Abstract
Gram-negative bacteria like Rhodobacter capsulatus use intertwined pathways to carry out the posttranslational maturation of c-type cytochromes (Cyts). This periplasmic process requires at least 10 essential components for apo-Cyt c chaperoning, thio-oxidoreduction, and the delivery of heme and its covalent ligation. One of these components, CcmI (also called CycH), is thought to act as an apo-Cyt c chaperone. In R. capsulatus, CcmI-null mutants are unable to produce c-type Cyts and thus sustain photosynthetic (Ps) growth. Previously, we have shown that overproduction of the putative heme ligation components CcmF and CcmHRc (also called Ccl1 and Ccl2) can partially bypass the function of CcmI on minimal, but not on enriched, media. Here, we demonstrate that either additional overproduction of CcmG (also called HelX) or hyperproduction of CcmF-CcmHRc is needed to completely overcome the role of CcmI during the biogenesis of c-type Cyts on both minimal and enriched media. These findings indicate that, in the absence of CcmI, interactions between the heme ligation and thioreduction pathways become restricted for sufficient Cyt c production. We therefore suggest that CcmI, along with its apo-Cyt chaperoning function, is also critical for the efficacy of holo-Cyt c formation, possibly via its close interactions with other components performing the final heme ligation steps during Cyt c biogenesis.
The c-type cytochromes (Cyts) are ubiquitous proteins that function as electron carriers between various energy transduction complexes in major metabolic pathways like photosynthesis (Ps) and respiration (Res). They contain heme (iron protoporphyrin IX) molecules as prosthetic groups that are covalently attached via thioether bonds formed between the vinyl groups of the heme moiety and the cysteine thiol groups of a conserved signature motif (almost exclusively Cys-Xxx-Yyy-Cys-His) within apo-Cyt c (46). Three distinct posttranslational maturation processes are known for the production of c-type holo-Cyts in prokaryotic and eukaryotic organisms (35, 43). Plant and most protozoan mitochondria, Archaea, and many gram-negative bacteria use system I, depicted in Fig. 1, top. It consists of at least 10 essential membrane-bound components that chaperone the c-type apo-Cyts to the maturation sites after their translocation across the cytoplasmic membrane, reduce the disulfide bonds between the cysteines of their heme binding motifs, export the heme cofactors into the periplasm, and attach them covalently and stereospecifically to the apo-Cyts (2, 35, 43, 60, 61).
FIG. 1.
Current model of Cyt c biogenesis in R. capsulatus. Pre-apo-Cyt c produced in the cytoplasm are translocated across the membrane via the Sec-dependent secretory pathway. Their signal peptides (SP) are either processed or used as N-terminal membrane anchors. Disulfide bonds are assumed to be formed within the cysteines of the conserved apo-Cyt c heme binding motif CxxCH and then possibly chaperoned via CcmI to the Cyt c maturation site. CcdA shuttles electrons across the membrane to the thioreduction component CcmHRc via CcmG. CcmHRc is thought to reduce the heme binding site disulfides prior to heme attachment. The heme cofactor (H) is synthesized in the cytoplasm from δ-amino levulinic acid (δ-ALA), putatively transported to the periplasm via the ATP-binding cassette (ABC) transporter CcmAB, and covalently ligated to CcmE via CcmCD, with WWD referring to the putative heme binding motif. CcmE is postulated to provide the heme to the putative heme lyase component CcmF, which is believed to attach it covalently and stereospecifically to the reduced apo-Cyt c. According to this model, CcmI-null mutants are not able to produce any holo-Cyt c, because apo-Cyt c is not delivered efficiently to the maturation site, as they probably are degraded rapidly. Hence, overproduction of CcmF and CcmHRc can alleviate the need for apo-Cyt c chaperoning and protection function of CcmI. Arrows refer to electron (e-) transfer steps, and ? indicates hypothetical steps. The bottom portion depicts the proposed interactions between CcmI, CcmG, CcmF, CcmHRc and CcmE, forming a hypothetical Cyt c biogenesis “core complex” in Rhodobacter capsulatus. This model proposes that apo-Cyt c interacts with CcmI as part of a multisubunit protein complex including CcmF and CcmHRc.
The gram-negative, purple nonsulfur facultatively photosynthetic bacterium Rhodobacter capsulatus produces various soluble or membrane-bound c-type Cyts, including Cyts c1, c2, c′, cy, co, and cp (14, 28, 33, 66), to sustain its versatile growth modes. Cyt c1 and either Cyt c2 or cy are required for Ps growth (66), whereas Cyt co and cp are essential subunits of the cbb3-type Cyt c oxidase involved in Res growth (33). The activity of the latter enzyme can be directly detected by monitoring the staining of colonies following the Nadi (α-naphthol + dimethylphenylenediamine → indophenol blue + H2O) reaction (32), Thus, R. capsulatus mutants unable to form c-type holo-Cyts can be identified by their Ps−/Nadi− dual phenotype (34), via which essential components of Cyt c biogenesis have been identified in this species (Fig. 1, top) (4, 5, 7, 15, 26, 38). As in Escherichia coli, the CcmABCD (also called HelABCD) components form an ATP-binding cassette (ABC)-containing transporter complex, thought to be involved in the export of the heme cofactor across the cytoplasmic membrane. The exported heme binds to CcmE (also called CycJ) (54), which conveys it to the putative apo-Cyt c heme lyase component CcmF (also called Ccl1) (25-27). Unlike E. coli, R. capsulatus and some other species contain two additional components: CcmI (also called CycH), which is proposed to chaperone the c-type apo-Cyts to the maturation site following their translocation to the periplasm (5, 38), and a membrane-bound thioredoxin, designated here as CcmHRc (also called Ccl2), which is involved in the apo-Cyt c thioreduction pathway (4, 5). In E. coli, the carboxyl-terminal part of CcmI is fused to the homologue of CcmHRc to form CcmH (35, 43, 59) (referred to as CcmHEc here).
As disulfide bonds are formed in the periplasm between the cysteines of translocated proteins via the DsbA/DsbB pathway for oxidative protein folding (11, 29), similar bonds are also assumed to be built between the heme binding cysteine thiols (39, 40, 50, 51). For periplasmic disulfide reduction, electrons originating from TrxA are thought to be shuttled across the cytoplasmic membrane via CcdA (homologous to the central domain of E. coli DsbD) to CcmG (also called HelX) and then to CcmHRc or CcmHEc depending on the species. The disulfide bonds at the heme binding sites of the c-type apo-Cyts are then believed to be reduced by CcmHRc (or CcmHEc) prior to covalent and stereo-specific heme ligation (Fig. 1, top) (15, 21-23, 30, 41, 44, 48, 56).
In R. capsulatus, CcmI is thought to act as a chaperone that delivers apo-Cyts to the maturation sites (38). Its sequence analysis indicates that it is a bipartite membrane protein that contains two amino-terminal transmembrane helices and a large periplasmic carboxyl-terminal extension of over 300 amino acids (38), designated CcmI-1 and CcmI-2, respectively. In R. capsulatus CcmI mutants lacking the CcmI-2 domain, c-type Cyt maturation becomes growth medium dependent. For example, they can still produce several c-type Cyts (e.g., Cyt c1, cp, and co) during growth in minimal medium but only Cyt c1, which is a subunit of the Cyt bc1 complex (14), in minimal and enriched media, indicating that CcmI-2 is not required for its maturation (38). Interestingly, E. coli, which does not contain a Cyt c1 homologue, naturally lacks an equivalent of CcmI-1.
Our previous analysis of Ps+/Nadi+ revertants of R. capsulatus CcmI-null mutants, such as MT-SRP1.r1, indicated that overexpression of ccmFHRc can bypass the function of CcmI during c-type Cyt maturation on minimal, but not on enriched, medium (16). Remarkably, these mutants can become fully proficient for Cyt c maturation on both media upon acquisition of additional mutations. In this work, we demonstrate that, in addition to the overexpression of ccmFHRc, either overexpression of ccmG or hyperexpression of ccmFHRc is required to fully overcome the role of CcmI in minimal and enriched growth media. These findings indicate that, in the absence of CcmI, interactions between the thioreduction and heme ligation components become restricted, limiting adequate c-type Cyt production. Therefore, although CcmI is not essential for the attachment of the heme cofactor per se, it is an important component for sufficient holo-Cyt c formation under Res and Ps growth on minimal and enriched media.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this work are described in Table 1. R. capsulatus strains were grown at 35°C on enriched (12) or minimal (57) medium, supplemented with appropriate antibiotics (tetracycline, kanamycin, and spectinomycin at 2.5, 10, and 10 μg/ml, respectively), either chemoheterotrophically (Res growth conditions) or photoheterotrophically in anaerobic jars with H2+CO2-generating gas packs from BBL Microbiology Systems (Cockeysville, MD) (Ps growth conditions). Plus and minus signs indicate the ability of the strains to form visible colonies after approximately 3 days of incubation under appropriate growth conditions. E. coli strains were grown on Luria broth (LB) supplemented as needed with appropriate antibiotics (tetracycline, kanamycin, spectinomycin, and ampicillin at 12.5, 50, 50, and 100 μg/ml, respectively).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Relevant phenotypeb | Source or reference |
|---|---|---|---|
| Strains | |||
| E. coli HB101 | F− Δ(gpt-proA)62 araC14 leuB6(Am) glnV44(AS) galK2(Oc) lacY1 Δ(mcrC-mrr) rpsL20(Strr) xylA5 mtl-1 thi-1 | Strr | 52 |
| R. capsulatus | |||
| SB1003a | Wild type; Res+/Nadi+ Ps+ Cyt c+ | 64 | |
| MT1131a | crtD121 Rifr | Wild type; Res+/Nadi+ Ps+ Cyt c+ | 55 |
| Y262 | GTA overproducer | 63 | |
| MT-SRP1 | Δ(ccmI::kan) | Res+/Nadi− Ps−; Cyt c− | 38 |
| MT-SRP1.r1 | Δ(ccmI::kan) G488A in promoter of ccmFHRc | Res+/Nadi+ Ps+ Cyt c+ on MedA | 16 |
| MT-SRP1.r1big | Δ(ccmI::kan) G488A in promoter of ccmFHRc unknown mutation | Res+/Nadi+ Ps+ Cyt c+ on MedA and MPYE | 16 |
| SB1003ΔhelD | Δ(ccmD::kan) | Res+/Nadi− Ps− Cyt c− | 26 |
| SB1003ΔhelX | Δ(ccmG::kan) | Res+/Nadi− Ps− Cyt c− | 4 |
| TCK2 | Δ(pyrE::kan), Δ(ccmC::pyrE) | Res+/Nadi− Ps− | 62 |
| KR319 | Δ(ccmA::kan) | Res+/Nadi− Ps− Cyt c− | 5 |
| MD12 | ccmF::spe (ccmHRc+) | Res+/Nadi− Ps− Cyts c− | 16 |
| MD14 | (ccmF+) ccmHRc::spe | Res+/Nadi− Ps− Cyt c− | 16 |
| Plasmids | |||
| pBluescript II SK(+) | Ampr | Stratagene | |
| pHP45Ω-Spec | Sper | 47 | |
| pRK2013 | Kanr, helper | 19 | |
| pRK404 | Broad host-range vectors, gene expression could be supported by E. coli lacZ promoter | Tetr | 19, 31, 36 |
| pRK415 | |||
| pUCA9 | |||
| pCHB500 | Broad host-range expression vector via R. capsulatus cycA promoter | Tetr | 6 |
| pDA5 | pRK415 derivative with 1.6- and 1.4-kb chromosomal EcoRI fragments from MT-SRP1.r1big | Tetr | This work |
| pDA10 | pRK415 derivative with 3.1-kb and 13.5-kb chromosomal EcoRI fragments from MT-SRP1.r1big | Tetr | This work |
| p2hel-404 | pRK404 derivative with ccmABCDG | Ampr Tetr | 38 |
| pCS1526 | 280-bp PCR product containing ccmD from MT-SRP1.r1big cloned into XbaI-KpnI sites of pRK415 | Tetr | This work |
| pCS1527 | 280-bp PCR product containing ccmD from MT-SRP1.r1big cloned into XbaI-KpnI sites of pCHB500 | Tetr | This work |
| pCS1530 | 830-bp PCR product containing ccmC from MT-SRP1.r1big cloned into XbaI-KpnI sites of pRK415 | Tetr | This work |
| pCS1531 | 1.01-kb PCR product containing ccmCD from MT-SRP1.r1big cloned into XbaI and KpnI sites of pRK415 | Tetr | This work |
| pCS1532 | 1.49-kb PCR product containing ccmCDG from MT-SRP1.r1big cloned into XbaI and KpnI sites of pRK415 | Tetr | This work |
| pCS1534 | 1.4-kb EcoRI fragment of pDA5 cloned into pRK415 | Tetr | This work |
| pCS1535 | 1.6-kb EcoRI fragment of pDA5 cloned into pRK415 | Tetr | This work |
| pCS1536 | 3.1-kb EcoRI fragment from pDA10 in pRK415 | Tetr | This work |
| pCS1537 | 13.5-kb EcoRI fragment from pDA10 in pRK415 | Tetr | This work |
| pCS1538 | 960-bp PCR product containing ubiE::ccmC from pDA5 cloned in XbaI-KpnI sites of pRK415 | Tetr | This work |
| pCS1539 | ΔccmC created by filling the unique BamHI site of pCS1532 | Tetr | This work |
| pCS1540 | 570-bp PCR product containing ccmG from MT-SRP1.r1big cloned into XbaI-KpnI sites of pCHB500 | Tetr | This work |
| pCS1541 | 750-bp PCR product containing ccmDG from MT-SRP1.r1big cloned into XbaI-KpnI sites of pCHB500 | Tetr | This work |
| pCS1542 | Sper cassette from pHP45Ω cloned into the BamHI site of ccmC gene in pCS1532 | Tetr Sper | This work |
| pCS1545 | Replacement of ccmG cysteines 75 and 78 with serines in pCS1540 | Tetr | This work |
| pMM401 | 7-kb fragment containing G488A promoter-up allele of ccmFHRc in pRK404 | Tetr | 16 |
| pMM403 | 7-kb fragment containing G488 wild-type promoter allele of ccmFHRc in pRK404 | Tetr | 16 |
| pMM500 | pMM401 derivative with a frameshift mutation in ccmF pYZ10 derivative with a nonpolar Sper cassette cloned into ccmF | Tetr, CcmF− CcmHRc+ Tetr Sper CcmF− CcmHRc+ | 16 |
| pYZ4 | 16 | ||
| pYZ10 | 2.8-kb fragment containing G488A promoter-up allele of ccmFHRc in pRK404 | Tetr | 16 |
| pYZ12 | pYZ10 derivative with a nonpolar Sper cassette cloned into ccmHRc | Tetr Sper, CcmF+ CcmHRc− | 16 |
| pK1 | ccdA in pRK404 | Tetr | 15 |
| pMD1 | ccmE in pRK404 | Tetr | 15 |
R. capsulatus SB1003 and MT1131 are both referred to as wild-type strains with respect to their Cyt c profile and growth properties. Y262, SB1003ΔhelD, and SB1003ΔhelX are derivatives of SB1003, and all other listed R. capsulatus strains are derivatives of MT1131.
MedA and MPYE refer to minimal and enriched growth media, respectively.
Preparation of enriched medium supplemented with redox-active chemicals or various nutritional compounds.
To characterize the medium-dependent phenotype of the ccmI suppressors like MT-SRP1.r1, enriched medium plates were supplemented at the same final concentrations with different nutritional compounds that make up the minimal medium (57). These were 0.2% (wt/vol) sodium succinate (pH 6.8), 0.01% (wt/vol) glutamic acid (pH 7.0), 0.004% (wt/vol) aspartic acid (pH 7.0), 0.05% (wt/vol) ammonium sulfate, 20 mM potassium phosphate (pH 6.8), 0.05% (wt/vol) sodium chloride, 0.2% (vol/vol) solution C consisting of 0.02% (wt/vol) nitrilotriacetic acid, 8.6 μM EDTA, 2.39 mM MgSO4, 0.45 mM CaCl2 · H2O, various trace elements [ZnSO4 · 7H2O, FeSO4 · 7H2O, MnSO4 · H2O, CuSO4 · 5H2O, Co(NO3)2 · 6H2O, and H3BO3] as in minimal medium, and a vitamin cocktail containing 0.1 mg/ml nicotinic acid, 0.05 mg/ml thiamine-HCl, and 1 μg/ml biotin. Enriched medium supplemented with various redox-active chemicals were prepared by adding cystine (0.05, 0.5, and 5 mM), diamide (N,N,N′,N′-tetramethylazodicarboxamide) (1, 10, and 100 μM), cysteine (0.1, 1, and 10 mM) and MESNA (2-mercaptoethanesulfonic acid) (0.1, 1, and 10 mM) or a mixture of cystine:cysteine at a 1:2 ratio (0.05:0.1, 0.5:1 and 5:10 mM) from appropriate fresh stock solutions. Plates were used immediately following solidification to determine the Res and Ps growth phenotypes of appropriate strains.
Molecular genetic techniques.
Standard molecular genetic techniques were performed as described by Sambrook and Russell (52). Conjugal transfer of plasmids from E. coli to R. capsulatus was performed as described previously (12). The R. capsulatus strain MT-SRP1.r1big was grown under Ps conditions on enriched medium and its chromosomal DNA isolated by using the QIAGEN DNeasy kit (Valencia, CA). A genomic library was prepared as described previously (16, 17) by digesting chromosomal DNA with EcoRI and cloning the resulting fragments into the broad-host-range vector pRK415 (19). A 280-bp PCR product containing ccmD was amplified by using the primers helD-Fwd (5′-GCT CTA GAA CCG AGA TCC GCG CCC GTC G-3′) and helD-Rev (5′-GGG GTA CCC CCG CGC CAA CAA AGC CCG C-3′) carrying the recognition sites for the restriction enzymes XbaI and KpnI at their 5′ ends and was then cloned into the respective sites of pRK415 and pCHB500 to yield pCS1526 and pCS1527, respectively. Similarly, pCS1530 was constructed by PCR amplification of an 830-bp fragment containing ccmC using the primers helC2 (5′-CTT ATT CTA GAT TGA TGC GAG TC-3′) and helC3 (5′-GAC GCA AGG TAC CTG ACG GCG TAT TTT C-3′); pCS1531 was constructed by PCR amplification of a 1,010-bp fragment containing ccmCD using the primers helC2 and helD-Rev; pCS1532 was constructed by PCR amplification of a 1,490-bp fragment containing ccmCDG using the primers helC2 and RchelX1 (5′-GGC CCC GGT ACC AAG ACG CAG GAT-3′); and pCS1538 was constructed by PCR amplification of a 960-bp fragment containing ubiE::ccmC using the primers helC1 (5′-GAC AGT TCT AGA TCG GCA AGG GCA G-3′) and helC3 following appropriate restriction digestion and ligation of the amplified DNA fragments into pRK415. For the construction of pCS1540 and pCS1541, a 570-bp PCR product containing ccmG was amplified by using the primers helXforward-XbaI (5′-CGG GTC TAG AAA GCC CAG GAA GAA CGG A-3′) and RchelX1, a 750-bp PCR product containing ccmDG was amplified by using the primers helD-Fwd and RchelX1, respectively, and a restriction digestion and ligation procedure similar to that mentioned above was carried out by using the broad-host-range vector pCHB500 (6). For all of these plasmids, MT-SRP1.r1big chromosomal DNA was used as a template. To create pCS1542, a spectinomycin resistance cassette (2.0 kb) from pHP45Ω-Spec (47) was inserted in reverse orientation into the unique BamHI site of pCS1532 located in ccmC. Plasmid pCS1539 was obtained by cutting pCS1530 at its unique BamHI site and blunting the ends with T4 DNA polymerase before ligation, resulting in a frameshift mutation in ccmC. For construction of pCS1545, both cysteine codons 75 and 78 of ccmG in pCS1540 were mutated to serine codons by using primers HelX-C78Ss (5′-CTT CTG GGC TTC CTG GAG CGC GCC CTG TCG-3′) and HelX-C78Sas (5′-ACC CGA CAG GGC GCG CTC CAG GAA GCC CAG-3′) following the instructions of the QuikChange XL site-directed mutagenesis kit from Stratagene (La Jolla, CA). All plasmids were verified by DNA sequence analyses using appropriate primers (see below).
DNA sequence analysis.
Automated DNA sequencing with the Big-Dye terminator cycle sequencing kit (Applied Biosystems) was performed according to the manufacturer by using the helC2, helC3, M13-forward, and M13-reverse primers to verify all pRK415 and pCHB500 derivatives constructed in this study. Chromosomal DNA of MT1131, MT-SRP1.r1, and MT-SRP1.r1big isolated with the DNeasy kit were used as templates, and the ORF124/ccmABCDG promoter region was amplified using the primers RcORF124down (5′-CTG GCG CTG GTG CCG ATG CTG GAT C-3′) and RchelCpromup (5′-CGG CTC TCC TCC CTC TCG CGT CCC G-3′) to generate a 2,070-bp PCR product, which was purified by using the QIAGEN PCR purification kit and sequenced using the same primers. Similarly, the 5′ region of ccmC including the intergenic space between ccmB and ccmC was amplified by using the same DNA template and the primers helC2 and helC3 to yield an 830-bp PCR fragment, which was purified and sequenced by using the primer helC2. DNA sequence analyses, homology searches, and genome sequence comparisons were done using MacVector (Kodak) and BLAST software packages (3).
Biochemical techniques.
Intracytoplasmic membrane vesicles, or chromatophores, were prepared in 20 mM MOPS (morpholinepropanesulfonic acid)-KOH buffer (pH 7.0) containing 1 mM KCl, 10 mM EDTA and a protease inhibitor cocktail (Complete, EDTA-free from Roche Inc., Indianapolis, IN) as recommended by the manufacturer using a French pressure cell (28). Protein concentrations were determined according to the method of Bradford (9) using the stain solution Bio-Safe Coomassie from Bio-Rad (Hercules, CA). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (16.5% T, 6% C) was used as described by Schaegger and von Jagow (53), and various c-type Cyts were revealed via their endogenous peroxidase activity using tetramethylbenzidine (TMBZ) and H2O2 (58). Cyt c oxidase activity of the colonies was detected using the Nadi reaction as described earlier (32). Antisera against CcmA and CcmG were produced, and immunoblot analyses were performed, as described elsewhere (26).
Chemicals.
All chemicals were of reagent grade and obtained from commercial sources.
RESULTS
Suppressors of CcmI-null mutants that are Ps+/Nadi+ on both minimal and enriched media.
R. capsulatus CcmI-null mutants, such as MT-SRP1, are Ps−/Nadi− on both minimal and enriched media due to their inability to produce c-type holo-Cyts (38). Ps+ revertants of MT-SRP1, such as MT-SRP1.r1, are readily isolated (at a frequency of approximately 10−6) on minimal, but not enriched, medium. These revertants overexpress ccmFHRc to bypass the need for CcmI during c-type Cyt biogenesis (16). However, they are Ps+ only on minimal medium, indicating that in such strains c-type Cyt production becomes restricted under different growth conditions. Interestingly, they acquire extra mutations (at a frequency of about 10−6) in addition to the overexpression of ccmFHRc (such as MT-SRP1.r1big) to become Ps+ on enriched medium (Table 2).
TABLE 2.
Phenotypes of various CcmI-null mutants and their derivatives
| Strain | Relevant characteristics | Phenotype ona:
|
|||
|---|---|---|---|---|---|
| MedA
|
MPYE
|
||||
| Res/Nadi | Ps | Res/Nadi | Ps | ||
| MT1131 | CcmI+ CcmF+ CcmHRc+ | +/+ | + | +/+ | + |
| MT-SRP1 | CcmI− CcmF+ CcmHRc+ | +/− | − | +/− | − |
| MT-SRP1.r1 | CcmI− CcmFup CcmHRcup | +/+ | + (slow) | +/+ | − |
| MT-SRP1.r1big | CcmI− CcmFup CcmHRcup BigXb | +/+ | + | +/+ | + |
MedA and MPYE refer to minimal and enriched growth media, respectively. For Res and Ps, + and − refer to the ability of the strains to grow under these conditions. For Nadi, + and − indicate the presence or absence of the Cyt c oxidase activity, respectively.
BigX refers to the unknown mutated allele that confers the Nadi+/Ps+ phenotype to MT-SRP1.r1 on enriched medium.
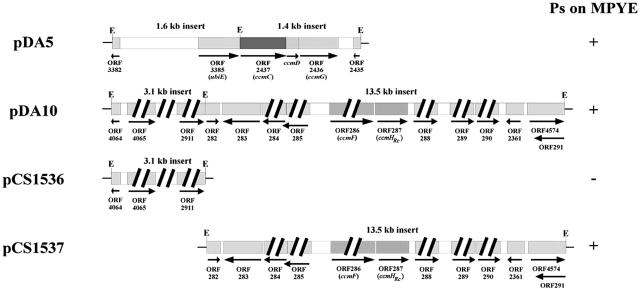
A conjugally transferable genomic library with EcoRI-digested chromosomal DNA from MT-SRP1.r1big was used to complement MT-SRP1.r1 for Ps+ growth on enriched medium and yielded the plasmids pDA5 and pDA10 (Fig. 2). pDA5 contained two EcoRI inserts of 1.6 and 1.4 kb, whose DNA sequence comparisons to the R. capsulatus genome sequence (http://ergo.integratedgenomics.com) indicated that they are not contiguous on the chromosome. The 1.6-kb insert consisted of two incomplete open reading frames (ORFs) (RRC03382 and RRC03385, which corresponds to ubiE (ubiquinone-menaquinone biosynthesis methyltransferase) of R. capsulatus. The 1.4-kb insert carried a 588-bp 3′ fragment of ccmC (RRC02437), the ccmDG (RRC02436) portion of the ORF124/ccmABCDG operon involved in Cyt c biogenesis (5), and an additional incomplete ORF (RRC02435). Similarly, pDA10 also contained two noncontiguous chromosomal inserts (3.1 and 13.5 kb). The 3.1-kb fragment consisted of one complete (RRC04065) and two incomplete (RRC04064 and RRC02911) ORFs (Fig. 2). On the other hand, the 13.5-kb fragment carried seven complete (RRC00283 through RRC00285, RRC00288 through RRC00290, and RRC02361) and three incomplete (RRC00282 and overlapping RRC00291 and RRC04574) ORFs, as well as the ccmFHRc gene cluster (RRC00286 and RRC00287), which is required for Cyt c maturation (5).
FIG. 2.
Plasmids pDA5 and pDA10, which confer the Ps+ growth phenotype on MT-SRP1.r1. The 1.4- and 1.6-kb EcoRI fragments of pDA5 are not contiguous with each other on the R. capsulatus chromosome. The 1.4-kb fragment carries two complete (ccmD and RRC02436 [ccmG]) and two incomplete (RRC02435 [hpt, hypoxanthine-guanine phosphoribosyltransferase] and RRC02437 [ccmC]) ORFs. The 1.6-kb fragment contains two incomplete ORFs, RRC03382 (ysiB, putative enoyl-CoA hydratase) and RRC03385 (ubiE, ubiquinone/menaquinone biosynthesis methyltransferase). The two EcoRI fragments of pDA10 are also noncontiguous on the R. capsulatus chromosome. The 3.1-kb fragment carries one complete (RRC04065 [serC]) and two incomplete (RRC04064 [serB] and RRC02911 [serA]) ORFs involved in serine biosynthesis. The 13.5-kb fragment contains nine complete ORFs (RRC00283 [entB, isochorismatase], RRC00284 and RRC00285 [argF and argD, respectively, involved in arginine biosynthesis], RRC00286 and RRC00287 [ccmF and ccmHRc, respectively, encoding the putative apo-Cyt c heme ligation complex CcmF and CcmHRc], RRC00288 [fadB, enoyl-CoA hydratase], RRC00289 [msbA, ATP binding lipid transporter], RRC00290 [acs, acetyl-CoA synthetase], and RRC002361 [cspA, cold shock DNA binding protein]), and three incomplete (RRC00282 [hrpB, ATP-dependent helicase], and overlapping RRC00291 [miaB, 2-methylthioadenine synthetase] and RRC04574 [a putative membrane protein with a possible oxidoreductase function]). The ability of pDA5, pDA10, pCS1536, and pCS1537 to confer on MT-SRP1.r1 a Ps+ phenotype on enriched medium is indicated to the right, E refers to EcoRI cleavage sites, and thick slashes indicate that the ORFs and the intergenic spaces are truncated as needed to fit the figure.
Hyperexpression of ccmFHRc can render MT-SRP1.r1 Ps+ on enriched medium.
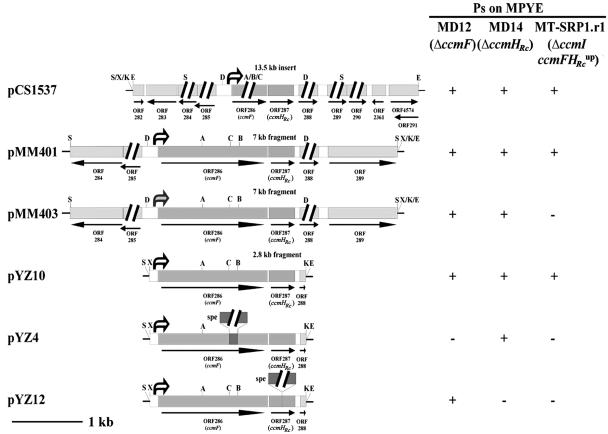
The plasmid pDA10 was analyzed by cloning its two EcoRI inserts separately into pRK415 to yield pCS1536 and pCS1537, respectively (Table 2). Of these plasmids, pCS1537 complemented MT-SRP1.r1 for Ps+ growth on enriched medium, whereas pCS1536 did not, indicating that the complementing activity was associated with the large EcoRI insert containing the ccmFHRc gene cluster (Fig. 2). As this fragment was originated from MT-SRP1.r1big, which is a derivative of MT-SRP1.r1 comprising a chromosomal overexpressing allele of ccmFHRc (16), cells harboring pDA10 were considered to hyperexpress ccmFHRc due to an increased copy number of these genes. Consistent with this assumption, it was observed that, while the plasmids pMM401 and pYZ10, which contain an overexpressing allele of ccmFHRc mediated the Ps+ growth of MT-SRP1.r1 on enriched medium, pMM403, which carries a wild-type allele of ccmFHRc, or pYZ4 and pYZ12, which are CcmF− CcmHRc+ and CcmF+ CcmHRc− derivatives of pYZ10, could not do so (Fig. 3). As both pYZ4 and pYZ12 complemented appropriate CcmF− and CcmHRc− mutants (16), these results indicated that not only hyperexpression of ccmFHRc but also intact ccmF and ccmHRc were required for the Ps+ phenotype of MT-SRP1.r1 on enriched medium.
FIG. 3.
Comparison of plasmids carrying various alleles of ccmF-ccmHRc. Plasmids pCS1537, pMM401, and pYZ10 carry a ccmF-ccmHRc up-promoter allele, whereas pMM403 has a wild-type ccmF-ccmHRc allele (Table 1). Plasmids pYZ4 and pYZ12 are CcmF− CcmHRc+ and CcmF+ CcmHRc− derivatives, respectively, of pYZ10. The ability of various plasmids to complement null mutants of CcmF (MD12), CcmHRc (MD14), and CcmI (MT-SRP1.r1) for Ps+ growth on enriched medium is indicated to the right. White arrows with black borders and the gray arrow refer to the promoter-up allele and wild type of ccmF-ccmHRc, respectively, and S, X, K, E, D, A, B, and C refer to cleavage sites of the restriction enzymes SphI, XbaI, KpnI, EcoRI, BclI, SacI, BamHI, and ClaI, respectively.
Chromatophore membranes and soluble fractions prepared from appropriate MT-SRP1.r1 (CcmI− CcmFHRcup) derivatives grown by Res in enriched medium were analyzed by TMBZ SDS-PAGE to confirm that overproduction of CcmFHRc correlated with increased production of c-type Cyts. The amounts of membrane-bound (Fig. 4) and soluble (data not shown) c-type Cyts present in derivatives of MT-SRP1.r1 containing pDA10 and pYZ10 (both ccmFHRcup) were higher than in those carrying pYZ4 (ccmF::spe ccmHRc) and pYZ12 (ccmF ccmHRc::spe), although still lower than in the wild-type strain MT1131. Therefore, whereas overexpression of ccmFHRc was sufficient to overcome the Ps− phenotype of a CcmI-null mutant on minimal medium, its additional hyperexpression via plasmid-borne copies was required for Ps+ growth on both media.
FIG. 4.
c-type Cyt profiles of various pDA10-related Ps+ derivatives of MT-SRP1.r1. Chromatophore membranes were prepared from R. capsulatus strains grown in enriched medium under respiratory condition and subjected to TMBZ SDS-PAGE analyses as described in Materials and Methods. Approximately 75 μg of proteins of strains MT1131 (wild type), MT-SRP1.r1 (CcmI− CcmFHRcup), and its derivatives containing pDA10 [p(ccmFHRcup)1], pYZ10 [p(ccmFHRcup)], pYZ4 [p(ΔccmF ccmHRcup)], and pYZ12 [p(ccmFup ΔccmHRc)], were loaded in each lane. Their respective Ps growth phenotypes on enriched medium are indicated at the bottom of the figure, and the molecular weight markers (in thousands) are shown on the left.
Multiple copies of ccmCDG can mediate Ps growth of MT-SRP1.r1 on enriched medium.
The two EcoRI inserts of pDA5 were also cloned separately into the conjugally transferable plasmid pRK415 to yield pCS1534 and pCS1535 (Fig. 5; Table 1). However, neither of these plasmids complemented MT-SRP1.r1 for Ps+ growth on enriched medium, even though both pCS1534 and pDA5 did so for CcmD-null (SB1003 ΔhelD) and CcmG-null (SB1003 ΔhelX) mutants (Fig. 5). This finding suggested that, although the 1.4-kb fragment (pCS1534) contained expressed wild-type copies of ccmD and ccmG, including a truncated form of ccmC (′ccmC), both of the EcoRI inserts were required for complementation of MT-SRP1.r1 by pDA5. On the other hand, the plasmids p2hel-404 containing ccmABCDG and pCS1532 carrying ccmCDG clusters conferred Ps+ growth ability on MT-SRP1.r1 on enriched medium like pDA5 (Fig. 5, rows 4 and 5). Thus, providing multiple copies of ccmCDG in trans was sufficient to render MT-SRP1.r1 Ps+ on enriched medium.
FIG. 5.
Restriction maps of pDA5 and its derivatives. Plasmid pDA5 carries a ubiE::ccmC fusion in frame spanning the internal EcoRI site joining the 1.6- and 1.4-kb fragments, which are cloned separately to yield pCS1534 and pCS1535, respectively. Plasmids p2hel-404 and pCS1532 carry ccmABCDG and ccmCDG, respectively. pCS1542 and pCS1539 are derivatives of pCS1532 with a ccmC::spe insertion polar on the downstream genes ccmDG and a frameshift mutation in ccmC (ccmΔCB), respectively. pCS1541, pCS1527 and pCS1540 are derivatives of pCHB500 and express ccmDG, ccmD and ccmG, respectively, via the cycA promoter. The ability of these plasmids to complement the CcmI-null mutant MT-SRP1.r1, the CcmD-null mutant SB1003 ΔhelD, the CcmG-null mutant SB1003 ΔhelX, and the CcmC-null mutant TCK2 for Ps+ growth on enriched medium is shown on the right. Curved arrows refer to various promoters mediating different levels of expression of downstream genes, as follows. Black arrow, E. coli lacZ promoter; dark gray arrow, ubiE promoter; light gray arrow, promoter downstream of the ccmB and upstream of the EcoRI site in ccmC; white arrow with black border, cycA promoter; gray arrow with black border, promoter downstream of the BamHI site in ccmC and upstream of ccmD). X, K, E, and B refer to cleavage sites of the restriction enzymes XbaI, KpnI, EcoRI, and BamHI, respectively.
Chromatophore membrane and supernatant fractions prepared from various MT-SRP1.r1 (CcmI− CcmFHRcup) derivatives grown by respiration in enriched medium were analyzed by TMBZ SDS-PAGE to correlate the growth phenotypes monitored here with the Cyt c profile changes. These analyses confirmed that the amounts of the membrane-bound Cyts cp, c1, cy, and co in MT-SRP1.r1big and MT-SRP1.r1 derivatives harboring p2hel-404 (ccmABCDG), pCS1532 (ccmCDG), and pDA5 (ubiE::ccmC ccmDG) (Fig, 6A, lanes 3 to 6) were similar to those seen in the wild-type strain MT1131 (Fig. 6A, lane 1), whereas those in MT-SRP1.r1 and its derivative containing pCS1534 (′ccmC ccmDG) were lower (Fig. 6A, lanes 2 and 7), which is in agreement with their respective Ps phenotypes. Likewise, the amounts of Cyt c2 and c′ in MT-SRP1.r1big and MT-SRP1.r1 derivatives carrying p2hel-404, pCS1532 and pDA5 were also similar to those in wild-type samples (Fig. 6B, lanes 3 to 6) but again lower in MT-SRP1.r1 and its derivative harboring pCS1534 (Fig. 6B, lanes 2 and 7). Thus, the Ps+ phenotypes on enriched medium accompanied increased steady-state amounts of various c-type Cyts.
FIG. 6.
c-type Cyt profiles of various pDA5-related Ps+ derivatives of MT-SRP1.r1. Chromatophore membranes (A and C) and soluble fractions (B) prepared from R. capsulatus strains grown in enriched medium under respiratory conditions were subjected to TMBZ SDS-PAGE analyses, as described in Material and Methods. Approximately 75 μg of proteins was loaded in each lane, and the Ps growth phenotypes on enriched medium of the strains used are indicated at the bottom of the panels B and C. Panels A and B correspond to the membrane and soluble fractions, respectively, of MT1131 (wild type), MT-SRP1.r1 (CcmI− CcmFHRcup), MT-SRP1.r1big (CcmI− CcmFHRcup BigX) and the MT-SRP1.r1 derivatives containing p2hel-404 [p(ccmABCDG)], pCS1532 [p(ccmCDG)], pDA5 [p(ubiE::ccmCDG)] and pCS1534 [p(′ccmC ccmDG)], and panel C corresponds to the membrane fractions of MT1131 (wild type), MT-SRP1.r1 (CcmI− CcmFHRcup) and its derivatives carrying pCS1541 [p(ccmDG)], pCS1527 [p(ccmD)], pCS1540 [p(ccmG)], pCS1542 [p(ccmC::spe ccmDG)], and pCS1539 [p(ccmΔCB ccmDG)], as listed in Table 1. The molecular weight markers (in thousands) are indicated on the left.
A promoter upstream of ccmC is responsible for Ps+ growth of MT-SRP1.r1 on enriched medium.
In order to understand how pDA5, which contained only a truncated form of ccmC, could still complement MT-SRP1.r1, the junction region spanning its two EcoRI fragments was examined. In pDA5, the 507-bp 5′ end of ubiE including a sequence that resembles a R. capsulatus promoter in correct orientation with respect to ccmDG carried by the 1.6-kb insert was fused in frame to the 588-bp 3′ end of ccmC located on the 1.4-kb insert. Thus, this fortuitous construction either created a functional ubiE::ccmC fusion product or provided an additional promoter, thereby increasing the expression of the downstream genes ccmD and ccmG. The first possibility was excluded, because pDA5, unlike pCS1532 or p2hel-404 carrying an intact ccmC, could not complement a nonpolar CcmC-null mutant like TCK2 (62) (Fig. 5, rows 1 to 5). In addition, neither pCS1530 carrying an intact ccmC nor pCS1538 containing the ubiE::ccmC fusion could mediate Ps+ growth of MT-SRP1.r1 on enriched medium (Table 1 and data not shown). On the other hand, the second possibility, that a promoter is located upstream of ccmC, was supported by the fact that pCS1539 carrying a frameshift mutation in ccmC (ccmΔC) but not pCS1542 containing a polar ccmC::spe insertion (47) was able to restore the Ps+ phenotype of MT-SRP1.r1 on enriched medium (Table 1). Indeed, control experiments indicated that both pCS1539 and pCS1542 complemented appropriate CcmD− (SB1003 ΔccmD) and CcmG− (SB1003 ΔccmG) mutants to Ps+/Nadi+ (Fig. 5, rows 6 and 7). Thus, it was concluded that the complementation seen with pDA5 was due to the ubiE promoter.
Overproduction of ccmG is sufficient for Ps+ growth of MT-SRP1.r1 on enriched medium.
The plasmids pCS1527, pCS1540, and pCS1541, overexpressing ccmD, ccmG, and ccmDG, respectively, via the R. capsulatus cycA promoter, were constructed to probe whether Ps+ growth of MT-SRP1.r1 on enriched medium (Table 1) required either or both of these genes. Upon conjugation into MT-SRP1.r1, both pCS1540 and pCS1541, but not pCS1527, were able to confer Ps+ growth on enriched medium, indicating that overexpression of only ccmG was sufficient for this phenotype (Fig. 5, rows 8 to 10). Appropriate MT-SRP1.r1 (CcmI− CcmFHRcup) derivatives were then subjected to immunoblot analyses using polyclonal antibodies against CcmG to confirm that overexpression of ccmG via additional ccmABCDG-endogenous or -exogenous promoters resulted in its overproduction. The amounts of the 18-kDa CcmG protein (41) were indeed higher in the Ps+ derivatives of MT-SRP1.r1 containing pCS1541 (ccmDG), pCS1540 (ccmG) and pCS1539 (ccmΔC and ccmDG) and thus overexpressing ccmG via an additional promoter than in the Ps− derivatives of the same strain carrying pCS1527 (ccmG) and pCS1542 (ccmC::spe and ccmDG) (Fig. 7A, compare lanes 3, 5, and 7 to 4 and 6). Equally, in chromatophore membranes of MT-SRP1.r1 derivatives carrying p2hel404 (ccmABCDG), pCS1532 (ccmCDG) and pDA5 (ubiE::ccmC and ccmDG) CcmG amounts were higher than those in derivatives harboring pCS1534 (′ccmC and ccmDG) and the wild-type strain MT1131 (Fig. 7B, compare lanes 5 to 7 to 2 and 8), which in turn were higher than those in MT-SRP1.r1 and MT-SRP1.r1big (Fig. 7B, lanes 3 and 4). As a control, the 25-kDa membrane-associated protein CcmA (26) was absent in a CcmA-null mutant (KR319) and not overproduced in MT-SRP1.r1, MT-SRP1.r1big, or MT-SRP1.r1 derivatives carrying additional copies of various portions of the ccmABCDG cluster (i.e., p2hel404, pCS1532, pDA5, or pCS1534) (Fig. 7C). Thus, in all instances, the increased amounts of CcmG correlated with the Ps+ phenotype of MT-SRP1.r1 derivatives, demonstrating that its overproduction in addition to that of CcmF and CcmHRc resulted in restoration of sufficient Cyt c biogenesis in the absence of CcmI.
FIG. 7.
Immunoblot analyses of CcmG and CcmA in MT-SRP1.r1 and its Ps+ derivatives. Chromatophore membranes were prepared from R. capsulatus strains grown in enriched medium under respiratory conditions and subjected to SDS-PAGE, approximately 25 μg of protein was loaded in each lane, and immunoblot analyses using polyclonal antibodies against CcmG (A and B) and CcmA (C) were performed as described in Material and Methods. Strains KR319 (CcmA−), MT1131 (wild type), MT-SRP1.r1 (CcmI− CcmFHRcup), and MT-SRP1.r1big (CcmI− CcmFHRcup BigX) and the MT-SRP1.r1 derivatives harboring p2hel-404 [p(ccmABCDG)], pCS1532 [p(ccmCDG)], pDA5 [p(ubiE::ccmCDG)] and pCS1534 [p(′ccmC ccmDG)], pCS1541 [p(ccmDG)], pCS1527 [p(ccmD)], pCS1540 [p(ccmG)], pCS1542 [p(ccmC::spe ccmDG)], pCS1539 [p(ccmΔCB ccmDG)] and pCS1545 [p(ccmG-C75S/C78S)] were used. Their respective Ps growth phenotypes on enriched medium are indicated at the bottom of panels A and C. Relevant molecular weight markers (in thousands) are also indicated.
Profiles of c-type Cyts of MT-SRP1.r1 (CcmI− CcmFHRcup) derivatives carrying various ccmG constructs were analyzed by TMBZ SDS-PAGE, using chromatophore membranes and soluble fractions from cells grown under Res conditions in enriched medium. In agreement with the overproduction of CcmG, the amounts of membrane-bound (Fig. 6C) and soluble (data not shown) c-type Cyts were larger in the Ps+ derivatives of MT-SRP1.r1 carrying pCS1541 (ccmDG), pCS1540 (ccmG) and pCS1539 (ccmΔC and ccmDG) (lanes 3, 5 and 7) than in those containing pCS1527 (ccmD) and pCS1542 (ccmC::spe and ccmDG) (lanes 2, 4, and 6). However, they were still lower than those in the wild-type strain MT1131 (lane 1).
To further probe whether the thioredoxin function of CcmG was indeed required for its ability to confer Ps+ growth on MT-SRP1.r1 in minimal and enriched media, its cysteine residues (Cys 75 and Cys 78) were replaced with serines in pCS1540 to yield pCS1545. This cysteineless CcmG derivative (CcmG-C75S/C78S) was unable to support Ps+ growth of MT-SRP1.r1 on enriched media, although it was produced at adequate amounts (Fig. 7A, compare lanes 5 and 8). Thus, the thioredoxin motif of CcmG was necessary for its suppressor ability under the Ps growth conditions tested here.
DISCUSSION
In this work, a detailed analysis of the Ps+ derivatives of CcmI-null suppressors that overproduce CcmF and CcmHRc and that can grow on both minimal and enriched medium was undertaken to further define the role of CcmI in Cyt c biogenesis. Fortuitous isolation of a ubiE::ccmCDG fusion plasmid led to the discovery that overproduction of CcmG, a component involved in apo-Cyt c thioreduction, was sufficient to remedy the Ps growth inability on enriched medium of CcmI-null suppressors overproducing CcmF and CcmHRc. Overexpression of ccmG could be achieved via various promoter activities, presumably by increasing its transcription. Positioning ccmG either artificially downstream of the exogenous ubiE or cycA promoters or using chromosomal fragments carrying promoter activities that are naturally located upstream of it yielded appreciable overproduction of CcmG.
The data obtained also pointed out that the transcriptional organization of the ccmABCDG cluster is complex. In addition to a promoter located upstream of ORF124/ccmA (5), a second promoter located between the stop codon of ccmB and the EcoRI site within ccmC is also present as a plasmid harboring only ccmCDG (Table 1) could render MT-SRP1.r1 Ps+ on enriched media. Moreover, since plasmids containing a polar insertion in ccmC or a fragment carrying a truncated ccmC upstream of intact ccmD and ccmG can complement appropriate CcmD− and CcmG− mutants as well (Fig. 5) but fail to support Ps+ growth of MT-SRP1.r1 on enriched medium, a third, weaker promoter appears to be located downstream from the BamHI site in ccmC and upstream of ccmD. The presence of a promoter within the ccmABCDG cluster endowing ccmCDG with the ability to be expressed independently of ccmAB raises the possibility that the products of these two clusters could be synthesized in different stoichiometries.
In addition to CcmG overproduction, CcmI− mutants containing both chromosomal and plasmid-borne copies of a promoter-up allele of ccmFHRcup cluster (16), hence highly overproducing CcmF and CcmHRc (data not shown), are also proficient for Cyt c maturation in minimal and enriched media. However, the photosynthetic growth of a CcmI− mutant carrying a chromosomal wild-type ccmFHRc allele (e.g., MT-SRP1) is much slower (requiring at least 7 days of incubation) than growth of a mutant with a similar promoter-up ccmFHRcup allele (e.g., MT-SRP1.r1). Furthermore, neither of these CcmI− mutants could be complemented for Ps+ growth on enriched medium upon introduction of a plasmid-borne wild-type copy of the ccmFHRc cluster (Fig. 3). Thus, a threshold level of CcmF and CcmHRc overproduction appears to be required to overcome the absence of CcmI during Cyt c biogenesis.
The fact that both CcmG and CcmHRc are known to be components of the thioreduction pathway (41), and that the thioredoxin motif of CcmG is required for suppression, raises the possibility that the growth defect of CcmI mutants on enriched medium might be due to a lack of adequate redox power to ensure sufficient holo-Cyt c formation. How the levels of CcmF, CcmHRc, and CcmG are regulated in R. capsulatus is not yet well known. Available data indicate that the amounts of CcmHRc and possibly of CcmF are dramatically increased during Res, compared to Ps, growth conditions, whereas those of CcmG remain almost unchanged. Similarly, the levels of CcmHRc and CcmG are higher in enriched than in minimal medium (24). These findings suggest that, while larger amounts of thioreduction components are required to support both Res and Ps growth in enriched medium, smaller amounts of CcmF and CcmHRc are sufficient under Ps than under Res conditions. Although the amounts of CcmF, CcmHRc and CcmG are regulated depending on the growth conditions, in the absence of CcmI, their levels might need to be adjusted to sustain productive Cyt c biogenesis. However, supplementation of enriched medium neither with different redox-active chemicals nor with the different nutritional constituents of the minimal medium (see Material and Methods) was sufficient to restore the Ps+ phenotype of MT-SRP1.r1. Thus, bypassing the absence of CcmI might not be mediated solely by the increased protein dithiol:disulfide oxidoreductase activities via the overproduction of CcmG and CcmHRc.
Remarkably, suppression of ccmI always requires both CcmF and CcmHRc, as if these two components act together. In E. coli, CcmF, CcmE, and CcmHEc coimmunoprecipitate, suggesting that they might form a heme ligation complex transferring heme from CcmE to apo-Cyt c (49). If a similar situation is also valid in R. capsulatus (16), then comparable interactions between CcmF, CcmHRc, CcmI, and CcmE might be expected (Fig. 1, bottom). Thus, a possibility is that overproduction of CcmG, CcmF, and CcmHRc in the absence of CcmI might enhance more efficient interactions of the remaining components so that adequate thioreduction of, and heme ligation to, the apo-Cyt c substrates can occur. Alternatively, overproduction of CcmF and CcmHRc could increase the capture rate of apo-Cyt c substrates by the heme ligation complex to compete out their proteolysis in the absence of specific protection factors like CcmI, as proposed earlier (16). In E. coli, the formation of specific disulfide intermediates between the thioredoxin motifs of CcmG and CcmHEc, respectively, as well as the heme binding motif of apo-Cyt c and the thioredoxin motif of CcmHEc, has been documented (23, 48). Conceivably, in the absence of CcmI, the rate of these interactions compared to that of apo-Cyt c proteolysis might be limiting.
Recently, it has been shown that expression in trans of the N-terminal transmembrane helix and the adjacent cytoplasmic loop of CcmI-1 is not sufficient for Cyt c maturation but alleviates the protoporphyrin IX (PPIX) accumulation phenotype of CcmI-null mutants in Sinorhizobium meliloti (10). This observation led to the proposal that in the absence of the CcmI-1 region, either heme synthesis via ferrochelatase or heme transport across the membrane is defective, leading to PPIX accumulation. However, the fact that CcmI-null mutants are fully proficient for c-type Cyt biogenesis by overproduction of CcmG in addition to CcmF and CcmHRc, as shown in this work, argues that the main function of CcmI might not be directly related to heme biosynthesis or delivery. It has been reported that CcmF in Paracoccus denitrificans, Pseudomonas putida, and Rhizobium etli, as well as CcmI in the latter species, is required not only for Cyt c maturation but also for siderophore synthesis and iron acquisition (18, 45, 65). In R. etli, CcmF− or CcmI− mutants also accumulate PPIX (65) raising the possibility that the CcmI-1 homologue of S. meliloti might be specifically involved in these periplasmic processes. Several R. capsulatus Cyt c biogenesis mutants exhibit a similar porphyrin excretion phenotype (7, 15, 26, 36); however, whether CcmI, CcmF and CcmHRc are necessary for iron acquisition in this species remains to be seen.
How CcmI interacts with the c-type apo-Cyts is unknown. Disulfide bonds are thought to be formed within the cysteines of the conserved heme-binding motif of the apo-Cyt c (22), possibly via the DsbA and DsbB components of the periplasmic protein disulfide oxidoreductase pathway (39, 40, 50, 51). However, it is now clear that this thio-oxidative pathway is not essential for Cyt c maturation (1, 17, 20). Nonetheless, when it is present, these bonds may need to be reduced via the thioreduction branch prior to heme attachment (15, 30, 41). It is thus tempting to speculate that CcmI might interact with other Cyt c biogenesis components, such as CcmF, CcmHRc, and CcmG, via the leucine zipper-like domain in CcmI-1 (38), while recognizing the disulfide bond containing c-type apo-Cyt substrates via its tetratricopeptide-like (TPR) motifs in CcmI-2 (10) (Fig. 1), or vice versa. Whereas leucine zipper motifs facilitate dimerization (37, 42), TPR-containing proteins interact with a partner protein(s) in multiprotein complexes (8, 13).
Finally, it should be mentioned that overexpression of ccmG is not the molecular basis of the Ps+ phenotype on enriched medium of the ccmI suppressor MT-SRP1.r1big (Table 2) (which was used to construct the initial chromosomal library), because similar amounts of CcmG are present in chromatophore membranes of MT-SRP1.r1 and MT-SRP1.r1big (Fig. 7B). Moreover, no DNA sequence difference in the 5′ upstream promoter region of the ORF124/ccmABCDG cluster or in the ccmB-ccmC intergenic region was found in MT-SRP1.r1big compared to MT-SRP1.r1 (data not shown). Thus, another mutation(s) in a currently unknown component(s), such as periplasmic proteases, might also confer Ps+ growth ability on a CcmI-null mutant.
In summary, it is clear that overproduction of CcmG in addition to that of CcmF and CcmHRc is necessary for full efficiency of Cyt c maturation in the absence of CcmI, suggesting specific interactions between these components. Future analyses of these interactions should provide further insights on the cross-point between the apo-Cyt c delivery and the thioreduction branches of Cyt c biogenesis.
Acknowledgments
This work was supported by grants DOE 91ER20052 and NIH GM 38237 to F.D. and GM 47909 to R.G.K.
We thank K. Wade for her help during this work and J. W. Cooley for his editorial comments.
REFERENCES
- 1.Allen, J. W., P. D. Barker, and S. J. Ferguson. 2003. A cytochrome b562 variant with a c-type cytochrome CXXCH heme-binding motif as a probe of the Escherichia coli cytochrome c maturation system. J. Biol. Chem. 278:52075-52083. [DOI] [PubMed] [Google Scholar]
- 2.Allen, J. W., O. Daltrop, J. M. Stevens, and S. J. Ferguson. 2003. c-type cytochromes: diverse structures and biogenesis systems pose evolutionary problems. Philos. Trans. R. Soc. Lond. B. 358:255-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Beckman, D. L., and R. G. Kranz. 1993. Cytochromes c biogenesis in a photosynthetic bacterium requires a periplasmic thioredoxin-like protein. Proc. Natl. Acad. Sci. USA 90:2179-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckman, D. L., D. R. Trawick, and R. G. Kranz. 1992. Bacterial cytochromes c biogenesis. Genes Dev. 6:268-283. [DOI] [PubMed] [Google Scholar]
- 6.Benning, C., and C. R. Somerville. 1992. Isolation and genetic complementation of a sulfolipid-deficient mutant of Rhodobacter sphaeroides. J. Bacteriol. 174:2352-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biel, S. W., and A. J. Biel. 1990. Isolation of a Rhodobacter capsulatus mutant that lacks c-type cytochromes and excretes porphyrins. J. Bacteriol. 172:1321-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blatch, G. L., and M. Lassle. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932-939. [DOI] [PubMed] [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 10.Cinege, G., A. Kereszt, S. Kertesz, G. Balogh, and I. Dusha. 2004. The roles of different regions of the CycH protein in c-type cytochrome biogenesis in Sinorhizobium meliloti. Mol. Genet. Genomics 271:171-179. [DOI] [PubMed] [Google Scholar]
- 11.Collet, J. F., and J. C. Bardwell. 2002. Oxidative protein folding in bacteria. Mol. Microbiol. 44:1-8. [DOI] [PubMed] [Google Scholar]
- 12.Daldal, F., S. Cheng, J. Applebaum, E. Davidson, and R. C. Prince. 1986. Cytochrome c2 is not essential for photosynthetic growth of Rhodopseudomonas capsulata. Proc. Natl. Acad. Sci. USA 83:2012-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Andrea, L. D., and L. Regan. 2003. TPR proteins: the versatile helix. Trends Biochem. Sci. 28:655-662. [DOI] [PubMed] [Google Scholar]
- 14.Davidson, E., and F. Daldal. 1987. Primary structure of the bc1 complex of Rhodopseudomonas capsulata. Nucleotide sequence of the pet operon encoding the Rieske cytochrome b, and cytochrome c1 apoproteins. J. Mol. Biol. 195:13-24. [DOI] [PubMed] [Google Scholar]
- 15.Deshmukh, M., G. Brasseur, and F. Daldal. 2000. Novel Rhodobacter capsulatus genes required for the biogenesis of various c-type cytochromes. Mol. Microbiol. 35:123-138. [DOI] [PubMed] [Google Scholar]
- 16.Deshmukh, M., M. May, Y. Zhang, K. K. Gabbert, K. A. Karberg, R. G. Kranz, and F. Daldal. 2002. Overexpression of ccl1-2 can bypass the need for the putative apocytochrome chaperone CycH during the biogenesis of c-type cytochromes. Mol. Microbiol. 46:1069-1080. [DOI] [PubMed] [Google Scholar]
- 17.Deshmukh, M., S. Turkarslan, D. Astor, M. Valkova-Valchanova, and F. Daldal. 2003. The dithiol:disulfide oxidoreductases DsbA and DsbB of Rhodobacter capsulatus are not directly involved in cytochrome c biogenesis, but their inactivation restores the cytochrome c biogenesis defect of CcdA-null mutants. J. Bacteriol. 185:3361-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Vrind, J. P., G. J. Brouwers, P. L. Corstjens, J. den Dulk, and E. W. de Vrind-de Jong. 1998. The cytochrome c maturation operon is involved in manganese oxidation in Pseudomonas putida GB-1. Appl. Environ. Microbiol. 64:3556-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ditta, G., T. Schmidhauser, E. Yakobson, P. Lu, X. W. Liang, D. R. Finlay, D. Guiney, and D. R. Helinski. 1985. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid 13:149-153. [DOI] [PubMed] [Google Scholar]
- 20.Erlendsson, L. S., and L. Hederstedt. 2002. Mutations in the thiol-disulfide oxidoreductases BdbC and BdbD can suppress cytochrome c deficiency of CcdA-defective Bacillus subtilis cells. J. Bacteriol. 184:1423-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fabianek, R. A., H. Hennecke, and L. Thony-Meyer. 1998. The active-site cysteines of the periplasmic thioredoxin-like protein CcmG of Escherichia coli are important but not essential for cytochrome c maturation in vivo. J. Bacteriol. 180:1947-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fabianek, R. A., H. Hennecke, and L. Thony-Meyer. 2000. Periplasmic protein thiol:disulfide oxidoreductases of Escherichia coli. FEMS Microbiol. Rev. 24:303-316. [DOI] [PubMed] [Google Scholar]
- 23.Fabianek, R. A., T. Hofer, and L. Thony-Meyer. 1999. Characterization of the Escherichia coli CcmH protein reveals new insights into the redox pathway required for cytochrome c maturation. Arch. Microbiol. 171:92-100. [DOI] [PubMed] [Google Scholar]
- 24.Gabbert, K. K., B. S. Goldman, and R. G. Kranz. 1997. Differential levels of specific cytochrome c biogenesis proteins in response to oxygen: analysis of the ccl operon in Rhodobacter capsulatus. J. Bacteriol. 179:5422-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldman, B. S., D. L. Beck, E. M. Monika, and R. G. Kranz. 1998. Transmembrane heme delivery systems. Proc. Natl. Acad. Sci. USA 95:5003-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldman, B. S., D. L. Beckman, A. Bali, E. M. Monika, K. K. Gabbert, and R. G. Kranz. 1997. Molecular and immunological analysis of an ABC transporter complex required for cytochrome c biogenesis. J. Mol. Biol. 268:724-738. [DOI] [PubMed] [Google Scholar]
- 27.Goldman, B. S., and R. G. Kranz. 2001. ABC transporters associated with cytochrome c biogenesis. Res. Microbiol. 152:323-329. [DOI] [PubMed] [Google Scholar]
- 28.Gray, K. A., M. Grooms, H. Myllykallio, C. Moomaw, C. Slaughter, and F. Daldal. 1994. Rhodobacter capsulatus contains a novel cb-type cytochrome c oxidase without a CuA center. Biochemistry 33:3120-3127. [DOI] [PubMed] [Google Scholar]
- 29.Kadokura, H., F. Katzen, and J. Beckwith. 2003. Protein disulfide bond formation in prokaryotes. Annu. Rev. Biochem. 72:111-135. [DOI] [PubMed] [Google Scholar]
- 30.Katzen, F., M. Deshmukh, F. Daldal, and J. Beckwith. 2002. Evolutionary domain fusion expanded the substrate specificity of the transmembrane electron transporter DsbD. EMBO J. 21:3960-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 32.Keilin, D. 1966. The history of cell respiration and cytochrome. Cambridge University Press, Cambridge, United Kingdom.
- 33.Koch, H. G., O. Hwang, and F. Daldal. 1998. Isolation and characterization of Rhodobacter capsulatus mutants affected in cytochrome cbb3 oxidase activity. J. Bacteriol. 180:969-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koch, H. G., H. Myllykallio, and F. Daldal. 1998. Using genetics to explore cytochrome function and structure in Rhodobacter. Methods Enzymol. 297:81-94. [Google Scholar]
- 35.Kranz, R., R. Lill, B. Goldman, G. Bonnard, and S. Merchant. 1998. Molecular mechanisms of cytochrome c biogenesis: three distinct systems. Mol. Microbiol. 29:383-396. [DOI] [PubMed] [Google Scholar]
- 36.Kranz, R. G. 1989. Isolation of mutants and genes involved in cytochromes c biosynthesis in Rhodobacter capsulatus. J. Bacteriol. 171:456-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Landschulz, W. H., P. F. Johnson, and S. L. McKnight. 1988. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759-1764. [DOI] [PubMed] [Google Scholar]
- 38.Lang, S. E., F. E. Jenney, Jr., and F. Daldal. 1996. Rhodobacter capsulatus CycH: a bipartite gene product with pleiotropic effects on the biogenesis of structurally different c-type cytochromes. J. Bacteriol. 178:5279-5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metheringham, R., L. Griffiths, H. Crooke, S. Forsythe, and J. Cole. 1995. An essential role for DsbA in cytochrome c synthesis and formate-dependent nitrite reduction by Escherichia coli K-12. Arch. Microbiol. 164:301-307. [DOI] [PubMed] [Google Scholar]
- 40.Metheringham, R., K. L. Tyson, H. Crooke, D. Missiakas, S. Raina, and J. A. Cole. 1996. Effects of mutations in genes for proteins involved in disulphide bond formation in the periplasm on the activities of anaerobically induced electron transfer chains in Escherichia coli K12. Mol. Gen. Genet. 253:95-102. [DOI] [PubMed] [Google Scholar]
- 41.Monika, E. M., B. S. Goldman, D. L. Beckman, and R. G. Kranz. 1997. A thioreduction pathway tethered to the membrane for periplasmic cytochromes c biogenesis; in vitro and in vivo studies. J. Mol. Biol. 271:679-692. [DOI] [PubMed] [Google Scholar]
- 42.O'Shea, E. K., J. D. Klemm, P. S. Kim, and T. Alber. 1991. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254:539-544. [DOI] [PubMed] [Google Scholar]
- 43.Page, M. D., Y. Sambongi, and S. J. Ferguson. 1998. Contrasting routes of c-type cytochrome assembly in mitochondria, chloroplasts and bacteria. Trends Biochem. Sci. 23:103-108. [DOI] [PubMed] [Google Scholar]
- 44.Page, M. D., N. F. Saunders, and S. J. Ferguson. 1997. Disruption of the Pseudomonas aeruginosa dipZ gene, encoding a putative protein-disulfide reductase, leads to partial pleiotropic deficiency in c-type cytochrome biogenesis. Microbiology 143:3111-3122. [DOI] [PubMed] [Google Scholar]
- 45.Pearce, D. A., M. D. Page, H. A. Norris, E. J. Tomlinson, and S. J. Ferguson. 1998. Identification of the contiguous Paracoccus denitrificans ccmF and ccmH genes: disruption of ccmF, encoding a putative transporter, results in formation of an unstable apocytochrome c and deficiency in siderophore production. Microbiology 144:467-477. [DOI] [PubMed] [Google Scholar]
- 46.Pettigrew, G. W., and G. R. Moore. 1987. Cytochromes c. Biological aspects. Springer Verlag, New York, N.Y.
- 47.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 48.Reid, E., J. Cole, and D. J. Eaves. 2001. The Escherichia coli CcmG protein fulfils a specific role in cytochrome c assembly. Biochem. J. 355:51-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren, Q., U. Ahuja, and L. Thony-Meyer. 2002. A bacterial cytochrome c heme lyase. CcmF forms a complex with the heme chaperone CcmE and CcmH but not with apocytochrome c. J. Biol. Chem. 277:7657-7663. [DOI] [PubMed] [Google Scholar]
- 50.Sambongi, Y., and S. J. Ferguson. 1996. Mutants of Escherichia coli lacking disulphide oxidoreductases DsbA and DsbB cannot synthesise an exogenous monohaem c-type cytochrome except in the presence of disulphide compounds. FEBS Lett. 398:265-268. [DOI] [PubMed] [Google Scholar]
- 51.Sambongi, Y., and S. J. Ferguson. 1994. Specific thiol compounds complement deficiency in c-type cytochrome biogenesis in Escherichia coli carrying a mutation in a membrane-bound disulphide isomerase-like protein. FEBS Lett. 353:235-238. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 53.Schaegger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1-100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 54.Schulz, H., H. Hennecke, and L. Thony-Meyer. 1998. Prototype of a heme chaperone essential for cytochrome c maturation. Science 281:1197-1200. [DOI] [PubMed] [Google Scholar]
- 55.Scolnik, P. A., M. A. Walker, and B. L. Marrs. 1980. Biosynthesis of carotenoids derived from neurosporene in Rhodopseudomonas capsulata. J. Biol. Chem. 255:2427-2432. [PubMed] [Google Scholar]
- 56.Setterdahl, A. T., B. S. Goldman, M. Hirasawa, P. Jacquot, A. J. Smith, R. G. Kranz, and D. B. Knaff. 2000. Oxidation-reduction properties of disulfide-containing proteins of the Rhodobacter capsulatus cytochrome c biogenesis system. Biochemistry 39:10172-10176. [DOI] [PubMed] [Google Scholar]
- 57.Sistrom, W. R. 1960. A requirement for sodium in the growth of Rhodopseudomonas sphaeroides. J. Gen. Microbiol. 22:778-785. [DOI] [PubMed] [Google Scholar]
- 58.Thomas, P. E., D. Ryan, and W. Levin. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75:168-176. [DOI] [PubMed] [Google Scholar]
- 59.Thony-Meyer, L. 1997. Biogenesis of respiratory cytochromes in bacteria. Microbiol. Mol. Biol. Rev. 61:337-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thony-Meyer, L. 2002. Cytochrome c maturation: a complex pathway for a simple task? Biochem. Soc. Trans. 30:633-638. [DOI] [PubMed] [Google Scholar]
- 61.Thony-Meyer, L. 2000. Haem-polypeptide interactions during cytochrome c maturation. Biochim. Biophys. Acta 1459:316-324. [DOI] [PubMed] [Google Scholar]
- 62.Yano, T., C. Sanders, J. Catalano, and F. Daldal. 2005. 5-FOA/pyrE based bi-directional auxotrophic selection system of Rhodobacter capsulatus: Construction of an unmarked mutant in the helC gene. Appl. Environ. Microbiol. 71:3014-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yen, H. C., N. T. Hu, and B. L. Marrs. 1979. Characterization of the gene transfer agent made by an overproducer mutant of Rhodopseudomonas capsulata. J. Mol. Biol. 131:157-168. [DOI] [PubMed] [Google Scholar]
- 64.Yen, H. C., and B. L. Marrs. 1976. Map of genes for the carotenoid and bacteriochlorophyll biosynthesis in Rhodopseudomonas capsulata. J. Bacteriol. 126:619-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yeoman, K. H., M. J. Delgado, M. Wexler, J. A. Downie, and A. W. Johnston. 1997. High affinity iron acquisition in Rhizobium leguminosarum requires the cycHJKL operon and the feuPQ gene products, which belong to the family of two-component transcriptional regulators. Microbiology 143:127-134. [DOI] [PubMed] [Google Scholar]
- 66.Zannoni, D., and F. Daldal. 1993. The role of c-type cytochromes in catalyzing oxidative and photosynthetic electron transport in the dual functional plasma membrane of facultative phototrophs. Arch. Microbiol. 160:413-423. [DOI] [PubMed] [Google Scholar]