Abstract
Oxidative stress in Bacillus subtilis results in the accumulation of Spx protein, which exerts both positive and negative transcriptional control over a genome-wide scale through its interaction with the RNA polymerase α subunit. Previous microarray transcriptome studies uncovered a unique class of genes that are controlled by Spx-RNA polymerase interaction under normal growth conditions that do not promote Spx overproduction. These genes were repressed by Spx when sulfate was present as a sole sulfur source. The genes include those of the ytmI, yxeI, and ssu operons, which encode products resembling proteins that function in the uptake and desulfurization of organic sulfur compounds. Primer extension and analysis of operon-lacZ fusion expression revealed that the operons are repressed by sulfate and cysteine; however, Spx functioned only in sulfate-dependent repression. Both the ytmI operon and the divergently transcribed ytlI, encoding a LysR-type regulator that positively controls ytmI operon transcription, are repressed by Spx in sulfate-containing media. The CXXC motif of Spx, which is necessary for redox sensitive control of Spx activity in response to oxidative stress, is not required for sulfate-dependent repression. The yxeL-lacZ and ssu-lacZ fusions were also repressed in an Spx-dependent manner in media containing sulfate as the sole sulfur source. This work uncovers a new role for Spx in the control of sulfur metabolism in a gram-positive bacterium under nonstressful growth conditions.
Spx is a global transcriptional regulator of the oxidative stress response in Bacillus subtilis and is highly conserved among low-G+C-content gram-positive bacteria (34, 35, 43). It was initially identified as a protein encoded by a gene that was the site of suppressor mutations in clpP and clpX mutants (29). Spx concentration is proteolytically controlled by the ATP-dependent protease ClpXP (35, 36). The high concentrations of Spx in clpX and clpP mutants result in poor growth and sporulation and the loss of competence development as well as reduced anaerobic metabolism. Spx was discovered to exert global negative transcriptional control by blocking activator-RNA polymerase (RNAP) interaction (35). This it does by binding to the C-terminal domain of the RNAP α subunit at a site bearing Tyr263, which is a highly conserved amino acid position in the RNAP α of gram-positive bacteria. This interaction was determined to be responsible for the defects in growth and development observed in clpX and clpP mutants. Spx exhibited no DNA-binding activity in vitro.
High concentrations of Spx also resulted in the induction of genes whose products function in thiol homeostasis and the biosynthesis of cysteine, as shown by probing B. subtilis RNA with genomic microarrays (34). The trxA and trxB genes, encoding thioredoxin and thioredoxin reductase, respectively, are transcriptionally activated under conditions of oxidative stress by a mechanism that involves Spx-RNAP interaction. Reconstitution of transcription initiation in vitro demonstrated that Spx could stimulate transcription from the trxA and -B promoters by direct interaction with RNAP (33). Spx-dependent activation of trxA and -B was observed only under oxidative conditions and does not involve initial binding of Spx to DNA. The oxidative activation of Spx activity is attributed to the CXXC motif at the N terminus of Spx protein, which constitutes a disulfide/thiol switch. High concentrations of Spx and the accompanying increase in Spx-dependent positive and negative transcriptional control were observed in cells treated with the thiol-specific oxidant diamide (34). Spx bears no resemblance to any other transcription factor but instead shows secondary structure similarity to the ArsC protein (arsenate reductase) of Escherichia coli plasmid R773 (25, 43).
The aforementioned microarray experiments uncovered a unique class of Spx-regulated genes whose products function in alternative sulfur source utilization (see Results). These are repressed by low concentrations of Spx under nonstressful growth conditions and include the ytmI, ssu, and yxeI operons, which encode highly conserved proteins involved in the transport and utilization of alternative sulfur sources such as organic sulfonates and sulfate esters (1, 3, 6, 17-21, 40, 41). The expression of the operons in diverse bacterial species is regulated in response to sulfur source availability (3-5, 7, 18, 22, 39, 41). Transcriptional repression is observed in the presence of the preferred sulfur sources, sulfate or cysteine, while the operons are derepressed in the presence of methionine or taurine (1). This pattern of control has been reported in the cases of the ytmI and ssu operons of B. subtilis (7, 39).
In E. coli, the regulation of transcription in response to sulfur source is attributed to two transcriptional regulators, CysB and Cbl (15, 23, 26, 28, 37, 42). Cbl is a LysR homolog that activates ssu operon transcription in the absence of sulfate and is inhibited by the anti-inducer adenosine 5′-phosphosulfate (APS), which is an intermediate in the assimilation of sulfate. CysB down-regulates ssu transcription in the presence of cysteine and is sensitive to the inducer N-acetyl-l-serine. The ssu and ytmI operons of B. subtilis appear to be similarly regulated, but until recently no regulator conferring Cys- or sulfate-dependent control had been identified. The ytmI operon is regulated by the product of the divergently transcribed gene ytlI, which encodes a LysR homolog dedicated to ytmI operon transcriptional control (7). YtlI is necessary for activation of ytmI transcription in the absence of Cys and sulfate, but the inducer and/or coactivator for YtlI have not been uncovered.
In this report, evidence is presented that Spx negatively controls ssu, yxeI, and ytmI operon gene transcription in response to the presence of sulfate but not cysteine. A null mutation of the spx gene and a mutation of rpoA that blocks Spx-RNA polymerase interaction result in the derepression of ssu, yxeI, and ytmI operon expression when sulfate is the sole source of sulfur. The ytlI gene is also under Spx negative control when sulfate is the sole sulfur source. This function of Spx is carried out under nonstressful growth conditions that do not result in elevated Spx concentration, unlike the situation in cells undergoing oxidative stress, in which Spx concentration and activity are upregulated (33, 34).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The B. subtilis strains used in this study are listed in Table 1 and are derivatives of strain JH642. E. coli cells were grown at 37°C in 2× yeast extract-tryptone (YT) liquid or on Luria-Bertani solid medium containing 1.2% agar (Difco). B. subtilis cells were grown at 37°C in 2× YT, Difco sporulation medium (13), or TSS minimal medium (10) with some modifications (37.4 mM NH4Cl, 2 mM K2HPO4, 49.5 mM Tris, 5 mM MgCl2, 0.5% glucose, 0.004% FeCl3, 0.004% citric acid, 0.1% l-glutamate, 0.005% auxotrophic requirements) and supplemented with one of the following sulfur sources at a concentration of 1 mM: l-cysteine, l-methionine, or MgSO4. The solid medium was Difco sporulation medium supplemented with 1.2% agar. Sulfate-free solid medium was prepared by adding 1% agarose to TSS. X-Gal was added to plates at a concentration of 40 μg/ml. When necessary, the following antibiotics were added (at the concentrations shown in parentheses): ampicillin (25 μg/ml), neomycin (5 μg/ml), chloramphenicol (5 μg/ml), erythromycin plus lincomycin (1 and 25 μg/ml, respectively), and spectinomycin (75 μg/ml).
TABLE 1.
Bacillus subtilis strains
| Strain | Relevant genotype | Source and/or reference |
|---|---|---|
| BFS71 | trpC2 ytmI′-lacZ erm ΔytmI | 7 |
| BSIP1214 | trpC2 ytlI::aphA3 | 7 |
| SB24 | trpC2 amyE::ssu21-289-lacZ | 39 |
| JH642 | trpC2 pheA1 | J. A. Hoch |
| ORB3621 | trpC2 pheA1 rpoAcxs-1 | 32 |
| ORB3834 | trpC2 pheA1 spx::aphA3 | 29 |
| ORB3909 | trpC2 pheA1 spx::spc | This study |
| ORB4269 | trpC2 pheA1 yxeL::pMMN519 (yxeL-lacZ) | M. M. Nakano |
| ORB4271 | trpC2 pheA1 amyE::Pspank-hy-spx | 34 |
| ORB4785 | trpC2 pheA1 yxeL::pMMN519 (yxeL-lacZ) rpoAcxs-1 | This study |
| ORB4786 | trpC2 pheA1 yxeL::pMMN519 (yxeL-lacZ) spx::aphA3 | This study |
| ORB4794 | trpC2 pheA1 ytmI′-lacZ erm ΔytmI | This study |
| ORB4795 | trpC2 pheA1 ytlI::aphA3 | This study |
| ORB4801 | trpC2 pheA1 ytmI′-lacZ erm ΔytmI rpoAcxs-1 | This study |
| ORB4802 | trpC2 pheA1 ytmI′-lacZ erm ΔytmI spx::aphA3 | This study |
| ORB4803 | trpC2 pheA1 ytmI′-lacZ erm ΔytmI spx::spc | This study |
| ORB4804 | trpC2 pheA1 ytmI′-lacZ ytlI::aphA3 | This study |
| ORB4807 | trpC2 pheA1 ytmI′-lacZ erm ΔytmI spx::aphA3 amyE::Pspank-hy-spx | This study |
| ORB4814 | trpC2 pheA1 amyE::Pspank-hy-spx(C10A) | This study |
| ORB4815 | trpC2 pheA1 amyE::Pspank-hy-spx(C13A) | This study |
| ORB4863 | trpC2 pheA1 ytmI′-lacZ erm ΔytmI spx::neo amyE::Pspank-hy-spx(C10A) | This study |
| ORB4864 | trpC2 pheA1 ytmI′-lacZ erm ΔytmI spx::neo amyE::Pspank-hy-spx(C13A) | This study |
| ORB4866 | trpC2 pheA1 amyE::ytlI-lacZ cat | This study |
| ORB4870 | trpC2 pheA1 amyE::ytlI-lacZ cat rpoAcxs-1 | This study |
| ORB4871 | trpC2 pheA1 amyE::ytlI-lacZ cat spx::aphA3 | This study |
| ORB4873 | trpC2 pheA1 amyE::ytlI-lacZ cat ytlI::aphA3 | This study |
| ORB4891 | trpC2 pheA1 amyE::ssu21-289-lacZ | This study |
| ORB4896 | trpC2 pheA1 amyE::ssu21-289-lacZ rpoAcxs-1 | This study |
| ORB4898 | trpC2 pheA1 amyE::ssu21-289-lacZ spx::neo | This study |
| ORB4962 | trpC2 pheA1 amyE::Pspac-ytlI | This study |
| ORB4977 | trpC2 pheA1 ytmI′-lacZ amyE::Pspac-ytlI | This study |
| ORB4979 | trpC2 pheA1 ytmI′-lacZ amyE::Pspac-ytlI ytlI::aphA3 | This study |
| ORB4983 | trpC2 pheA1 ytmI′-lacZ amyE::Pspac-ytlI ytlI::aphA3 spx::spc | This study |
Plasmid and strain construction.
Plasmids constructed in this study are listed in Table 2. Primers used in this study are listed in Table 3.
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant genotype or property | Source or reference |
|---|---|---|
| pDG1727 | Contains Spr cassette | 11 |
| pDH32 | Creates lacZ fusions | D. Henner |
| pDR66 | Allows IPTG-dependent gene expression | 14 |
| pDR111 | Allows IPTG-dependent gene expression | 2 |
| pFH3 | pUC18 with spx | 29 |
| pKE17 | pDR66 with ytlI under control of Pspac | This study |
| pSN95 | pDR111 with spx(C10A) under control of Pspank-hy | This study |
| pSN96 | pDR111 with spx(C13A) under control of Pspank-hy | This study |
| pSN104 | pUC19 with ytlI | This study |
| pSN105 | pDH32 with ytlI | This study |
| pYZ81 | pFH3 with spx::spc | This study |
TABLE 3.
Primers used in this study
| Primer | Sequence |
|---|---|
| oKE18 | 5′-ACGCGTCGACGCATACTCTGATCTTTGACTAATC-3′ |
| oKE19 | 5′-ACATGCATGCCGTGAAACCATCTCCTGATGAATC-3′ |
| oSN03-66 | 5′-CACATCACCAAGCGCGACTTCATGCAGAA-3′ |
| oSN03-67 | 5′-TTCTGCATGAAGTCGCGCTTGGTGATGTG-3′ |
| oSN03-68 | 5′-CAAGCTGTACTTCAGCGAGAAAGGCGAGAG-3′ |
| oSN03-69 | 5′-CTCTCGCCTTTCTCGCTGAAGTACAGCTTG-3′ |
| oSN03-70 | 5′-CACCAAGCGCGACTTCAGCGAGAAAGGCGAG-3′ |
| oSN03-71 | 5′-CTCGCCTTTCTCGCTGAAGTCGCGCTTGGTG-3′ |
| oSN03-73 | 5′-GGAATTCACCCTCTCCTTCTGTACATGTCAGC-3′ |
| oSN03-74 | 5′-CCGGATCCGTTGTGTTTAAGCGCCTCGTATTGC-3′ |
| Pspac-up | 5′-GACTTTATCTACAAGGTGTG-3′ |
| Pspac-down | 5′-AAATGATGACCTCGTTTCCA-3′ |
| oMN01-173 | 5′-CGAGGAAGCTTAGATGTTCATCCTACTA-3′ |
| oMN01-174 | 5′-TACCAGCAGGTCGACAAATAAAAGAAGG-3′ |
| o-sn04-90 | 5′-GAGTATTTCTGCAGCTTTATAGAAGCTTCC-3′ |
| o-sn04-91 | 5′-AGCTCCGATGCATCTTCAACAGTTGCC-3′ |
B. subtilis strain ORB3909 (spx::spc) was constructed as follows. A spectinomycin resistance cassette (Spr) was isolated from pDG1727 (11) by digestion with BamHI and XhoI. The fragment was filled in to create blunt ends and inserted into pFH3 digested with BclI, creating a product that was also filled in. The ligated product was pYZ81, which was introduced by transformation into JH642.
The strain ORB4269 contained yxeL-lacZ integrated at the yxeI operon locus of the chromosome. A 600-bp fragment that contains the 3′ end of yxeK and the 5′ end of yxeL was amplified by PCR using oligonucleotides oMN02-210 (5′-CTGCGGAATTCGGACGTGAT-3′) and oMN02-211 (5′-AACTGCAAGGCGGATCCGTGGCTT CA-3′) and chromosomal DNA prepared from JH642. The PCR product, after being digested with EcoRI and BamHI, was inserted into pTKlac (16). The resultant plasmid, pMMN519, was used to transform JH642 to generate ORB4269. The yxeL gene is the fourth open reading frame of the yxeI operon, and the introduction of the yxeL-lacZ plasmid by a single recombination event disrupts the operon.
B. subtilis strain ORB4814 [Pspank-hy-spx(C10A)] was constructed as follows. Two PCRs were performed to create a site-directed mutation. Primers oMN01-173 and oSN03-67 generated the first fragment, and oMN01-174 and oSN03-66 generated the second fragment. The fragments were ligated, and primers oMN01-173 and oMN01-174 were used to amplify the full-length mutagenized fragment. The resultant fragment was digested with HindIII and SalI and subsequently ligated to pDR111 (2) digested with the same enzymes, giving pSN95, which was introduced by transformation into strain JH642.
B. subtilis strain ORB4815[Pspank-hy-spx(C13A)] was constructed in a manner that was similar to that used for the construction of ORB4814, except that the first PCR was performed with mutagenic primers oSN03-69 and oSN03-68 replacing oSN03-67 and oSN03-66, respectively. The plasmid constructed was pSN96, which was introduced by transformation into JH642.
B. subtilis strain ORB4866 carrying a ytlI-lacZ fusion was constructed as follows. With oligonucleotides oSN03-73 and oSN03-74, a PCR fragment was obtained by using JH642 DNA as the template. The PCR product was digested with EcoRI and BamHI and ligated with pUC19, which was digested with the same enzymes, forming plasmid pSN104. Plasmid pSN104 was then digested with EcoRI and BamHI, and the resultant fragment was subcloned to plasmid pDH32, which had been digested with the same enzymes, forming pSN105. Plasmid pSN105 was then introduced by transformation into JH642, and chloramphenicol-resistant (Cmr) transformants were tested for loss of amylase activity on LB-starch agar plates.
B. subtilis strain ORB4962 carries the ytlI coding sequence under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Pspac promoter. To construct the strain, oligonucleotides oKE18 and oKE19 were used to perform PCR on JH642 chromosomal DNA previously digested with EcoRI and BamHI. The resultant 1,056-bp fragment corresponding to positions −81 to + 975 relative to the ytlI translational start site was digested with SalI and SphI. The digested fragment was then ligated to pDR66 (14) digested with the same enzymes, forming pKE17. The sequence of ytlI was verified by sequencing the plasmid with both Pspac-up and Pspac-down primers (Table 3). Plasmid pKE17 was transformed into JH642, resulting in recombination at amyE. Cmr clones were picked and checked for loss of amylase activity.
β-Galactosidase assays.
Cells from a frozen stock were used to inoculate to 1 or 2 ml of 2× YT plus antibiotics. The culture was allowed to grow for several hours, after which it was diluted 100-fold into 2 ml of TSS supplemented with 1 mM of different sulfur sources. After overnight growth, the culture was inoculated to 20 ml of TSS containing the same sulfur source to a starting optical density at 600 nm (OD600) of 0.05. After growth with agitation to an OD600 of 0.7 to 0.8 (or 0.7 to 1.0 in the case of the experiment with cells bearing the ytmI-lacZ fusion [see Fig. 3A]), two 1-ml samples were taken. Cells were pelleted by centrifugation and stored at −80°C until assayed for β-galactosidase activity. When required, 1 mM IPTG was added at the initial OD600 of 0.05.
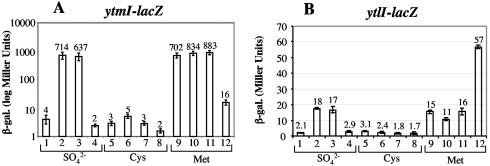
FIG. 3.
Expression of ytmI- and ytlI-lacZ fusions in wild-type, ytlI, spx, and rpoAcxs-1 strains. Cells of each fusion-bearing strain were grown in TSS media containing sulfate, cysteine, or methionine as the sole sulfur source. Samples were collected at late log phase, where maximum β-galactosidase activity was observed. The samples were assayed for β-galactosidase activity, which was expressed in Miller units (27). Experiments were performed in triplicate. (A) Bar graph of β-galactosidase activity in cells bearing the ytmI-lacZ fusion. Lanes 1, 5, and 9, strain ORB4794 (ytmI-lacZ); lanes. 2, 6, and 10, ORB4801 (ytmI-lacZ rpoAcxs-1); lanes 3, 7, and 11, ORB4802 (ytmI-lacZ spx); lanes 4, 8, and 12, ORB4804 (ytmI-lacZ ytlI). (B) Bar graph of β-galactosidase activity in cells bearing the ytlI-lacZ fusion. Lanes 1, 5, and 9, ORB4866 (ytlI-lacZ); lanes 2, 6, and 10, ORB4870 (ytlI-lacZ rpoAcxs-1); lanes 3, 7, and 11, ORB4871 (ytlI-lacZ spx); lanes 4, 8, and 12, ORB4873 (ytl-lacZ ytlI).
β-Galactosidase activity was determined as previously described (30) (27). The two samples from a single time point were averaged to give the Miller units. Experiments were repeated three times, and the data are presented as the mean of three independent experiments ± one standard deviation.
RESULTS
Genes that are derepressed in the rpoAcxs-1 mutant background.
The mutant rpoAcxs-1 allele of the RNAP α subunit gene, rpoA, encodes a product bearing a Y263C substitution in the C-terminal domain of the RNAP α subunit that was shown to impair interaction with the Spx protein (35). Previous microarray studies (34) were conducted that were designed to identify genes whose expression was affected by the overproduction of a protease-resistant form of Spx, SpxDD, but was not so affected in the rpoAcxs-1 background. This strategy succeeded in the discovery of a number of genes controlled by the Spx-RNAP interaction.
The microarray experiment uncovered a class of Spx-controlled genes in B. subtilis that was not expressed in a rpoA+ genetic background but was derepressed in an rpoAcxs-1 mutant. While derepression of these genes was observed in the rpoAcxs-1 background, repression occurred when the protease-resistant form of Spx, SpxDD, was produced by IPTG-dependent induction of a Pspank-hy-spxDD construct despite the weakened Spx interaction with the RpoAcxs-1 RNAP subunit. The unique class of Spx-controlled genes was not expressed in rpoA+ cells incubated in minimal medium; hence, the production of SpxDD had no observable effect on expression in the wild-type background.
Nearly all of the genes identified as members of this unique class encode products that function in alternative sulfur source utilization (Table 4). The ytmI, ssu, and yxeI operons encode ABC transport components and products showing homology to flavin mononucleotide or F420-dependent monooxygenases that function in the desulfurization of sulfur-containing organic compounds (18). Products of the ssu genes had also been implicated in dimethyl sulfide utilization in Pseudomonas (8).
TABLE 4.
Genes derepressed in an rpoAcxs-1 mutant background and negatively controlled by Spxa
| Gene (operon) | Product(s) |
|---|---|
| cssS | kinase, protein secretion stress |
| ytmI, ytmO, ytmJ, ytmN, | sulfonate utilization |
| ytmL, ytmK, ytmM, ytnI, ytnJ, ytnM, ribR (ytmI operon) | |
| ytlI | LysR-like regulator of ytmI operon |
| yxeK, yxeO, yxeP, yxeN, | sulfonate utilization |
| yxeL, yxeQ, yxeR, yxeD | |
| (yxe operon) | |
| ssuD, ssuA, ssuC, ssuB, | sulfonate; DMS utilization |
| ygaN, yhzA (ssu operon) | |
| yjnA | permease |
| cysC, cysP, sat, ylnE, ylnD, | sulfate transport, APS, sulfite |
| ylnF (cysP operon) | reduction |
| yubD | MDR transporter |
| metE | cobalamin-independent methionine synthase |
From data from reference 34.
Spx represses YtlI-dependent transcription of the ytmI operon in cultures containing sulfate as the sole sulfur source.
One of the Spx-controlled operons identified was the ytmI (ytmIJKLMNOytnIJribRytnLM) operon (7). The operon is induced in the absence of cysteine and sulfate and in the presence of methionine (7). It encodes products resembling components of an ABC-type transport system, two FMN-dependent monooxygenases, and proteins that appear to function in flavin cofactor synthesis. A low-molecular-weight glutaredoxin-like protein, YtnI, may function as a monooxygenase-protecting antioxidant to eliminate reactive oxygen species, which are by-products of the FMN reductase-catalyzed reaction (19).
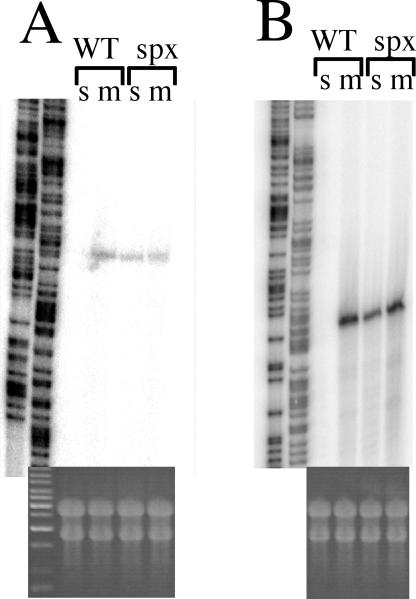
A recent report provides evidence that the divergently transcribed ytlI gene encodes a LysR-like product that functions in ytmI operon transcriptional activation (7). Primer extension analysis (Fig. 1) shows that the start sites of transcription for the ytmI operon and the ytlI gene are only 15 bp apart, with their −10 regions overlapping and with opposite orientations (Fig. 2). The primer extension shows that ytmI and ytlI are transcribed when methionine is present but are repressed by sulfate. Figure 1 also shows that SO42−-dependent repression of ytmI and ytlI is not observed in cells of an spx mutant.
FIG. 1.
Primer extension analysis of ytmI and ytlI RNA in wild-type and spx mutant cells. Cultures were grown in TSS medium containing either MgSO4 or methionine (see Materials and Methods) to OD600s of 0.7. RNA was harvested as described previously (35). Primer extension reactions were performed using oligonucleotides oSN04-90 (for ytlI) and oSN04-91 (for ytmI). Sanger dideoxynucleotide termination sequence reactions were performed using the same primers. Products were resolved by polyacrylamide-urea gel electrophoresis and visualized by phosphorimaging. WT, wild type (JH642); spx, spx mutant (ORB3834); s, sulfate; m, methionine. (A) Primer extension products of ytlI RNA. Sequencing reactions are shown on the left. (B) Primer extension products of ytmI RNA. At the bottoms of the primer extension product lanes are photographs of formaldehyde agarose gels of total RNA from each RNA sample.
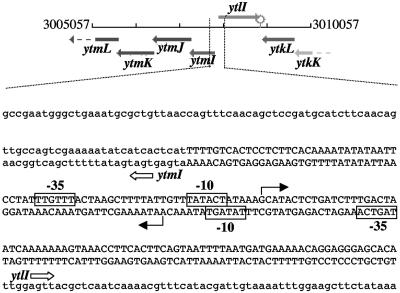
FIG. 2.
Diagram and nucleotide sequence of the ytmI-ytlI promoter region. At the top is a diagram of the ytmI-ytlI locus from the SubtiList website (http://genolist.pasteur.fr/SubtiList/). The numbers at either end refer to the nucleotide positions in the chromosome. At the bottom is the nucleotide sequence of the ytmI-ytlI promoter region. Uppercase letters denote the noncoding, intergenic region between ytmI and ytlI. Lowercase letters specify the coding sequences of ytmI and ytlI. The ATG and TTG start codons are labeled with gene names and arrows for ytmI and ytlI. Solid arrows mark the transcriptional start sites for both genes as identified by primer extension analysis (Fig. 1). The −10 regions of both promoters are labeled, and the sequences are enclosed in rectangles.
Spx-dependent repression of ytmI and ytlI in the presence of sulfate is not the result of Spx overproduction of the kind that characterizes cells of a clpX or clpP mutant or cells treated with the thiol-specific oxidant diamide. Western analysis with anti-Spx antiserum failed to detect Spx protein from extracts of cells grown in sulfate, cysteine, or methionine media (data not shown), indicating that changing the sulfur source does not significantly affect spx expression.
Spx is required for sulfate- but not cysteine-dependent repression.
The expression of the ytmI-lacZ fusion of strains ORB4794, ORB4804 (ytlI::aphA3), ORB4801 (rpoAcxs-1), and ORB4802 (spx::aphA3) was examined in cultures containing sulfate, cysteine, or methionine as sole sulfur sources. As shown in Fig. 3, ytmI-lacZ expression was repressed in cells of cultures grown in sulfate or cysteine (Fig. 3A, lanes 1 and 5). Strains bearing either the rpoAcxs-1 or the spx null mutation had higher levels of ytmI-lacZ activity in sulfate-containing medium (Fig. 3A, lanes 2 and 3), but this activity was still repressed in cysteine medium (Fig. 3A, lanes 6 and 7). Cells of cultures grown in medium containing methionine exhibited derepressed ytmI-lacZ expression, and the introduction of the spx or the rpoAcxs-1 mutation resulted in no further increase in expression (Fig. 3A, lanes 9 to 11). A mutation in the ytlI gene resulted in reduced expression in the presence of each of the three sulfur sources, although expression is relatively high in the methionine-grown cultures (Fig. 3A, lanes 4, 8, and 12).
As reported previously (7), YtlI exerts positive control over the ytmI operon. It is therefore possible that Spx indirectly affects ytmI expression by controlling the expression of the divergently transcribed ytlI gene. To examine this possibility, a ytlI-lacZ fusion was constructed and introduced into the ytlI, spx, and rpoAcxs-1 mutants. The expression of the fusion was repressed in media containing either sulfate or cysteine (Fig. 3B, lanes 1 and 5) but was derepressed in medium containing methionine (Fig. 3B, lane 9). Thus, the expression patterns of ytlI with respect to the sulfur sources resembled those of the ytmI-lacZ fusion. Introduction of the rpoAcxs-1 or spx mutations resulted in derepression in sulfate-containing medium (Fig. 3B, lanes 2 and 3) but not in medium containing cysteine (Fig. 3B, lanes 6 and 7), which again is similar to the pattern of ytmI operon regulation. As with ytmI, ytlI-lacZ was derepressed in medium containing methionine, and this expression was not elevated further by the introduction of the rpoAcxs-1 and spx mutations (Fig. 3B, lanes 9 to 11). Unlike ytmI expression, however, a ytlI mutation resulted in overexpression in methionine-containing medium (Fig. 3B, lane 12), which is suggestive of YtlI-mediated negative autoregulation. This negative autoregulation is not involved with cysteine-dependent repression of ytlI, since repression is observed in wild-type and ytlI mutant cells. The results show that there are two modes of negative control, one involving Spx in sulfate medium and one that is dependent on exogenous cysteine.
Mutations in the CXXC motif of Spx do not eliminate sulfate-dependent negative control.
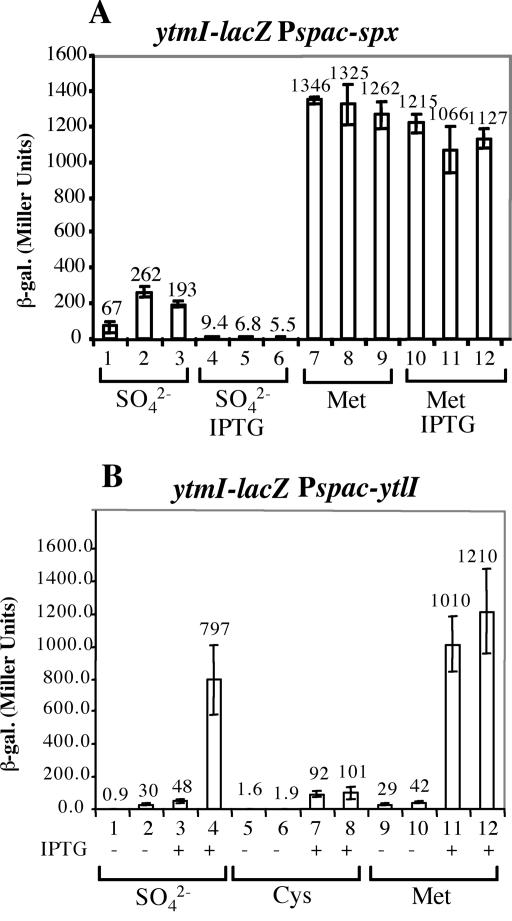
The CXXC motif of Spx is required for redox sensitive control of Spx activity (33). The transcription of the trxA and trxB genes requires Spx in its oxidized form, in which it bears a disulfide linkage between the cysteine residues of the redox active site. The requirement of the CXXC motif in sulfate-dependent repression was tested by expression in an spx mutant background of the wild-type and mutant alleles of Spx, the latter bearing C-to-A codon substitutions at C10 and C13. The spx alleles were expressed as IPTG-inducible constructs from the amyE locus of the B. subtilis chromosome. In the presence of SO42−, somewhat elevated ytmI-lacZ expression is observed in cells in the absence of IPTG (Fig. 4A, lanes 1 to 3). The induction of wild-type and mutant spx resulted in the complete repression of ytmI in the presence of sulfate (Fig. 4A, lanes 4 to 6). When the cells were grown in methionine, complete derepression was observed in both the presence and the absence of IPTG, whether or not the wild-type or mutant alleles were expressed. The low-level derepression in the absence of IPTG in sulfate medium suggests that the IPTG-inducible constructs are leaky, allowing some expression in the absence of inducer. This derepression was higher in the case of the mutant alleles, suggesting that these encode slightly defective Spx proteins. However, the mutants are capable of mediating SO42−-dependent repression of ytmI.
FIG. 4.
(A) Complementation of spx with wild-type and CXXC mutant alleles of spx. Cells were grown in TSS media containing either sulfate or methionine as sole sulfur sources in the absence (lanes 1 to 3 and 7 to 9) and the presence (lanes 4 to 6 and 10 to 12) of IPTG. Samples were collected as described in the legend to Fig. 3. β-Galactosidase activity was expressed in Miller units. Experiments were performed in triplicate. Lanes 1, 4, 7 and 10, ORB4807 (ytmI-lacZ spx amyE::Pspank-hy-spx); lanes 2, 5, 8, and 11, ORB4863 [ytmI-lacZ spx amyE::Pspank-hy-spx(C10A)]; lanes 3, 6, 9, and 12, ORB4864 [ytmI-lacZ spx amyE::Pspank-hy-spx(C13A)]. (B) The ytmI-lacZ fusion is still controlled by Spx when ytlI is constitutively expressed. Strains containing ytmI-lacZ and Pspank-hy-ytlI with or without an spx mutation were grown in TSS containing either sulfate, cysteine, or methionine as the sole sulfur source. The absence (−) and the presence (+) of IPTG is indicated. Lanes 1, 3, 5, 7, 9, and 11, ORB4979 (ytmI-lacZ amyE::Pspac-ytlI ytlI::aphA3); lanes 2, 4, 6, 8, 10 and 12, ORB4983 (ytmI-lacZ amyE::Pspac-ytlI ytlI::aphA3 spx::spc).
Constitutive expression of ytlI does not render ytmI transcription independently of Spx-mediated repression in sulfate medium.
Spx could regulate ytmI only indirectly, by controlling the expression of its positive regulator, YtlI. To determine if this was true, we expressed the ytlI gene from an IPTG-inducible Pspac promoter so as to render the ytlI expression independent of Spx. The expression of ytmI-lacZ was then examined in both the presence and the absence of IPTG and sulfate. As shown in Fig. 4B, lanes 1 and 3, in spx+ cells, the expression of ytmI-lacZ is repressed in sulfate medium in the presence or the absence of IPTG induction of ytlI. A much higher level of ytmI-lacZ expression is observed upon IPTG addition to cultures of the spx mutant (Fig. 4B, lanes 2 and 4). Repression of ytmI by cysteine is retained in the presence or the absence of IPTG in spx and spx+ strains (Fig. 4B, lanes 5 to 8), showing again that cysteine-dependent repression does not involve Spx. In methionine medium (Fig. 4B, lanes 9 to 12), IPTG addition results in high-level ytmI-lacZ expression in spx+ and spx mutant backgrounds. Thus, Spx was shown to exert negative control at the ytlI and the ytmI promoters when cells are grown in medium containing sulfate.
Spx exerts negative control of the ssu and yxeI operons in cells grown in sulfate medium.
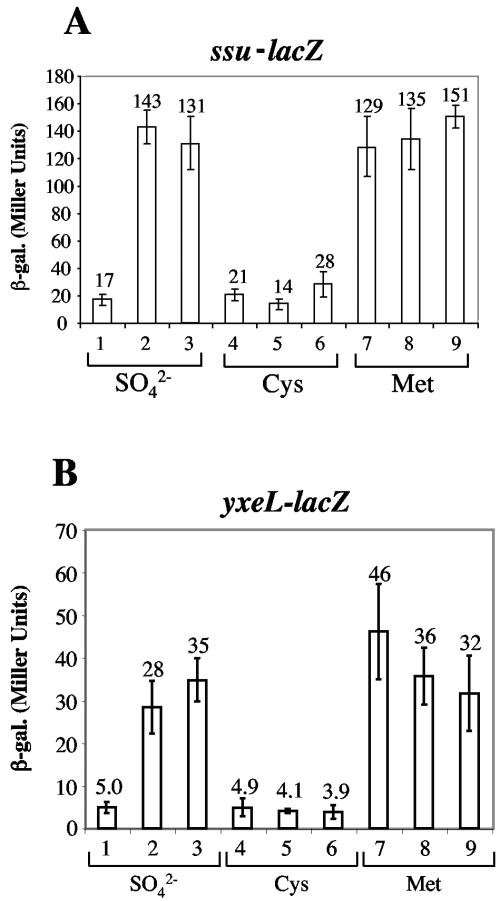
Like the ytmI operon, the ssu and yxeI operons encode products resembling ABC-type transport components and FMN-dependent monooxygenases, characteristics of operons whose products function in the metabolism of organosulfur compounds. Both fall into the class of operons that are derepressed in the rpoAcxs-1 mutant but repressed by Spx overproduction. The expression of an ssu-lacZ fusion (obtained from J. R. van der Ploeg) and that of a yxeL-lacZ fusion (constructed by M. M. Nakano; unpublished) was examined in wild-type, spx, and rpoAcxs-1 cells grown in sulfate, cysteine, or methionine media. Both fusions are repressed by sulfate and cysteine but are expressed to their highest levels in methionine medium (Fig. 5). Sulfate-dependent repression requires the spx gene and the wild-type allele of rpoA (Fig. 5A and B, lanes 1 to 3). As was the case with ytmI and ytlI expression, the spx and rpoAcxs-1 mutations do not confer any increase in expression further than that observed in wild-type cells incubated in methionine medium.
FIG. 5.
(A) The expression of ssu-lacZ is negatively controlled by Spx in the presence of sulfate. The strains bearing amyE::ssu21-289-lacZ (39) in a wild-type, spx, or rpoAcxs-1 background were grown in TSS containing either sulfate, cysteine, or methionine as the sole sulfur source. Samples were collected and assayed for β-galactosidase activity as described for Fig. 3. Lanes 1, 4, and 7, ORB4891 (ssu21-289-lacZ); lanes 2, 5, and 8, ORB4896 (ssu21-289-lacZ rpoAcxs-1); lanes 3, 6, and 9, ORB4898 (ssu21-289-lacZ spx). (B) The expression of yxeL-lacZ is negatively controlled by Spx in the presence of sulfate. The strains bearing yxeL-lacZ in a wild-type, spx, or rpoAcxs-1 background were grown in TSS containing either sulfate, cysteine, or methionine as the sole sulfur source. Samples were collected and assayed for β-galactosidase activity as for Fig. 3. Lanes 1, 4, and 7, ORB4269 (yxeL-lacZ); lanes 2, 5, and 8, ORB4785 (yxeL-lacZ spx); lanes 3, 6, and 9, ORB4786 (yxeL-lacZ rpoAcxs-1).
DISCUSSION
The ytmI, ssu, and yxeI operons encode products resembling proteins that function in organic sulfonate uptake and desulfonation. The ssu operon of B. subtilis has been reported to be required for growth in medium containing short-chain alkanesulfonates, sulfoacetate, taurine, or isethionate as sole sulfur sources (40). Each of the operons encodes not only the components of organic sulfonate uptake but also the FMNH2- or coenzyme F420-requiring monooxygenases that catalyze the oxygenolytic desulfonation of the substrate. The desulfonation of organic sulfonates yields sulfite, which is subsequently reduced to sulfide, a substrate for cysteine synthase. The transport components of the ytmI operon constitute one of three systems that can function in the uptake of cystine (4). All three operons are induced by sulfur limitation, which is a consequence of low exogenous concentrations of the preferred sulfur sources sulfate or cysteine. Sulfate is normally transported by the sulfate/thiosulfate transporter encoded by cysP. Sulfate sulfur assimilation proceeds with the formation of adenosine 5′-phosphosulfate (APS), which is generated by ATP sulfurylase using SO42− and ATP followed by the phosphorylation of APS by APS kinase to yield PAPS (phosphoadenosine phosphosulfate). The product of the cysH gene, PAPS sulfotransferase (or phosphoadenylyl sulfate reductase), generates sulfite, which is reduced by sulfite reductase to sulfide for cysteine synthesis. In organic sulfonate utilization, sulfite is generated by a process that bypasses the sulfate reduction pathway (18).
Sulfate-dependent control of sulfonate utilization genes is accomplished through the LysR-like transcriptional activator Cbl in E. coli (15). APS, an intermediate in sulfate sulfur assimilation, serves as an antiactivator preventing the transcription of the ssu operon (5). The ssu operon is also under negative control by CysB, another LysR-type regulator that is sensitive to O-acetyl serine, a precursor of cysteine biosynthesis that accumulates under low cysteine concentrations (5). Until recently, there have been no such regulators identified in gram-positive bacteria. The evidence presented herein indicates that Spx is at least a component of the sulfate-dependent control system. This brings the number of regulatory factors controlling sulfur metabolism at the level of transcription initiation in gram-positive bacteria to five, with the others being CmbR of Lactococcus lactis (9), CysL (12), YtlI, and, putatively, YrzC of B. subtilis (38). CmbR is a LysR homolog that activates the metC (cystathionine β-lyase) cysK (cysteine synthase) operon in response to low cysteine and high o-acetylserine (OAS) concentrations. CmbR was reported to be a functional homolog of CysB. While there are many LysR-type regulators encoded in the B. subtilis genome, there are none that are close homologs of CmbR. OAS has been reported to exert a positive influence on expression of the cysH operon in B. subtilis (24). The specific regulator that mediates this control, the hypothetical CysR factor, has not been uncovered. CysL, another LysR-like regulator, is required for transcription of the cysIJ dicistronic operon, which encodes sulfite reductase. CysL-dependent transcription is activated in response to the presence of sulfate as the sole sulfur source and is repressed by thiosulfate (12). YtlI and YrzC have been implicated in the control of the ytmI operon. At present, it is not known if YtlI activity is controlled in response to cysteine or sulfate availability. The yrzC gene is the site of mutations that lead to the derepression of ribR, a gene that resides within the ytmI operon and encodes riboflavin kinase (38). It is presumed that YrzC is a negative regulator of ytmI operon expression, but here too it is not known how YrzC is controlled in response to sulfur source availability. Spx exerts a negative effect on both the ytlI gene and the ytmI operon and is responsible for sulfate-dependent repression. It is not clear if this is accomplished in conjunction with or independently of YrzC.
The finding that Spx can function in the control of genes involved in sulfur metabolism was not entirely unexpected, since microarray studies have shown that genes whose products function directly in cysteine biosynthesis are induced when Spx is overproduced as a protease-resistant variant (34). However, this and other Spx activities have been heretofore observed only when Spx is present in a high concentration (29, 31, 34, 35). Spx functions in sulfate-dependent control under normal laboratory culture growth and at concentrations far below the levels at which Spx was previously observed to activate the trxA gene and repress activator-stimulated transcription. Western blot analysis using anti-Spx antiserum did not result in the detection of Spx in cells of cultures grown in sulfate, cysteine, or methionine (data not shown), while Spx is easily detected after diamide treatment or in a clpX mutant. One could imagine that the ytmI operon is repressed by Spx interaction with the RNA polymerase α subunit, which prevents YtlI-RNA polymerase contact. But this kind of activation interference had been observed only when Spx concentration was high. Further complicating attempts to explain Spx function in controlling ytmI operon transcription is the finding that the divergently transcribed ytlI gene is also under sulfate-dependent Spx control.
At present, it is not known in what capacity Spx functions in the negative control of ytmI and ytlI. Even less is known about the role Spx plays in the sulfate-dependent transcriptional control of the yxeI and ssu operons, as there have been no regulators, aside from Spx, that have been uncovered. It is not clear how sulfur availability might influence the activity of Spx. The CXXC motif which constitutes the thiol/disulfide switch governing redox sensitive control does not play a significant role in sulfate-dependent control. The recently solved crystal structure of Spx has revealed two sulfate ions that are coordinated in the vicinity of the Cys10 and C13 positions (K. Newberry, S. Nakano, P. Zuber, and R. Brennan; unpublished data). It is possible that Spx directly senses the presence of sulfate and responds through a conformational change that affects Spx activity.
Acknowledgments
We thank I. Martin-Verstraete for the ytmI′-lacZ fusion and the ytlI null mutant, J. R. van der Ploeg for the ssu-lacZ fusion construct, and Michiko M. Nakano for reagents and helpful discussions and for reading the manuscript.
The research presented herein was supported by grant GM45898 from the National Institutes of Health and a grant from the Oregon Medical Research Foundation.
REFERENCES
- 1.Auger, S., A. Danchin, and I. Martin-Verstraete. 2002. Global expression profile of Bacillus subtilis grown in the presence of sulfate or methionine. J. Bacteriol. 184:5179-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Britton, R. A., P. Eichenberger, J. E. Gonzalez-Pastor, P. Fawcett, R. Monson, R. Losick, and A. D. Grossman. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 184:4881-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruggemann, C., K. Denger, A. M. Cook, and J. Ruff. 2004. Enzymes and genes of taurine and isethionate dissimilation in Paracoccus denitrificans. Microbiology 150:805-816. [DOI] [PubMed] [Google Scholar]
- 4.Burguiere, P., S. Auger, M. F. Hullo, A. Danchin, and I. Martin-Verstraete. 2004. Three different systems participate in l-cystine uptake in Bacillus subtilis. J. Bacteriol. 186:4875-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bykowski, T., J. R. van der Ploeg, R. Iwanicka-Nowicka, and M. M. Hryniewicz. 2002. The switch from inorganic to organic sulphur assimilation in Escherichia coli: adenosine 5′-phosphosulphate (APS) as a signalling molecule for sulphate excess. Mol. Microbiol. 43:1347-1358. [DOI] [PubMed] [Google Scholar]
- 6.Cook, A. M., H. Laue, and F. Junker. 1999. Microbial desulfonation. FEMS Microbiol. Rev. 22:399-419. [DOI] [PubMed] [Google Scholar]
- 7.Coppee, J. Y., S. Auger, E. Turlin, A. Sekowska, J. P. Le Caer, V. Labas, V. Vagner, A. Danchin, and I. Martin-Verstraete. 2001. Sulfur-limitation-regulated proteins in Bacillus subtilis: a two-dimensional gel electrophoresis study. Microbiology 147:1631-1640. [DOI] [PubMed] [Google Scholar]
- 8.Endoh, T., K. Kasuga, M. Horinouchi, T. Yoshida, H. Habe, H. Nojiri, and T. Omori. 2003. Characterization and identification of genes essential for dimethyl sulfide utilization in Pseudomonas putida strain DS1. Appl. Microbiol. Biotechnol. 62:83-91. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez, M., M. Kleerebezem, O. P. Kuipers, R. J. Siezen, and R. van Kranenburg. 2002. Regulation of the metC-cysK operon, involved in sulfur metabolism in Lactococcus lactis. J. Bacteriol. 184:82-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fouet, A., S. F. Jin, G. Raffel, and A. L. Sonenshein. 1990. Multiple regulatory sites in the Bacillus subtilis citB promoter region. J. Bacteriol. 172:5408-5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 12.Guillouard, I., S. Auger, M. F. Hullo, F. Chetouani, A. Danchin, and I. Martin-Verstraete. 2002. Identification of Bacillus subtilis CysL, a regulator of the cysJI operon, which encodes sulfite reductase. J. Bacteriol. 184:4681-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harwood, C. R., and S. M. Cutting. 1990. Molecular biological methods for bacillus. John Wiley & Sons, Chichester, United Kingdom.
- 14.Ireton, K., D. Z. Rudner, K. J. Siranosian, and A. D. Grossman. 1993. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev. 7:283-294. [DOI] [PubMed] [Google Scholar]
- 15.Iwanicka-Nowicka, R., and M. M. Hryniewicz. 1995. A new gene, cbl, encoding a member of the LysR family of transcriptional regulators belongs to Escherichia coli cys regulon. Gene 166:11-17. [DOI] [PubMed] [Google Scholar]
- 16.Kenney, T. J., and C. P. Moran, Jr. 1991. Genetic evidence for interaction of σA with two promoters in Bacillus subtilis. J. Bacteriol. 173:3282-3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kertesz, M. A. 2001. Bacterial transporters for sulfate and organosulfur compounds. Res. Microbiol. 152:279-290. [DOI] [PubMed] [Google Scholar]
- 18.Kertesz, M. A. 2000. Riding the sulfur cycle—metabolism of sulfonates and sulfate esters in gram-negative bacteria. FEMS Microbiol. Rev. 24:135-175. [DOI] [PubMed] [Google Scholar]
- 19.Kertesz, M. A., and C. Wietek. 2001. Desulfurization and desulfonation: applications of sulfur-controlled gene expression in bacteria. Appl. Microbiol. Biotechnol. 57:460-466. [DOI] [PubMed] [Google Scholar]
- 20.Kirimura, K., K. Harada, H. Iwasawa, T. Tanaka, Y. Iwasaki, T. Furuya, Y. Ishii, and K. Kino. 2004. Identification and functional analysis of the genes encoding dibenzothiophene-desulfurizing enzymes from thermophilic bacteria. Appl. Microbiol. Biotechnol. 65:703-713. [DOI] [PubMed] [Google Scholar]
- 21.Lei, B., and S. C. Tu. 1996. Gene overexpression, purification, and identification of a desulfurization enzyme from Rhodococcus sp. strain IGTS8 as a sulfide/sulfoxide monooxygenase. J. Bacteriol. 178:5699-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, M. Z., C. H. Squires, D. J. Monticello, and J. D. Childs. 1996. Genetic analysis of the dsz promoter and associated regulatory regions of Rhodococcus erythropolis IGTS8. J. Bacteriol. 178:6409-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lochowska, A., R. Iwanicka-Nowicka, D. Plochocka, and M. M. Hryniewicz. 2001. Functional dissection of the LysR-type CysB transcriptional regulator. Regions important for DNA binding, inducer response, oligomerization, and positive control. J. Biol. Chem. 276:2098-2107. [DOI] [PubMed] [Google Scholar]
- 24.Mansilla, M. C., and D. de Mendoza. 1997. l-Cysteine biosynthesis in Bacillus subtilis: identification, sequencing, and functional characterization of the gene coding for phosphoadenylylsulfate sulfotransferase. J. Bacteriol. 179:976-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin, P., S. DeMel, J. Shi, T. Gladysheva, D. L. Gatti, B. P. Rosen, and B. F. Edwards. 2001. Insights into the structure, solvation, and mechanism of ArsC arsenate reductase, a novel arsenic detoxification enzyme. Structure (Cambridge) 9:1071-1081. [DOI] [PubMed] [Google Scholar]
- 26.Mascarenhas, D. M., and M. D. Yudkin. 1980. Identification of a positive regulatory protein in Escherichia coli: the product of the cysB gene. Mol. Gen. Genet. 177:535-539. [DOI] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 28.Monroe, R. S., J. Ostrowski, M. M. Hryniewicz, and N. M. Kredich. 1990. In vitro interactions of CysB protein with the cysK and cysJIH promoter regions of Salmonella typhimurium. J. Bacteriol. 172:6919-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakano, M. M., F. Hajarizadeh, Y. Zhu, and P. Zuber. 2001. Loss-of-function mutations in yjbD result in ClpX- and ClpP-independent competence development of Bacillus subtilis. Mol. Microbiol. 42:383-394. [DOI] [PubMed] [Google Scholar]
- 30.Nakano, M. M., M. A. Marahiel, and P. Zuber. 1988. Identification of a genetic locus required for biosynthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis. J. Bacteriol. 170:5662-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakano, M. M., S. Nakano, and P. Zuber. 2002. Spx (YjbD), a negative effector of competence in Bacillus subtilis, enhances ClpC-MecA-ComK interaction. Mol. Microbiol. 44:1341-1349. [DOI] [PubMed] [Google Scholar]
- 32.Nakano, M. M., Y. Zhu, J. Liu, D. Y. Reyes, H. Yoshikawa, and P. Zuber. 2000. Mutations conferring amino acid residue substitutions in the carboxy-terminal domain of RNA polymerase α can suppress clpX and clpP with respect to developmentally regulated transcription in Bacillus subtilis. Mol. Microbiol. 37:869-884. [DOI] [PubMed] [Google Scholar]
- 33.Nakano, S., K. N. Erwin, M. Ralle, and P. Zuber. 2005. Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol. Microbiol. 55:498-510. [DOI] [PubMed] [Google Scholar]
- 34.Nakano, S., E. Küster-Schöck, A. D. Grossman, and P. Zuber. 2003. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 100:13603-13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakano, S., M. M. Nakano, Y. Zhang, M. Leelakriangsak, and P. Zuber. 2003. A regulatory protein that interferes with activator-stimulated transcription in bacteria. Proc. Natl. Acad. Sci. USA 100:4233-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakano, S., G. Zheng, M. M. Nakano, and P. Zuber. 2002. Multiple pathways of Spx (YjbD) proteolysis in Bacillus subtilis. J. Bacteriol. 184:3664-3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ostrowski, J., and N. M. Kredich. 1990. In vitro interactions of CysB protein with the cysJIH promoter of Salmonella typhimurium: inhibitory effects of sulfide. J. Bacteriol. 172:779-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Solovieva, I. M., R. A. Kreneva, L. E. Lopes, and D. A. Perumov. 2005. The riboflavin kinase encoding gene ribR of Bacillus subtilis is a part of a 10 kb operon, which is negatively regulated by the yrzC gene product. FEMS Microbiol. Lett. 243:51-58. (First published 30 November 2004; 10.1016/j.femsle.2004.11.038.) [DOI] [PubMed] [Google Scholar]
- 39.van der Ploeg, J. R., M. Barone, and T. Leisinger. 2001. Expression of the Bacillus subtilis sulphonate-sulphur utilization genes is regulated at the levels of transcription initiation and termination. Mol. Microbiol. 39:1356-1365. [PubMed] [Google Scholar]
- 40.van der Ploeg, J. R., N. J. Cummings, T. Leisinger, and I. F. Connerton. 1998. Bacillus subtilis genes for the utilization of sulfur from aliphatic sulfonates. Microbiology 144:2555-2561. [DOI] [PubMed] [Google Scholar]
- 41.van der Ploeg, J. R., R. Iwanicka-Nowicka, T. Bykowski, M. M. Hryniewicz, and T. Leisinger. 1999. The Escherichia coli ssuEADCB gene cluster is required for the utilization of sulfur from aliphatic sulfonates and is regulated by the transcriptional activator Cbl. J. Biol. Chem. 274:29358-29365. [DOI] [PubMed] [Google Scholar]
- 42.van der Ploeg, J. R., R. Iwanicka-Nowicka, M. A. Kertesz, T. Leisinger, and M. M. Hryniewicz. 1997. Involvement of CysB and Cbl regulatory proteins in expression of the tauABCD operon and other sulfate starvation-inducible genes in Escherichia coli. J. Bacteriol. 179:7671-7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuber, P. 2004. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J. Bacteriol. 186:1911-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]