Abstract
Protein carbonylation is an irreversible oxidative modification that increases during organism aging and bacterial growth arrest. We analyzed whether the heat shock regulon has a role in defending Escherichia coli cells against this deleterious modification upon entry into stationary phase. Providing the cell with ectopically elevated levels of the heat shock transcription factor, σ32, effectively reduced stasis-induced carbonylation. Separate overproduction of the major chaperone systems, DnaK/DnaJ and GroEL/GroES, established that the former of these is more important in counteracting protein carbonylation. Deletion of the heat shock proteases Lon and HslVU enhanced carbonylation whereas a clpP deletion alone had no effect. However, ClpP appears to have a role in reducing protein carbonyls in cells lacking Lon and HslVU. Proteomic immunodetection of carbonylated proteins in the wild-type, lon, and hslVU strains demonstrated that the same spectrum of proteins displayed a higher load of carbonyl groups in the lon and hslVU mutants. These proteins included the β-subunit of RNA polymerase, elongation factors Tu and G, the E1 subunit of the pyruvate dehydrogenase complex, isocitrate dehydrogenase, 6-phosphogluconate dehydrogenase, and serine hydroxymethyltranferase.
Protein carbonylation has become a commonly used biomarker of severe oxidative damage to proteins, and many diseases have been associated with this modification, including Parkinson's disease, Alzheimer's disease, cancer, cataractogenesis, diabetes, and sepsis (6, 17). Carbonylation increases with the age of cells, organelles, and tissues of various species and has been linked to age-dependent deterioration of specific enzymes, e.g., the aconitase and the adenine nucleotide translocator ANT (29, 30). Carbonyl derivatives can be formed by a direct metal-catalyzed oxidative attack on the amino acid side chains of proline, arginine, lysine, and threonine. The quantitatively most important products of the carbonylation reaction are glutamic semialdehyde from arginine and proline and aminoadipic semialdehyde from lysine (21). Compared to other oxidative modifications, carbonyls are relatively difficult to induce and are irreversible modifications (6).
The levels of carbonylated proteins increase rapidly as Escherichia coli cells enter stationary phase as a result of carbon/energy (3, 9) or nitrogen (3) starvation. This modification has been associated with the reduced plating efficiency of stationary phase cells (7). Specifically, using an in situ detection of protein carbonyls in single cells and a density gradient centrifugation technique to separate culturable and nonculturable cells of the same chronological age it was demonstrated that proteins of the nonculturable cell population exhibited markedly higher levels of irreversible carbonylation (7). Apart from the fact that the RpoS and OxyR regulons and the superoxide dismutases and catalases are important in mitigating protein carbonylation (10), little is known about the function and identity of cellular defenses against this oxidation. Since heat shock genes have a role in cellular resistance against oxidative stress (14, 24) and are increasingly expressed during oxidant exposure (2, 26), we tested whether the heat shock regulon is involved in attenuating stasis-induced carbonylation. We report that several members of this regulon, both chaperones and proteases, are key factors in the cellular defense against this deleterious oxidative modification.
MATERIALS AND METHODS
Chemicals and reagents
Detection of carbonylated proteins was performed using the chemical and immunological reagents of an OxyBlot oxidized protein detection kit (Intergen Company). Anti-GroEL and anti-DnaK mouse monoclonal antibodies were purchased from Stressgen Bioreagents (Biosite). Anti-DnaK mouse polyclonal antibodies and anti-σ32 mouse monoclonal antibodies were from Neoclone. Anti-mouse immunoglobulin G peroxidase conjugates, trypsin, and isopropyl-β-galactopyranoside (IPTG) were from Sigma. The chemiluminescence blotting substrate (ECL+) was obtained from Amersham Corp., and the Immobilon-P polyvinylidene difluoride (PVDF) membrane was from Millipore. Bicinchoninic protein assay reagents were purchased from Pierce. The precast polyacrylamide gels used for one-dimensional (1-D) electrophoresis were either Criterion 10% Tris-HCl (Bio-Rad) or NuPAGE 12% Bis-Tris gel (Invitrogen). The ampholines (Resolyte 4-8) used for two-dimensional electrophoresis were from BDH (VWR International). The LIVE/DEAD BacLight bacterial viability kit was from Molecular Probe. All chemicals and reagents were used according to instructions provided by the manufacturer.
Bacterial strains, plasmids, and growth conditions
The E. coli K-12 strains and plasmids used in this study are listed in Table 1. Cultures were grown aerobically at 37°C in minimal M9 medium (18) supplemented with thiamine (10 mM), all 20 amino acids, and glucose (0.2%) in Erlenmeyer flasks in a rotary shaker. When appropriate, the medium was supplemented with carbenicillin (100 μg/ml), tetracycline (20 μg/ml), kanamycin (50 μg/ml) or spectinomycin (100 μg/ml). To overproduce Hsps, IPTG was added to the exponentially growing cultures at an optical density at 420 nm (OD420) of 0.05 to induce expression from the plasmid pKV1278 (σ32), pBB535 (DnaK/DnaJ), or pBB541 (GroES/GroEL). Early stationary phase samples were collected 30 min after exponential growth (measured as OD420) ceased due to glucose depletion in the medium. For protein stability measurements, cells were grown exponentially at 37°C and at an OD420 of 0.1 and IPTG was added to induce expression from the plasmid pKV1278 (σ32). After 3 h, protein synthesis was blocked by addition of spectinomycin (400 μg/ml).
TABLE 1.
E. coli strains and plasmids used in this work
| Strain | Plasmid | Genotype | Origin |
|---|---|---|---|
| MJ252 | pKV1278(Plac-rpoH) | MG1655 λ−ilvG-rfb-50 rph-1 | M. Jishage |
| PhB1465 | MG1655 Δ(clpPX-lon)1196::Catr | P. Bouloc | |
| PhB1466 | MG1655 Δ(clpPX-lon)1196::Catr ΔhslVU 1172::Tetr | P. Bouloc | |
| PhB1907 | MG1655 ΔclpPX | P. Bouloc | |
| ÅF38 | pBB535(PAl/lacO-1-dnaKdnaJ) | MG1655 Δlac | This work |
| ÅF41 | pKV1278(Ptrc-rpoH) | MG1655 Δlac | This work |
| ÅF42 | ptrc99A | MG1655 Δlac | This work |
| ÅF49 | MG1655 Δlac ΔhslVU::Tetr | This work | |
| ÅF50 | MG1655 Δlac ΔclpP::Camr | This work | |
| ÅF64 | pBB541(groESL) | MG1655 Δlac | This work |
| pBB528(laclq) | |||
| ÅF66 | MG1655 Δlac Δlon146::Tetr | This work | |
| MG1655Δlac | F− | MG1655 λ− Δlac ilvG-rfb-50 rph-1 | Lab stock |
General methods.
Crude cell extracts were obtained using a 20 K French pressure cell (Spectronic Instruments) for all experiments. Derivatized protein extracts were processed for resolution on two-dimensional polyacrylamide (2-D) gels by the methods of O′Farrell (20) with modifications (25). Isoelectric focusing was performed as described in reference 25. Gel electrophoresis using 11.5% sodium dodecyl sulfate-polyacrylamide gels and immunoblotting with mouse-monoclonal or mouse-polyclonal antibodies was carried out according to standard procedures. For detection, the ECL+ blotting kit was used with horseradish peroxidase-conjugated anti-mouse immunoglobulin G as secondary antibody. Blots were subsequently exposed in a charge-coupled device camera (FUJIFILM Image Reader LAS-1000Pro). For quantitative analyses of the blots, Image Gauge 3.46, Science Lab 99 software was used.
Protein identification.
Proteins separated on two-dimensional gels were blotted onto PVDF membranes and used for immunodetection of carbonylated proteins. The same membranes were stained with Coomassie brilliant blue after exposure in the charge-coupled device camera and were used as references when matching the carbonylated proteins to the spots on the 2-D gels. The matched protein spots were subsequently cut out and used for mass-spectrometric analysis. Samples were analyzed using a matrix-assisted laser desorption ionization-linear reflection mass spectrometer (Micromass, Manchester, UK) in reflection mode. Tryptic digest (0.5 μl) was mixed with 0.5 μl matrix solution (12 mg/ml α-cyano-4-OH-cinnamic acid in acetonitrile/water [1:1]-0.1% trifluoroacetic acid) directly on the matrix-assisted laser desorption ionization probe and allowed to dry at ambient conditions. ACTH (18-39) MH+ 2465.199 was used as an external and tryptic autodigest MH+ 2211.105 as an internal lock mass. Monoisotopic mass values were used for peptide mass mapping using MASCOT (available at www.matrixscience.com) against the nr database at the National Center for Biotechnology Information. All identifications had a score equivalent to a 95% level of confidence. When necessary, samples were purified and concentrated with ZipTips containing C18 resin according to the manufacturer's instructions. The peptides eluted in 3.5 μl of acetonitrile/water (1:1) containing 0.1% formic acid were then analyzed by electrospray ionization mass spectrometry/mass spectrometry (MS/MS) on a quadrupole time-of-flight ultima atmospheric pressure ionization apparatus (Micromass, Manchester, UK). Fragment ion data were acquired by nanoflow electrospray using argon as the collision gas. The MS/MS data were used as input for protein identification by MASCOT (www.matrixscience.com). No restrictions in species, molecular weight, or pI were applied when searching against the nr database of the National Center for Biotechnology Information. Proteins identified all had a score equivalent to a 95% confidence level or higher.
Viability assays.
Cells collected after 3 h in stationary phase were serially diluted in M9 medium lacking glucose and amino acids prior to plating onto solid LB medium in the presence and absence of the appropriate antibiotic selection. To induce expression from plasmid pBB535 (ÅF38), IPTG was added at an OD420 of 1.5 before transition into stationary phase. Viability was also determined by microscopic analyses using BacLight LIVE/DEAD methodology, whereby living cells fluoresce in green and dead cells in red when excited at 590 nm (12).
Carbonylation assays.
Analyses of carbonylated proteins were performed using the chemical and immunological reagents of an OxyBlot oxidized protein detection kit. The carbonyl groups in the protein side chains were derivatized to 2,4-dinitrophenylhydrazone (DNP-hydrazone) by reaction with 2,4-dinitrophenylhydrazine. The DNP-derivatized proteins were analyzed immunochemically on one- or two-dimensional Western blots or directly dot blotted onto PVDF membranes as described previously (9). In general, 1 μg of protein was loaded into the slot-blot apparatus and 10 μg onto 1- or 2-D gels.
RESULTS AND DISCUSSION
Overproduction of σ32 and DnaK/DnaJ prevents stasis-induced oxidation of proteins
Previous studies show that certain proteins are specifically susceptible to stasis-induced oxidation (9), and we determined the specificity of protein carbonylation at the time when this is maximal upon growth arrest (early stationary phase; see Fig. 1). A number of carbonylated proteins, some of which have not previously been shown to be carbonylated, were identified by mass spectrometry (Table 2). The identification suggests that several different processes, including, e.g., information transfer (transcription and translation), central metabolism (glycolysis and TCA cycle), protein folding, and fatty acid biosynthesis, are subjected to stasis-induced deterioration. Some of the identified carbonylated proteins have been demonstrated previously to be targets for the DnaK chaperone, i.e., these proteins are prone to aggregate in the absence of DnaK (19). Based on these results and the fact that aberrant proteins are more susceptible to carbonylation than the native counterparts (3, 8), we entertained the idea that stationary phase carbonylation of proteins may, in part, be a consequence of stasis-induced mistranslation (3) overwhelming the heat shock chaperones. If so, elevated levels of heat shock chaperones could partly counteract the effect of such mistranslation and attenuate stasis-induced carbonylation.
FIG. 1.
Protein carbonylation during growth and in stationary phase. Relative protein carbonylation levels (black squares) and the optical density of the wild-type culture (MG1655Δlac) (filled circles) are shown during growth and growth arrest caused by glucose depletion. Carbonyl levels were determined by one-dimensional Western blot immunoassays and quantified using Image Gauge software. Carbonyl levels were related to that obtained during exponential growth, which was assigned a value of 1.0. The arrow indicates the time at which samples were obtained for identification of carbonylated proteins (see Table 2). The experiment was repeated at least three times. Representative results are shown, and there was always less than 15% variation between experiments.
TABLE 2.
Proteins subjected to stasis-induced carbonylation at the time when carbonyl content is maximal. Proteins in bold have been demonstrated to be substrates for DnaK (19).
| Protein no. | Gene | Size (kDa) | Protein name |
|---|---|---|---|
| 1 | rpoB | 151 | RNA polymerase β-subunita |
| 2 | purL | 141 | Phosphoribosyl formylglycinamide synthase |
| 3 | carA | 119 | Carbamoylphosphate synthase |
| 4 | aceE | 100 | E1 component of pyruvate dehydrogenase |
| 5 | metE | 85 | Tetrahydropteroyltriglutamate methyltransferase |
| 6 | fus | 78 | Protein chain elongation factor G |
| 7 | dnaK | 69 | Molecular chaperone DnaK |
| 8 | aceF | 66 | E2 component of pyruvate dehydrogenase |
| 9 | ptsI | 64 | Phosphoenolpyruvate-protein phosphotransferase |
| 10 | 61 | Polyamine-induced protein precursor | |
| 11 | groEL | 57 | Molecular chaperone GroEL |
| 12 | glnA | 52 | Glutamine synthetase |
| 13 | gltD | 52 | Glutamate synthase |
| 14 | pyk | 51 | Pyruvate kinase |
| 15 | gnd | 49 | 6-Phosphogluconate dehydrogenase |
| 16 | icd | 46 | Isocitrate dehydrogenase |
| 17 | glyA | 46 | Serine hydroxymethyltransferase |
| 18 | serA | 44 | d-3-phosphoglycerate dehydrogenase |
| 19 | sucB | 44 | Dihydrolipoamide succinyltransferase |
| 20 | fabB | 43 | β-Keto-[acyl-carrier-protein] synthetase |
| 21 | tufA | 42 | Protein chain elongation factor Tu |
| 22 | sucC | 41 | Succinyl CoA ligase |
| 23 | znuA | 34 | High-affinity zinc uptake system protein precursor |
| 24 | mdh | 32 | Malate dehydrogenase |
Proteins that have been demonstrated to be substrates for DnaK (19) are shown in boldface characters.
We determined whether overproduction of the Hsp transcription factor σ32 mitigated protein carbonylation in cells entering a growth-arrested state. To confirm that overproduction of σ32 resulted in significantly elevated concentrations of Hsps, we checked the levels of DnaK and GroEL (Fig. 2A). As seen in Fig. 2A, overproduction of the heat shock regulon drastically reduced (3.5-fold lower than the control) stasis-induced carbonylation. In fact, carbonylation did not increase upon growth arrest in cells with ectopically elevated Hsp levels.
FIG. 2.
Effects of overproducing the Hsp regulon on protein oxidation. Severalfold change in the levels of DnaK, GroEL, and carbonylated proteins as a consequence of overproducing σ32 (ÅF41) (A), DnaK/DnaJ (ÅF38) (B), and GroEL/GroES (ÅF64) (C) in cells entering a growth arrested state (30 min after growth ceased) is shown. “1” on the y axis means no change compared to wild type, while “2” means a twofold increase and “−2” means a twofold decrease. The cells were grown with (100 μM) and without IPTG prior to starvation. Protein and carbonyl levels were determined by one-dimensional Western blot immunoassays and quantified using Image Gauge software. All levels were related to that obtained in the control culture without overproduction, which was assigned a value of 1.0.
Next, we determined the effect of overproducing the DnaK/DnaJ chaperone system alone. DnaK/DnaJ overproduction markedly reduced the levels of other Hsps, such as GroEL (Fig. 2B), consistent with the role of DnaK/DnaJ as negative modulators of the Hsp regulon. Nevertheless, despite the reduced levels of other Hsps, overproduction of the DnaK/DnaJ chaperones counteracted protein carbonylation to the same extent as overproduction of σ32 (Fig. 2B). This was due not to a reduced production of proteins that happen to be carbonylation sensitive but rather to a reduction in such proteins' carbonylation load. For example, the carbonyl level of elongation factor EF-Tu, when normalized to the total levels of EF-Tu present, was substantially reduced by DnaK/DnaJ overproduction (Fig. 3A). As depicted (Fig. 3B), the total amount of carbonylated proteins was generally lower upon DnaK/DnaJ overproduction, indicating a general protective function of this chaperone system.
FIG. 3.
Effects of overproducing DnaK/DnaJ (ÅF38) on EF-Tu carbonylation during glucose starvation (A). Open squares, no IPTG; gray squares; 100 μM IPTG; black squares, 250 μM IPTG. Carbonyl levels are normalized to EF-Tu levels. (B) General pattern and level of carbonylated proteins (equal amounts of total cellular proteins were loaded/well) in growth-arrested (2.5 h after cell division ceased) wild-type (ÅF38) cells harboring the PA1/lacO-1- dnaK dnaJ construct. Cells were grown with (250 μM) and without IPTG prior to starvation. Arrow indicates EF-Tu. Protein and carbonyl levels were determined by one-dimensional Western blot immunoassays and quantified using Image Gauge software. All experiments were repeated at least three times. Representative results are shown, and there was always less than 15% variation between experiments.
Overproduction of the GroEL/GroES chaperone system affected carbonylation marginally (Fig. 2C), suggesting that the DnaK/DnaJ system is more important in the cellular defense against this oxidative modification. This is consistent also with the fact that DnaK/DnaJ overproduction reduced carbonylation despite a concomitantly reduced level of GroEL (Fig. 2B). A role for DnaK in protecting against protein oxidation is in line with results demonstrating that aerobic growth of E. coli on ethanol depends on DnaK protection of a mutant ethanol oxidoreductase (AdhE) against metal-catalyzed oxidation (11), but a global role for DnaK/DnaJ in mitigating stasis-induced carbonylation has not been reported previously.
Defense against carbonylation damage requires the Lon and HslVU (ClpQY) proteases.
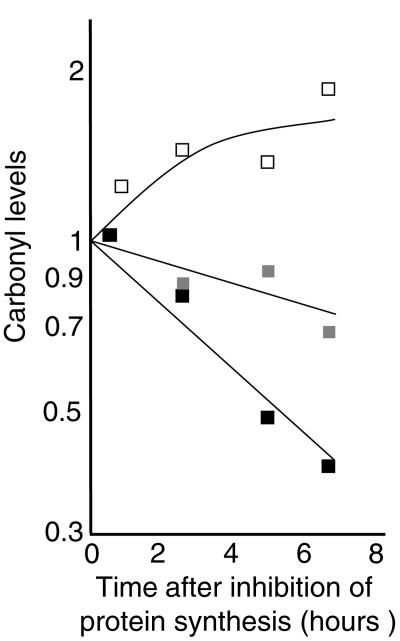
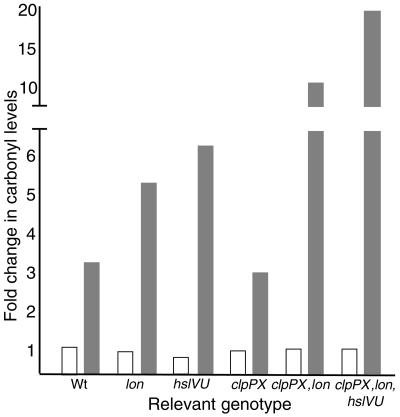
Oxidized aconitase has been shown to be recognized and degraded by mitochondrial Lon in mammalian cells (4). Since the bacterial Lon protease is a member of the heat shock regulon, we investigated whether attenuated carbonylation caused by Hsp overproduction resulted, in part, from increased degradation of carbonylated proteins. To approach this, we blocked protein synthesis with spectinomycin in cells with and without overproduced levels of σ32 and subsequently analyzed protein carbonyl levels at time intervals. As depicted (Fig. 4A), protein carbonyl levels decreased when σ32 had been overproduced, in stark contrast to the vector control results. This provided the impetus to analyze carbonylation levels in mutants lacking Lon and other Hsp proteases. Deletion of Lon increased protein carbonylation upon entry of cells into stationary phase, and deletion of the HslVU (ClpQY) protease had an even more pronounced effect (Fig. 5). However, a clpP deletion alone did not affect total protein carbonyl levels (Fig. 5). It is possible that increased levels of σS in the clpP mutant (22) may mask a possible role for ClpP in degrading carbonylated proteins, since σS-dependent genes have been shown to counteract protein oxidation (10). However, the clpP mutations had no significant effect on total protein carbonyl levels even in a background lacking rpoS (not shown). In contrast, the lack of ClpP further elevated carbonylation in cells lacking Lon, suggesting that ClpP has a role in the defense against protein carbonylation but that Lon can fully compensate for the absence of ClpP (Fig. 5). In addition, HslVU appears to partly compensate for the lack of Lon and ClpP since carbonylation was further increased by introducing an hslVU deletion (Fig. 5).
FIG. 4.
Relative stability of carbonylated proteins determined by one-dimensional Western blot immunoassays in cells with different levels of σ32. Protein synthesis was inhibited with spectinomycin during entry to stationary phase (30 min after growth ceased) in cells containing either the empty vector control (ÅF42; open squares) or the Ptrc- rpoH construct (ÅF41) grown in the presence (black squares) or absence (gray squares) of 100 μM IPTG.
FIG. 5.
Severalfold change in the levels of carbonylated proteins in wild-type (MG1655Δlac) cells and in cells lacking the proteases Lon (ÅF66), HslVU (ÅF49), and ClpPX (PhB1907), ClpPX and Lon (PhB1465), and ClpPX, Lon, and HslVU (PhB1466). Protein extracts were from exponentially growing cells (open bars) and cells from an overnight stationary phase sample (grey bars). Carbonyl levels were determined by a slot-blot immunoassay and quantified using Image Gauge software. All levels were related to that obtained for the wild type during exponential growth, which was assigned a value of 1.0.
Proteomic immunodetection of carbonylated proteins in the wild-type, lon, and hslVU strains demonstrated that the increased levels of protein carbonyls were, for the most part, nonspecific in the sense that the same spectrum of oxidized proteins exhibited a higher load of carbonyl groups in both mutants (Fig. 6). These results are consistent with previous results demonstrating that the function and substrate recognition of Lon and HslVU overlap (28). Several proteins being increasingly carbonylated in the mutants were identified by mass spectrometry; they included the β-subunit of RNA polymerase (RpoB), elongation factors Tu and G (EF-Tu, EF-G), the E1 component (AceE) of the pyruvate dehydrogenase complex, isocitrate dehydrogenase (Idh), 6-phosphogluconate dehydrogenase (Gnd), and serine hydroxymethyltranferase (GlyA) (Fig. 6). Interestingly, GroEL and DnaK were two of only a few proteins whose carbonylation load did not increase by deleting lon or hslVU (Fig. 6). At present, it is not clear whether carbonylated proteins are targets for proteases because they contain oxidation cues for protease recognition or whether oxidation-induced unfolding is the primary cause of increased attack by heat shock proteases.
FIG. 6.
Proteomic immunodetection of carbonylated proteins in wild type (MG1655Δlac), Δlon (ÅF66), and ΔhslVU (ÅF49) cells entering a growth-arrested state due to glucose starvation. The major target proteins of carbonylation were identified by mass spectrometry and included the β-subunit of RNA polymerase (RpoB), elongation factors Tu and G (EF-Tu, EF-G), the E1 component (AceE) of the pyruvate dehydrogenase complex, isocitrate dehydrogenase (Icd), 6-phosphogluconate dehydrogenase (Gnd), and Serine hydroxymethyltranferase (GlyA). The boxed proteins are GroEL and DnaK. The experiment was repeated three times, and the patterns of carbonylated proteins were the same in each experiment. Representative results are shown.
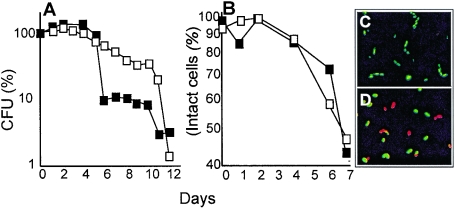
Overproduction of DnaK/DnaJ does not prolong the average life span of stationary phase cells.
The DnaK/DnaJ system is not expected to repair or refold carbonylated proteins, since carbonylation is an irreversible modification. Reduced carbonylation in cells overproducing this chaperone system may instead result from (i) a reduced abundance of aberrant proteins and/or (ii) an increased DnaK/DnaJ-dependent solubility of carbonylated proteins. The former of these suggestions stems from data demonstrating that aberrant forms of proteins are more susceptible to oxidative carbonylation than their native counterpart (8). Thus, any condition reducing the levels of such aberrant proteins, such as increased ribosomal proofreading (3) or elevated levels of DnaK/DnaJ (this work), may be expected to also reduce cellular carbonyl levels. The second idea is based on the fact that carbonylated proteins are susceptible to proteolysis as long as they remain soluble (13). However, heavily carbonylated proteins are prone to aggregate and such high-level molecular aggregates escape proteolysis (4). Increased levels of DnaK/DnaJ may keep carbonylated proteins in a soluble, protease-susceptible form and thereby contribute to their degradation by, e.g., Lon and HslVU. It has been suggested that the decline in proteosomal activity during aging (1, 5) may be closely connected to a gradual accumulation of proteolysis-resistant aggregates of carbonylated proteins that bind and inhibit proteosomal function (13). Thus, it may be worth considering that the effects of overproducing Hsp70 (DnaK family proteins) in retarding aging (31) and delaying the onset of age-related protein aggregation (14) in eukaryotes may be linked to its role in reducing the level of carbonylated and aggregation-prone proteins. However, overproduction of DnaK/DnaJ did not extend the average life span of stationary phase cells (Fig. 7A and B). Both plating efficiency and membrane integrity were lost with the same rate, or at a higher rate, in cells with ectopically elevated levels of DnaK/DnaJ (Fig. 7). Thus, it may be that increased carbonylation is a diagnostic marker of deterioration (7) but is not the limiting factor in stationary phase survival of E. coli cells. Alternatively, it is possible that the reduced concentration of other important proteins of the heat shock regulon resulting from DnaK/DnaJ overproduction masks potential benefits of reducing carbonylation. In addition, elevating the levels of the entire complement of heat shock proteins by overproducing σ32 is not informative with respect to stasis survival since it drastically reduces expression, by sigma factor competition (15), of σS- and σE-dependent genes, which are essential for stasis survival. Thus, while it is clear that mutant cells lacking specific members of the heat shock regulon (e.g., DnaK, ClpP, HslVU, and Lon) (16, 23, 27) die at an accelerated rate in stationary phase, it is uncertain whether the level of heat shock proteins constitutes a bottleneck in stationary phase survival of wild-type E. coli cells.
FIG. 7.
Stasis-survival of cells overproducing DnaK/DnaJ (ÅF38). Open squares, no IPTG; black squares, 250 μM IPTG. Plating efficiency (A) and membrane integrity (B) was monitored in cells growth arrested due to glucose starvation. Membrane integrity of cells was analyzed using the BacLight LIVE/DEAD methodology (12). Intact cells appear fluorescent green (C; 1 h starvation), whereas cells with a debilitated and leaky membrane appear red (D; 7 days of starvation). Representative results are shown, and there was always less than 15% variation between experiments.
Acknowledgments
We thank B. Bukau, C. Gross, K. Gerdes, M. Kanemori, S. Gottesmann, T Yura, J. Urbonavicius, P. Bouloc, and M. P. Mayer for providing strains, plasmids, and information necessary for this work, Thomas Larsson for help with the mass-spectrometric analyses, and also the people in the Nyström laboratory for valuable comments on the manuscript.
This work was sponsored by grants from the Swedish Natural Research Council and the Foundation for Strategic Research in Sweden and an award from the Göran Gustafsson Foundation for Scientific Research in Molecular Biology.
REFERENCES
- 1.Agarwal, S., and R. S. Sohal. 1994. Aging and proteolysis of oxidized proteins. Arch. Biochem. Biophys. 309:24-28. [DOI] [PubMed] [Google Scholar]
- 2.Amoros, M., and F. Estruch. 2001. Hsf1p and Msn2/4p cooperate in the expression of Saccharomyces cerevisiae genes HSP26 and HSP104 in a gene- and stress type-dependent manner. Mol. Microbiol. 39:1523-1532. [DOI] [PubMed] [Google Scholar]
- 3.Ballesteros, M., A. Fredriksson, J. Henriksson, and T. Nyström. 2001. Bacterial senescence: protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes. EMBO J. 20:5280-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bota, D. A., and K. J. Davies. 2002. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat Cell Biol. 4:674-680. [DOI] [PubMed] [Google Scholar]
- 5.Bota, D. A., H. Van Remmen, and K. J. Davies. 2002. Modulation of Lon protease activity and aconitase turnover during aging and oxidative stress. FEBS Lett. 532:103-106. [DOI] [PubMed] [Google Scholar]
- 6.Dalle-Donne, I., D. Giustarini, R. Colombo, R. Rossi, and A. Milzani. 2003. Protein carbonylation in human diseases. Trends Mol. Med. 9:169-176. [DOI] [PubMed] [Google Scholar]
- 7.Desnues, B., C. Cuny, G. Gregori, S. Dukan, H. Aguilaniu, and T. Nyström. 2003. Differential oxidative damage and expression of stress defence regulons in culturable and non-culturable Escherichia coli cells. EMBO Rep. 4:400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dukan, S., A. Farewell, M. Ballesteros, F. Taddei, M. Radman, and T. Nyström. 2000. Protein oxidation in response to increased transcriptional or translational errors. Proc. Natl. Acad. Sci. USA 97:5746-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dukan, S., and T. Nyström. 1998. Bacterial senescence: stasis results in increased and differential oxidation of cytoplasmic proteins leading to developmental induction of the heat shock regulon. Genes Dev. 12:3431-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dukan, S., and T. Nyström. 1999. Oxidative stress defense and deterioration of growth-arrested Escherichia coli cells. J. Biol. Chem. 274:26027-26032. [DOI] [PubMed] [Google Scholar]
- 11.Echave, P., M. A. Esparza-Ceron, E. Cabiscol, J. Tamarit, J. Ros, J. Membrillo-Hernandez, and E. C. Lin. 2002. DnaK dependence of mutant ethanol oxidoreductases evolved for aerobic function and protective role of the chaperone against protein oxidative damage in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:4626-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ericsson, M., D. Hanstorp, P. Hagberg, J. Enger, and T. Nyström. 2000. Sorting out bacterial viability with optical tweezers. J. Bacteriol. 182:5551-5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grune, T., T. Jung, K. Merker, and K. J. Davies. 2004. Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and ‘aggresomes’ during oxidative stress, aging, and disease. Int. J. Biochem. Cell Biol. 36:2519-2530. [DOI] [PubMed] [Google Scholar]
- 14.Hsu, A. L., C. T. Murphy, and C. Kenyon. 2003. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300:1142-1145. [DOI] [PubMed] [Google Scholar]
- 15.Jishage, M., K. Kvint, V. Shingler, and T. Nyström. 2002. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 16:1260-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jurkiewicz, D., and K. I. Wolska. 1999. Effect of DnaK and DnaJ proteins deprivation on Escherichia coli response to starvation. Acta Microbiol. Pol. 48:197-201. [PubMed] [Google Scholar]
- 17.Levine, R. L. 2002. Carbonyl modified proteins in cellular regulation, aging, and disease. Free Radic. Biol. Med. 32:790-796. [DOI] [PubMed] [Google Scholar]
- 18.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 19.Mogk, A., T. Tomoyasu, P. Goloubinoff, S. Rudiger, D. Roder, H. Langen, and B. Bukau. 1999. Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 18:6934-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 21.Requena, J. R., C. C. Chao, R. L. Levine, and E. R. Stadtman. 2001. Glutamic and aminoadipic semialdehydes are the main carbonyl products of metal-catalyzed oxidation of proteins. Proc. Natl. Acad. Sci. USA 98:69-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schweder, T., K. H. Lee, O. Lomovskaya, and A. Matin. 1996. Regulation of Escherichia coli starvation sigma factor (sigma s) by ClpXP protease. J. Bacteriol. 178:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spence, J., A. Cegielska, and C. Georgopoulos. 1990. Role of Escherichia coli heat shock proteins DnaK and HtpG (C62.5) in response to nutritional deprivation. J. Bacteriol. 172:7157-7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ueom, J., S. Kwon, S. Kim, Y. Chae, and K. Lee. 2003. Acquisition of heat shock tolerance by regulation of intracellular redox states. Biochim. Biophys. Acta 1642:9-16. [DOI] [PubMed] [Google Scholar]
- 25.VanBogelen, R. A., M. E. Hutton, and F. C. Neidhardt. 1990. Gene-protein database of Escherichia coli K-12: edition 3. Electrophoresis 11:1131-1166. [DOI] [PubMed] [Google Scholar]
- 26.VanBogelen, R. A., P. M. Kelley, and F. C. Neidhardt. 1987. Differential induction of heat shock, SOS, and oxidation stress regulons and accumulation of nucleotides in Escherichia coli. J. Bacteriol. 169:26-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weichart, D., N. Querfurth, M. Dreger, and R. Hengge-Aronis. 2003. Global role for ClpP-containing proteases in stationary-phase adaptation of Escherichia coli. J. Bacteriol. 185:115-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu, W. F., Y. Zhou, and S. Gottesman. 1999. Redundant in vivo proteolytic activities of Escherichia coli Lon and the ClpYQ (HslUV) protease. J. Bacteriol. 181:3681-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan, L. J., R. L. Levine, and R. S. Sohal. 1997. Oxidative damage during aging targets mitochondrial aconitase. Proc. Natl. Acad. Sci. USA 94:11168-11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan, L. J., and R. S. Sohal. 1998. Mitochondrial adenine nucleotide translocase is modified oxidatively during aging. Proc. Natl. Acad. Sci. USA 95:12896-12901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yokoyama, K., K. Fukumoto, T. Murakami, S. Harada, R. Hosono, R. Wadhwa, Y. Mitsui, and S. Ohkuma. 2002. Extended longevity of Caenorhabditis elegans by knocking in extra copies of hsp70F, a homolog of mot-2 (mortalin)/mthsp70/Grp75. FEBS Lett. 516:53-57. [DOI] [PubMed] [Google Scholar]