Abstract
Buchnera aphidicola, the obligate symbiont of aphids, has an extremely reduced genome, of which about 10% is devoted to the biosynthesis of essential amino acids needed by its hosts. Most regulatory genes for these pathways are absent, raising the question of whether and how transcription of these genes responds to the major shifts in dietary amino acid content encountered by aphids. Using full-genome microarrays for B. aphidicola of the host Schizaphis graminum, we examined transcriptome responses to changes in dietary amino acid content and then verified behavior of individual transcripts using quantitative reverse transcriptase PCR. The only gene showing a consistent and substantial (>twofold) response was metE, which underlies methionine biosynthesis and which is the only amino acid biosynthetic gene retaining its ancestral regulator (metR). In another aphid host, Acyrthosiphon pisum, B. aphidicola has no functional metR and shows no response in metE transcript levels to changes in amino acid concentrations. Thus, the only substantial transcriptional response involves the one gene for which an ancestral regulator is retained. This result parallels that from a previous study on heat stress, in which only the few genes retaining the global heat shock promoter showed responses in transcript abundance. The irreversible losses of transcriptional regulators constrain ability to alter gene expression in the context of environmental fluctuations affecting the symbiotic partners.
In diverse bacteria, as well as archaea and eukaryotes, a drastic reduction in overall genome size is associated with obligate dependence on a host (e.g., see references 1, 10, 17, and 34). One of the best-studied cases of genome reduction is that of the intracellular bacterial symbiont Buchnera aphidicola, which shows a long evolutionary history of strict maternal transmission in its aphid hosts (Insecta: Homoptera: Aphidoidea) (18). Genomes of B. aphidicola are only 500 to 640 kb in length and contain 450 to 580 protein-coding genes, essentially all of which have orthologs in related bacteria, including Escherichia coli (12, 28-30, 33). These related lineages typically contain 3,000 to 6,000 genes. Reconstructions of the gene set of the shared ancestor of the γ-Proteobacteria provide strong evidence that B. aphidicola genomes are derived from larger genomes through loss of most genes (12, 19).
Despite this massive gene loss, the three completed genome sequences for B. aphidicola of different aphid species show that each retains the enzyme-encoding genes for the biosynthesis of all or most essential amino acids. These 45 to 55 genes comprise approximately 10% of the genome. Retention of these genes fits with a major nutritional role of B. aphidicola, as hypothesized by early investigators (3) and as supported by experimental evidence (5-7, 26). Using relatively abundant nonessential amino acids and sugar derived from the aphid's diet of phloem sap, B. aphidicola synthesizes essential amino acids required by animals but rare in phloem (23). The mutualistic role of these pathways is highlighted by the observations that genes underlying synthesis of the nonessential amino acids have been eliminated from B. aphidicola genomes and that all amino acid biosynthetic pathways are typically missing from similarly small genomes of pathogenic bacteria, which obtain the needed nutrients from hosts. Since persistence of B. aphidicola depends on host reproduction, natural selection should favor the ability to regulate these genes appropriately in the context of the symbiosis.
Reduced bacterial genomes are particularly lacking in genes regulating transcription. For example, the E. coli genome encodes 233 experimentally verified regulatory proteins, including those targeting individual pathways and those acting as global regulatory proteins (27), but B. aphidicola contains only about five such genes, as inferred from homology. Most of the difference reflects gene loss in the B. aphidicola lineage. With regard to amino acid biosynthesis, B. aphidicola retains many of the structural genes encoding the enzymes but has lost almost all genes regulating their expression. For example, in E. coli and some other bacteria, transcription of argA, underlying the rate-limiting step in arginine biosynthesis, is repressed by argR, with repression enhanced in the presence of arginine (14). Two of the sequenced B. aphidicola genomes retain argA (and other genes for arginine biosynthesis) but have lost argR. Of the 16 E. coli genes known to encode regulators affecting transcription of amino acid biosynthetic genes (Table 1), only two are present in the sequenced genome of B. aphidicola of Schizaphis graminum [Buchnera(Sg)]. Only one of these, metR, has regulation of amino acid synthesis as a primary role; MetR regulates methionine production in E. coli (15, 32). The other, himA, encodes a protein that affects transcription of numerous E. coli genes, including those encoding carbamoyl phosphate synthetase, which underlies steps in both arginine and pyrimidine biosynthesis (9). In E. coli, most regulatory genes affecting amino acid biosynthesis act by repressing transcription in the presence of the amino acid end product. In the evolution of the Buchnera-aphid symbiosis, selection for overproduction of amino acids may have favored the inactivation or elimination of repressors. Indeed, metR, the only gene encoding a specific regulator of amino acid biosynthesis that is retained in Buchnera(Sg), is unusual in acting as a transcriptional activator. Evidence from E. coli and other bacteria shows that MetR enhances the transcription of metE especially when bound to the MetE substrate, homocysteine (32, 35).
TABLE 1.
Regulatory genes for essential amino acid biosynthetic pathways in E. coli, with those retained in the genomes of Buchnera(Sg) and Buchnera(Ap) indicated
| Amino acid | Loci regulated | Regulatory locus | Regulatory complex | Type of regulation | Present in Buchnera (Sg)/(Ap)? |
|---|---|---|---|---|---|
| Arg | argCBH | argR | ArgR + Arg | Repressor | −/− |
| argE | argR | ArgR + Arg | Repressor | −/− | |
| argF | argR | ArgR + Arg | Repressor | −/− | |
| carAB | argR | ArgR + Arg | Repressor | −/− | |
| carAB | himA | IHFa | Activator/repressor | +/+ | |
| Lys | lysA | lysR | LysR | Activator | −/− |
| Thr | thrABC | thrL | ThrL | Attenuation | −/− |
| Val/Ile/Leu | ilvC | ilvY | IlvY | Activator | −/− |
| ilvIH | lrp | Lrp | Activator | −/− | |
| ilvD | lrp | Lrp | Activator | −/− | |
| ilvD | himA | IHF | Activator/repressor | +/+ | |
| ilvD | ilvL | IlvL | Attenuation | −/− | |
| Leu | leuABCD | leuO | LeuO | Activator | −/− |
| leuABCD | lrp | Lrp | Activator | −/− | |
| leuABCD | leuL | LeuL | Attenuation | −/− | |
| Trp | trpE, trpDCBA | trpR | TrpR + Trp | Repressor | −/− |
| trpL | TrpL | Attenuation | −/− | ||
| aroH | trpR | TrpR + Trp | Repressor | −/− | |
| aroA | lrp | Lrp | Activator | −/− | |
| Phe | pheA | pheL | PheL | Attenuation | −/− |
| His | hisGDCBHAFI | hisL | HisL | Attenuation | −/− |
| Met | metE | metJ | MetJ + SAMb | Repressor | −/− |
| metE | metR | MetR + Hcn | Activator | +/− | |
| Gly | glyA | metR | MetR + Hcn | Activator | +/− |
| Cys | cysK | cysB | CysB | Activator | −/− |
IHF, integration host factor.
SAM, S-adenogylmethionine.
Because of variation in plants, aphids do encounter wide fluctuations in quantity and composition of dietary amino acids (e.g., see references 23, 24, and 31). Without some kind of transcriptional control, B. aphidicola would continue to transcribe and translate genes even when the end product is abundant or when substrate for these pathways is limiting, as occurs when the total amino acid content of phloem sap is low. The resulting metabolic waste (transcription, translation, synthesis of unneeded end products) would be expected to adversely affect aphid growth efficiency and reproduction, to the detriment of both symbionts and hosts.
Although B. aphidicola has clearly lost most ancestral regulatory elements, inspection of the genome sequence cannot rule out the possibility that novel mechanisms for transcriptional regulation have evolved. For example, modifications of individual genes or binding sites may yield novel control mechanisms suited to the symbiotic lifestyle. That B. aphidicola may have novel mechanisms for controlling amino acid biosynthetic genes is strongly suggested by the relocation of genes for biosynthesis of leucine and tryptophan to plasmids (2, 11, 22).
In this paper, we present a first attempt to determine the capacity of B. aphidicola for controlling the expression of amino acid biosynthetic genes in response to changes in nutrition. Initially we employed a full-genome microarray based on the genome sequence of Buchnera(Sg) (20, 30, 36) to identify transcripts that respond to experimentally induced dietary changes. Quantitative reverse transcriptase PCR (qRT-PCR) was used to verify microarray findings and to examine consequences of interspecies differences in the presence of intact metR.
MATERIALS AND METHODS
Aphid stock and rearing conditions.
We used two study insect species, S. graminum and Acyrthosiphon pisum; for both, the full genomic sequence of the corresponding Buchnera isolate is available (28, 30) (Table 2). Clonal stocks of both species were maintained in the laboratory (aphids reproduce parthenogenetically). The S. graminum genotype was “biotype E” and was the same as that used for the genomic sequence determination (30) and for the microarray construction (36). It was maintained on barley (Hordeum vulgare cv. “Golf”). The A. pisum strain “7-2-1,” which lacks secondary symbionts, was grown on fava bean; it was used in qRT-PCR experiments. All stock cultures and experimental treatments were subjected to a constant 20°C and a photocycle of 14 h of light:10 h of dark.
TABLE 2.
Aphids, symbiotic bacteria, and corresponding food plants for organisms used in this study
Experimental treatments.
For microarray analyses of whole transcriptome responses to nutritional changes, we manipulated the nutritional status of aphids by supplementing host plants with several amino acids and by starvation. Replicates were performed in plastic chambers (12 × 2 × 8 cm) with fine mesh screen windows for ventilation and with a well into which leaves were inserted. The wells contained water supplemented with single amino acids or with no added amino acid in the case of controls. All experiments were conducted at the same temperature and photoperiod as the stock conditions (20°C and 14 h of light:10 h of dark). Plants were cut and placed in the solutions 24 h before aphids were introduced to the chambers. The control treatment consisted of cut leaves of seedling barley (of the same variety as that used for rearing) in tap water. Amino acid treatments consisted of similar barley leaves inserted into solutions of single amino acids, including arginine (10 mM), glutamine (39 mM), glutamic acid (75 mM), aspartic acid (51 mM), leucine (42 mM), and tryptophan (10 mM). The solutions were made with tap water and brought to the same pH as the control tap water. The starvation treatment consisted of aphids kept at high humidity in Petri dishes with no plants. Aphids were nymphs of synchronized age (born within 24 h of one another), transferred from small colonies 5 days after birth. These individuals were allowed to feed (or starve) for 24 h, harvested by gently brushing from plant leaves, and frozen at −80°C within 2 min of harvest. For the microarray experiments, three treatment sets were performed at different times. These represent biological replicates and were used in separate microarray hybridizations to identify responses to dietary changes.
Two additional sets of experiments were performed for use in qRT-PCR assays aimed at examining relative expression of metE. Treatments consisted of a control (tap water), tap water supplemented with glutamine (100 mM), and tap water supplemented with homocysteine (20 mM), with each solution placed in the reservoirs containing plant leaves, as described above. As in the treatments for the microarray experiments, plants were left in the treatment conditions for 24 h, and then aphids were added and allowed to feed for 24 to 30 h. In these treatments, aphids were mixed-age individuals transferred from a large colony. Each condition was replicated four times within each experimental set in separate chambers.
To compare the changes in metE transcript levels of Buchnera from A. pisum [Buchnera(Ap)] to those of Buchnera(Sg), we performed similar amino acid manipulations for A. pisum using cut fava bean seedlings in tap water and glutamine and homocysteine solutions (same concentrations as above). Aphids were mixed-age individuals from a large colony on fava bean. These manipulations were performed synchronously with the treatments for the qRT-PCR experiments for S. graminum. These samples were analyzed using qRT-PCR only.
When S. graminum was harvested from barley leaves, each sample was separated into two subsamples. One was used for microarray hybridizations, with RNA extraction and labeling protocols as described by Wilcox et al. (36). The other subsample was used to verify that the treatments did affect amino acid profiles in aphid body fluids. Free amino acids were extracted and individual concentrations determined using high-performance liquid chromatography, as described by Telang et al. (31). In addition, for some of the amino acid treatments and controls, we collected leaf exudates in order to assess treatment effects on amino acid profiles in phloem sap, as described by Telang et al. (31).
As for S. graminum, portions of the A. pisum experimental samples were also reserved for free amino acid determinations of aphid body fluids.
Microarrays and hybridization.
The design and construction of the microarrays are described by Wilcox et al. (36) and only briefly summarized here. Individual genes were represented by PCR-amplified fragments obtained using primers designed using the Buchnera(Sg) genome sequence (30). The amplicons corresponded to 95% of coding genes, plus numerous spacers that were longer than 100 nucleotides, stamped onto glass slides, with each gene fragment present as eight copies per slide, as paired spots within four subgrids. All amino acid biosynthetic genes were represented. RNA extraction, labeling of cDNA with Cy3/Cy5, hybridization, and image scoring were carried out as described by Wilcox et al. (36). Total extracted RNA from the aphid sample was used, including host RNA and stable RNAs. Each slide was hybridized with transcriptomes resulting from two treatments, labeled with separate dyes. Each treatment was hybridized a total of two times, labeled once with Cy3 and once with Cy5 (arginine, glutamic acid, and leucine treatments), or three times, including labeling with both dyes (control, glutamine, tryptophan, and starvation treatments). Different hybridizations represented separate treatment sets; therefore, hybridizations are both biological and technical replicates. Slides were scanned using an Applied Precision ArrayWorx scanner, and spot intensity data were extracted using Digital Genome Version 2.19 (MolecularWare). Hybridization with extracted DNA was also carried out to determine differences in spot intensities arising from differential binding or fragment length of individual genes.
Microarray data analyses.
Spot signal data were imported into SAS version 9 (25). Analyses followed the procedure described by Wilcox et al. (36) for assessing signals for individual genes. Briefly, after log2 transformation of the raw signal data for individual spots, we applied a mixed-model ANOVA to remove systematic effects attributable to individual slides, to dye, and to interactions. Average spot intensities from the DNA hybridization were used to remove gene-specific effects such as those reflecting individual fragment length, DNA concentration in the probe spots, or sequence features. The signal was normalized for each slide, and we obtained the residuals for each spot. We used the microarrays to explore the overall patterns and magnitudes of response, and we did not select a particular P value as a threshold for significance. However, we considered those with P < 0.0001 to be candidates for further examination. Our focus in these experiments was on the 55 genes underlying amino acid biosynthesis.
qRT-PCR.
To verify expression changes in metE, the gene appearing to be most responsive to any treatments based on the microarray hybridizations, we performed qRT-PCR. These tests were performed for glutamine and homocysteine treatments relative to the control and were conducted for both Buchnera(Sg) and Buchnera(Ap). Primers were designed to amplify short regions of these genes, using the determined genome sequences for these two B. aphidicola isolates (28, 30). Oligonucleotide primers (5′→3′) for metE were AAAAGCGATGCTAACTGGACCA and CCAACCCGATTTGTTTAGCAA for Buchnera(Sg) and TAAAGGGATGTTAACAGGACCG and CTAAAGCAATTTGTGTTGCAA for Buchnera(Ap), with both reactions yielding 95-bp products. In addition to the target gene, we included two control genes for each comparison. The first, rpsL, encodes a ribosomal protein; the second, atpE, encodes a subunit of ATP synthase. Both control genes showed near-constant relative transcript abundance under different treatments based on the microarray results, and neither is directly related to amino acid synthesis. Primers (5′→3′) for rpsL wereGGCCACGGTTAATCAATTGGT and AACGCCTCTTTTTTGCGGAC[Buchnera(Sg)] and CTACACCTAAAAAGCCTAATTCAGC and CCTGCACAATCTAAGGAGCC [Buchnera(Ap)], giving products of 97 and 199, bp respectively. Primers (5′→3′) for rpsL were TAGAAGGAGCTGCAAGGC and AGCCTAATCCAACTGCAATC [Buchnera(Sg)] and AAGTTTTTAGAGGGAGCCGC and CAGCAATCATTGGAATCGC [Buchnera(Ap)], yielding products of 108 and 103 bp, respectively. qRT-PCR was carried out with a Roche Lightcycler and Roche reagents. We used the same “touchdown” reaction protocol for all reactions, as follows: 95°C for 10 min and then 40 cycles of 95°C for 5 s, variable annealing temperature for 15 s, and 72°C for 5 s. The annealing temperature was 68°C for the first three cycles and was then lowered by 1°C each cycle until it reached 55°C, with remaining cycles at 55°C. An internal standard curve was generated for each of the reactions, using serial dilutions of the cloned template. Each experimental sample consisted of several pooled aphids from a treatment replicate. The same RNA extraction was used for control and test genes, and template copy number was estimated from the standard curve for each gene. Therefore, variation in quantity of sample or in efficiency of an extraction would affect control and test genes equally.
For metE, the ratio of its copy number to the copy number for each of the two control genes was calculated to yield a measure of relative transcript level for a particular treatment. Within each experimental set, all treatments were run synchronously, and analyses were performed separately for each set. Data were categorized according to treatment (control, glutamine, homocysteine). The ratios of transcript copy numbers (metE/control gene) were normalized by a factor that set the lowest value at 1. For parametric tests (one-way ANOVA and t test), results were log transformed to meet criteria of normality and equality of variances; nonparametric tests were also performed (Mann-Whitney for set 1, Kruskal-Wallis for sets 2 and 3). Statistical tests were carried out using the InStat (GraphPad) software package.
RESULTS
Effectiveness of dietary manipulations.
The amino acid treatments resulted in large changes in the amino acid content of both phloem sap and aphid body fluids. To determine the effects of amino acid additions on phloem sap composition, we determined the content of exudates from the treatment plants; representative results are shown in Fig. 1A. This approach to altering amino acid profile dramatically affects the phloem sap contents, but due to transamination in the plant (4), the changes are not always a simple increase in the added amino acid. For example, addition of tryptophan has little effect on tryptophan content in sap but results in elevation of proline (Fig. 1A). In contrast, addition of leucine results in a large spike in leucine in both phloem (Fig. 1A) and the aphid body content (data not shown), indicating that leucine is transported through the phloem and is not readily catabolized in the plant or aphid. Addition of arginine results in more arginine and also elevates relative levels of other amino acids such as proline and aspartic acid (Fig. 1A).
FIG. 1.
Effects of homocysteine (Hcn) and glutamine (Gln) treatments on relative concentrations of amino acids in phloem sap of barley (A), body fluids of S. granimum (B), and body fluids of A. pisum (C). Data are presented as proportions of the total amino acid pool.
In general, treatments had large effects on profiles of free amino acids in the aphids; representative results for glutamine and homocysteine manipulations are shown in Fig. 1. For glutamine treatments, the free amino acid most elevated in both plant sap and aphid tissues was glutamine itself (Fig. 1). In animals, plants, and bacteria, glutamine can be converted to other nonessential amino acids such as aspartic acid and glutamic acid, the substrates for the essential amino acid biosynthetic pathways. The most elevated amino acid in aphid tissues for homocysteine treatments was methionine (Fig. 1B), which may be produced from homocysteine within plant tissues or by Buchnera within the aphid.
The results in Fig. 1 depict relative abundances. We presume that adding an amino acid did not result in absolute decreases in concentrations of any amino acids (i.e., decreased proportions of some amino acids in treatment samples must reflect elevation of other amino acids). Overall, the evidence consistently showed that adding amino acids to the phloem resulted in large effects on amino acid content of the aphid diet and on free amino acid levels in aphid body fluids. Similar changes in body fluid composition occurred in both S. graminum and A. pisum (Fig. 1B and C).
Microarray results.
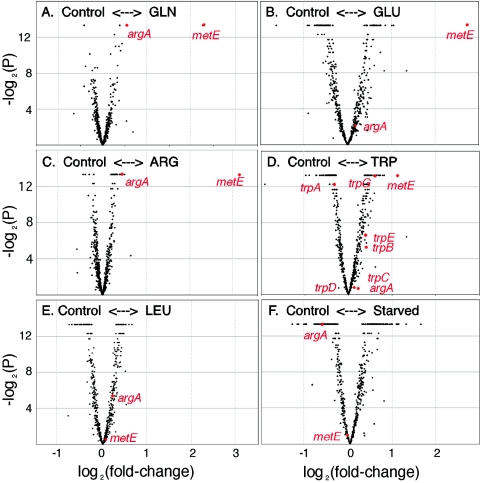
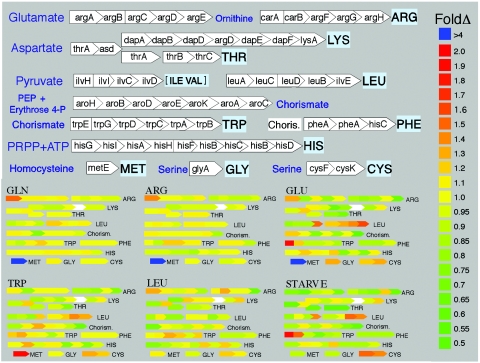
Overviews of gene expression changes in response to dietary manipulations are shown in Fig. 2, which contrasts the control treatments with the various experimental treatments, in the form of “volcano” plots of fold change and statistical significance, and Fig. 3, which shows the average fold changes for genes underlying different enzymatic steps in amino acid biosynthesis. Only one gene, metE, shows a strong response to any of the treatments; transcripts of metE changed >fourfold in response to two nonessential amino acids (glutamic acid and glutamine), >fourfold in response to the essential amino acid arginine, and about twofold in response to tryptophan (Fig. 2A to D and 3). In contrast, metE showed no response to leucine or to starvation (Fig. 2E and F). After metE, the most dramatic change in an amino acid biosynthetic gene involved argA, which showed a slight but statistically robust increase in relative transcript abundance in response to glutamine and arginine and a decrease in response to starvation but no response to leucine or glutamic acid (Fig. 2 and 3). In all cases, changes in argA transcript levels were less than twofold. The only other amino acid biosynthetic gene that showed a possibly significant response was trpA, which showed a minor decrease in tryptophan-treated samples (Fig. 2D). The starvation treatment, compared to controls, showed a decrease in transcripts of argA and some increase in the plasmid-encoded genes (trpEG and leuABCD). Other than the change in metE transcript abundance, all of the responses were small in magnitude (<twofold), and most did not approach statistical significance.
FIG. 2.
Volcano plots showing the statistical significance (y axis) and the fold change (x axis) for differences between controls and treatments involving the experimental addition of single amino acids to the host plants of S. graminum. Statistical significance is based on a mixed-model ANOVA and reflects both biological and technical replication. Significance levels less than 0.0001 were rounded to that value.
FIG. 3.
Average effects of treatments on transcript levels of amino acid biosynthetic genes of Buchnera(Sg), based on microarray results.
qRT-PCR experiments.
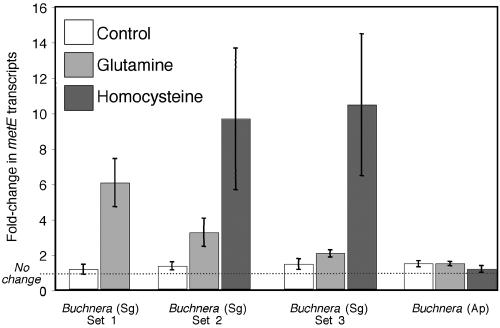
Initially, qRT-PCR was performed to verify the microarray results indicating an increase in metE transcript with glutamine supplementation (Fig. 4, set 1). Using either atpE or rpsL as a control gene, these assays confirmed an increase in relative abundance of metE transcripts of Buchnera(Sg) under treatments with glutamine (Mann-Whitney U test = 0.0, P = 0.028; t test on log-transformed data, t = 9.77, degrees of freedom = 6, P < 0.0001). For this assay, the average difference was about fivefold in relative expression between treatments (Fig. 4, set 1).
FIG. 4.
Transcript abundance of metE following control, glutamine, and homocysteine treatments. Values are based on estimates from qRT-PCR. The three sets for Buchnera(Sg) were performed at separate times, with the third set of bars based on the glutamine and control treatments performed for the microarrays. Each value represents four replicates, with replicates derived from aphids on different plants in different growth chambers.
Roles of MetR and homocysteine.
Transcription of metE is activated by the complex of MetR plus homocysteine in E. coli (32, 35). To address whether homocysteine affected the metE response in Buchnera(Sg), we expanded the qRT-PCR assays to include a treatment involving experimentally elevated homocysteine. In these experiments (Fig. 4, sets 2 and 3), homocysteine treatment had an even greater effect on metE transcript levels than did glutamine treatments, resulting in an average 10-fold increase in metE transcript abundance compared to a 0.5- to 3-fold increase in response to glutamine (Fig. 4, sets 2 and 3; Kruskal-Wallis test for set 2, KW = 9.88, P = 0.0072; for set 3, KW = 7.56, P = 0.023). (The large variation in fold-change response to homocysteine and the smaller response to glutamine in sets 2 and 3 compared to set 1 probably reflect the use of mixed-age aphids in sets 2 and 3; the dietary methionine is probably delayed in reaching the bacteriocytes within embryos which make up most of the symbionts in the older aphid individuals). Overall, these results suggest that the mechanism underlying the effect on metE expression involves MetR and that repression by the MetR plus homocysteine complex is especially strong, as in E. coli (32).
Buchnera(Ap), which diverged from Buchnera(Sg) about 60 million years ago (18), lacks metR and retains metE. Thus, if MetR is the basis for the major changes in metE transcript abundance in Buchnera(Sg), Buchnera(Ap) is predicted to lack the metE response. Treatments with glutamine and homocysteine had similar effects on amino acid profiles of aphid body fluids in both species (Fig. 1). But, as predicted if MetR is involved in metE repression, these treatments failed to induce a metE response in Buchnera(Ap) (Fig. 4, P > 0.8 for both Kruskal-Wallis and ANOVA on log-transformed data). Thus, MetR appears to be the major regulator underlying the large responses of metE to changes in amino acid pools in Buchnera(Sg), and loss of metR in the lineage leading to Buchnera(Ap) corresponds to a lack of regulation of metE expression.
DISCUSSION
The main question motivating this study was whether the elimination of ancestral regulatory genes in reduced and rearranged symbiont genomes corresponds to less responsive transcriptional control or whether these organisms have novel mechanisms for regulating gene expression. Specifically, we addressed whether amino acid biosynthetic genes are subject to transcriptional regulation in response to changes in amino acid concentrations encountered by the hosts. The amino acid biosynthetic pathways, some of the best-studied systems of transcriptional regulation, show dramatic responses in E. coli and related bacteria; for example, the combined mechanisms of trp operon regulation in E. coli can effect a change in transcription rates of up to 500-fold (38). In B. aphidicola, amino acid biosynthesis is central to its symbiotic role, yet the underlying genes have lost most of their ancestral regulatory systems (Table 1). Expression of these genes is relevant to the ecology of aphids/B. aphidicola. For S. graminum in particular, amino acid content of dietary phloem sap varies at different stages of the growing season, at different stages of colonization of an individual plant, and on different host plant species (e.g., see references 23 and 24).
These results for transcript abundances under different treatments would reflect any regulatory responses affecting either transcription rates or transcript longevity. Based on observations of other bacteria, transcriptional control is expected to be central in regulating these pathways, but our study does not exclude the possibility of regulatory changes involving the translation, persistence, or activity of enzymes.
Our results indicate that strong shifts in transcriptional patterns occur solely when the ancestral regulator is retained: metE is the only gene showing a strong response in response to the numerous dietary manipulations that we applied. Furthermore, this response occurs only when metR, the known regulator of metE, is present in the genome. Buchnera(Ap), lacking metR, shows no such response. Interestingly, metR is one of the few regulatory genes that acts as an activator of transcription rather than as a repressor. Therefore, the immediate effect of its loss would be to lower rather than increase transcription of its target gene.
Another complicating factor in understanding the nutrient exchange underlying this symbiosis is that not all amino acids enter the bacteriome and the symbiont cells. Compartmentalization of the symbionts, with barriers to the free exchange of materials with the host, means that excess amounts of a particular essential amino acid in hemolymph or in host cells may not be detectable or useable by the symbionts. Previous nutritional studies using labeled N sources provided evidence that many amino acids are not taken up by B. aphidicola (6, 7, 13). Possibly, only nonessential amino acids are imported; two of these, glutamic acid and aspartic acid, serve as substrates for the pathways producing the essential amino acids. Arginine is readily broken down into glutamic acid plus glutamine in animal tissues, probably explaining why arginine and glutamine treatments had similar effects on transcript profiles (Figs. 2 and 3). In our treatments, tryptophan was also metabolized, at least partly in the plant, to give an increase in glutamine in hemolymph (data not shown), consistent with the elevated expression of metE and mildly elevated argA, as observed for glutamine and glutamic acid treatments. In contrast, the addition of leucine, which appears not to be catabolized in plants or aphids, did not result in elevated glutamine (Fig. 1) and had no effect on metE transcript levels (Fig. 2 and 3). It is possible that manipulation of glutamine, glutamic acid, arginine, and tryptophan all increased homocysteine levels within B. aphidicola cells. In plant and insect samples, the levels of homocysteine were always low relative to other amino acids; we did not assay absolute concentrations.
This study, as well as a previous study on transcriptome responses to heat stress (36), point to a lack of novel mechanisms for regulating transcript levels in B. aphidicola. Buchnera(Sg) does mount a heat shock response, but compared to free-living bacteria, it is relatively feeble in terms of numbers of loci involved and magnitude of changes (36). For variability in amino acid pools and in temperature, the only strong responses are those expected on the basis of the few ancestral regulatory mechanisms that are retained. Furthermore, these appear to function in a fashion similar to that in ancestors. For example, the consensus sequence of the global heat shock promoter is exactly the same in Buchnera(Sg) and in E. coli, and only the loci retaining this promoter show a response to heat stress (36). Likewise, homocysteine appears to be the main molecule affecting MetR activation of metE promoters, as in E. coli and other enterics. Most “heat shock” genes and most genes for amino acid biosynthesis show little or no transcriptional responses to these variables, in contrast to the case for free-living, related bacteria.
The difference between Buchnera(Sg) and Buchnera(Ap) in the presence and action of metR indicates that loss of regulatory genes is ongoing during Buchnera evolution. It is possible that differences in content of sulfur-containing organic compounds between grasses (the host plants of S. graminum) and herbaceous legumes (used by A. pisum) affect the need for regulating methionine production and thus underlie the differential retention of metR by these two species. Based on the genome sequences, the two species also differ in ability to utilize sulfate; the genes underlying reduction to sulfide are intact in Buchnera(Ap) but are inactivated as pseudogenes in Buchnera(Sg), possibly due to the presence of more sulfur-containing organic compounds in grasses (30).
Distantly related bacteria often display completely different regulatory mechanisms for the expression of amino acid biosynthetic pathways, indicating that new regulatory systems often can replace old ones while the structural genes encoding the biosynthetic enzymes are continuously maintained (e.g., see references 37 and 38). In light of this lability in regulatory mechanisms and of the great age of the Buchnera symbiosis (>100 million years [18]), the lack of novel transcriptional responses distinctive to the symbiotic lifestyle is somewhat surprising. Among the possible reasons that B. aphidicola lacks novel regulatory systems is the remarkable stasis of its genome contents and arrangement (30), which may have limited its ability to acquire novel regulatory elements. Another explanation may lie in its small genetic population size, which may limit the effectiveness of selection for more fine-tuned transcriptional regulation (8, 16, 21).
Acknowledgments
We thank Gordon Plague for substantial help with the quantitative PCR assays and for helpful comments on experimental design, Howard Ochman for comments, Susan Miller for advice on analyses, and Becky Nankivell for assisting with preparation of the manuscript and figures.
Funding was from an NSF postdoctoral fellowship to J.L.W. and from grant NSF-0313737 to N.A.M.
REFERENCES
- 1.Akman, L., A. Yamashita, H. Watanabe, K. Oshima, T. Shiba, M. Hattori, and S. Aksoy. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32:402-407. [DOI] [PubMed] [Google Scholar]
- 2.Bracho, A. M., D. Martinez-Torres, A. Moya, and A. Latorre. 1995. Discovery and molecular characterization of a plasmid localized in Buchnera sp. bacterial endosymbiont of the aphid Rhopalosiphum padi. J. Mol. Evol. 41:67-73. [DOI] [PubMed] [Google Scholar]
- 3.Buchner, P. 1965. Endosymbiosis of animals with plant microorganisms. Wiley and Sons, New York, N.Y.
- 4.Corruzi, G., and R. Last. 2000. Amino acids, p. 358-411. In B. B. Buchnanan, W. Gruissem, and R. L. Jones (ed.), Biochemistry and molecular biology of plants. American Society of Plant Biology, Rockville, Md.
- 5.Douglas, A. E., and W. A. Prosser. 1992. Synthesis of the essential amino acid tryptophan in the pea aphid (Acyrthosiphon pisum) symbiosis. J. Insect Physiol. 38:565-568. [Google Scholar]
- 6.Febvay, G., Y. Rahbé, M. Rynkiewiecz, J. Guillard, and G. Bonnot. 1999. Fate of dietary sucrose and neosynthesis of amino acids in the pea aphid, Acyrthosiphon pisum, reared on different diets. J. Exp. Biol. 202:2639-2652. [DOI] [PubMed] [Google Scholar]
- 7.Febvay, G., I., Liadouze, J. Guillaud, and G. Bonnot. 1995. Analysis of energetic amino acid metabolism in Acyrthosiphon pisum—a multidimensional approach to amino acid metabolism in aphids. Arch. Insect Biochem. Physiol. 29:45-69. [Google Scholar]
- 8.Funk, D. J., J. J. Wernegreen, and N. A. Moran. 2001. Intraspecific variation in symbiont genomes: bottlenecks and the aphid-Buchnera association. Genetics 157:477-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goosen, N., and P. van de Putte. 1995. The regulation of transcription initiation by integration host factor. Mol. Microbiol. 16:1-7. [DOI] [PubMed] [Google Scholar]
- 10.Keeling, P. J., and N. M. Fast. 2002. Microsporidia: biology and evolution of highly reduced intracellular parasites. Annu. Rev. Microbiol. 56:93-116. [DOI] [PubMed] [Google Scholar]
- 11.Lai, C. Y., L. Baumann, and P. Baumann. 1994. Amplification of trpEG: adaptation of Buchnera aphidicola to an endosymbiotic association with aphids. Proc. Natl. Acad. Sci. USA 91:3819-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lerat, E., V. Daubin, and N. A. Moran. 2003. From gene trees to organismal phylogeny in prokaryotes: the case of the γ-Proteobacteria. PLOS Biol. 1:101-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liadouze, I., G. Febvay, J. Guillaud, and G. Bonnot. 1995. Effect of diet on the free amino-acid pools of symbiotic and aposymbiotic pea aphids, Acyrthosiphon pisum. J. Insect Physiol. 41:33-40. [Google Scholar]
- 14.Lim, D. B., J. D. Oppenheim, T. Eckhardt, and W. K. Maas. 1987. Nucleotide sequence of the argR gene of Escherichia coli K-12 and isolation of its product, the arginine repressor. Proc. Natl. Acad. Sci. USA 84:6697-6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maxson, M. E., B. Redfield, X. Y. Cai, R. Shoeman, K. Fujita, W. Fisher, G. Stauffer, H. Weissback, and N. Bron. 1989. Regulation of methionine synthesis in Escherichia coli: effect of the MetR protein on the expression of the metE and metR genes. Proc. Natl. Acad. Sci. USA 86:85-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moran, N. A. 1996. Accelerated evolution and Muller's rachet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. USA 93:2873-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moran, N. A. 2002. Microbial minimalism: genome reduction in bacterial pathogens Cell 108:583-586. [DOI] [PubMed] [Google Scholar]
- 18.Moran, N. A., M. A. Munson, P. Baumann, and H. Ishikawa. 1993. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc. R. Soc. Lond. B Biol. Sci. 253:167-171. [Google Scholar]
- 19.Moran, N. A., and A. Mira. 2001. The process of genome shrinkage in the obligate symbiont, Buchnera aphidicola. Genome Biol. 2:research0054.1-0054.12. [Online.] http://genomebiology.com/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moran, N. A., G. Plague, J. P. Sandström, and J. A. Wilcox. 2003. A genomic perspective on nutrient-provisioning by bacterial symbionts of insects. Proc. Natl. Acad. Sci. USA 100:14543-14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rispe, C., and N. A. Moran. 2000. Accumulation of deleterious mutations in endosymbionts: Muller's ratchet with two levels of selection. Am. Nat. 156:425-441. [DOI] [PubMed] [Google Scholar]
- 22.Rouhbakhsh, D., C.-Y. Lai, C. D. von Dohlen, L. Baumann, P. Baumann, N. A. Moran, and D. J. Voegtlin. 1996. The tryptophan biosynthetic pathway of aphid endosymbionts (Buchnera): genetics and evolution of plasmid-associated trpEG within the Aphididae. J. Mol. Evol. 42:414-421. [DOI] [PubMed] [Google Scholar]
- 23.Sandström, J., and N. Moran. 1999. How nutritionally imbalanced is phloem sap for aphids? Entomol. Exp. Appl. 91:203-210. [Google Scholar]
- 24.Sandström, J. P., A. Telang, and N. A. Moran. 2000. Nutritional enhancement of host plants by aphid—a comparison of three aphid species on grasses. J. Insect Physiol. 46:33-40. [DOI] [PubMed] [Google Scholar]
- 25.SAS Institute Inc. 1996. SAS/STAT software version 6.12. SAS Institute Inc., Cary, N.C.
- 26.Sasaki, T., and H. Ishikawa. 1995. Production of essential amino-acids from glutamate by mycetocyte symbionts of the pea aphid, Acyrthosiphon pisum J. Insect Physiol. 41:41-46. [Google Scholar]
- 27.Serres, M. H., S. Goswami, and M. Riley. 2004. GenProtEC: an updated and improved analysis of functions of Escherichia coli K-12 proteins. Nucleic Acids Res. 32:D300-D302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakaki, and H. Ishikawa. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature (London) 407:81-86. [DOI] [PubMed] [Google Scholar]
- 29.Silva, F. J., A. Latorre, and A. Moya. 2001. Genome size reduction through multiple events of gene disintegration in Buchnera APS. Trends Genet. 17:615-618. [DOI] [PubMed] [Google Scholar]
- 30.Tamas, I., L. Klasson, B. Canbäck, A. K. Näslund, A. S. Eriksson, J. J. Wernegreen, J. P. Sandström, N. A. Moran, and S. G. Andersson. 2002. 50 million years of genomic stasis in endosymbiotic bacteria. Science 296:2376-2379. [DOI] [PubMed] [Google Scholar]
- 31.Telang, A., J. Sandström, E. Dyreson, and N. A. Moran. 1999. Feeding damage by Diuraphis noxia results in a nutritionally enhanced phloem diet. Entomol. Exp. Appl. 91:403-412. [Google Scholar]
- 32.Urbanowski, M. L., and G. V. Stauffer. 1989. Role of homocysteine in metR-mediated activation of the metE and metH genes in Salmonella typhimurium and Escherichia coli. J. Bacteriol. 171:3277-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Ham, R. C., J. Kamerbeek, C. Palacios, C. Rausell, F. Abascal, U. Bastolla, J. M. Fernandez, L. Jimenez, M. Postigo, F. J. Silva, J. Tamames, E. Viguera, A. Latorre, A. Valencia, F. Moran, and A. Moya. 2003. Reductive genome evolution in Buchnera aphidicola. Proc. Natl. Acad. Sci. USA 100:581-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waters, E., M. J. Hohn, I. Ahel, D. E. Graham, M. D. Adams, M. Barnstead, K. Y. Beeson, L. Bibbs, R. Bolanos, M. Keller, K. Kretz, X. Y. Lin, E. Mathur, J. W. Ni, M. Podar, T. Richardson, G. G. Sutton, M. Simon, D. Soll, K. O. Stetter, J. M. Short, and M. Noordewier. 2003. The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc. Natl. Acad. Sci. USA 100:12984-12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weissbach, H., and N. Brot. 1991. Regulation of methionine synthesis in Escherichia coli. Mol. Microbiol. 5:1593-1597. [DOI] [PubMed] [Google Scholar]
- 36.Wilcox, J. L., H. E. Dunbar, R. D. Wolfinger, and N. A. Moran. 2003. Consequences of reductive evolution for gene expression in an obligate endosymbiont. Mol. Microbiol. 48:1491-1500. [DOI] [PubMed] [Google Scholar]
- 37.Xie, G., N. O. Keyhani, C. A. Bonner, and R. A. Jensen. 2002. Ancient origin of the tryptophan operon and the dynamics of evolutionary change. Microbiol. Mol. Biol. Rev. 67:303-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yanovsky, C. 2004. The different roles of tryptophan transfer RNA in regulating trp operon expression in E. coli versus B. subtilis. Trends Genet. 20:367-374. [DOI] [PubMed] [Google Scholar]