Abstract
Expression of the type III protein secretion system (TTSS), encoded in the locus of enterocyte effacement (LEE) of enterohemorrhagic Escherichia coli (EHEC), has been shown to be controlled by various regulators. In a search for additional regulatory genes, we identified a DNA fragment containing clpX and clpP that has a positive regulatory effect on LEE expression in EHEC O157. The expression of LEE-encoded Esp proteins was significantly reduced in a clpXP deletion mutant. Deletion of grlR, a negative regulatory gene within LEE, markedly increased LEE expression even in the clpXP mutant. To verify the regulatory mechanism of GrlR expression, a chromosomal epitope-tagged allele of grlR (grlR-FLAG) was constructed. GrlR-FLAG expression was increased significantly in the clpXP deletion mutant, suggesting that the GrlR level is under the control of ClpXP, and this regulation is critical for the ClpXP-dependent expression of LEE in EHEC. Deletion of rpoS, the gene encoding a stationary-phase-inducing sigma factor that is a substrate for ClpXP protease, partially restored LEE expression in the clpXP mutant. A multicopy plasmid carrying rpoS strongly repressed expression of Esp proteins, suggesting that positive regulation by ClpXP is partially mediated through a negative effect of RpoS on LEE expression. We also found that rpoS deletion induces transcription of pchA, which encodes one of the positive regulators for LEE expression in EHEC. These results suggest that ClpXP controls expression of LEE through the regulation of RpoS and GrlR levels in EHEC.
Enterohemorrhagic Escherichia coli (EHEC) strains are life-threatening human pathogens and cause hemorrhagic colitis, bloody diarrhea, and hemolytic uremic syndrome (40). EHEC is a member of the attaching and effacing pathogens (40, 56), a group that includes enteropathogenic E. coli (EPEC) (39) and the mouse pathogen Citrobacter rodentium (45). The attaching/effacing lesion on intestinal epithelial cells is characterized by destruction of microvilli and formation of a pedestal-like structure, triggered by rearrangement of cytoskeletal proteins (30, 42). The genes essential for causing the attaching/effacing lesion are encoded in a pathogenicity island designated the locus of enterocyte effacement (LEE). LEE consists of more than 40 genes organized into five major operons, designated LEE1 to LEE5 (6, 8, 43). LEE1, -2 and -3 operons contain mostly genes encoding the structural and auxiliary proteins necessary for formation of a dedicated type III protein secretion system (TTSS) (22). The LEE4 operon encodes several secreted proteins (e.g., EspA, EspB, EspD, and EspF), all of which are secreted through TTSS (28, 29, 34, 37). The LEE5 operon encodes an adhesion factor, designated intimin (23, 24), and Tir, a protein that is also translocated through TTSS and acts as a receptor for intimin at the host cell membrane (27).
A transcriptional activator, Ler, is encoded by the first gene of the LEE1 operon and is essential for the expression of almost all LEE genes (9, 38). Deng et al. (5) reported that LEE-encoded Orf10 (GrlR) and Orf11 (GrlA) are positive and negative regulators, respectively. Among regulatory proteins encoded outside LEE, nucleoid-associated proteins such as Fis (10), H-NS (2, 57), integration host factor (9), and Hha (46) have been shown to be involved in positive or negative control of LEE expression. Several quorum-sensing-related genes also are important for regulation of LEE expression (25, 49, 50). Other regulatory factors such as YhiE (GadR), YhiF, YhiX (GadE), EtrA, EivF, and BipA are involved in regulation of LEE gene expression in EPEC and/or EHEC (15, 47, 52, 61).
Although most of these regulatory mechanisms are thought to be common in both EHEC and EPEC, there are some differences in LEE regulation between these two organisms. For example, the EPEC adherence factor plasmid, which is widely distributed in EPEC strains (48, 51) but is not found in EHEC, contains perA, B, and C (11), also designated bfpT, V, and W (53). The functions of perA and perC have been found to be important for full activation of LEE genes (11, 38, 41). We previously reported that perC homologues A, B, and C (renamed pchA, B, and C) were identified by screening an EHEC O157 genomic library, and that pchA, B, and C positively regulate LEE transcription in EHEC and are essential for EHEC adhesion to HEp-2 cells (20).
In the present study, we have identified clpX and clpP as positive regulatory genes for LEE expression in EHEC O157 and found that this positive regulation partially depends on negative regulation by RpoS, a stationary-phase sigma factor and a substrate for the ClpXP protease. We further investigated ClpXP down-regulation of expression of the LEE-encoded negative regulator GrlR in strains with or without rpoS deletion. These results suggest that RpoS and GrlR are involved in the regulation of ClpXP-dependent expression of LEE in EHEC.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are summarized in Table 1. The wild-type EHEC O157:H7 strain used was Sakai (16). E. coli K-12 strain JM109 (60) was used for DNA procedures. Unless otherwise specified, bacteria were grown in Luria broth (LB). Antibiotics were added, as required, at these final concentrations: ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; kanamycin, 50 μg/ml; and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), 50 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant phenotype | Source or reference |
|---|---|---|
| Strains | ||
| EDL933-1 | EDL933 Δ(ler)::kan | 20 |
| EDL933-5141 | EDL933 Δ(lac) | 20 |
| Sakai | Wild-type EHEC O157:H7 (same as RIMD0509952) | 16 |
| SKI-5142 | Sakai Δ(lacZYA) | This study |
| SKI-5143 | SKI-5142 Δ(ler)::kan | This study |
| SKI-5144 | SKI-5142 Δ(pchA)::kan | This study |
| SKI-5147 | SKI-5142 Δ(clpX)::kan | This study |
| SKI-5148 | SKI-5142 Δ(clpP)::kan | This study |
| SKI-5149 | SKI-5142 Δ(clpXP)::kan | This study |
| SKI-5151 | SKI-5142 Δ(rpoS)::cat | This study |
| SKI-5152 | SKI-5142 Δ(grlR)::cat | This study |
| SKI-5153 | SKI-5142 Δ(grlA)::cat | This study |
| SKI-5154 | SKI-5142 Δ(grlRA)::cat | This study |
| SKI-5155 | SKI-5149 Δ(rpoS)::cat | This study |
| SKI-5156 | SKI-5149 Δ(grlR)::cat | This study |
| SKI-5180 | SKI-5142 grlR::FLAG Kmr | This study |
| SKI-5181 | SKI-5180 Δ(ler)::cat | This study |
| SKI-5182 | SKI-5180 Δ(clpXP)::cat | This study |
| SKI-5183 | SKI-5180 Δ(rpoS)::cat | This study |
| SKI-5188 | SKI-5183 Kms | This study |
| SKI-5189 | SKI-5188 Δ(clpXP)::kan | This study |
| SKI-5190 | SKI-5142 grlA::FLAG Kmr | This study |
| SKI-5191 | SKI-5190 Δ(ler)::cat | This study |
| SKI-5192 | SKI-5190 Δ(clpXP)::cat | This study |
| SKI-5193 | SKI-5190 Δ(rpoS)::cat | This study |
| Plasmids | ||
| pKD3, pKD4, pKD13 | Template plasmid for lambda Red recombination system, Apr | 4 |
| pKD46 | Red recombinase expression plasmid, Apr | 4 |
| pCP20 | FLP recombinase expression plasmid, Apr | 4 |
| pSUB11 | Template plasmid for FLAG epitope tagging, Apr | 57 |
| pLEE19 | pSSVI215-LEE, espB::Tn3-lacZYA Kmr Apr | 20 |
| pGEM-T-Easy | TA cloning vector, Apr | Promega |
| pGEM-self | Self-ligated pGEM-T-Easy | A. Iguchi, unpublished |
| pGEMLER | pGEM-T-Easy-ler | 20 |
| pGEMRS | pGEM-T-Easy-rpoS | This study |
| pGEMGA | pGEM-T-Easy-grlA | This study |
| pGEMPA | pGEM-T-Easy-pchA | This study |
| pACYC184 | Cloning vector, Cmr Tcr | 3 |
| pSPH4 | pACYC184 carrying a 6.29-kbp SphI fragment containing clpP and clpX from EDL933-1 | This study |
| pACXP | pACYC184-clpXP | This study |
| pRL124 | Promoter probe vector, Apr | 36 |
| pRLLER | pRL124-ler promoter region | This study |
| pRLSL | pRL124-sepL promoter region | This study |
| pRLPA | pRL124-pchA promoter region | This study |
DNA procedures.
Standard DNA procedures, including DNA sequencing and PCR, were performed as described previously (17-19, 33).
Shotgun cloning.
A genomic library of strain EDL933-1 (a ler derivative of wild-type EHEC O157 strain EDL933) (20) was screened to identify regulatory genes for LEE expression in EHEC O157 as described previously (20).
Construction of plasmids.
The reporter plasmid pLEE19 and ler-expressing plasmid pACLER were described previously (20). Plasmids pACXP, pGEMGA, pGEMPA, and pGEMRS were constructed by cloning amplified PCR fragments into pACYC184 (3) or pGEM-T-Easy (Promega). The DNA sequences of the PCR primers used in these studies are shown in Table 2.
TABLE 2.
Oligonucleotide primers used in this studya
| Primer | Nucleotide sequence (5′-3′) |
|---|---|
| LACIP1 | GCGGTATGGCATGATAGCGCCCGGAAGAGAGTCAATTCAGGGTGGTGAATGTGTAGGCTGGAGCTGCTTC |
| LACAP2 | CATATCGGTAAATAGCTTGCCTGCTTTTATTCTTTCTGTCATCGACATGTCATATGAATATCCTCCTTAGT |
| CLPPP1 | AGGTTACAATCGGTACAGCAGGTTTTTTCAATTTTATCCAGGAGACGGAAGTGTAGGCTGGAGCTGCTTC |
| CLPPP2 | AGCGTTGTGCCGCCCTGGATAAGTATAGCGGCACAGTTGCGCCTCTGGCACATATGAATATCCTCCTTAGT |
| CLPXP1 | CATTTGCGTCGTCGTGTGCGGCACAAAGAACAAAGAAGAGGTTTTGACCCGTGTAGGCTGGAGCTGCTTC |
| CLPXP2 | GGAGATAAAATCCCCCCTTTTTGGTTAACTAATTGTATGGGAATGGTTAACATATGAATATCCTCCTTAGT |
| RPOSP52 | AAGAAAAAGGCCAGCCTCGCTTGAGACTGGCCTTTCTGACAGATGCTTACCTTACGCCCCGCCCTGCCAC |
| RPOSP6 | AGGCTTTTGCTTGAATGTTCCGTCAAGGGATCACGGGTAGGAGCCACCTTCTACCTGTGACGGAAGATCA |
| ORF10P5 | TTTTAAATAAACTTGTGGCATTCCTGTGTTTTTAATAACTAATGGCTTACGCCCCGCCCTGCCAC |
| ORF10P6 | ATTATGAAGGATGGCATCTATAGCATTATATTTATTAGCAATGAACTACCTGTGACGGAAGATCA |
| ORF11P5 | ACTCTCCTTTTTCCGCCTCATGATCATTTCGTTCCAGATATTAGACTTACGCCCCGCCCTGCCAC |
| ORF11P6 | GAATCTAAAAATAAAAATGGCGACTATGTAATTCCTGACTCAGTACTACCTGTGACGGAAGATCA |
| ORF10P5FF | CCATTAGTTATTAAAAACACAGGAATGCCACAAGTTTATTTAAAAGACTACAAAGACCATGACGG |
| ORF10FR | TCTTTTTATTTGATAAATAAATCGACATAAAAAACATACATAAAACATATGAATATCCTCCTTAG |
| ORF11P5FF | TCTAATATCTGGAACGAAATGATCATGAGGCGGAAAAAGGAGAGTGACTACAAAGACCATGACGG |
| ORF11FR | GAGAAAAAGGCTTACCCTGGAAAACAAAACCCTTAAATATAGCTTCATATGAATATCCTCCTTAG |
| CLPBHI | CGGGATCCCGTTTACGCAGCATAACGCGCT |
| CLPSPH | CATGCATGCATGATCCCCCCTTTTTGGTTAAC |
| ORF11BHI | CGGGATCCCGTGTATGTTTTTTATGTCGAT |
| ORF11STU | GAAGGCCTTCACCCTAAATATAGCTTTAT |
| RPOSSTU | GAAGGCCTTCACTGGCCTTTCTGACAGATG |
| RPOSBHI | CGGGATCCCGATCGGCGGAACCAGGCTTTT |
| LERKPN | GGGGTACCCCTCTTTATAGAGGGGCGCATT |
| LERERI2 | CGGAATTCCGTGCTTCCTGCTGTAGAACTG |
| LEE4KPN | GGGGTACCCCGTCGACTTTTGGCAGAGAAC |
| LEE4ERI | CGGAATTCCGTGCGATATCCCAGGCTTAGC |
| PERCKPN | GGGGTACCCCGTACCCAGTGTATACCGTGA |
| PERCERI | CGGAATTCCGTTTCTCCAGACATTCTGCCA |
Priming sites on pKD3, pKD4, pKD13, and pSUB11 are underlined. Restriction sites are shown in bold type.
To construct pACXP, a 2.31-kbp DNA fragment carrying clpX and clpP (including 244 bp upstream from the clpP initiation codon) that had been amplified with CLPBHI/CLPSPH primers was digested with BamHI and SphI and inserted into the corresponding sites of pACYC184. To construct pGEMGA, a 0.52-kbp DNA fragment containing the grlA sequence (including 49 bp upstream from the grlA initiation codon) that had been amplified with ORF11BHI/ORF11STU primers was cloned into pGEM-T-Easy. To construct pGEMPA, a 0.41-kbp DNA fragment containing the pchA sequence (including 38 bp upstream from the pchA initiation codon) that had been amplified with PERC4STU/PERC3BHI primers (20) was cloned into pGEM-T-Easy. To construct pGEMRS, a 1.08-kbp DNA fragment containing the rpoS sequence (including 62 bp upstream from the rpoS initiation codon) that had been amplified with RPOSSTU/RPOSBHI primers was cloned into pGEM-T-Easy. The transcriptional direction of grlA, pchA, and rpoS was the same as that of the lac promoter in pGEM-T-Easy.
Plasmids pRLLER, pRLSL, and pRLPA carry PCR-amplified DNA fragments containing the putative transcriptional regulatory regions of ler, sepL, and pchA, respectively, inserted into the promoter-probe vector pRL124 (36). To construct pRLLER, a 0.98-kbp DNA fragment (−904 to +80 relative to the ler initiation codon) that had been amplified with LERKPN (20) and LERERI2 primers was digested with KpnI and EcoRI and inserted into the corresponding sites of pRL124. To obtain pRLSL, a 0.69-kbp DNA fragment (−488 to +205 relative to the sepL initiation codon) that had been amplified with LEE4KPN/LEE4ERI primers was cloned into pRL124. To construct pRLPA, a 0.52-kbp DNA fragment (−489 to + 36, relative to the pchA initiation codon) that had been amplified with PERCKPN/PERCERI primers was cloned into pRL124. Though the exact promoter structures are still not known for the pchA gene, we believe that all pRL124-derived plasmids contain functional promoters, because cells carrying these plasmids had much higher levels of β-galactosidase than cells carrying pRL124 (Table 3, data not shown). Plasmid pGEM-self is self-ligated pGEM-T-Easy (A. Iguchi, unpublished).
TABLE 3.
Effect of clpXP deletion on transcription of LEE regulator genes
| Strain | β-Galactosidase activity, Miller unitsa (SD)
|
||
|---|---|---|---|
| pRL124 | pRLLER | pRLPA | |
| SKI-5142 | 63.7 (4.74) | 43,100 (4,510) | 12,500 (1,280) |
| SKI-5149 | 56 (5.43) | 11,100 (1,190) | 5,950 (499) |
β-Galactosidase activity was measured in late-log-phase cells grown in DMEM plus 0.5% glycerol.
Construction of mutant strains.
A one-step inactivation method (4) was used to construct mutant strains. PCR products containing kanamycin or chloramphenicol resistance cassettes flanked by 45 or 50 bp of homology to the 5′ and 3′ termini of each gene, were electroporated into competent cells of parent strains carrying pKD46. To construct strain SKI-5142, PCR fragments were amplified from pKD4 using primers LACIP1 and LACAP2. The resultant kanamycin-resistant colonies were examined for the Lac-negative phenotype on LB agar plates containing X-Gal. The FRT-flanked kanamycin cassette was removed after transformation with pCP20 as described previously (4).
We confirmed that the expression levels of LEE-encoded Esp proteins in wild-type Sakai and SKI-5142 cells were comparable (data not shown). To construct strains SKI-5143 and SKI-5144, PCR products amplified from pKD13 and pKD4 using primers LERP1/LERP4 and PERCP1/PERCP2, respectively, were electroporated into strain SKI-5142 carrying pKD46, as described previously (20). To construct strains SKI-5147, SKI-5148, and SKI-5149, PCR products amplified from pKD4 with primers CLPXP1/CLPXP2, CLPPP1/CLPPP2, and CLPPP1/CLPXP2, respectively, were electroporated into SKI-5142 carrying pKD46. To construct strains SKI-5151, SKI-5152, SKI-5153, and SKI-5154, PCR products amplified from pKD3 with primers RPOSP52/RPOSP6, ORF10P5/ORF10P6, ORF11P5/ORF11P6, and ORF11P5/ORF10P6, respectively, were electroporated into SKI-5142 carrying pKD46. To construct SKI-5155 and SKI-5156, PCR products amplified from pKD3 with primers RPOSP52/RPOSP6 and ORF10P5/ORF10P6, respectively, were electroporated into SKI-5149 carrying pKD46.
To tag the chromosomal grlR and grlA genes with the DNA sequence encoding triple FLAG epitopes, the modified lambda Red recombination system was used as described previously (58). Briefly, forward primers ORF10P5FF and ORF11P5FF containing the 3′-terminal sequence (without a stop codon) of grlR and grlA, respectively, followed by a sequence encoding triple FLAG epitopes, and reverse primers ORF10FR and ORF11FR, corresponding to a chromosomal region downstream from grlR and grlA, respectively, were used for PCR amplification. PCR products containing the kanamycin resistance cassette were amplified from pSUB11 with each primer set as described above and electroporated into SKI-5142 carrying pKD46 to construct strains SKI-5180 and SKI-5190. To construct strains SKI-5181 (or SKI-5191), SKI-5182 (or SKI-5192), and SKI-5183 (or SKI-5193), the parent strain SKI-5180 (or SKI-5190) carrying pKD46 was electroporated with PCR products amplified from pKD3 with primers LERP5/LERP6 (20), CLPXP1/CLPPP2, and RPOSP52/RPOSP6, respectively. To construct strain SKI-5188, a kanamycin-sensitive derivative of SKI-5183, the FRT-flanked kanamycin cassette was removed after transformation with pCP20 as described previously (4). To construct strain SKI-5189, PCR products amplified from pKD4 with CLPPP1/CLPXP2 primers were electroporated into SKI-5188 carrying pKD46.
All the intact and mutant loci described above were verified by PCR and/or DNA sequence analysis (data not shown).
β-Galactosidase assay.
β-Galactosidase activity was assayed as described previously (17, 20). Bacteria were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco/Invitrogen) supplemented with 0.5% glycerol and appropriate antibiotics, at 37°C with shaking until they reached an optical density at 600 nm of 0.6. To evaluate the effect of an rpoS deletion on pchA transcription, stationary-phase cultures, grown as above to an optical density at 600 nm of 1.0, were used. All assays were done in triplicate and repeated at least three times.
Analysis of proteins in culture supernatants and whole-cell lysates.
Proteins in culture supernatants and whole-cell lysates from stationary-phase cultures, grown as above, were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and Western blotting as described previously (20). Western blotting was performed with polyclonal anti-EspB (20) or monoclonal anti-FLAG M2 (SIGMA) antibodies to probe EspB or FLAG tag, respectively. Binding of secondary anti-mouse immunoglobulin G antibody conjugated to horseradish peroxidase was detected using ECL Western blotting detection reagents (Amersham). All assays were performed in duplicate and repeated at least three times. The quantification of Orf10-FLAG levels in log-phase and stationary-phase cultures of strains with or without clpXP deletion was performed as follows: both spectinomycin (200 μg/ml) and tetracycline (20 μg/ml) were added to late-log-phase (5 h after initiation of culture growth) and stationary-phase (9 h after initiation of culture growth) cultures of bacteria to inhibit further protein synthesis. Proteins in whole-cell lysates sampled at 0.5, 1, 2, and 4 h after addition of antibiotics were probed with anti-FLAG antibody as described above. All assays were performed in duplicate and repeated at least three times.
RESULTS
ClpXP protease positively regulates expression of LEE-encoded TTSS.
In our previous study, the pchB gene was identified by shotgun cloning as one of the positive regulatory gene for LEE expression (20). Using the same strategy, another putative LEE regulator was found on a 6.29-kbp SphI fragment of pACYC184 (designated plasmid pSPH4). Introduction of pSPH4 into EDL933-5141 resulted in about a twofold increase in espB-lacZ expression on plasmid pLEE19 (data not shown). Sequence analysis showed that pSPH4 contains intact clpX and clpP genes (data not shown, Table 1), which encode a heteromultimeric ATP-dependent protease.
To examine the effect of ClpXP on LEE expression, we isolated clpXP mutants. For this purpose, Sakai was used as the wild-type EHEC O157 strain because of the availability of sequence information for its pchA and pchB genes, which are important for positive regulation of LEE expression (20). We constructed clpX, clpP, and clpXP deletion mutants of strain SKI-5142, as described in Materials and Methods. Proteins from equal amounts of culture supernatants from strains carrying pACYC184 or pACXP were analyzed by SDS-PAGE. All clpX and/or clpP deletions abolished the expression of LEE-encoded secreted proteins, such as EspA, -B, and -D (data not shown, Fig. 1A). Introduction of pACXP restored the amount of Esp proteins to that of the wild-type strain (Fig. 1A).
FIG. 1.
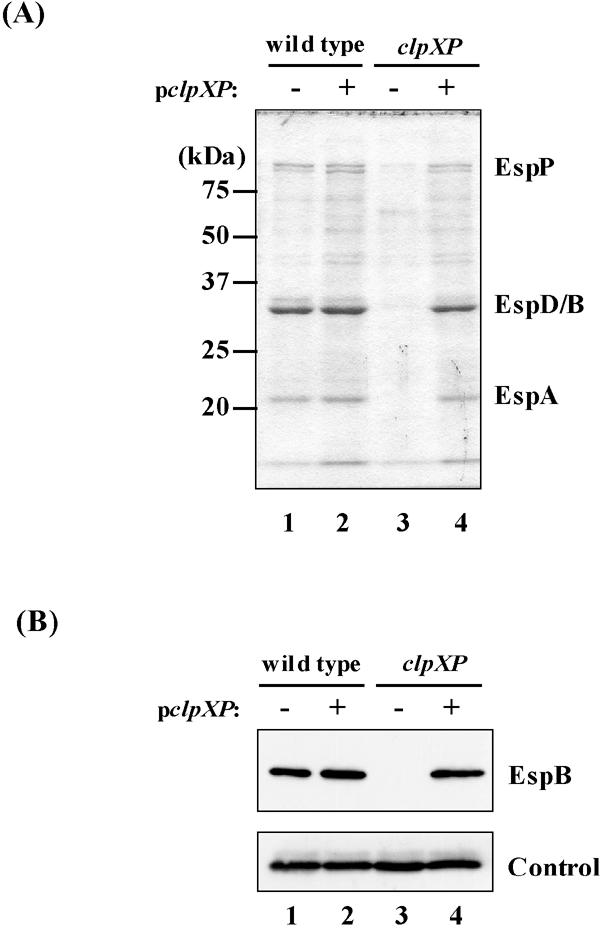
Effect of clpXP deletion and of a multicopy plasmid carrying clpXP on expression of LEE-encoded Esp proteins. (A) Coomassie brilliant blue-stained SDS-PAGE profiles of Esp proteins secreted in culture supernatants. (B) Western blotting analysis of EspB in whole-cell lysates from equal amounts of wild-type (SKI-5142) and clpXP (SKI-5149) strains. Cultures were sampled at stationary phase. Strains used: lanes 1, SKI-5142/pACYC184; lanes 2, SKI-5142/pACXP; lanes 3, SKI-5149/pACYC184; lanes 4, SKI-5149/pACXP. The control shows a cross-reacting protein used to normalize loading.
The amount of EspB was examined further by Western blotting using a polyclonal anti-EspB antibody. These results confirmed that the clpXP deletion impaired EspB expression but did not change the amount of the cross-reacting control band (Fig. 1B, upper and lower panels). We further confirmed these results by determining the transcriptional activity of genes in the LEE4 operon. Transcriptional activities of sepL and espB decreased more than eightfold in the clpXP deletion strain as well as in the ler deletion strain (Fig. 2).
FIG. 2.
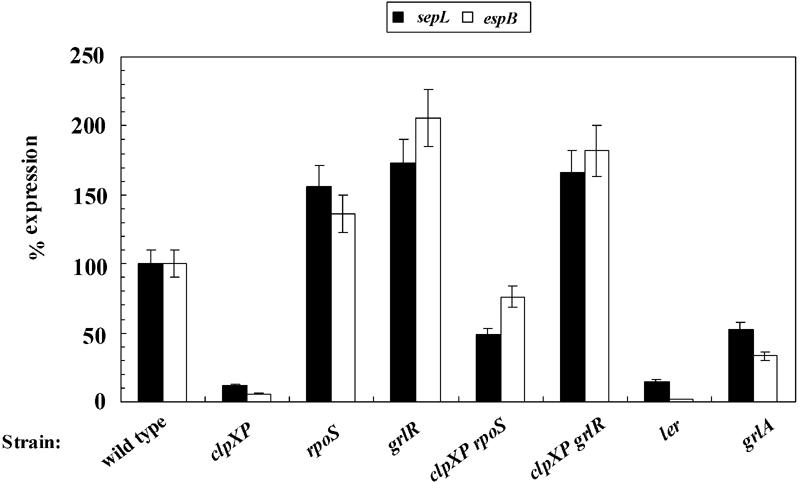
Transcriptional activity of LEE4 in various mutants. β-Galactosidase activities of espB-lacZ in pLEE19 (open bars) and sepL-lacZ on pRLSL (solid bars) were calculated as percentages relative to those in wild-type SKI-5142. Cultures were sampled at late log phase. Average percentages were calculated using three or more measurements from at least five different experiments. Error bars represent standard errors of the normalized values. Typical β-galactosidase activities in Miller units (with standard deviation) of each fusion in SKI-5142 were pLEE19, 66.5 (7.11) and pRLSL, 11,900 (798). Strains used: wild type, SKI-5142; clpXP, SKI-5149; rpoS, SKI-5151; grlR, SKI-5152; clpXP rpoS, SKI-5155; clpXP grlR, SKI-5156; ler, SKI-5143; grlA, SKI-5153.
grlR deletion suppresses the negative effect of clpXP deletion on expression of the LEE-encoded TTSS.
As suggested above, ATP-dependent protease ClpXP positively regulates the expression of LEE-encoded TTSS in EHEC. One explanation for this would be if ClpXP protease controls the expression level of a negative regulator for LEE expression. Since grlR has been shown to encode a negative regulator for LEE expression (5, 35), we examined the effect of grlR deletion on LEE expression. The grlR deletion mutant showed constitutive expression of EspB even in repressed culture conditions (Fig. 3B), confirming a previous report (35). We also constructed a grlR clpXP double mutant to examine whether the negative effect of a clpXP deletion can be suppressed by an additional grlR deletion. The amount of EspB in the double mutant was comparable to that in the strain carrying the grlR deletion alone (Fig. 3), suggesting that ClpXP may regulate the expression level of GrlR.
FIG. 3.
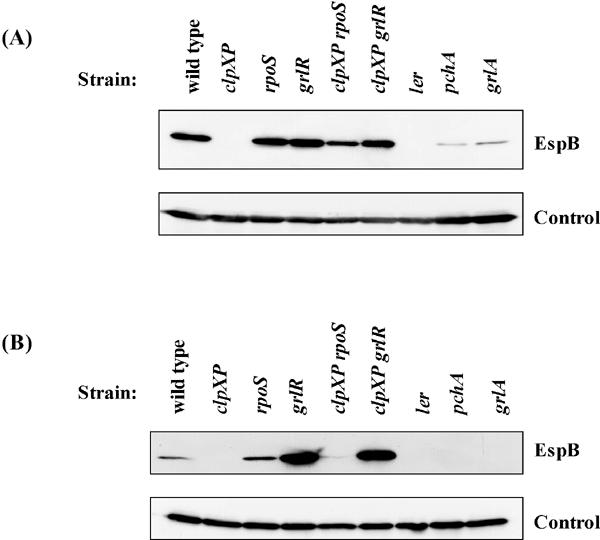
Expression of EspB in whole-cell lysates from stationary-phase cultures of mutants grown in (A) DMEM and (B) LB. Strains used: wild type, SKI-5142; clpXP, SKI-5149; rpoS, SKI-5151; grlR, SKI-5152; clpXP rpoS, SKI-5155; clpXP grlR, SKI-5156; ler, SKI-5143; pchA, SKI-5144; grlA, SKI-5153. The control is as described for Fig. 1.
Amount of GrlR increases in the clpXP deletion strain.
To examine the effect of the clpXP deletion on the GrlR level, we inserted FLAG epitope sequences into the chromosomal grlR at the 3′ terminus, as described in Materials and Methods. We also constructed a control strain, which carries a chromosomal grlA-FLAG fusion gene. An additional ler, clpXP, or rpoS deletion was introduced into these strains. The amounts of GrlA-FLAG and GrlR-FLAG were found to be decreased in the ler mutant (Fig. 4A and B), as also shown in a previous study (7). Although the clpXP deletion markedly reduced GrlA-FLAG expression, it conversely increased the amount of GrlR-FLAG (Fig. 4AB). These results suggest that ClpXP negatively controls the GrlR level.
FIG. 4.
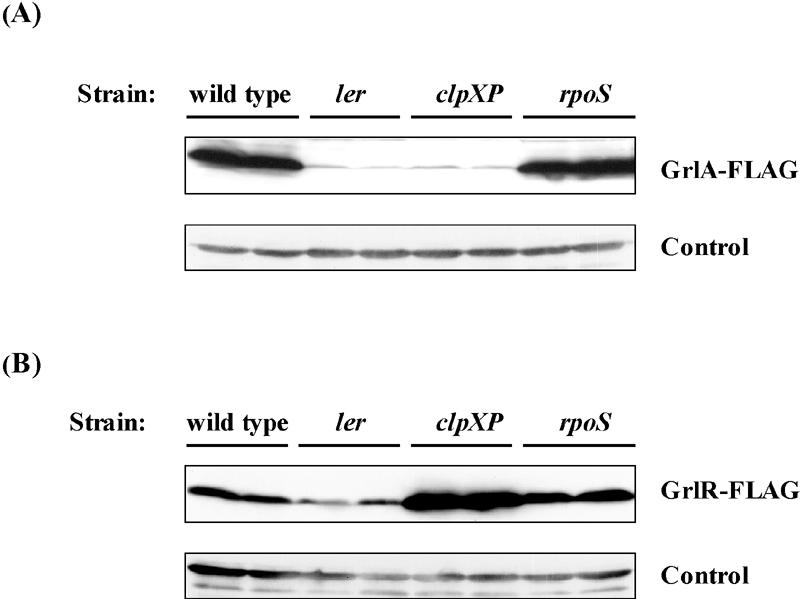
Expression of GrlA-FLAG and GrlR-FLAG. The amount of (A) GrlA-FLAG and (B) GrlR-FLAG in whole-cell lysates was examined by Western blotting using monoclonal anti-FLAG antibody. Lanes received independent samples from each strain. Cultures were sampled at stationary phase. Strains used in A: wild type, SKI-5190; ler, SKI-5191; clpXP, SKI-5192; rpoS, SKI-5193. Strains used in B: wild type, SKI-5180; ler, SKI-5181; clpXP, SKI-5182; rpoS, SKI-5183. The control is as described for Fig. 1.
Growth-phase-dependent expression of GrlR.
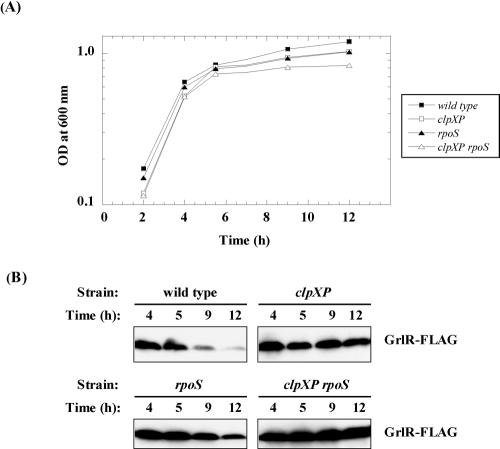
We also examined the growth-phase-dependent expression of GrlR. The GrlR-FLAG level in wild-type background was minimal in stationary-phase cultures (12 h after initiation of culture growth), while the level in the clpXP deletion strain was constitutive (Fig. 5B, upper two panels). To investigate this further, we constructed an rpoS clpXP double mutant in a grlR-FLAG-carrying strain, as described in Materials and Methods. More GrlR-FLAG was found in the stationary-phase culture of rpoS clpXP mutant than in that of the rpoS mutant (Fig. 5B, lower two panels). These results indicate that ClpXP controls GrlR expression even in an rpoS deletion strain.
FIG. 5.
Growth phase-dependent expression of GrlR-FLAG. (A) Growth curves of SKI-5180 (wild type), SKI-5182 (clpXP), SKI-5188 (rpoS), and SKI-5189 (rpoS clpXP) in DMEM plus 0.5% glycerol. (B) Western blots of GrlR-FLAG from whole-cell lysates of each mutant sampled at 4, 5, 9, and 12 h during the growth cycle.
We also examined the GrlR-FLAG levels at different times after addition of spectinomycin and tetracycline in late-log-phase and stationary-phase cultures (Fig. 6). Although the GrlR-FLAG was quite stable in log-phase cultures of strains with or without clpXP deletion even 4 hours after addition of antibiotics, that in stationary-phase cultures of a strain without clpXP deletion was gradually decreased and minimal at 4 h after antibiotic addition. These results suggest that ClpXP may be directly involved in controlling the GrlR levels and thereby regulate LEE expression in stationary phase.
FIG. 6.
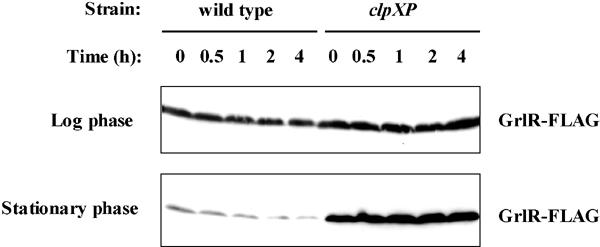
Stability of GrlR-FLAG in log-phase (upper panel) and stationary-phase (lower panel) cultures of the wild type (SKI-5180) and clpXP mutant (SKI-5182). Western blots of GrlR-FLAG from whole-cell lysates of each strain were sampled at 0, 0.5, 1, 2, and 4 h after antibiotic addition.
Deletion in rpoS partially suppresses the negative effect of the clpXP deletion on LEE expression.
The ClpXP protease has been shown to be responsible for degradation of various regulatory proteins (12, 13). Therefore, we examined the RpoS sigma factor, which is one of these regulatory proteins and is responsible for transcription of stationary-phase-induced genes. To analyze the effect of an rpoS deletion on LEE expression, with or without clpXP mutation, we constructed an rpoS deletion strain and an rpoS clpXP double deletion strain and compared the amount of EspB in whole-cell lysates from DMEM (LEE-activated) and LB (LEE-repressed) cultures with those of wild-type, ΔclpXP, Δler, ΔpchA, and ΔgrlA strains. As shown previously (5, 7, 20), the level of LEE expression was reduced by ler, pchA, and grlA deletions (Fig. 3). The loss of EspB observed in the clpXP deletion mutant was partially suppressed by introduction of an additional deletion in rpoS in both DMEM and LB cultures (Fig. 3). These results suggest that RpoS negatively regulates LEE expression in EHEC. In agreement with these results, the transcriptional activities of sepL and espB were increased by an rpoS deletion, with or without a clpXP mutation (Fig. 2).
Multicopy rpoS significantly reduces expression of LEE-encoded TTSS.
To confirm these results, we examined the effect of multicopy rpoS on expression of Esp proteins. As shown in Fig. 7, multicopy rpoS significantly reduced the amount of Esp proteins in culture supernatants and whole-cell lysates in wild-type and rpoS mutant strains. These results indicate that RpoS negatively regulates LEE expression.
FIG. 7.
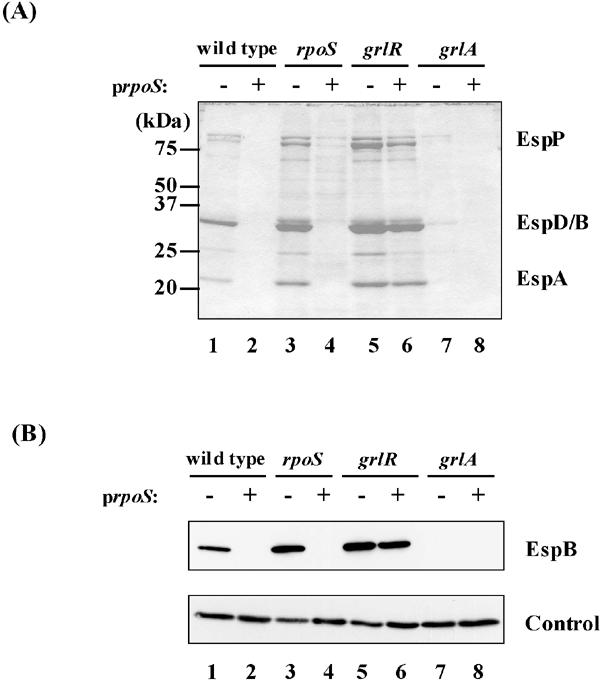
Effect of rpoS overexpression on expression of LEE-encoded secreted proteins. (A) Coomassie brilliant blue-stained SDS-PAGE profiles of Esp proteins secreted in the culture supernatants. (B) Western blotting analysis of EspB in whole-cell lysates. Cultures were sampled at stationary phase. Strains used: wild type, SKI-5142; rpoS, SKI-5151; grlR, SKI-5152; grlA, SKI-5153; lane 1, SKI-5142/pGEM-self; lane 2, SKI-5142/pGEMRS; lane 3, SKI-5151/pGEM-self; lane 4, SKI-5151/pGEMRS; lane 5, SKI-5152/pGEM-self; lane 6, SKI-5152/pGEMRS; lane 7, SKI-5153/pGEM-self; lane 8, SKI-5153/pGEMRS. The control is as described for Fig. 1.
Effect of clpXP deletion on LEE expression is mediated through regulation of the pchA gene.
As described previously, LEE-encoded GrlR and GrlA have been shown to act as negative and positive regulators, respectively, for LEE expression (5, 35). Those studies also showed that the positive effect of GrlA was not observed in a ler mutant. Therefore, we investigated whether the negative effect of GrlR on LEE expression only occurred in the presence of GrlA.
We showed that derepression of LEE expression by the grlR deletion did not occur if the strain also had a grlA deletion (Fig. 8A). In the clpXP deletion mutant, a multicopy plasmid carrying ler, pchA, or grlA restored LEE expression to the wild-type level (Fig. 8B), suggesting that clpXP may also regulate LEE expression by modulating the expression of pchA or ler. Therefore, we analyzed transcriptional regulation of the pchA and ler genes. The β-galactosidase activities of pchA and ler promoters decreased more than twofold in the clpXP deletion strain, while that of pRL124 (vector control) did not change (Table 3). These results indicate that transcription of pchA as well as of ler is under the positive control of ClpXP. We also examined the effect of rpoS deletion on pchA transcription. As shown in Table 4, transcriptional activity of pchA in cells from stationary-phase cultures was increased about twofold due to the rpoS deletion. These results suggest that ClpXP regulates LEE expression via controlling RpoS and GrlR levels (Fig. 9).
FIG. 8.
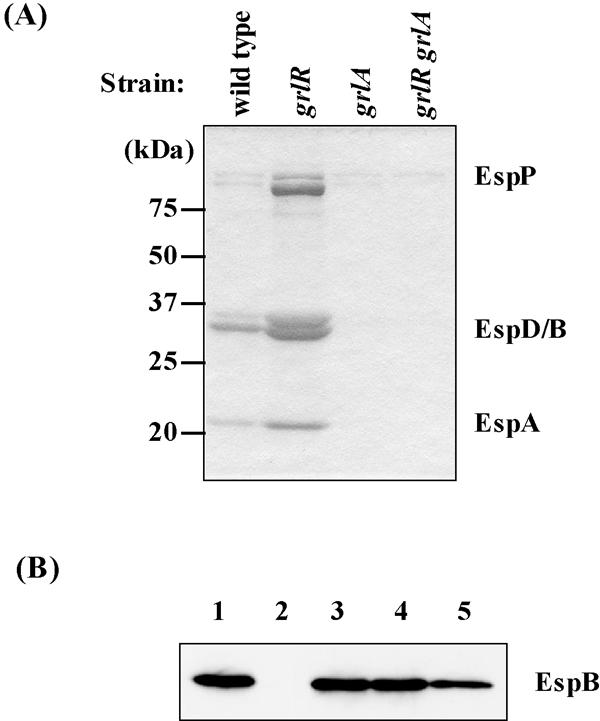
Effect of grlR and/or grlA deletions on expression of secreted proteins. (A) Coomassie brilliant blue-stained SDS-PAGE profiles of Esp proteins secreted in culture supernatants. (B) Complementation of clpXP mutant (SKI-5149) by ler-, pchA-, or grlA-expressing plasmid (pGEMLER, pGEMPA, and pGEMGA, respectively) monitored by the amount of EspB in whole-cell lysates. Cultures were sampled at stationary phase. Strains used in A: wild type, SKI-5142; grlR, SKI-5152; grlA, SKI-5153; grlR grlA, SKI-5154; Strains used in B: lane 1, SKI-5142/pGEM-self; lanes 2, SKI-5149/pGEM-self; lane 3, SKI-5149/pGEMGA; lane 4, SKI-5149/pGEMPA; lane 5, SKI-5149/pGEMLER.
TABLE 4.
Effect of rpoS deletion on pchA transcriptiona
| Strain | β-Galactosidase activity, Miller units (SD)
|
||
|---|---|---|---|
| pRL124 | pRLPA | Ratio (pRLPA/pRL124) | |
| SKI-5142 | 60.4 (8.63) | 9,940 (913) | 165 |
| SKI-5151 | 49.5 (4.00) | 15,600 (1,340) | 315 |
β-Galactosidase activity was measured in late-log-phase cells grown in DMEM plus 0.5% glycerol
FIG. 9.
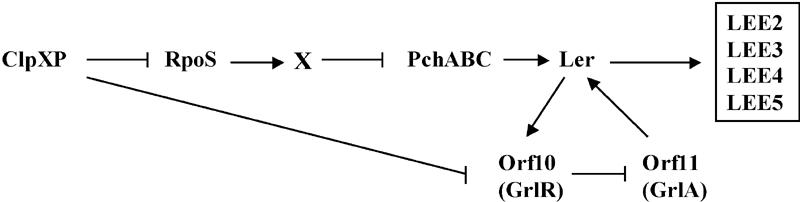
Model for ClpXP-mediated regulation of LEE expression in EHEC. RpoS-dependent regulation by ClpXP is hypothesized to occur through an unidentified factor, X, which negatively regulates pch transcription. RpoS-independent regulation by ClpXP could be mediated through direct or indirect effects of the ClpXP protease on the GrlR level. Ler positively regulates transcription of grlR, whose gene product may antagonize GrlA, thereby facilitating Ler-dependent expression of other LEE operons. As described in the text, additional regulatory elements such as Fis, H-NS, integration host factor Hha, GadR, YhiF, GadE, EtrA, EivF, BipA, and quorum sensing have also been implicated in regulation of the LEE.
DISCUSSION
Regulation of LEE expression is induced in cells under conditions similar to those in the gastrointestinal tract, but repressed in cells grown in rich culture media such as LB (1, 26). In the studies reported here, we confirmed previous findings (35) that deletion of grlR overcomes the repression of LEE-encoded TTSS expression in LB (Fig. 3B). Deng et al. (5) showed that mutation of grlA caused a significant reduction in the expression of LEE-encoded secreted proteins, and introduction of a plasmid carrying grlA could complement this phenotype only in the presence of Ler.
In this report, we found that the negative effect of the grlR deletion on LEE expression was observed only in the presence of grlA (Fig. 8). Since grlA is immediately downstream of grlR and these two genes probably form an operon, GrlR may directly repress GrlA by protein-protein interactions that down-regulate Ler expression. Since transcriptional expression of grlR is under the control of Ler (7), interactions among these regulators are summarized in the model shown in Fig. 9. Ler is central for the regulatory cascade, negative regulation by GrlR of LEE expression depends on the presence of GrlA, and GrlA activates Ler, which can induce expression of other LEE genes.
The level of GrlR-FLAG in the wild-type strain was markedly decreased in stationary-phase cultures, while the level in the clpXP mutant was constitutive (Fig. 4B and 5B). Therefore, ClpXP is involved in regulation of the GrlR level. One explanation for this would be if the ClpXP level is induced in stationary-phase cultures. However, this is unlikely because the ClpXP level is not significantly different in exponential and stationary-phase E. coli K-12 cultures (44). Another possibility may be that GrlR is a substrate for ClpXP. To test this possibility, the level of GrlR-FLAG was compared at different times after addition of spectinomycin and tetracycline in strains with and without a clpXP mutation (Fig. 6). The GrlR-FLAG level of a strain without clpXP deletion was unstable in stationary-phase cultures for several hours after antibiotic addition, suggesting that ClpXP may directly be involved in controlling the stability of GrlR. However, further experiments should be carried out to confirm this conclusion. Since we measured the level of the FLAG-tagged allele of GrlR in this study, the level of native GrlR needs to be determined to rule out the possibility that the FLAG tag may influence the stability of GrlR. Therefore, it is premature to conclude that GrlR is a direct substrate of ClpXP.
The ClpXP protease plays diverse physiological roles in degrading damaged and incomplete proteins (14, 59) and regulatory proteins in response to various stresses (12, 13). Recently, Jackson et al. (21) demonstrated that ClpXP and another ATP-dependent protease, Lon, together control TTSS expression by regulating the stability of a small histone-like protein, YmoA, in Yersinia pestis. YmoA is a homologue of E. coli Hha protein, which has been reported to regulate LEE expression through Ler in EHEC (46). Therefore, it is possible that ClpXP controls Hha stability and thereby regulates LEE expression in EHEC. It will be important to learn whether Hha is a substrate for ClpXP in EHEC and whether Hha controls other LEE regulators such as GrlR, GrlA, and PchA/B/C.
We also found that abolishment of LEE expression in the clpXP deletion mutant was partially suppressed by an rpoS deletion. A previous report showed that expression of the LEE3 operon is positively regulated by RpoS in E. coli K-12 (50). We have not examined LEE3 transcription in the EHEC rpoS mutant used in our studies. However, we have shown that RpoS negatively regulates ler transcription through reduction of pchA transcription in EHEC (Table 4). Therefore, RpoS may also negatively regulate LEE3 transcription in EHEC O157. Because rpoS encodes a stationary-phase alternative sigma factor, an additional regulator, tentatively designated X in Fig. 9, should exist downstream of RpoS and negatively regulate Pch expression. Further study to identify this regulator is now in progress.
ClpXP has been shown to negatively regulate flagellar synthesis in Salmonella enterica serovar Typhimurium by modulating turnover of FlhD and FlhC, encoded by the flagellar master operon (54, 55). We also observed that clpXP deletion in strain O157 Sakai causes overproduction of flagellin, the subunit of the flagellar filament (Iyoda and Watanabe, unpublished). Therefore, clpXP-dependent repression of flagellar synthesis is also observed in EHEC O157. The supramolecular structure of bacterial flagella resembles that of TTSS, and they are considered phylogenetically related (31). As shown in Salmonella, flagellar synthesis is an energy-consuming process, and overproduced flagellin causes significant growth retardation in vitro (32). Therefore, it is possible that ClpXP controls expression of both the flagellar system and other TTSS (in this case, LEE-encoded TTSS) to minimize their expression when they are not needed by the cell.
We previously have found that the pchABC genes encode positive transcriptional regulators that induce ler transcription in EHEC (20). The molecular mechanism by which other regulators, encoded outside LEE, control LEE expression is largely unknown. However, some mechanisms may directly affect Pch expression. Analysis of the regulatory mechanisms of Pch as well as GrlR will be important for a better understanding of the regulation of LEE expression in EHEC.
Acknowledgments
We are grateful to Hiromi Sato and Hitomi Sato for technical assistance. We thank Toru Tobe and Hiroyuki Abe (Osaka University), Tetsuya Hayashi (Miyazaki University), and Kazuhiro Kutsukake (Okayama University) for helpful discussions. We also thank Sergio Uzzau (Sassari University) for providing pSUB11 plasmid.
This work was supported by grants-in-aid for scientific research from the Ministry of Education, Science and Culture of Japan, the Ministry of Health and Welfare of Japan, and the Japan Health Science Foundation.
REFERENCES
- 1.Abe, H., I. Tatsuno, T. Tobe, A. Okutani, and C. Sasakawa. 2002. Bicarbonate ion stimulates the expression of locus of enterocyte effacement-encoded genes in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 70:3500-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bustamante, V. H., F. J. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39:664-678. [DOI] [PubMed] [Google Scholar]
- 3.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng, W., J. L. Puente, S. Gruenheid, Y. Li, B. A. Vallance, A. Vazquez, J. Barba, J. A. Ibarra, P. O'Donnell, P. Metalnikov, K. Ashman, S. Lee, D. Goode, T. Pawson, and B. B. Finlay. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. USA 101:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott, S. J., S. W. Hutcheson, M. S. Dubois, J. L. Mellies, L. A. Wainwright, M. Batchelor, G. Frankel, S. Knutton, and J. B. Kaper. 1999. Identification of CesT, a chaperone for the type III secretion of Tir in enteropathogenic Escherichia coli. Mol. Microbiol. 33:1176-1188. [DOI] [PubMed] [Google Scholar]
- 7.Elliott, S. J., V. Sperandio, J. A. Giron, S. Shin, J. L. Mellies, L. Wainwright, S. W. Hutcheson, T. K. McDaniel, and J. B. Kaper. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliott, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. K. Deng, L. C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 9.Friedberg, D., T. Umanski, Y. Fang, and I. Rosenshine. 1999. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol. Microbiol. 34:941-952. [DOI] [PubMed] [Google Scholar]
- 10.Goldberg, M. D., M. Johnson, J. C. Hinton, and P. H. Williams. 2001. Role of the nucleoid-associated protein Fis in the regulation of virulence properties of enteropathogenic Escherichia coli. Mol. Microbiol. 41:549-559. [DOI] [PubMed] [Google Scholar]
- 11.Gómez-Duarte, O. G., and J. B. Kaper. 1995. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect. Immun. 63:1767-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottesman, S. 1996. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30:465-506. [DOI] [PubMed] [Google Scholar]
- 13.Gottesman, S. 2003. Proteolysis in bacterial regulatory circuits. Annu. Rev. Cell. Dev. Biol. 19:565-587. [DOI] [PubMed] [Google Scholar]
- 14.Gottesman, S., S. Wickner, and M. R. Maurizi. 1997. Protein quality control: triage by chaperones and proteases. Genes Dev. 11:815-823. [DOI] [PubMed] [Google Scholar]
- 15.Grant, A. J., M. Farris, P. Alefounder, P. H. Williams, M. J. Woodward, and C. D. O'Connor. 2003. Co-ordination of pathogenicity island expression by the BipA GTPase in enteropathogenic Escherichia coli (EPEC). Mol. Microbiol. 48:507-521. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 17.Iyoda, S., T. Kamidoi, K. Hirose, K. Kutsukake, and H. Watanabe. 2001. A flagellar gene fliZ regulates the expression of invasion genes and virulence phenotype in Salmonella enterica serovar Typhimurium. Microb. Pathog. 30:81-90. [DOI] [PubMed] [Google Scholar]
- 18.Iyoda, S., and K. Kutsukake. 1995. Molecular dissection of the flagellum-specific anti-sigma factor, FlgM, of Salmonella typhimurium. Mol. Gen. Genet. 249:417-424. [DOI] [PubMed] [Google Scholar]
- 19.Iyoda, S., K. Tamura, K. Itoh, H. Izumiya, N. Ueno, K. Nagata, M. Togo, J. Terajima, N. Ueno, K. Nagata, M. Togo, J. Terajima, and H. Watanabe. 2000. Inducible stx2 phages are lysogenized in the enteroaggregative and other phenotypic Escherichia coli O86:HNM isolated from patients. FEMS Microbiol. Lett. 191:7-10. [DOI] [PubMed] [Google Scholar]
- 20.Iyoda, S., and H. Watanabe. 2004. Positive effects of multiple pch genes on expression of the locus of enterocyte effacement genes and adherence of enterohaemorrhagic Escherichia coli O157:H7 to Hep-2 cells. Microbiology 150:2357-2371. [DOI] [PubMed] [Google Scholar]
- 21.Jackson, M. W., E. Silva-Herzog, and G. V. Plano. 2004. The ATP-dependent ClpXP and Lon proteases regulate expression of the Yersinia pestis type III secretion system via regulated proteolysis of YmoA, a small histone-like protein. Mol. Microbiol. 54:1364-1378. [DOI] [PubMed] [Google Scholar]
- 22.Jarvis, K. G., J. A. Girón, A. E. Jerse, T. K. McDaniel, M. S. Donnenberg, and J. B. Kaper. 1995. Enteropathogenic Escherichia coli contains a specialized secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. USA 92:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jerse, A. E., and J. B. Kaper. 1991. The eae gene of enteropathogenic Escherichia coli encodes a 94-kilodalton membrane protein, the expression of which is influenced by the EAF plasmid. Infect. Immun. 59:4302-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanamaru, K., K. Kanamaru, I. Tatsuno, T. Tobe, and C. Sasakawa. 2000. SdiA, an Escherichia coli homologue of quorum-sensing regulators, controls the expression of virulence factors in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 38:805-816. [DOI] [PubMed] [Google Scholar]
- 26.Kenny, B., A. Abe, M. Stein, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) protein secretion is induced in response to factors similar to those of the gastrointestinal tract. Infect. Immun. 65:2606-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 28.Kenny, B., and B. B. Finlay. 1995. Protein secretion by enteropathogenic Escherichia coli is essential for transducing signals to epithelial cells. Proc. Natl. Acad. Sci. USA 92:7991-7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kenny, B., L. C. Lai, B. B. Finlay, and M. S. Donnenberg. 1996. EspA, a protein secreted by enteropathogenic Escherichia coli, is required to induce signals in epithelial cells. Mol. Microbiol. 20:313-323. [DOI] [PubMed] [Google Scholar]
- 30.Knutton, S., D. R. Lloyd, and A. S. McNeish. 1987. Adhesion of enteropathogenic Escherichia coli to human intestinal enterocytes and cultured human intestinal mucosa. Infect. Immun. 55:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kubori, T., Y. Matsushima, D. Nakamura, J. Uralil, M. Lara-Tejero, A. Sukhan, J. E. Galan, and S. I. Aizawa. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602-605. [DOI] [PubMed] [Google Scholar]
- 32.Kutsukake, K., and T. Iino. 1994. Role of FliA-FlgM regulatory system on the transcriptional control of the flagellar regulon and flagella formation in Salmonella typhimurium. J. Bacteriol. 176:3598-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kutsukake, K., S. Iyoda, K. Ohnishi, and T. Iino. 1994. Genetic and molecular analyses of the interaction between the flagellum-specific sigma and anti-sigma factors in Salmonella typhimurium. EMBO J. 13:4568-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai, L. C., L. A. Wainwright, K. D. Stone, and M. S. Donnenberg. 1997. A third secreted protein that is encoded by the enteropathogenic Escherichia coli pathogenicity island is required for transduction of signals and for attaching and effacing activities in host cells. Infect. Immun. 65:2211-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lio, C.-W. J., and W.-J. Syu. 2004. Identification of a negative regulator for the pathogenicity island of enterohemorrhagic Escherichia coli O157:H7. J. Biomed. Sci. 11:855-863. [DOI] [PubMed] [Google Scholar]
- 36.Malo, M. S., and R. E. Loughlin. 1988. Promoter-detection vectors for Escherichia coli with multiple useful features. Gene 64:207-215. [DOI] [PubMed] [Google Scholar]
- 37.McNamara, B. P., and M. S. Donnenberg. 1998. A novel proline-rich protein, EspF, is secreted from enteropathogenic Escherichia coli via the type III export pathway. FEMS Microbiol. Lett. 166:71-78. [DOI] [PubMed] [Google Scholar]
- 38.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 39.Moon, H. W., S. C. Whipp, R. A. Argenzio, M. M. Levine, and R. A. Giannella. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porter, M. E., P. Mitchell, A. J. Roe, A. Free, D. G. E. Smith, and D. L. Gally. 2004. Direct and indirect transcriptional activation of virulence genes by an AraC-like protein, PerA from enteropathogenic Escherichia coli. Mol. Microbiol. 54:1117-1133. [DOI] [PubMed] [Google Scholar]
- 42.Rosenshine, I., S. Ruschkowski, M. Stein, D. J. Reinscheid, S. D. Mills, and B. B. Finlay. 1996. A pathogenic bacterium triggers epithelial signals to form a functional bacterial receptor that mediates actin pseudopod formation. EMBO J. 15:2613-2624. [PMC free article] [PubMed] [Google Scholar]
- 43.Sánchez-SanMartin, C., V. H. Bustamante, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of the orf19 gene and the tir-cesT-eae operon of enteropathogenic Escherichia coli. J. Bacteriol. 183:2823-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schweder, T., K.-Y. Lee, O. Lomovskaya, and A. Matin. 1996. Regulation of Escherichia coli starvation sigma factor (sigma s) by ClpXP protease. J. Bacteriol. 178:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schauer, D. B., and S. Falkow. 1993. Attaching and effacing locus of a Citrobacter freundii biotype that causes transmissible murine colonic hyperplasia. Infect. Immun. 61:2486-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma, V. K., and R. L. Zuerner. 2004. Role of hha and ler in transcriptional regulation of the esp operon of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 186:7290-7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin, S., M. P. Castanie-Cornet, J. W. Foster, J. A. Crawford, C. Brinkley, and J. B. Kaper. 2001. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol. Microbiol. 41:1133-1150. [DOI] [PubMed] [Google Scholar]
- 48.Sohel, I., J. L. Puente, S. W. Ramer, D. Bieber, C.-Y. Wu, and G. K. Schoolnik. 1996. Enteropathogenic Escherichia coli: identification of a gene cluster coding for bundle-forming pilus morphogenesis. J. Bacteriol. 178:2613-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sperandio, V., C. C. Li, and J. B. Kaper. 2002. Quorum-sensing Escherichia coli regulator A: a regulator of the LysR family involved in the regulation of the locus of enterocyte effacement pathogenicity island in enterohemorrhagic E. coli. Infect. Immun. 70:3085-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stone, K. D., H. Z. Zhang, L. K. Carlson, and M. S. Donnenberg. 1996. A cluster of fourteen genes from enteropathogenic Escherichia coli is sufficient for the biogenesis of a type IV pilus. Mol. Microbiol. 20:325-337. [DOI] [PubMed] [Google Scholar]
- 52.Tatsuno, I., K. Nagano, K. Taguchi, L. Rong, H. Mori, and C. Sasakawa. 2003. Increased adherence to Caco-2 cells caused by disruption of the yhiE and yhiF genes in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 71:2598-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tobe, T., G. K. Schoolnik, I. Sohel, V. H. Bustamante, and J. L. Puente. 1996. Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 21:963-975. [DOI] [PubMed] [Google Scholar]
- 54.Tomoyasu, T., T. Ohkishi, Y. Ukyo, A. Tokumitsu, A. Takaya, M. Suzuki, K. Sekiya, H. Matsui, K. Kutsukake, and T. Yamamoto. 2002. The ClpXP ATP-dependent protease regulates flagellum synthesis in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:645-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomoyasu, T., A. Takaya, E. Isogai, and T. Yamamoto. 2003. Turnover of FlhD and FlhC, master regulator proteins for Salmonella flagellum biogenesis, by the ATP-dependent ClpXP protease. Mol. Microbiol. 48:443-452. [DOI] [PubMed] [Google Scholar]
- 56.Tzipori, S., F. Gunzer, M. S. Donnenberg, L. de Montigny, J. B. Kaper, and A. Donohue-Rolfe. 1995. The role of the eaeA gene in diarrhea and neurological complications in a gnotobiotic piglet model of enterohemorrhagic Escherichia coli infection. Infect. Immun. 63:3621-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Umanski, T., I. Rosenshine, and D. Friedberg. 2002. Thermoregulated expression of virulence genes in enteropathogenic Escherichia coli. Microbiology 148:2735-2744. [DOI] [PubMed] [Google Scholar]
- 58.Uzzau, S., N. Figueroa-Bossi, S. Rubino, and L. Bossi. 2001. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. USA 98:15264-15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wickner, S., M. R. Maurizi, and S. Gottesman. 1999. Post-translational quality control: folding, refolding, and degrading proteins. Science 286:1888-1893. [DOI] [PubMed] [Google Scholar]
- 60.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, L., R. R. Chaudhuri, C. Constantinidou, J. L. Hobman, M. D. Patel, A. C. Jones, D. Sarti, A. J. Roe, I. Vlisidou, R. K. Shaw, F. Falciani, M. P. Stevens, D. L. Gally, S. Knutton, G. Frankel, C. W. Penn, and M. J. Pallen. 2004. Regulators encoded in the Escherichia coli type III secretion system 2 gene cluster influence expression of genes within the locus for enterocyte effacement in enterohemorrhagic E. coli O157:H7. Infect. Immun. 72:7282-7293. [DOI] [PMC free article] [PubMed] [Google Scholar]