimental data are available concerning its physiological functions in B. subtilis. Although there are certainly drawbacks to the use of transcriptome analyses for the study of posttranslational effects, like the ones caused by thioredoxin A depletion, this type of analysis gives valuable insights in the resulting pleiotropic effects on transcription. Importantly, the results of the present transcriptome analyses will serve as leads to identify relevant targets for further research on thioredoxin A function in B. subtilis.
Supplementary Material
Acknowledgments
We thank Isabelle Martin-Verstraete for communicating results prior to publication and helpful suggestions with respect to the sulfur metabolism in B. subtilis.
W.K.S. was supported by grant 811.35.002 from NWO-ALW. J.Y.D., J.M.V.D. and S.B. were supported by European Union project grants QLK3-CT-1999-00413, QLRT-CT1999-01455, LSHC-CT-2004-503468, and LSHG-CT-2004-005257.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arner, E. S., and A. Holmgren. 2000. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267:6102-6109. [DOI] [PubMed] [Google Scholar]
- 2.Baerends, R. J., W. K. Smits, A. de Jong, L. W. Hamoen, J. Kok, and O. P. Kuipers. 2004. Genome2D: a visualization tool for the rapid analysis of bacterial transcriptome data. Genome Biol. 5:R37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baichoo, N., T. Wang, R. Ye, and J. D. Helmann. 2002. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol. Microbiol. 45:1613-1629. [DOI] [PubMed] [Google Scholar]
- 4.Balmer, Y., A. Koller, G. del Val, W. Manieri, P. Schurmann, and B. B. Buchanan. 2003. Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc. Natl. Acad. Sci. USA 100:370-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balmer, Y., W. H. Vensel, C. K. Tanaka, W. J. Hurkman, E. Gelhaye, N. Rouhier, J. P. Jacquot, W. Manieri, P. Schurmann, M. Droux, and B. B. Buchanan. 2004. Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc. Natl. Acad. Sci. USA 101:2642-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benov, L., and I. Fridovich. 1999. Why superoxide imposes an aromatic amino acid auxotrophy on Escherichia coli. The transketolase connection. J. Biol. Chem. 274:4202-4206. [DOI] [PubMed] [Google Scholar]
- 7.Benov, L., and I. Fridovich. 1997. Superoxide imposes leakage of sulfite from Escherichia coli. Arch. Biochem. Biophys. 347:271-274. [DOI] [PubMed] [Google Scholar]
- 8.Benov, L., N. M. Kredich, and I. Fridovich. 1996. The mechanism of the auxotrophy for sulfur-containing amino acids imposed upon Escherichia coli by superoxide. J. Biol. Chem. 271:21037-21040. [DOI] [PubMed] [Google Scholar]
- 9.Berndt, C., C. H. Lillig, M. Wollenberg, E. Bill, M. C. Mansilla, D. de Mendoza, A. Seidler, and J. D. Schwenn. 2004. Characterization and reconstitution of a 4Fe-4S adenylyl sulfate/phosphoadenylyl sulfate reductase from Bacillus subtilis. J. Biol. Chem. 279:7850-7855. [DOI] [PubMed] [Google Scholar]
- 10.Bloomfield, K. L., S. A. Osborne, D. D. Kennedy, F. M. Clarke, and K. F. Tonissen. 2003. Thioredoxin-mediated redox control of the transcription factor Sp1 and regulation of the thioredoxin gene promoter. Gene 319:107-116. [DOI] [PubMed] [Google Scholar]
- 11.Burguière, P., S. Auger, M.-F. Hullo, A. Danchin, and I. Martin-Verstraete. 2004. Three different systems participate in l-cystine uptake in Bacillus subtilis. J. Bacteriol. 186:4875-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlioz, A., and D. Touati. 1986. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 5:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, L., L. Keramati, and J. D. Helmann. 1995. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc. Natl. Acad. Sci. USA 92:8190-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danon, A. 2002. Redox reactions of regulatory proteins: do kinetics promote specificity? Trends Biochem. Sci. 27:197-203. [DOI] [PubMed] [Google Scholar]
- 15.Dooley, C. T., T. M. Dore, G. T. Hanson, W. C. Jackson, S. J. Remington, and R. Y. Tsien. 2004. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J. Biol. Chem. 279:22284-22293. [DOI] [PubMed] [Google Scholar]
- 16.Erlendsson, L. S., M. Moller, and L. Hederstedt. 2004. Bacillus subtilis StoA is a thiol-disulfide oxidoreductase important for spore cortex synthesis. J. Bacteriol. 186:6230-6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eymann, C., G. Homuth, C. Scharf, and M. Hecker. 2002. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J. Bacteriol. 184:2500-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuangthong, M., S. Atichartpongkul, S. Mongkolsuk, and J. D. Helmann. 2001. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J. Bacteriol. 183:4134-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuangthong, M., A. F. Herbig, N. Bsat, and J. D. Helmann. 2002. Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J. Bacteriol. 184:3276-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaballa, A., and J. D. Helmann. 2002. A peroxide-induced zinc uptake system plays an important role in protection against oxidative stress in Bacillus subtilis. Mol. Microbiol. 45:997-1005. [DOI] [PubMed] [Google Scholar]
- 21.Gaballa, A., T. Wang, R. W. Ye, and J. D. Helmann. 2002. Functional analysis of the Bacillus subtilis Zur regulon. J. Bacteriol. 184:6508-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gleason, F. K., and A. Holmgren. 1988. Thioredoxin and related proteins in procaryotes. FEMS Microbiol. Rev. 4:271-297. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez-Pastor, J. E., E. C. Hobbs, and R. Losick. 2003. Cannibalism by sporulating bacteria. Science 301:510-513. [DOI] [PubMed] [Google Scholar]
- 24.Grundy, F. J., and T. M. Henkin. 2001. Synthesis of serine, glycine, cysteine, and methionine, p. 245-254. In A. L. Sonenshein, J. A. Hoch, and R. M. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 25.Guillouard, I., S. Auger, M. F. Hullo, F. Chetouani, A. Danchin, and I. Martin-Verstraete. 2002. Identification of Bacillus subtilis CysL, a regulator of the cysJI operon, which encodes sulfite reductase. J. Bacteriol. 184:4681-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamoen, L. W., W. K. Smits, A. de Jong, S. Holsappel, and O. P. Kuipers. 2002. Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach. Nucleic Acids Res. 30:5517-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanson, G. T., R. Aggeler, D. Oglesbee, M. Cannon, R. A. Capaldi, R. Y. Tsien, and S. J. Remington. 2004. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J. Biol. Chem. 279:13044-13053. [DOI] [PubMed] [Google Scholar]
- 28.Hayes, C. S., B. Illades-Aguiar, L. Casillas-Martinez, and P. Setlow. 1998. In vitro and in vivo oxidation of methionine residues in small, acid-soluble spore proteins from Bacillus species. J. Bacteriol. 180:2694-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imamura, D., K. Kobayashi, J. Sekiguchi, N. Ogasawara, M. Takeuchi, and T. Sato. 2004. spoIVH (ykvV), a requisite cortex formation gene, is expressed in both sporulating compartments of Bacillus subtilis. J. Bacteriol. 186:5450-5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson, D. E., and C. C. Richardson. 2003. A covalent linkage between the gene 5 DNA polymerase of bacteriophage T7 and Escherichia coli thioredoxin, the processivity factor: fate of thioredoxin during DNA synthesis. J. Biol. Chem. 278:23762-23772. [DOI] [PubMed] [Google Scholar]
- 31.Jonas, R. M., E. A. Weaver, T. J. Kenney, C. P. Moran, Jr., and W. G. Haldenwang. 1988. The Bacillus subtilis spoIIG operon encodes both sigma E and a gene necessary for sigma E activation. J. Bacteriol. 170:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, L., A. Mogk, and W. Schumann. 1996. A xylose-inducible Bacillus subtilis integration vector and its application. Gene 181:71-76. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouille, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauel, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. Seegers, J. Sekiguchi, A. Sekowska, S. J. Seror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar, J. K., S. Tabor, and C. C. Richardson. 2004. Proteomic analysis of thioredoxin-targeted proteins in Escherichia coli. Proc. Natl. Acad. Sci. USA 101:3759-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 36.Leichert, L. I., and U. Jakob. 2004. Protein thiol modifications visualized in vivo. PLoS Biol. 2:e333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leichert, L. I., C. Scharf, and M. Hecker. 2003. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 185:1967-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemaire, S. D., B. Guillon, P. Le Marechal, E. Keryer, M. Miginiac-Maslow, and P. Decottignies. 2004. New thioredoxin targets in the unicellular photosynthetic eukaryote Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 101:7475-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, Y., J. Hugenholtz, T. Abee, and D. Molenaar. 2003. Glutathione protects Lactococcus lactis against oxidative stress. Appl. Environ. Microbiol. 69:5739-5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindahl, M., and F. J. Florencio. 2003. Thioredoxin-linked processes in cyanobacteria are as numerous as in chloroplasts, but targets are different. Proc. Natl. Acad. Sci. USA 100:16107-16112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mansilla, M. C., and D. de Mendoza. 1997. l-Cysteine biosynthesis in Bacillus subtilis: identification, sequencing, and functional characterization of the gene coding for phosphoadenylylsulfate sulfotransferase. J. Bacteriol. 179:976-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mansilla, M. C., and D. de Mendoza. 2000. The Bacillus subtilis cysP gene encodes a novel sulphate permease related to the inorganic phosphate transporter (Pit) family. Microbiology 146:815-821. [DOI] [PubMed] [Google Scholar]
- 43.McDonnell, G. E., and D. J. McConnell. 1994. Overproduction, isolation, and DNA-binding characteristics of Xre, the repressor protein from the Bacillus subtilis defective prophage PBSX. J. Bacteriol. 176:5831-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDonnell, G. E., H. Wood, K. M. Devine, and D. J. McConnell. 1994. Genetic control of bacterial suicide: regulation of the induction of PBSX in Bacillus subtilis. J. Bacteriol. 176:5820-5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mostertz, J., C. Scharf, M. Hecker, and G. Homuth. 2004. Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology 150:497-512. [DOI] [PubMed] [Google Scholar]
- 46.Muller, E. G. 1991. Thioredoxin deficiency in yeast prolongs S phase and shortens the G1 interval of the cell cycle. J. Biol. Chem. 266:9194-9202. [PubMed] [Google Scholar]
- 47.Nakano, M. M., F. Hajarizadeh, Y. Zhu, and P. Zuber. 2001. Loss-of-function mutations in yjbD result in ClpX- and ClpP-independent competence development of Bacillus subtilis. Mol. Microbiol. 42:383-394. [DOI] [PubMed] [Google Scholar]
- 48.Nakano, S., K. N. Erwin, M. Ralle, and P. Zuber. 2005. Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol. Microbiol. 55:498-510. [DOI] [PubMed] [Google Scholar]
- 49.Nakano, S., E. Kuster-Schock, A. D. Grossman, and P. Zuber. 2003. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 100:13603-13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakano, S., M. M. Nakano, Y. Zhang, M. Leelakriangsak, and P. Zuber. 2003. A regulatory protein that interferes with activator-stimulated transcription in bacteria. Proc. Natl. Acad. Sci. USA 100:4233-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nanamiya, H., G. Akanuma, Y. Natori, R. Murayama, S. Kosono, T. Kudo, K. Kobayashi, N. Ogasawara, S. M. Park, K. Ochi, and F. Kawamura. 2004. Zinc is a key factor in controlling alternation of two types of L31 protein in the Bacillus subtilis ribosome. Mol. Microbiol. 52:273-283. [DOI] [PubMed] [Google Scholar]
- 52.Paget, M. S., V. Molle, G. Cohen, Y. Aharonowitz, and M. J. Buttner. 2001. Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the sigmaR regulon. Mol. Microbiol. 42:1007-1020. [DOI] [PubMed] [Google Scholar]
- 53.Prinz, W. A., F. Aslund, A. Holmgren, and J. Beckwith. 1997. The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J. Biol. Chem. 272:15661-15667. [DOI] [PubMed] [Google Scholar]
- 54.Scharf, C., S. Riethdorf, H. Ernst, S. Engelmann, U. Volker, and M. Hecker. 1998. Thioredoxin is an essential protein induced by multiple stresses in Bacillus subtilis. J. Bacteriol. 180:1869-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scotti, C., A. Valbuzzi, M. Perego, A. Galizzi, and A. M. Albertini. 1996. The Bacillus subtilis genes for ribonucleotide reductase are similar to the genes for the second class I NrdE/NrdF enzymes of Enterobacteriaceae. Microbiology 142:2995-3004. [DOI] [PubMed] [Google Scholar]
- 56.Sekowska, A., S. Robin, J. J. Daudin, A. Henaut, and A. Danchin. 2001. Extracting biological information from DNA arrays: an unexpected link between arginine and methionine metabolism in Bacillus subtilis. Genome Biol. 2:RESEARCH0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singha, N. C., A. Vlamis-Gardikas, and A. Holmgren. 2003. Real-time kinetics of the interaction between the two subunits, Escherichia coli thioredoxin and gene 5 protein of phage T7 DNA polymerase. J. Biol. Chem. 278:21421-21428. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka, R., Y. Araki, M. Mizukami, A. Miyauchi, M. Ishibashi, H. Tokunaga, and M. Tokunaga. 2004. Expression and purification of the Bacillus subtilis thioredoxin superfamily protein YkvV. Biosci. Biotechnol. Biochem. 68:1801-1804. [DOI] [PubMed] [Google Scholar]
- 59.van der Ploeg, M., Jr., Barone, and T. Leisinger. 2001. Functional analysis of the Bacillus subtilis cysK and cysJI genes. FEMS Microbiol. Lett. 201:29-35. [DOI] [PubMed] [Google Scholar]
- 60.van Sinderen, D., A. Luttinger, L. Kong, D. Dubnau, G. Venema, and L. Hamoen. 1995. comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis. Mol. Microbiol. 15:455-462. [DOI] [PubMed] [Google Scholar]
- 61.Venema, G., R. H. Pritchard, and T. Venema-Schroeder. 1965. Fate of transforming deoxyribonucleic acid in Bacillus subtilis. J. Bacteriol. 89:1250-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wong, J. H., Y. Balmer, N. Cai, C. K. Tanaka, W. H. Vensel, W. J. Hurkman, and B. B. Buchanan. 2003. Unraveling thioredoxin-linked metabolic processes of cereal starchy endosperm using proteomics. FEBS Lett. 547:151-156. [DOI] [PubMed] [Google Scholar]
- 63.Wood, H. E., M. T. Dawson, K. M. Devine, and D. J. McConnell. 1990. Characterization of PBSX, a defective prophage of Bacillus subtilis. J. Bacteriol. 172:2667-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamazaki, D., K. Motohashi, T. Kasama, Y. Hara, and T. Hisabori. 2004. Target proteins of the cytosolic thioredoxins in Arabidopsis thaliana. Plant Cell Physiol. 45:18-27. [DOI] [PubMed] [Google Scholar]
- 65.Yasbin, R. E., D. Cheo, and K. W. Bayles. 1991. The SOB system of Bacillus subtilis: a global regulon involved in DNA repair and differentiation. Res. Microbiol. 142:885-892. [DOI] [PubMed] [Google Scholar]
- 66.Yasueda, H., Y. Kawahara, and S. Sugimoto. 1999. Bacillus subtilis yckG and yckF encode two key enzymes of the ribulose monophosphate pathway used by methylotrophs, and yckH is required for their expression. J. Bacteriol. 181:7154-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.