Abstract
The Pseudomonas syringae type III secretion system (TTSS) translocates effector proteins into plant cells. Several P. syringae effectors require accessory proteins called type III chaperones (TTCs) to be secreted via the TTSS. We characterized the hopO1-1, hopS1, and hopS2 operons in P. syringae pv. tomato DC3000; these operons encode three homologous TTCs, ShcO1, ShcS1, and ShcS2. ShcO1, ShcS1, and ShcS2 facilitated the type III secretion and/or translocation of their cognate effectors HopO1-1, HopS1, and HopS2, respectively. ShcO1 and HopO1-1 interacted with each other in yeast two-hybrid and coimmunoprecipitation assays. Interestingly, ShcS1 and ShcS2 were capable of substituting for ShcO1 in facilitating HopO1-1 secretion and translocation and each TTC was able to bind the other's cognate effectors in yeast two-hybrid assays. Moreover, ShcO1, ShcS1, and ShcS2 all bound to the middle-third region of HopO1-1. The HopS2 effector possessed atypical P. syringae TTSS N-terminal characteristics and was translocated in low amounts. A site-directed HopS2 mutation that introduced a common N-terminal characteristic from other P. syringae type III secreted substrates increased HopS2 translocation, supporting the idea that this characteristic functions as a secretion signal. Additionally, hopO1-2 and hopT1-2 were shown to encode effectors secreted via the DC3000 TTSS. Finally, a DC3000 hopO1-1 operon deletion mutant produced disease symptoms similar to those seen with wild-type DC3000 but was reduced in its ability to multiply in Arabidopsis thaliana. The existence of TTCs that can bind to dissimilar effectors and that can substitute for each other in effector secretion provides insights into the nature of how TTCs function.
Pseudomonas syringae is a gram-negative bacterial pathogen that is capable of causing disease in many different plants (4, 38). The complete genome sequence of P. syringae pv. tomato DC3000, a pathogen of tomato and the model plant Arabidopsis thaliana, has been determined (15). To be pathogenic, P. syringae requires a type III protein secretion system (TTSS) known as the Hrp system, which is encoded by the Hrp pathogenicity island (3, 5). TTSSs “inject” or translocate bacterial proteins called effectors into host cells and are present in many gram-negative pathogens of plants, animals, and insects and other bacteria closely associated with eukaryotes (22, 39).
In addition to translocating effectors, the P. syringae TTSS also secretes extracellular accessory proteins or helper proteins in the extracellular spaces of plant leaves (i.e., the apoplast), which help in the translocation of effectors into plant cells. P. syringae effectors and helper proteins have been named Hop proteins (for “Hrp outer proteins”). The secretion signals or recognition motifs for proteins secreted by TTSSs are encoded within the N-terminal half of TTSS substrates, with the N-terminal 15 to 20 amino acids being particularly important (62, 63, 66). Specifically, amphipathicity within the N-terminal 15 amino acids appears to be an important characteristic for secretion of Yersinia TTSS substrates (51, 52). P. syringae Hops also share biochemical characteristics that may be part of an N-terminal secretion signal recognized by the TTSS apparatus. For example, the first 50 amino acids are rich in Ser residues and other polar amino acids; position 3 or 4 in most Hops is occupied by aliphatic amino acids (Ile, Leu, Val) or Pro; and the first 12 amino acids rarely have any acidic amino acids (Glu and Asp) (34, 59). However, it remains unknown whether these shared biochemical characteristics are functionally important for the type III secretion of P. syringae Hops.
The availability of the P. syringae pv. tomato DC3000 genome sequence has spurred several bioinformatic and functional genomic searches for additional DC3000 hop genes (21, 33). These studies have exploited the fact that most TTSS-related genes in P. syringae have promoters that contain a conserved sequence called a Hrp box, which is the presumed binding site for HrpL, an alternate sigma factor (30, 73, 75). Additional hop genes were identified based on the biochemical characteristics in the N termini of their predicted products (34, 59). The confirmed Hop inventory of DC3000 is now greater than 40 and is likely to become even larger as additional putative Hops are shown to be secreted or translocated through the TTSS (6). The pathogenic roles for the majority of these Hops are poorly understood, although recently many P. syringae effectors have been implicated in suppression of plant innate immunity (1, 2, 7, 14, 25-27, 36, 42, 55). Recently, a unified naming system for P. syringae Hops has been established and this naming system will be used for the effectors described in this paper (50).
Many TTSS substrates utilize intracellular accessory proteins called type III chaperones (TTCs) that facilitate their secretion and translocation via the TTSS (6, 28, 58). TTCs generally do not share significant sequence similarity with each other unless they are homologous TTCs from different bacterial strains. But they do share the following general characteristics: a low molecular mass (about 10 to 20 kDa); an acidic isoelectric point (pI < 6); and a C-terminal amphipathic region. Several classes of TTCs have been described (58). Class IA TTCs associate with one effector and are usually encoded by TTC genes that are next to their cognate effector gene; class 1B TTCs associate with multiple effectors and are encoded by genes within the DNA regions encoding the TTSS apparatus; class II TTCs assist in the type III secretion of translocators, a class of helper proteins that assist effectors in crossing the eukaryotic plasma membrane; and class III TTCs are utilized in the related flagellar biogenesis TTSS.
The roles of TTCs appear to be multidimensional. TTCs have been shown to prevent interactions between TTSS substrates in the bacterial cell (57, 74) and to stabilize effectors (28, 31). In addition, TTCs may help establish an order of effector secretion (12, 69) and several TTCs play a regulatory role (17, 24, 56, 72). The binding sites for most TTCs are on the N-terminal half of TTSS substrates and may provide a second secretion signal for effectors to be recognized by the secretion apparatus (20). Recently, it has been shown that TTCs in Salmonella spp. are involved in TTSS specificity by directing substrates to a specific TTSS (49). The crystal structure of several TTCs has been determined, including several bound to binding domains of their cognate effectors (68). The majority of these TTCs possess a similar structure and bind to their substrate binding sites as a homodimer (9, 10, 54, 65). Interestingly, the crystal structure of an Escherichia coli TTC substrate pair, CesA-EspA, revealed a distinct structure and CesA was shown to bind EspA as a monomer (74).
TTCs have been confirmed to function in the TTSSs of the bacterial plant pathogens Erwinia amylovora (DspB) (31) and Xanthomonas campestris pv. vesicatoria (HpaB) (16), as well as several in P. syringae (ShcA, ShcF, ShcM, ShcN, and ShcV) (8, 53, 64, 70, 71). All of the TTCs identified in plant pathogens thus far belong to class IA, with the exception of HpaB, a class IB member. Of the TTCs confirmed in P. syringae, all but one, ShcA, is from DC3000, though an uncharacterized ShcA is also present in DC3000 (15, 71). Several other candidate TTCs have been identified in DC3000 on the basis of the general TTC characteristics and gene location (15, 34, 70, 71). One of the DC3000 TTC genes, shcO1, is in the hopO1-1 operon and is likely a TTC for the HopO1-1 effector (formerly known as HopPtoO or HopPtoS1) (34, 59). The hopO1-1 operon has been shown to be actively transcribed (11, 75). Two other DC3000 operons have TTC genes homologous to shcO1 (33). One of these operons, the hopS1 operon, contains shcS1, encoding a putative TTC for the confirmed effector HopS1 (formerly HopPtoS4′) (62). The other operon, the hopS2 operon, contains a TTC gene homologous to shcO1 and is an apparent two-gene operon, which contains the putative TTC gene shcS2 and putative effector gene hopS2. Recent translocation experiments suggested that HopS2 was not translocated (62). The hopO1-1, hopS1, and hopS2 operons also share homologies between their confirmed and/or putative effector genes (33).
In this study we characterized the hopO1-1, hopS1, and hopS2 operons from DC3000 and confirmed that two novel Hops, HopO1-2 and HopT1-2, are secreted via the TTSS. We showed that ShcO1, ShcS1, and ShcS2 act as TTCs for their cognate effectors, HopO1-1, HopS1, and HopS2, respectively. ShcS1 and ShcS2 can substitute for ShcO1 and enhance the type III secretion and translocation of the HopO1-1 effector. We delimited the TTC binding site within HopO1-1 for each of these TTCs and found that each interacted with the middle third of HopO1-1 in yeast two-hybrid assays. Interestingly, we found HopS2 was translocated at a low level into plant cells and when a common N-terminal characteristic present in other Hops was introduced into HopS2, the translocation of HopS2 increased, suggesting that this characteristic is functionally important for TTSS substrate secretion.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used are listed in Table 1. Escherichia coli strains were grown routinely in LM (35) or Terrific (61) broth at 37°C. P. syringae strains were grown in KB broth (46) at 30°C. For type III secretion assays, P. syringae pv. tomato DC3000 strains were grown in hrp-inducing fructose medium at 22°C (41). Antibiotics were used at the following concentrations: ampicillin (Ap), 100 μg ml−1; chloramphenicol (Cm), 30 μg ml−1; gentamicin (Gm), 10 μg ml−1; kanamycin (Km), 50 μg ml−1; rifampin (Rif), 100 μg ml−1; spectinomycin (Sp), 50 μg ml−1; and tetracycline (Tc), 20 μg ml−1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristics | Reference and/or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | supE44 ΔlacU169(φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1, Nalr | 35; Life Technologies |
| E. coli DB3.1 | F gyrA462 endA1 Δ(sr1-recA) mcrB mrr hsdS20 (rB−, mB−) supE44 ara-14 galK2 lacY1 proA2 rpsL20 (Smr) xyl-5 λ−leu mtl-1 | Invitrogen |
| P. syringae pv. tomato DC3000 | Wild type; spontaneous Rifr | 23 |
| P. syringae pv. syringae B728A | Wild type; spontaneous Rifr | 37 |
| P. syringae pv. tomato UNL137 | DC3000 hopO1-1 operon deletion mutant, Rifr Spr | This work |
| Plasmids | ||
| pBluescript-II KS+ | Cloning vector, Apr | Stratagene |
| pML123 | Broad-host-range cloning vector, Gmr Kmr | 48 |
| pBBR1MCS1 | Broad-host-range vector, Cmr | 47 |
| pRK415 | Broad-host-range vector, unstable in absence of selection, Tcr | 45 |
| pEG202 | Yeast two-hybrid vector, creates fusion to LexA DNA binding domain, Apr | 32 |
| pJG4-5 | Yeast two-hybrid vector, creates fusion to B42 activation domain, Apr | 32 |
| pENTR/D-TOPO | Gateway system donor vector, Kmr | Invitrogen |
| pCPP2318 | pCPP30 derivative carrying blaM lacking signal peptide sequences, Tcr | 19 |
| pCPP3234 | Gateway destination vector containing the adenylate cyclase (cyaA) gene for C-terminal fusions, Spr/Smr Cmr | 62 |
| pLN142 | pML123 derivative carrying hopO1-1-flag, Gmr | 59 |
| pLN241 | pJG4-5 derivative carrying hopT1-1, Apr | This work |
| pLN248 | pJG4-5 derivative carrying hopO1-1, Apr | This work |
| pLN249 | pEG202 derivative carrying shcO1, Apr | This work |
| pLN250 | pJG4-5 derivative carrying shcO1, Apr | This work |
| pLN256 | pML123 derivative carrying hopT1-1-flag, Gmr | This work |
| pLN314 | pBluescript-II KS+ derivative containing 1.6 kb DNA region upstream of shcO1, omega cassette, and 2.0 kb DNA region downstream of hopT1-1, Apr Spr | This work |
| pLN366 | pENTR/D-TOPO derivative carrying hopS2, Kmr | This work |
| pLN367 | pENTR/D-TOPO derivative carrying hopS1, Kmr | This work |
| pLN370 | pRK415 derivative containing upstream and downstream flanking sequences of hopO1-1 operon with omega cassette in between, Tcr Spr | This work |
| pLN411 | pML123 derivative carrying shcO1/hopO1-1-flag, Gmr | This work |
| pLN428 | pJG4-5 derivative carrying hopO1-2, Apr | This work |
| pLN454 | pBBR1MCS5 derivative carrying hopO1-1 operon, Gmr | This work |
| pLN521 | pML123 derivative carrying hopO1-2-flag, Gmr | This work |
| pLN567 | pML123 derivative carrying hopT1-2-flag, Gmr | This work |
| pLN568 | pEG202 derivative carrying shcS1, Apr | This work |
| pLN622 | pENTR/D-TOPO derivative carrying shcS2lhopS2, Kmr | This work |
| pLN625 | pJG4-5 derivative carrying hopO1-1 fragment corresponding to HopO1-11-140, Apr | This work |
| pLN627 | pJG4-5 derivative carrying hopO1-1 fragment corresponding to HopO1-1141-283, Apr | This work |
| pLN705 | pBBR1MCS1 Gateway destination vector containing a hemagglutinin tag for C-terminal fusions, Cmr | This work |
| pLN716 | pJG4-5 derivative carrying hopO1-1 fragment corresponding to HopO1-1190-283, Apr | This work |
| pLN750 | pJG4-5 derivative carrying shcS1, Apr | This work |
| pLN752 | pJG4-5 derivative carrying hopT1-2, Apr | This work |
| pLN753 | pJG4-5 derivative carrying hopS1, Apr | This work |
| pLN786 | pENTR/D-TOPO derivative carrying shcO1, Kmr | This work |
| pLN814 | pENTR/D-TOPO derivative carrying hopO1-1, Kmr | This work |
| pLN898 | Gateway construct made by recombining pLN705 and pLN786, Cmr | This work |
| pLN966 | pEG202 derivative carrying shcS2, Apr | This work |
| pLN967 | pJG4-5 derivative carrying hopS2, Apr | This work |
| pLN980 | pJG4-5 derivative carrying shcS2, Apr | This work |
| pLN998 | pJG4-5 derivative carrying hopO1-1 fragment corresponding to HopO1-1111-175, Apr | This work |
| pLN999 | pJG4-5 derivative carrying hopO1-1 fragment corresponding to HopO1-171-189, Apr | This work |
| pLN1003 | pENTR/D-TOPO derivative carrying shcS1/hopS1, Kmr | This work |
| pLN1005 | Gateway construct made by recombining pCPP3234 and pLN1003, Spr | This work |
| pLN1006 | Gateway construct made by recombining pCPP3234 and pLN367, Spr | This work |
| pLN1018 | pML123 derivative carrying shcS2-ha/hopO1-1-flag, Gmr | This work |
| pLN1019 | pML123 derivative carrying shcS1-ha/hopO1-1-flag, Gmr | This work |
| pLN1022 | pJG4-5 derivative carrying hopO1-1 fragment corresponding to HopO1-11-125, Apr | This work |
| pLN1023 | pJG4-5 derivative carrying hopO1-1 fragment corresponding to HopO1-11-110, Apr | This work |
| pLN1027 | pJG4-5 derivative carrying hopO1-1 fragment corresponding to HopO1-1161-283, Apr | This work |
| pLN1063 | pENTR/D-TOPO derivative carrying shcO1/hopO1-1, Kmr | This work |
| pLN1064 | pENTR/D-TOPO derivative carrying shcS2-ha/hopO1-1, Kmr | This work |
| pLN1065 | pENTR/D-TOPO derivative carrying shcS1-ha/hopO1-1, Kmr | This work |
| pLN1074 | Gateway construct made by recombining pCPP3234 and pLN1063, Spr | This work |
| pLN1075 | Gateway construct made by recombining pCPP3234 and pLN1064, Spr | This work |
| pLN1076 | Gateway construct made by recombining pCPP3234 and pLN1065, Spr | This work |
| pLN1077 | Gateway construct made by recombining pCPP3234 and pLN814, Spr | This work |
| pLN1090 | pENTR/D-TOPO derivative carrying shcS2/hopS2 containing an S4I site-directed mutation in HopS2, Kmr | This work |
| pLN1138 | Gateway construct made by recombining pCPP3234 and pLN622, Spr | This work |
| pLN1140 | Gateway construct made by recombining pCPP3234 and pLN1090, Spr | This work |
| pLN1233 | Gateway construct made by recombining pCPP3234 and pLN366, Spr | This work |
| pLN1234 | pENTR/D-TOPO derivative carrying shcS2/hopS2 from P. syringae pv. tomato PT23 | This work |
| pLN1235 | pENTR/D-TOPO derivative carrying shcS2/hopS2 from P. syringae pv. maculicola 84-67 | This work |
| pLN1236 | pENTR/D-TOPO derivative carrying shcS2/hopS2 from P. syringae pv. maculicola NCPPB1886 | This work |
| pLN1243 | Gateway construct made by recombining pCPP3234 and pLN1245, Spr | This work |
| pLN1245 | pENTR/D-TOPO derivative carrying hopS2 containing an S4I site-directed mutation in HopS2, Kmr | This work |
| pLN1460 | pJG4-5 derivative carrying hopO1-1 fragment corresponding to HopO1-11-70, Apr | This work |
General DNA manipulations.
For standard molecular biological manipulations, well-described protocols were used (61). Restriction enzymes, T4 ligase, and DNA polymerase were purchased from New England Biolabs (Beverly, Mass.). The thermostable DNA polymerase used in PCRs was Pfu polymerase (Stratagene, La Jolla, Calif.). Oligonucleotide primers were ordered from Integrated DNA Technologies (Coralville, Iowa). For cloning using Gateway technology, we amplified desired target genes using PCR and Pfu polymerase and cloned the amplified fragments into the pENTR/D-TOPO vector (Invitrogen, Carlsbad, Calif.). The resulting pENTR constructs were recombined with Gateway destination vectors by LR reaction using LR clonase (Invitrogen) following the manufacturer's instructions. Information on the primers used for PCR cloning is provided at the following URL: http://psiweb.unl.edu/alfano/tables/Table51.pdf. The standard cycling conditions used for PCRs were as follows: 2 min at 94°C; 30 cycles of 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C; and 1 cycle for 10 min at 72°C. Plasmids were introduced into P. syringae strains by electroporation. General DNA sequence analysis was performed with Lasergene software (DNAstar Inc., Madison, Wis.). Database searches were done with BLASTN, BLASTP, BLASTX (http://www.ncbi.nlm.nih.gov/BLAST/index.html), and 3D-PSSM (http://www.sbg.bio.ic.ac.uk/∼3dpssm/).
Type III protein secretion assays.
To clone shcS1 and shcS2 separately into the BamHI site of pLN142 we used primers sets P1308/1309 and P1306/P1307, respectively, resulting in constructs pLN1019 and pLN1018. To clone hopO1-2-flag into pML123 we used primer set P1093/P821 (which contained either a ClaI or a XhoI site), resulting in pLN521. Primer set P823/P824 (which contained either a XbaI or BamHI site) was used to clone hopT1-2-flag, resulting in construct pLN567. These constructs were transformed into DC3000(pCPP2318) or UNL137(pCPP2318) by electroporation and used for secretion assays. pCPP2318 encodes a mature β-lactamase and should remain cell bound and was used as a lysis control. In other secretion assays, NPTII was used as a lysis control. Secretion assays were performed as previously described (59). Briefly, bacterial strains were grown at 22°C in hrp-inducing conditions for 4 to 6 h. Cell and supernatant fractions were separated with centrifugation. Proteins in the supernatants were precipitated with trichloroacetic acid and resuspended in 1× sample sodium dodecyl sulfate (SDS) buffer. The cell and supernatant fraction preparations resulted in 13.3- and 133-fold concentrations, respectively, relative to initial volumes. Equal volumes of each sample were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) using standard procedures (61), transferred to polyvinylidene difluoride membranes, and immunoblotted with anti-FLAG (Sigma Chemical Co., St. Louis, Mo.), anti-NPTII (Cortex Biochem, San Leandro, Calif.), or anti-β-lactamase or anti-hemagglutinin (HA) (Roche Diagnostics Corp., Indianapolis, Ind.) primary antibodies. Anti-β-lactamase and anti-NPTII antibodies were recognized with goat anti-rabbit immunoglobulin G-alkaline phosphatase conjugate (Sigma Chemical Co., St. Louis, Mo.), anti-FLAG antibodies were recognized with goat anti-mouse immunoglobulin G-alkaline phosphatase conjugate (Sigma Chemical Co.), and anti-HA antibodies were recognized with goat anti-rat immunoglobulin G-alkaline phosphatase conjugate (Sigma Chemical Co.). Membrane-bound secondary antibodies were visualized on autoradiograms with a Western Star chemiluminescence detection kit (Applied Biosystems, Foster City, Calif.).
Coimmunoprecipitation assays with ShcO1 and HopO1-1.
A Gateway entry construct pLN786 containing shcO1 was recombined with pLN705 (a broad-host-range Gateway destination vector), resulting in pLN898, which encodes ShcO1 fused with a C-terminal HA epitope. We electroporated a control vector or pLN898 into DC3000(pLN142) and pLN898 into DC3000(pML123) and used these strains to perform coimmunoprecipitation experiments. Briefly, saturated overnight cultures were inoculated in 500 ml KB cultures and grown to an optical density at 600 nm (OD600) of 0.8. The cells were harvested by centrifugation for 30 m at 5,000 × g, and the pelleted cells were frozen at −20°C until needed. The cells were thawed at room temperature for 30 m. The cells were resuspended in 10 ml of TBS (50 mM Tris-HCl, 150 mM NaCl, pH 7.6) with complete protease inhibitor cocktails (Roche Diagnostics Corp., Indianapolis, Ind.). The cell suspensions were lysed on ice by sonicating three times for 30 s each time and centrifuged. Cell-free supernatants were transferred to fresh tubes, and the aliquots were stored at −80°C until needed. Total protein (500 μg) from each sample was incubated with 100 μl M2 anti-FLAG resin (Sigma Chemical Co.) and rocked at 4°C overnight. The resin was briefly centrifuged, washed twice with TBS, and resuspended in 100 μl 1× SDS sample buffer. Samples from different stages of the procedure along with final eluate were analyzed with immunoblots using either anti-HA or anti-FLAG antibodies.
RNA isolation and RT-PCR analysis.
DC3000 was grown in hrp-inducing minimal or KB media to an OD600 of 0.6. Cells were harvested and lysed with RLT buffer (QIAGEN, Valencia, CA) containing guanidinium thiocyanate (GITC) and 1% β-mercaptoethanol followed by an incubation with GITC-saturated hot phenol (pH 5 to 6) at 65°C for 10 min. Total RNA was precipitated with ammonium acetate and ethanol and resuspended in water. RNA was treated with DNase I (Ambion, Austin, TX) as recommended by the manufacturer. Reverse transcription was performed with 2 μg of total RNA using gene specific primers and a RETROScript reverse transcriptase PCR (RT-PCR) kit (Ambion). The fragments corresponding to shcS1/hopS1, hopO1-2, and shcS2/hopS2 were amplified from the resulting cDNA by PCR with the primer sets P933/P949, P1260/P615, and P763/P745, respectively, using the following cycling conditions: 5 min at 94°C; 30 cycles of 30 s at 94°C, 1 min at 52°C, and 3 min at 72°C; and 1 cycle at 10 min at 72°C.
Site-directed mutagenesis of PSPTO4588 (HopS2).
Site-directed mutagenesis of hopS2 was performed using a QuikChange mutagenesis kit (Stratagene) according to the manufacturer's instructions. Primers P1279 and P1280 were used in PCRs to change the fourth residue of HopS2 from a Ser to Ile (S4I). The template DNA used was construct pLN622, which contained hopS2 with its chaperone gene shcS2, and resulted in Gateway entry construct pLN1090. This Gateway entry construct was recombined into pCPP3234, a broad-host-range Gateway destination vector (62). The resulting construct, pLN1140, contained a site-directed mutation in hopS2 which altered residue 4 (S4I) of HopS2. Additionally, another Gateway entry construct, pLN1245, was made using pLN316 as the template DNA and primer set P1279/P1280, which lacked shcS2 and encoded a HopS2 derivative that had the S4I mutation. This construct was recombined with destination vector pCPP3234, resulting in construct pLN1243.
Adenylate cyclase (cyaA) translocation assay.
pENTR/D-TOPO plasmids containing hopO1-1 (pLN814), hopS1 (pLN367), shcS1/hopS1 (pLN1003), hopS2 (pLN366), or shcS2/hopS2 (pLN622) were recombined with pCPP3234, which resulted in the construct pLN1077, pLN1006, pLN1005, pLN1233, or pLN1138, respectively. To clone shcO1/hopO1-1 we used primer set P1102 and P0814 and pLN411 as the template DNA. The amplified product was recombined into pENTR/D-TOPO, resulting in construct pLN1063. Similarly, to clone shcS1/hopO1-1 into pENTR/D-TOPO we used primer set P1104 and P0814 and pLN1018 as the template, resulting in construct pLN1065. Finally, to clone shcS2/hopO1-1, we used primer set P1106/P0814 and pLN1018 as the template DNA, resulting in construct pLN1064. These constructs were used in Gateway LR reactions with pCPP3234, generating pLN1074 (shcO1/hopO1-1), pLN1075 (shcS2/hopO1-1), and pLN1076 (shcS1/hopO1-1).
Each construct that encoded CyaA fusions was introduced into P. syringae pv. syringae B728a or DC3000 by electroporation. For CyaA assays we followed a protocol recently described (62). Briefly, bacterial suspensions in 5 mM MES (morpholineethanesulfonic acid) (pH 5.6, containing100 μM IPTG [isopropyl-β-d-thiogalactopyranoside]) at an OD600 of 0.4 were infiltrated into Nicotiana benthamiana leaves. Disks of leaf tissue 0.9 cm in diameter were harvested 6 h after infiltration and ground with a mortar and pestle in liquid nitrogen and resuspended in 0.1 M HCl. A direct cyclic AMP (cAMP) correlate enzyme immunoassay kit (Assay Designs, Ann Abor, MI) was used to measure cAMP concentrations in each sample by following the manufacturer's instructions.
DNA gel blots.
Total DNAs (3 μg) from different bacterial strains were digested with EcoRI, separated in a 1% agarose gel, and transferred to an Immobilon-Ny+ membrane (Millipore, Bedford, MA) by following a standard protocol (61). hopS2 DNA was PCR amplified with primers P615 and P617 and used for a probe prepared with a Prime-It random priming labeling kit (Stratagene, La Jolla, CA). Hybridization was carried out at 55°C in hybridization buffer (7% SDS, 2 mM EDTA, and 0.5 M Na2HPO4) for 18 h. After hybridization the membranes were washed twice with wash solution (0.1% SDS and 1× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) and exposed to X-ray films for 12 h at −80°C.
Construction of the DC3000 hopO1-1 operon mutant UNL137.
DC3000 deletion mutants lacking the hopO1-1 operon were made using homologous recombination techniques, which replaced the operon with an antibiotic marker. To do this, we cloned a 1.6 kb DNA region upstream of shcO1 (with primers P0574 and P0575) and a 1.6 kb DNA region downstream of hopT1-1 (with primers P0576 and P0577) adjacent to each other in the same orientation into pBluescript SK+ (Stratagene). A 2 kb Sp/Smr Ω fragment from pHP45 (29) was cloned into a BamHI site in between the above-described fragments, resulting in construct pLN314. The 5.2 kb fragment containing the hopO1-1 operon flanking regions with the Ω fragment was cut out with restriction enzymes and cloned into the XbaI and KpnI sites of pRK415 (45), resulting in construct pLN370. pLN370 was transformed into DC3000 by electroporation and retention of the Ω fragment, and loss of the plasmid was selected by growing DC3000(pLN370) cultures for 5 consecutive days in which each day a new culture was inoculated in fresh KB broth containing Sp. On the 5th day, the culture was plated onto KB plates containing Sp, and Spr colonies were screened for Tc sensitivity, which would indicate that these colonies lost the plasmid. A Tcs Spr colony confirmed with PCR to have an Ω fragment substituted for the hopO1-1 operon was designated UNL137.
Yeast two-hybrid analysis.
Protein-protein interactions were determined with yeast two-hybrid assays that used pEG202, a vector that encodes a LexA DNA binding domain (DBD) (i.e., the bait vector), and pJG4-5, a vector encoding an activation domain (AD) (i.e., the fish vector) (13). We made constructs containing shcO1, shcS1, shcS2, hopO1-1, hopO1-2, hopT1-1, hopT1-2, hopS1, or hopS2 in pEG202 or pJG4-5. The pEG202 constructs, cloned genes, and PCR primer sets used for cloning are as follows: pLN249, shcO1, and P568/P569; pLN568, shcS1, and P933/P934; and pLN966, shcS2, and P1260/P951. The pJG4-5 constructs, the genes, and the primer sets used are as follows: pLN241, hopT1, and P570/P571; pLN248, hopO1-1, and P722/P723; pLN250, shcO1, and P568/P569; pLN428, hopO1-2, and P763/P764; pLN750, shcS1, and P933/P934; pLN752, hopT1-2, and P1117/P1118; pLN753, hopS1, and P948/P949; pLN967, hopS2, and P952/P953; and pLN980, shcS2, and P1260/P951. Additional pJG4-5 constructs were made that corresponded to different regions of HopO1-1. The construct names, the corresponding HopO1-1 amino acids, and the primer sets used are the following: pLN625, HopO1-1 amino acids (aa) 1 to 140, and P722/P975; pLN627, HopO1-1 aa 141 to 283, and P976/P723; pLN716, HopO1-1 aa 190 to 283, and P1036/P723; pLN998, HopO1-1 aa 111 to 175, and P1298/P1299; pLN999, HopO1-1 aa 71 to 189, and P1034/P1297; pLN1022, HopO1-1 aa 1 to 125, and P722/P1351; pLN1023, HopO1-1 aa 1 to 110, and P722/P1355; pLN1027, HopO1-1 aa 161 to 283, and P1353/P723; and pLN1460, HopO1-1 aa 1 to 70, and P722/P1677. To test interactions, different combinations of plasmids were cotransformed into Saccharomyces cerevisiae strain EGY48(pSH18-34) and the interactions were analyzed using published protocols (32). Briefly, different combinations of plasmids carried in EGY48(pSH18-34) were analyzed on galactose-containing plates that lacked leucine and contained X-gal. Interactions were detected based on the ability to rescue leucine auxotrophy and β-galactosidase activity.
Pathogenicity assays.
Arabidopsis thaliana Col-0 plants used in pathogenicity assays were grown in a growth chamber at 23°C with 8 h of light per day. The ability of DC3000 and derivative strains to grow in planta in Col-0 plants was determined as described previously (27). Briefly, bacteria were grown at 28°C overnight on KB agar plates and suspended at an OD600 of 0.2 in 10 mM MgCl2 containing 0.02% Silwet L-77 (Lehle Seeds, Round Rock, TX). Four-week-old Col-0 plants were dip-inoculated into cell suspensions for 1 m and allowed to dry. Four leaf disks 0.4 cm2 in size were sampled for each treatment at days 0, 2, and 4 postinoculation and macerated in 1 ml sterile water. Serial dilutions were plated on KB plates containing the appropriate antibiotics. Plates were incubated at 28°C for 48 h, after which the colonies were enumerated. The following strains were used in pathogenicity assays: wild-type DC3000, the DC3000 hrcC mutant, and the DC3000 hopO1-1 operon mutant UNL137. Complementation of UNL137 was performed using subclones of the hopO1-1 operon, which included the following: pLN454 contained the complete hopO1-1 operon in pBBR1MCS5, which was cloned using primer set P939/P583; pLN256, which contained hopT1-1 and was made with primer set P824/P583; and pLN411, which contained shcO1 and hopO1-1.
RESULTS
ShcO1 facilitates the type III secretion of HopO1-1 and interacts with HopO1-1 in vivo.
HopO1-1 (formerly known as HopPtoS1 or HopPtoO) is a DC3000 effector that shares similarity to ADP ribosyltransferases (34, 59). It was shown to be secreted via the DC3000 TTSS (59) and translocated into plant cells (34). hopO1-1 is located on pDC3000A, one of the two plasmids identified in P. syringae pv. tomato DC3000 (15). The hopO1-1 operon contains two other genes (Fig. 1A). hopT1-1, located downstream of hopO1-1, encodes an effector demonstrated to be translocated (62). We identified an open reading frame (ORF) upstream of hopO1-1, PSPTOA0017, which shared the general characteristics of type III chaperones (TTCs) (71). These characteristics include its gene location, a small molecular mass (15.6 kDa), and an acidic isoelectric point (5.5). Moreover, PSPTOA0017 shows a predicted 3-D structure that is similar to several animal pathogen class 1A TTCs as determined based on the protein fold recognition program 3D-PSSM.
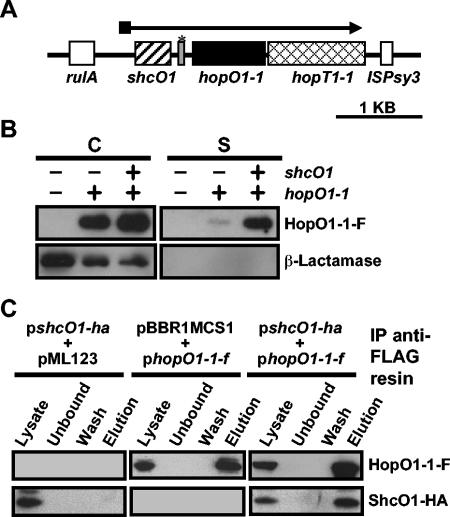
FIG. 1.
ShcO1 facilitates the type III secretion of HopO1-1 and interacts with HopO1-1 in vivo. (A) Genetic organization of the HopO1-1 operon. The operon is located on plasmid pDC3000A in DC3000. The TTC gene shcO1 is depicted with a striped box. The hopO1-1 and hopT1-1 genes are depicted with a black box and a hatched box, respectively. The open boxes show the genes flanking the hopO1-1 operon. The gray box marked with an asterisk between shcO1 and hopO1-1 represents a gene fragment which is homologous to hopS1, a gene shown in Fig. 2A. (B) Secretion of HopO1-1 in culture via the DC3000 TTSS is enhanced in the presence of the ShcO1 TTC. Constructs containing hopO1-1-flag (pLN142) or shcO1/hopO1-1-flag (pLN411) were electroporated into the DC3000 hopO1-1 operon deletion mutant UNL137, which also carried pCPP2318 encoding β-lactamase lacking its export signal. Cultures of the above strains were grown in hrp-inducing fructose medium and separated into cell (C) and supernatant (S) fractions. Proteins were resolved with SDS-PAGE and transferred to polyvinylidene difluoride membranes. HopO1-1-F and β-lactamase were detected with anti-FLAG and anti-β-lactamase antibodies, respectively. (C) ShcO1 coimmunoprecipitates with HopO1-1. DC3000(pLN142, pLN898) cultures, which expressed ShcO1-HA and HopO1-1-F, were sonicated, and soluble protein samples were obtained by centrifugation. Soluble proteins were mixed with anti-FLAG resin to immunoprecipitate (IP) HopO1-1-F as described in Materials and Methods. DC3000(pLN898, pML123) and DC3000(pBBR1MCS1, pLN142) were used as vector controls. Fractions representing the total soluble protein (Lysate), total soluble protein after treatment with resin (Unbound), the final wash (Wash), and the proteins that were eluted from the resin (Elution) were subjected to SDS-PAGE and immunoblotted. ShcO1-HA and HopO1-1-F were detected with anti-HA and anti-FLAG antibodies, respectively.
To determine whether PSPTOA0017 encoded a TTC that facilitated the type III secretion of HopO1-1 we made a DC3000 mutant, UNL137, lacking the complete hopO1-1 operon as described in Materials and Methods. We grew UNL137 cultures carrying plasmids that contained hopO1-1 (fused to a flag epitope to allow for detection of HopO1-1) with and without PSPTOA0017 and performed type III secretion assays to determine whether HopO1-1 was secreted in higher amounts when PSPTOA0017 was present. The cultures were separated into cell and supernatant fractions, and these samples were subjected to SDS-PAGE and analyzed for the presence of HopO1-1-FLAG with immunoblots. We found that a much greater amount of HopO1-1 was found in the supernatant fraction when PSPTOA0017 was present, as is consistent with the presumed function of PSPTOA0017 as a TTC for HopO1-1 (Fig. 1B). In the absence of PSPTOA0017, only a small amount of HopO1-1 was secreted, which may have been due to the presence of other homologous TTC genes present in the genome of DC3000. Hereafter, we refer to PSPTOA0017 as ShcO1. Similar secretion experiments were carried out for HopT1-1, and we found that this effector was secreted in high amounts in the absence of ShcO1, suggesting that ShcO1 is not a TTC for HopT1-1 (data not shown). In addition, secretion assays were done to determine whether ShcO1 was type III secreted and we were unable to detect any ShcO1 in supernatant fractions, suggesting that it is not secreted (data not shown).
To facilitate secretion, TTCs interact directly with their cognate effectors. To determine whether ShcO1 interacted with HopO1-1, we did coimmunoprecipitation experiments with DC3000 extracts from cultures expressing ShcO1 fused to an hemagglutinin (HA) epitope (ShcO1-HA) and/or HopO1-1 fused to the FLAG epitope (HopO1-1-FLAG). When anti-FLAG resin was incubated with these extracts, ShcO1-HA was immunoprecipitated along with HopO1-1-FLAG (Fig. 1C). Importantly, ShcO1-HA did not associate with the anti-FLAG resin in the absence of HopO1-1-FLAG (Fig. 1C). This indicates that ShcO1 and HopO1-1 interact in vivo, as is consistent with ShcO1 acting as a TTC for HopO1-1.
Characterization of a chromosomal region in DC3000 that shares similarity to the hopO1-1 operon.
Several genomic investigations have identified a DC3000 chromosomal region that contains ORFs homologous to the genes in the hopO1-1 operon (15, 34, 59). This region includes two predicted transcriptional units that have putative promoters containing hrp boxes, suggesting that they are regulated by the HrpL alternative sigma factor used in the transcription of many P. syringae type III genes (40) (Fig. 2A).
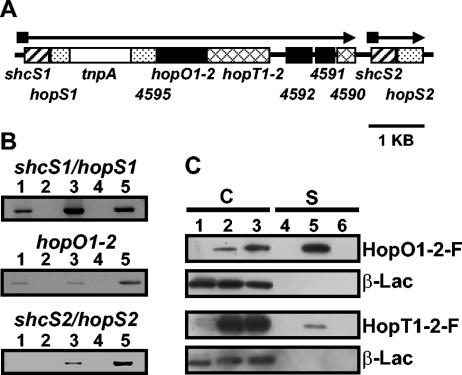
FIG. 2.
RT-PCR experiments indicate that shcS1/hopS1 and shcS2/hopS2 are transcribed in a type III-dependent manner and that HopO1-2 and HopT1-2 are both secreted in culture via the DC3000 TTSS. (A) The genetic organization of a cluster of genes in the DC3000 chromosome which are homologous to shcO1, hopO1-1, or hopT1-2. The homologous TTC genes, shcS1 and shcS2, are depicted with striped boxes. The homologous effector genes hopS1 and hopS2 are depicted with stippled boxes. Black boxes depict genes or ORFs that are homologous to hopO1-2 and hatched boxes depict hopT1-2 and a nearby homologous ORF, PSPTO4590. The open box represents a transposase that interrupts hopS1 and PSPTO4595. The ORFs with number designations represent the PSPTO numbers from the databases. The arrows show the predicted orientation of transcription and the black squares indicate the presence of hrp boxes, which are often found associated with type III-related promoters. (B) RT-PCR analysis of the DNA region shown in panel A. RNA was isolated from DC3000 cultures grown in either rich KB medium or hrp-inducing minimal medium. RT-PCR using RNA from KB-grown cultures with reverse transcriptase (RT) (lane 1) or without RT (lane 2); RT-PCR using RNA from hrp-inducing grown cultures with RT (lane 3) or without RT (lane 4); PCR using DC3000 genomic DNA (lane 5). The primer sets used were complementary to sequences within the coding regions of shcS1/hopS1, hopO1-2, or shcS2/hopS2. (C) Immunoblot analyses were performed cultures of DC3000 or a DC3000 hrcC mutant disabled in type III secretion, carrying either pLN521 (encoding HopO1-2-FLAG) or pLN567 (encoding HopT1-2-FLAG). Cultures were separated into cell-bound (C, lanes 1 to 3) or supernatant (S, lanes 4 to 6) fractions. Lanes 1 and 4, wild-type DC3000; lanes 2 and 5, DC3000(pLN521) (top two blots) or DC3000(pLN567) (bottom two blots); lanes 3 and 6, DC3000 hrcC(pLN521) (top two blots) or DC3000 hrcC(pLN567) (bottom two blots). Strains also carried pCPP2318, which encoded a mature form of β-lactamase that should remain cell bound unless significant nonspecific cell lysis occurred. Cell-bound and supernatant fractions were concentrated 13.3- and 133-fold, respectively. Anti-FLAG antibodies were used to visualize HopO1-2-F and HopT1-2-F and β-lactamase (β-Lac) was detected with anti-β-lactamase antibodies.
One predicted transcriptional unit, referred to here as the hopS1 operon, contained a putative TTC gene, shcS1, which encodes a protein that is 79% identical to ShcO1 (Fig. 2A). The next gene in the operon, hopS1 (formerly named hopPtoS4′), has been shown to encode a translocated effector (62). A transposon is located downstream of hopS1, followed by an ORF, PSPTO4595, that shares similarity with the 3′ half of hopO1-1. Another ORF overlapping with PSPTO4595 also shares similarity with hopO1-1, suggesting that this region has incurred frameshift mutations and that hopS1 likely represents a truncated gene. The adjacent downstream ORF, now designated hopO1-2 (formerly designated hopPtoS3 or hopPtoO2) (34, 59), encodes a protein that is 75.3% identical to HopO1-1. Downstream of hopO1-2 is a putative effector gene, hopT1-2, which would encode a protein that is 71% identical to HopT1-1. The rest of the putative transcriptional unit contains PSPTO4592, PSPTO4591, and PSPTO4590. The predicted protein products of PSPTO4592 and PSPTO4591 are homologous to different regions of HopO1-1 and HopO1-2 and likely represent a single coding sequence interrupted with a nonsense mutation. PSPTO4590 appears to be a truncated hopT1-2 homolog and is probably nonfunctional. The other transcriptional unit in this DNA region, referred to here as the hopS2 operon (Fig. 2A), is an apparent two-gene operon that contains a putative chaperone gene, shcS2, encoding a protein that has 69.2% and 70.3% identity with ShcO1 and ShcS1, respectively, and a putative effector gene, hopS2, encoding a protein 35.3% identical to HopS1 (59).
We sought to determine the transcriptional status of the hopS1 operon, before and after the transposon insertion, as well as the hopS2 operon. To do so we did RT-PCR experiments with three different primer sets. The first primer set spanned the first two genes of the hopS1 operon, shcS1 and hopS1, the second primer set annealed within the coding region of hopO1-2, and the third primer set spanned shcS2 and hopS2 in the hopS2 operon. The results clearly showed that both shcS1/hopS1 and shcS2/hopS2 were transcribed (Fig. 2B). We detected RNA for shcS1/hopS1 even in rich medium, although the amount of RNA appeared greater from cultures grown in hrp-inducing conditions. We detected reduced amounts of RNA from hopO1-2 in a hrp-independent manner (Fig. 2B). The transposon may have polar effects and/or there may be an out-reading promoter within the transposon, suggesting that HopO1-2 and HopT1-2 may be made constitutively in low amounts in DC3000.
To determine whether the HopO1-2 and HopT1-2 proteins could be secreted via the DC3000 TTSS, we separately cloned these genes into a broad-host-range plasmid and electroporated the resulting constructs, pLN521 and pLN567, into DC3000. DC3000(pLN521), expressing HopO1-2, and DC3000(pLN567), expressing HopT1-2 (which both produced proteins fused to a C-terminal FLAG epitope), were grown in hrp-inducing minimal medium and separated into cell and supernatant fractions. Immunoblots using anti-FLAG antibodies showed that both HopO1-2-FLAG and HopT1-2-FLAG were found in the supernatant fraction, indicating that both were secreted via the DC3000 TTSS (Fig. 2C). Therefore, even though the transcription of these genes is likely to be at a low level due to the polar effects of the upstream transposon, their protein products are secreted via the DC3000 TTSS.
HopS2 is well distributed in different P. syringae strains and is translocated into plant cells.
A recent report concluded on the basis of adenylate cyclase (CyaA) reporter translocation assays (62) that HopS2 (PSPTO4588) was not translocated into plant cells. Because our RT-PCR data suggested that the hopS2 operon was active we were interested in experiments that further investigated this operon. To determine how common hopS2 was in other P. syringae strains and other plant pathogens we did DNA gel blot analyses with total DNA from different bacterial strains and probed with hopS2. In our survey we found that hopS2 was present in all the P. syringae pv. tomato strains (DC3000, Pt23, 3357, and 133) and both P. syringae pv. maculicola strains tested (84-67 and NCPPB1886) and in the lone strains tested from P. syringae pathovars phaseolicola (HB10Y) and tabaci (11528) (Fig. 3A). But hopS2 was not detected in several P. syringae strains, including pathovar syringae, glycinea, or pisi (Fig. 3A).
FIG. 3.
HopS2 and HopS2 secretion signal mutation derivatives are not detectably secreted in culture via the DC3000 TTSS, but are translocated into plant cells. (A) Distribution of hopS2 in different P. syringae strains and other bacterial plant pathogens. Total DNA from different strains of bacteria was digested with EcoRI and transferred to nylon membrane and probed with a PCR fragment corresponding to a region within hopS2. Numbered lanes correspond to total DNA from the following bacteria: 1, P. syringae pv. tomato DC3000; 2, P. syringae pv. syringae 61; 3, P. syringae pv. glycinea race 4; P. syringae pv. tomato Pt23; 5, P. syringae pv. syringae B728a; 6, Erwinia chrysanthemi 3937; 7, P. syringae pv. maculicola 84-67; 8, P. syringae pv. maculicola NCPPB1886; 9, P. syringae pv. phaseolicola HB10Y; 10, P. syringae pv. tabaci 11528; 11, P. syringae pv. tomato 3357; 12, P. syringae pv. tomato 133; 13, Xanthomomas campestris pv. vesicatoria 82-8; 14, P. syringae pv. pisi 11. Molecular mass markers are indicated in kB on the left. (B) HopS2-CyaA translocation assays with wild-type HopS2 or a HopS2 missense mutation derivative altered in its N-terminal secretion signal in the presence or absence of the ShcS2 TTC. DC3000 cultures expressing different HopS2-CyaA fusions were infiltrated into Nicotiana benthamiana and assayed for cAMP production 6 h after infiltration as described in Material and Methods. cAMP levels were reported in picomoles of cAMP per micrograms of protein with standard errors. The experiment was performed three times with similar results. Results indicate that wild-type HopS2 is translocated into plant cells in a ShcS2-dependent manner and that a HopS2 derivative that has a more typical N-terminal secretion signal was translocated into plant cells in higher amounts.
One possible reason that HopS2 was not translocated in the previous report (62) was that the biochemical characteristics of its N terminus, which may represent a secretion signal, were different from the majority of other P. syringae effectors. Specifically, one difference was that HopS2 lacked a leucine, isoleucine, valine, or proline in position 3 or 4 which is present in other P. syringae effectors (59). Instead, position 3 and 4 are occupied with a lysine or serine in HopS2 from DC3000. To determine whether hopS2 alleles from other strains encoded proteins with similar N-terminal characteristics, we PCR-cloned hopS2 from the P. syringae pv. maculicola strains 84-67 and NCPPB1886 and P. syringae pv. tomato Pt23 and completely sequenced both nucleotide strands of each allele. While there were several conservative amino acid changes within the predicted HopS2 products, the first 50 amino acids encompassing the N-terminal secretion signal characteristics were identical, including positions 3 and 4 (data not shown). This suggests that HopS2 may possess different N-terminal characteristics than other P. syringae effectors and led us to retest whether HopS2 was translocated into plant cells.
The earlier report that indicated that HopS2 was not translocated into plant cells was done only in the presence of its putative TTC ShcS2 (62). We rationalized that HopS2 was possibly translocated at a low level into plant cells and not easily detected. Hence, we retested its translocation in the absence and presence of ShcS2 and assessed HopS2 translocation. We made constructs that expressed HopS2 fused to a calmodulin-dependent adenylate cyclase (CyaA) and performed CyaA translocation assays (18, 67). In this assay, an effector-CyaA fusion will produce cyclic AMP (cAMP) only if it is translocated into eukaryotic cells because they contain calmodulin. Thus, production of significant levels of cAMP indicates that the effector-CyaA fusion was translocated into plant cells. We made constructs that encoded HopS2-CyaA with and without the candidate ShcS2 TTC. DC3000 strains expressing ShcS2 and HopS2-CyaA (pLN623) or HopS2-CyaA alone (pLN624) were infiltrated into N. benthamiana leaves, and cAMP levels were detected as described in Materials and Methods. The cAMP levels were significantly higher in leaf tissue infiltrated with DC3000(pLN623) expressing both ShcS2 and HopS2-CyaA than they were with DC3000(pLN624) expressing only HopS2-CyaA (Fig. 3B). This indicates on the basis of CyaA assays (60, 62) that HopS2 is translocated into plant cells, albeit at a relatively low level compared to amounts of cAMP produced by other P. syringae effectors. These results are also consistent with ShcS2 acting as a TTC for HopS2.
To test whether the atypical N-terminal characteristics of HopS2 contributed to its low level of translocation and whether the N terminus represented a type III secretion signal, we made CyaA fusions to a hopS2 derivative that encoded a HopS2 protein that substituted the fourth serine residue with isoleucine (S4I). DC3000 carrying constructs that made ShcS2 and HopS2S4I-CyaA were infiltrated into N. benthamiana leaf tissue, and cAMP assays were performed. In the presence of ShcS2, the HopS2S4I-CyaA carrying strain produced high levels of cAMP, indicating that this effector-CyaA fusion was translocated into plant cells in much higher amounts than the wild-type HopS2-CyaA fusion (Fig. 3B). The infiltration of DC3000 expressing HopS2S4I-CyaA in the absence of ShcS2 resulted in only a small amount of cAMP produced (Fig. 3B), which demonstrated that the HopS2 mutation derivative still maintained its dependence on ShcS2 for translocation.
ShcO1, ShcS1, and ShcS2 interact with each other and with each other's cognate effectors in yeast two-hybrid assays.
Because the hopO1-1, hopS1, and hopS2 operons contain TTC and effector genes that encoded homologous TTCs and effectors, we were interested in determining whether the TTCs could interact with each other and which effectors interacted with each TTC. To determine this, we cloned shcO1, shcS1, and shcS2 into the LexA-based yeast two-hybrid vector pEG202, resulting in constructs pLN249, pLN568, and pLN966, respectively. Each construct produced a TTC fused to the LexA DNA binding domain (DBD). We also cloned TTC genes and effector genes into an activation domain (AD) vector, pJG4-5, which produced proteins fused to the B42 AD. The DBD and AD constructs were expressed in yeast cells with every possible DBD and AD combination, and the yeast strains were screened for β-galactosidase activity and leucine prototrophy. We found that each TTC interacted with itself as well as with each of the others (Table 2), suggesting that these TTCs can form homodimers and heterodimers. Each TTC interacted with its most likely cognate effector (based on the location of the TTC and effector gene). That is, ShcO1 interacted with HopO1-1, ShcS1 interacted with HopS1, and ShcS2 interacted with HopS2 (Table 2). Each TTC also interacted with cognate effector of each of the others (Table 2). For example, ShcO1 interacted with both HopS1 and HopS2. This was unanticipated, since HopO1-1 is not homologous to HopS1 and HopS2. Each TTC also interacted with HopO1-2 (Table 2), suggesting that this effector may also be dependent on TTC for secretion. None of the TTCs interacted with HopT1-1 or HopT1-2, which both appeared to be secreted via the TTSS independent of TTCs (Fig. 2C) (62).
TABLE 2.
Interactions between ShcO1- and HopO1-1-related TTCs and effectors in yeast two-hybrid assaysa
| DBD-ShcO1 | DBD-ShcS1 | DBD-ShcS2 | |
|---|---|---|---|
| AD-vector | − | − | − |
| AD-ShcO1 | + | + | + |
| AD-ShcS1 | + | + | + |
| AD-ShcS2 | + | + | + |
| AD-HopO1-1 | + | + | + |
| AD-HopO1-2 | + | + | + |
| AD-HopS1 | + | + | + |
| AD-HopS2 | + | + | + |
| AD-HopT1-1 | − | − | − |
| AD-HopT1-2 | − | − | − |
−, no interaction; +, positive interaction.
ShcS1 and ShcS2 can substitute for ShcO1 in the type III secretion and translocation of the HopO1-1 effector.
To determine the relevance of the interaction of ShcO1 TTC homologs with multiple effectors, we tested whether ShcS1 or ShcS2 could substitute for ShcO1 in the type III secretion and translocation of HopO1-1. We separately PCR-cloned shcS1 and shcS2 into a broad-host-range plasmid which already contained hopO1-1-flag, resulting in constructs pLN1019 and pLN1018, respectively. When the DC3000 hopO1-1 operon deletion mutant UNL137 expressing HopO1-1-FLAG alone was grown, we noticed that in addition to full-length HopO1-1-FLAG, a smaller molecular mass protein band, which likely represented a HopO1-1-FLAG degradation product, was detected on immunoblots using anti-FLAG antibodies (Fig. 4A). This degradation product was not detected in E. coli DH5α expressing HopO1-1 from the same construct, suggesting that the apparent proteolysis was specific to P. syringae (Fig. 4A). Interestingly, the apparent HopO1-1-FLAG degradation product was no longer detectable in samples from UNL137 cultures expressing HopO1-1-FLAG with ShcO1 or ShcS2 (Fig. 4A). Moreover, the degradation product was reduced in samples from UNL137 expressing ShcS1 and HopO1-1-FLAG (Fig. 4A). These data suggest that these TTCs stabilize HopO1-1-FLAG.
FIG. 4.
The ShcO1 homologs ShcS1 and ShcS2 enhanced HopO1-1 type III secretion in culture, protected HopO1-1 from degradation, and enhanced HopO1-1 translocation into plant cells. (A) A HopO1-1 degradation product is made in the absence of ShcO1 and ShcS2 in the DC3000 hopO1-1 deletion mutant UNL137 but not in E. coli DH5α. Bacterial strains carrying either pLN142 (hopO1-1-flag), pLN411 (shcO1, hopO1-1-flag), pLN1018 (shcS2, hopO1-1-flag), or pLN1019 (shcS1, hopO1-1-flag) were lysed in SDS sample buffer and subjected to SDS-PAGE and immunoblot analysis. Anti-FLAG antibodies were used to detect HopO1-1-FLAG and the HopO1-1-related degradation product. In UNL137 lacking ShcO1, a small degradation product of about 10 kDa in mass was detectable. When ShcO1, ShcS1, or ShcS2 was present in UNL137, the degradation product was no longer detected or was reduced. However, the HopO1-1-related degradation product was not detected in E. coli DH5α. (B) Type III secretion assays of HopO1-1 in the absence or presence of ShcO1, ShcS1, or ShcS2. The DC3000 hopO1-1 deletion mutant UNL137 containing the following constructs were grown in hrp-inducing medium for type III secretion assays: pLN142 (hopO1-1-flag); pLN411 (shcO1, hopO1-1-flag); pLN1018 (shcS2, hopO1-1-flag); or pLN1019 (shcS1, hopO1-1-flag). Cultures were separated into cell-bound (C) and supernatant (S) fractions, which were concentrated 13.3- and 133-fold, respectively. NPTII, which should remain in the cell fraction, was used as a lysis control. Samples were separated with SDS-PAGE and transferred to a nylon membrane for immunoblot analysis. HopO1-1-FLAG and NPTII were detected with anti-FLAG and anti-NPTII antibodies, respectively. (C) HopO1-1-CyaA translocation assays in the presence or absence of the homologous TTCs ShcO1, ShcS1, or ShcS2. B72Ba cultures containing different constructs that made HopO1-1 alone (pLN1077) or with ShcO1 (pLN1074), with ShcS1 (pLN1076), or with ShcS2 (pLN1075) were infiltrated into N. benthamiana and assayed for cAMP production 7 h after infiltration as described in Material and Methods. cAMP levels were reported in picomoles of cAMP per micrograms of protein with standard errors. The experiment was per-formed three times with similar results. Results indicate that HopO1-1 is translocated into plant cells without a plasmid encoded TTC. However, when any of the three homologous chaperones were present, higher amounts of HopO1-1-CyaA were translocated into plant cells.
To determine whether ShcS1 and ShcS2 facilitated the type III secretion of HopO1-1 in culture we performed type III secretion assays with UNL137 containing different TTC/effector combinations. Cultures of UNL137 containing constructs that encoded HopO1-1-FLAG alone (pLN142), ShcO1 and HopO1-1-FLAG (pLN411), ShcS1 and HopO1-1-FLAG (pLN1019), or ShcS2 and HopO1-1-FLAG (pLN1018) were grown in hrp-inducing minimal medium and analyzed for the secretion of HopO1-1-FLAG. We detected higher levels of secreted HopO1-1-FLAG from cultures that contained ShcO1, ShcS1, or ShcS2 than in samples containing only HopO1-1-FLAG (Fig. 4B). This indicates that ShcS1 and ShcS2 can function as a TTC for HopO1-1 and is consistent with their interaction shown in the yeast two-hybrid assays.
We further tested whether each of the TTCs could enhance the translocation of HopO1-1 into plant cells by use of CyaA translocation assays. Constructs were made that contained hopO1-1 (pLN1077), shcO1 and hopO1-1 (pLN1074), shcS1 and hopO1-1 (pLN1076), or shcS2 and hopO1-1 (pLN1075) such that the hopO1-1 gene on each construct was in-frame with cyaA. Thus, each construct when expressed by P. syringae made a HopO1-1-CyaA fusion alone or with one of the ShcO1 TTC homologs. On the basis of the available draft genome information for P. syringae pv. syringae B728a, this strain does not contain any shcO1 homologs; therefore, it was used as a delivery strain to test whether the translocation of HopO1-1-CyaA fusions was facilitated by the presence of plasmid-encoded ShcO1, ShcS1, or ShcS2. B728a expressing HopO1-1-CyaA alone had relatively high cAMP levels, suggesting that this fusion protein was translocated in the absence of TTCs (Fig. 4C). However, cAMP levels were about three times greater when ShcO1, ShcS1, or ShcS2 was also present (Fig. 4C), clearly demonstrating that each TTC acted as a TTC for HopO1-1. We also confirmed that HopS1-CyaA used ShcS1 as a TTC on the basis of higher cAMP levels when plant tissue was infiltrated with B728a expressing both ShcS1 and HopS1-CyaA (108.1 ± 5.8 pmol/cAMP/μg) compared to cAMP levels in plant tissue infiltrated with B728a expressing only HopS1-CyaA (60.1 ± 7.8 pmol/cAMP/μg), clearly demonstrating that ShcS1 acts as a TTC for HopS1.
ShcO1, ShcS1, and ShcS2 bind to the same middle-third region of HopO1-1.
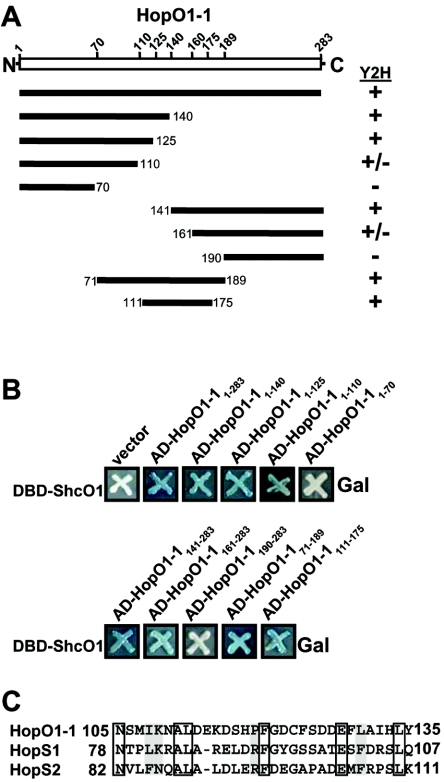
To determine the TTC binding site within HopO1-1 we made a series of yeast two-hybrid AD constructs that contained inserts corresponding to different regions of the HopO1-1 protein (Fig. 5A). These constructs were transformed into yeast strains that contained constructs that encoded ShcO1, ShcS1, or ShcS2 fused to the DBD. Yeast two-hybrid studies were carried out for both β-galactosidase activity and the rescue of leucine auxotrophy. All TTCs interacted with the different HopO1-1 truncations similarly. The yeast two-hybrid results for ShcO1-DBD with different HopO1-1-AD truncations are shown (Fig. 5A and B). ShcO1-DBD did not interact with the N-terminal 70 amino acids of HopO1-1 or the C-terminal region spanning amino acids 190 to 283 (Fig. 5A and B). All HopO1-1-AD fusion proteins were stably expressed, as determined on the basis of their detection in yeast extracts by use of immunoblot analysis with anti-HA antibodies (data not shown), excluding the possibility that the lack of interaction was due to the instability of the proteins. The HopO1-1-AD fusion corresponding to the first 125 amino acids interacted with ShcO1-DBD, as did the fusion corresponding to the last 142 amino acids (amino acids 141 to 283), suggesting that there are ShcO1 binding sites in both of these HopO1-1 truncations (Fig. 5A and B). The smallest HopO1-1-AD fusion that showed a strong interaction with ShcO1 was the HopO1-1 region from amino acids 111 to 175. Thus, the ShcO1 binding sites within HopO1-1 reside in the middle third of HopO1-1. The binding pattern of ShcS1 and ShcS2 to HopO1-1 was the same as seen in the above-described results, indicating that they bind to the same regions of HopO1-1 (data not shown).
FIG. 5.
ShcO1 interacts with a middle region of HopO1-1 in a LexA-based yeast two-hybrid assay. (A) Schematic representation of different fragments of HopO1-1 fused to the transcriptional activation domain (AD). The upper open box represents full-length HopO1-1 protein (283 amino acids). The lower bars represent the series of the DNA fragments that were cloned into pJG4-5 to construct AD-fusions and used in yeast two-hybrid assays with ShcO1 fused to the DNA-binding domain (DBD). The results are represented as follows: +, strong interaction; −, no detectable interaction; +/−, weak interaction. (B) Yeast strains carrying pEG202::shcO1 (producing DBD-ShcO1) and pJG4-5 carrying different fragments of hopO1-1 were grown 2 days on the appropriate medium containing X-Gal. Interactions of different AD-HopO1-1 fusions with DBD-ShcO1 were scored as follows: dark blue, strong interaction; pale blue, weak interaction; white, no interaction. Analogous experiments measuring the rescue of leucine auxotrophy showed similar results (data not shown). (C) The ShcO1 binding region of HopO1-1 was compared to HopS1 and HopS2 with DNAstar Megalign program to identify regions of HopS1 and HopS2 that may represent a TTC binding region. The regions that aligned the best are shown. Boxed amino acids represent amino acids that are identical in each protein and shaded amino acids are conservative changes.
Because ShcO1, ShcS1, and ShcS2 bind to the same region on HopO1-1 and because HopO1-1 does not share high similarity or identity with HopS1 or HopS2, we wanted to identify similar regions between these effectors that might represent a conserved TTC binding site within these effectors. Multiple protein alignments were done between the smallest region of HopO1-1 that interacted with ShcO1, ShcS1, and ShcS2 and regions within HopS1 and HopS2. The HopO1-1 region spanning amino acids 105 to 135, which is within the region that interacted strongly with each TTC (Fig. 5A and B), aligned best with region spanning amino acids 78 to 107 of HopS1 and amino acids 82 to 111 of HopS2 (Fig. 5C). These regions were predicted to have α-helices and amphipathic regions, which would be consistent with the TTC interacting with hydrophobic and polar amino acids exposed on the surface of effectors as shown in structural studies with other TTC/effector pairs (10, 65, 68). Additional experiments are in progress to determine the TTC binding regions within HopS1 and HopS2.
A DC3000 hopO1-1 operon mutant is reduced in its ability to multiply in plant tissue but produced disease symptoms similar to those seen with wild-type DC3000.
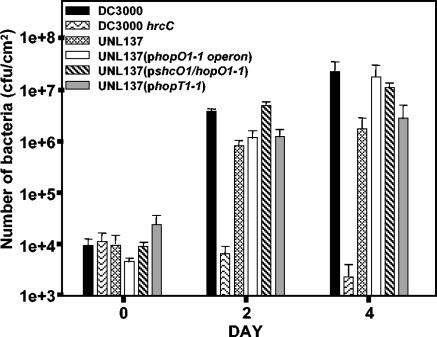
To determine the contribution of HopO1-1 and HopT1-1 to pathogenicity, A. thaliana ecotype Col-0 plants were dip-inoculated with the DC3000 hopO1-1 operon mutant UNL137 as described in Materials and Methods. The disease symptoms produced by UNL137 were not significantly different from those seen with the wild-type DC3000 controls (data not shown). However, bacterial multiplication was reduced in UNL137 compared to the in planta growth exhibited by wild-type DC3000 (Fig. 6). The reduction in bacterial growth was complemented on day 4 when the complete hopO1-1 operon was provided in trans on a broad-host-range plasmid in UNL137. When shcO1 and hopO1-1 or hopT1-1 alone was provided in trans to UNL137, shcO1 and hopO1-1 restored bacterial multiplication to a higher level than hopT1-1, suggesting that HopO1-1 contributes more to pathogenicity than HopT1-1.
FIG. 6.
The DC3000 hopO1-1 operon deletion mutant UNL137 is significantly reduced in its ability to grow in Arabidopsis leaf tissue. The following strains were dip-inoculated into A. thaliana Col-0 leaves as described in the Materials and Methods: Wild-type DC3000, the DC3000 hrcC mutant defective in the TTSS, the hopO1-1 operon deletion mutant UNL137, UNL137 carrying the complete hopO1-1 operon (pLN454), UNL137 carrying shcO1 and hopO1-1 (pLN411), and UNL137 carrying hopT1-1 (pLN256). The results suggest that the hopO1-1 operon does contribute to bacterial multiplication in planta and, based on the mutant complementation, HopO1-1 contributes more to in planta growth than HopT1-1. Disease symptoms produced by UNL137 were indistinguishable from disease symptoms produced by wild-type DC3000 (data not shown).
DISCUSSION
We have shown that ShcO1, ShcS1, and ShcS2 act as TTCs and facilitate the type III secretion and/or translocation of their cognate effectors HopO1-1, HopS1, and HopS2, respectively (Figs. 1, 3 and 4). Additional experiments showed that ShcO1 coimmunoprecipitated with HopO1-1 (Fig. 1C) and that all three TTCs stabilized HopO1-1 and enhanced the secretion and translocation of HopO1-1 (Fig. 4). Yeast two-hybrid assays showed that each of these TTCs interacted with the effectors of each of the other as well as the HopO1-1 homolog, HopO1-2 (Table 2). This was surprising since class 1A TTCs are known to interact with only one effector, and because HopS1 and HopS2 do not share high similarity with HopO1-1 and HopO1-2, it implied that dissimilar effectors could maintain TTC specificity. Each TTC also interacted with the other homologous TTCs consistent with these forming as either homo- or heterodimers. We used yeast two-hybrid assays to define the TTC binding site within HopO1-1 and found that each TTC bound to the middle third of HopO1-1 (Fig. 5).
Recently, a related study, which was focused on the ShcS1/HopS1 interaction, showed that ShcO1 could substitute for ShcS1 in HopS1 translocation (44). This paper also reports that ShcS1 and ShcO1 interact with each other's respective effectors HopO1-1 and HopS1. Additionally, they also showed that ShcO1 and ShcS1 interacted with themselves as well as with each other in yeast two-hybrid assays. Our results confirm these interactions. One curious difference between our results and theirs is that they were unable to show that ShcS2 interacted with its own effector (HopS2), HopS1, or HopO1-1 in yeast two-hybrid assays, whereas we found that ShcS2 interacted strongly with these three effectors and HopO1-2. Indeed, they reported that ShcS2 was unable to substitute for ShcS1 in HopS1 translocation (44). In contrast, we found that ShcS2 facilitated the translocation of HopS2 and protected HopO1-1 from proteolysis, enhanced secretion of HopO1-1 via the TTSS in culture, and enhanced the translocation of HopO1-1 into plant cells (Figs. 3 and 4). Thus, we conclude that ShcS2 is an active TTC.
HopO1-1 and HopS1 are known to be translocated effectors (34, 59, 62). However, prior to this report, HopS2 was not detectably translocated (62). It was speculated that the reason for this was that HopS2 had atypical N-terminal biochemical characteristics (62). Most P. syringae TTSS substrates have N termini that share biochemical characteristics, and these may be an important part of a type III secretion signal. The DC3000 HopS2 lacked one of these characteristics—the presence of an aliphatic amino acid (Leu, Ile, or Val) or a Pro in position 3 or 4. Interestingly, we found that if we introduced an aliphatic amino acid in position 4 of HopS2, much more HopS2 was translocated into plant cells, as determined on the basis of the translocation of HopS2-CyaA fusions (Fig. 3B). This supports the idea that this N-terminal characteristic does function as a type III secretion signal. It is important to note that the translocation of the HopS2 mutation derivatives retained dependence on the TTC ShcS2. This suggests that the N-terminal secretion signals in P. syringae TTSS substrates are not independent of TTC-mediated secretion signals as indicated for Yersinia TTSS substrates (20).
Transcriptional analyses indicated that the hopO1-1 and hopS1 operons were transcribed (11, 75). We revisited the expression of the hopS1 operon because we were interested in whether the putative effector genes downstream of the transposon in the shcS1 operon were transcribed. We confirmed that the hrp promoter upstream of the operon is active (Fig. 2B). Importantly, we also found that the hopO1-2 gene downstream of the transposon was transcribed, indicating that this gene is expressed, albeit at apparent lower levels. Additionally, we also found that the hopS2 operon was transcribed in an hrp-dependent manner (Fig. 2B), further supporting the idea that ShcS2 and HopS2 are functional.
We found that hopO1-2, hopS2, and hopT1-2 encoded proteins that were secreted in culture or translocated via the DC3000 TTSS (Fig. 2C and 3B). This increases the confirmed inventory of DC3000 Hops to 43. It is likely that the inventory will reach about 50 Hops, because several other candidate Hops await testing (21, 43). We have used several methods to identify hop genes, including searches for hrp promoter and N-terminal secretion signal characteristics (30, 59). Currently, the most efficient approach to identify hop genes is to use both criteria together. Even though the DC3000 Hop inventory is large, we know little about the effector activities and their plant targets. The effectors discussed here—HopO1-1 and HopO1-2—are putative ADP ribosyltransferases, while HopS1, HopS2, HopT1-1, and HopT1-2 have no significant similarities to any known genes or proteins in the databases. Of the other DC3000 effectors, only HopPtoD2 and HopPtoN have confirmed enzymatic activities (14, 27, 53).
The amount of redundancy of the genes in the hopO1-1, hopS1, and hopS2 operons is quite substantial. For example, there are three sets of effectors encoded that are clearly homologous (HopS1/HopS2, HopO1-1/HopO1-2, and HopT1-1/HopT1-2). And HopO1-1, HopS1, and HopS2 are further linked by utilizing the same TTCs for their secretion. At first glance, it is surprising how dissimilar effectors could evolve to use homologous TTCs. One possible explanation is that these effectors arose from gene duplication events and have since diverged. Supporting this notion is that the ORF immediately downstream of the transposon (and other overlapping ORFs) actually has similarity to hopO1-1 and encodes a protein domain for an active site of ADP ribosyltransferases similar to HopO1-1 and HopO1-2. Therefore, hopS1 may have previously encoded an effector more similar to HopO1-1 than it does currently. Further supporting this is that, in between shcO1 and hopO1-1 on the plasmid, there is a gene fragment with homology to hopS1. Therefore, it seems likely that the hopO1-1 operon was in the chromosome near the hopS1 operon or that the hopS1 operon was carried on the plasmid near hopO1-1.
Once a gene duplication event occurred, mutations may have arisen in regions important for TTC binding that still permitted TTC binding. The fact that the regions are dissimilar seems to suggest that the primary amino acid sequence is not important for TTC binding. Rather, it may be the mixture of hydrophobic patches and the polar amino acid residues that are important for TTC binding, which is supported by the structural studies of TTCs bound to their effectors (10, 65, 68). We have defined the TTC binding site within HopO1-1 and this region has predicted α-helices and exposed hydrophobic and polar amino acids consistent with those found in the structural studies of other TTC/effector pairs. Currently, we are defining the TTC binding regions within HopS1 and HopS2 to define the properties of these regions that provide TTC specificity. We hope these studies will allow us to exploit the redundancy of P. syringae effectors and TTCs to explore the molecular details of these interactions.
Acknowledgments
We thank Alan Collmer for providing us with plasmid pCPP3234 and Meredith Hanks and Misty D. Wehling for help with several of the experiments. We also thank members of the Alfano laboratory for reviewing the manuscript.
This research was supported by grant 03-35319-13862 from the National Research Initiative Competitive Grants Program of the U.S. Department of Agriculture (J.R.A.) and by National Science Foundation Plant Genome Research Program Cooperative Agreement DBI-0077622 (J.R.A.).
REFERENCES
- 1.Abramovitch, R. B., Y. J. Kim, S. Chen, M. B. Dickman, and G. B. Martin. 2003. Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J. 22:60-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abramovitch, R. B., and G. B. Martin. 2004. Strategies used by bacterial pathogens to suppress plant defenses. Curr. Opin. Plant Biol. 7:356-364. [DOI] [PubMed] [Google Scholar]
- 3.Alfano, J. R., A. O. Charkowski, W. Deng, J. L. Badel, T. Petnicki-Ocwieja, K. van Dijk, and A. Collmer. 2000. The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretions genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc. Natl. Acad. Sci. USA 97:4856-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alfano, J. R., and A. Collmer. 1996. Bacterial pathogens in plants: life up against the wall. Plant Cell 8:1683-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alfano, J. R., and A. Collmer. 1997. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J. Bacteriol. 179:5655-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alfano, J. R., and A. Collmer. 2004. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42:385-414. [DOI] [PubMed] [Google Scholar]
- 7.Axtell, M. J., and B. J. Staskawicz. 2003. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112:369-377. [DOI] [PubMed] [Google Scholar]
- 8.Badel, J. L., K. Nomura, S. Bandyopadhyay, R. Shimizu, A. Collmer, and S. Y. He. 2003. Pseudomonas syringae pv. tomato DC3000 HopPtoM (CEL ORF3) is important for lesion formation but not growth in tomato and is secreted and translocated by the Hrp type III secretion system in a chaperone-dependent manner. Mol. Microbiol. 49:1239-1251. [DOI] [PubMed] [Google Scholar]
- 9.Birtalan, S., and P. Ghosh. 2001. Structure of the Yersinia type III secretory system chaperone SycE. Nat. Struct. Biol. 8:974-978. [DOI] [PubMed] [Google Scholar]
- 10.Birtalan, S. C., R. M. Phillips, and P. Ghosh. 2002. Three-dimensional secretion signals in chaperone-effector complexes of bacterial pathogens. Mol. Cell 9:971-980. [DOI] [PubMed] [Google Scholar]
- 11.Boch, J., V. Joardar, L. Gao, T. L. Robertson, M. Lim, and B. N. Kunkel. 2002. Identification of Pseudomonas syringae pv. tomato genes induced during infection of Arabidopsis thaliana. Mol. Microbiol. 44:73-88. [DOI] [PubMed] [Google Scholar]
- 12.Boyd, A. P., I. Lambermont, and G. Cornelis. 2000. Competition between the Yops of Yersinia enterocolitica for delivery into eukaryotic cells: role of the SycE chaperone binding domain of YopE. J. Bacteriol. 182:4811-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brent, R., and R. L. Finley, Jr. 1997. Understanding gene and allele function with two-hybrid methods. Annu. Rev. Genet. 31:663-704. [DOI] [PubMed] [Google Scholar]
- 14.Bretz, J. R., N. M. Mock, J. C. Charity, S. Zeyad, C. J. Baker, and S. W. Hutcheson. 2003. A translocated protein tyrosine phosphatase of Pseudomonas syringae pv. tomato DC3000 modulates plant defence response to infection. Mol. Microbiol. 49:389-400. [DOI] [PubMed] [Google Scholar]
- 15.Buell, C. R., V. Joardar, M. Lindeberg, S. J., I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. S. Durkin, and J. F. Kolonay. 2003. The complete sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100:10181-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buttner, D., D. Gurlebeck, L. D. Noel, and U. Bonas. 2004. HpaB from Xanthomonas campestris pv. vesicatoria acts as an exit control protein in type III-dependent protein secretion. Mol. Microbiol. 54:755-768. [DOI] [PubMed] [Google Scholar]
- 17.Cambronne, E. D., J. A. Sorg, and O. Schneewind. 2004. Binding of SycH chaperone to YscM1 and YscM2 activates effector yop expression in Yersinia enterocolitica. J. Bacteriol. 186:829-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casper-Lindley, C., D. Dahlbeck, E. T. Clark, and B. J. Staskawicz. 2002. Direct biochemical evidence for type III secretion-dependent translocation of the AvrBs2 effector protein into plant cells. Proc. Natl. Acad. Sci. USA 99:8336-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Charkowski, A. O., H.-C. Huang, and A. Collmer. 1997. Altered localization of HrpZ in Pseudomonas syringae pv. syringae hrp mutants suggests that different components of the type III secretion pathway control protein translocation across the inner and outer membranes of gram-negative bacteria. J. Bacteriol. 179:3866-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng, L. W., D. M. Anderson, and O. Schneewind. 1997. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol. Microbiol. 24:757-765. [DOI] [PubMed] [Google Scholar]
- 21.Collmer, A., M. Lindeberg, T. Petnicki-Ocwieja, D. Schneider, and J. R. Alfano. 2002. Genomic mining type III secretion system effectors in Pseudomonas syringae yield new picks for all TTSS prospectors. Trends Microbiol. 10:462-469. [DOI] [PubMed] [Google Scholar]
- 22.Cornelis, G., and F. van Gijsegem. 2000. Assembly and function of type III secretion systems. Annu. Rev. Microbiol. 54:734-774. [DOI] [PubMed] [Google Scholar]
- 23.Cuppels, D. A. 1986. Generation and characterization of Tn5 insertion mutations in Pseudomonas syringae pv. tomato. Appl. Environ. Microbiol. 51:323-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darwin, K. H., and V. L. Miller. 2001. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 20:1850-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DebRoy, S., R. Thilmony, Y. B. Kwack, K. Nomura, and S. Y. He. 2004. A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc. Natl. Acad. Sci. USA 101:9927-9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Espinosa, A., and J. R. Alfano. 2004. Disabling surveillance: bacterial type III secretion system effectors that suppress innate immunity. Cell. Microbiol. 6:1027-1040. [DOI] [PubMed] [Google Scholar]
- 27.Espinosa, A., M. Guo, V. C. Tam, Z. Q. Fu, and J. R. Alfano. 2003. The Pseudomonas syringae type III-secreted protein HopPtoD2 possesses protein tyrosine phosphatase activity and suppresses programmed cell death in plants. Mol. Microbiol. 49:377-387. [DOI] [PubMed] [Google Scholar]
- 28.Feldman, M. F., and G. R. Cornelis. 2003. The multitalented type III chaperones: all you can do with 15 kDa. FEMS Microbiol. Lett. 219:151-158. [DOI] [PubMed] [Google Scholar]
- 29.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 30.Fouts, D. E., R. B. Abramovitch, J. R. Alfano, A. M. Baldo, C. R. Buell, S. Cartinhour, A. K. Chatterjee, M. D'Ascenzo, M. Gwinn, S. G. Lazarowitz, N.-C. Lin, G. B. Martin, A. H. Rehm, D. J. Schneider, K. van Dijk, X. Tang, and A. Collmer. 2002. Genomewide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proc. Natl. Acad. Sci. USA 99:2275-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaudriault, S., J. P. Paulin, and M. A. Barny. 2002. The DspB/F protein of Erwinina amylovora is a type III secretion chaperone ensuring efficient intrabacterial production of the Hrp-secreted DspA/E pathogenicity factor. Mol. Plant Pathol. 3:313-320. [DOI] [PubMed] [Google Scholar]
- 32.Golemis, E. A., J. Gyuris, and R. Brent. 1994. Interaction trap/two-hybrid system to identify interacting proteins, p. 13.14.1-13.14.17. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Greene Publishing Associates/John Wiley & Sons, New York, N.Y.
- 33.Greenberg, J. T., and B. A. Vinatzer. 2003. Identifying type III effectors of plant pathogens and analyzing their interaction with plant cells. Curr. Opin. Microbiol. 6:20-28. [DOI] [PubMed] [Google Scholar]
- 34.Guttman, D. S., B. A. Vinatzer, S. F. Sarkar, M. V. Ranall, G. Kettler, and J. T. Greenberg. 2002. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295:1722-1726. [DOI] [PubMed] [Google Scholar]
- 35.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 36.Hauck, P., R. Thilmony, and S. Y. He. 2003. A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. USA 100:8577-8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirano, S. S., A. O. Charkowski, A. Collmer, W. D. K., and C. D. Upper. 1999. Role of the Hrp type III protein secretion system in growth of Pseudomonas syringae pv. syringae B728a on host plants in the field. Proc. Natl. Acad. Sci. USA 96:9851-9856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirano, S. S., and C. D. Upper. 2000. Bacterial in the leaf ecosystem with emphasis on Pseudomonas syringae—a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64:624-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hutcheson, S. W., S. Heu, S. Jin, M. C. Lidell, M. U. Pirhonen, and D. L. Rowley. 1996. Function and regulation of Pseudomonas syringae hrp genes, p. 512-521. In T. Nakazawa, K. Furakawa, D. Haas, and S. Silver (ed.), Molecular biology of pseudomonads. ASM Press, Washington, D.C.
- 41.Huynh, T. V., D. Dahlbeck, and B. J. Staskawicz. 1989. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245:1374-1377. [DOI] [PubMed] [Google Scholar]
- 42.Jamir, Y., M. Guo, H.-S. Oh, T. Petnicki-Ocwieja, S. Chen, X. Tang, M. B. Dickman, A. Collmer, and J. R. Alfano. 2004. Identification of Pseudomonas syringae type III effectors that suppress programmed cell death in plants and yeast. Plant J. 37:554-565. [DOI] [PubMed] [Google Scholar]
- 43.Jamir, Y., X. Tang, and J. R. Alfano. 2004. The genome of Pseudomonas syringae pv. tomato DC3000 and functional genomic studies to better understand plant pathogenesis, p. 113-138. In J. L. Ramos (ed.), Pseudomonas: genomics, life style and molecular architecture, vol. 1. Kluwer/Plenum, London, United Kingdom.
- 44.Kabisch, U., A. Landgraf, J. Krause, U. Bonas, and J. Boch. 2005. Type III secretion chaperones ShcS1 and ShcO1 from Pseudomonas syringae pv. tomato DC3000 bind more than one effector. Microbiology 151:269-280. [DOI] [PubMed] [Google Scholar]
- 45.Keen, N. T., S. Tamaski, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 46.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Med. 22:301-307. [PubMed] [Google Scholar]
- 47.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 48.Labes, M., A. Puhler, and R. Simon. 1990. A new family of RSF1010-derived expression and lac-fusion broad-host-range vectors for gram-negative bacteria. Gene 89:37-46. [DOI] [PubMed] [Google Scholar]
- 49.Lee, S. H., and J. E. Galan. 2004. Salmonella type III secretion-associated chaperones confer secretion-pathway specificity. Mol. Microbiol. 51:483-495. [DOI] [PubMed] [Google Scholar]
- 50.Lindeberg, M., J. Stavrinides, J. H. Chang, J. R. Alfano, A. Collmer, J. L. Dangl, J. T. Greenberg, J. W. Mansfield, and D. S. Guttman. 2005. Proposed guidelines for a unified nomenclature and phylogenetic analysis of type III Hop effector proteins in the plant pathogen Pseudomonas syringae. Mol. Plant-Microbe Interact. 18:275-282. [DOI] [PubMed]
- 51.Lloyd, S. A., A. Forsberg, H. Wolf-Watz, and M. S. Francis. 2001. Targeting exported substrates to the Yersinia TTSS: different functions for different signals? Trends Microbiol. 9:367-371. [DOI] [PubMed] [Google Scholar]
- 52.Lloyd, S. A., M. Sjostrom, S. Andersson, and H. Wolf-Watz. 2002. Molecular characterization of type III secretion signals via analysis of synthetic N-terminal amino acid sequences. Mol. Microbiol. 43:51-59. [DOI] [PubMed] [Google Scholar]
- 53.Lopez-Solanilla, E., P. A. Bronstein, A. R. Schneider, and A. Collmer. 2004. HopPtoN is a Pseudomonas syringae Hrp (type III secretion system) cysteine protease effector that suppresses pathogen-induced necrosis associated with both compatible and incompatible plant interactions. Mol. Microbiol. 54:353-365. [DOI] [PubMed] [Google Scholar]
- 54.Luo, Y., M. G. Bertero, E. A. Frey, R. A. Pfuetzner, M. R. Wenk, L. Creagh, S. L. Marcus, D. Lim, F. Sicheri, C. Kay, C. Haynes, B. B. Finlay, and N. C. Strynadka. 2001. Structural and biochemical characterization of the type III secretion chaperones CesT and SigE. Nat. Struct. Biol. 8:1031-1036. [DOI] [PubMed] [Google Scholar]
- 55.Mackey, D., Y. Belkhadir, J. M. Alonso, J. R. Ecker, and J. L. Dangl. 2003. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112:379-389. [DOI] [PubMed] [Google Scholar]
- 56.Mavris, M., A. L. Page, R. Tournebize, B. Demers, P. Sansonetti, and C. Parsot. 2002. Regulation of transcription by the activity of the Shigella flexneri type III secretion apparatus. Mol. Microbiol. 43:1543-1553. [DOI] [PubMed] [Google Scholar]
- 57.Ménard, R., P. Sansonetti, C. Parsot, and T. Vasselon. 1994. Extracellular association and cytoplasmic partitioning of the IpaB and IpaC invasins of S. flexneri. Cell 79:515-525. [DOI] [PubMed] [Google Scholar]
- 58.Parsot, C., C. Hamiaux, and A. L. Page. 2003. The various and varying roles of specific chaperones in type III secretion systems. Curr. Opin. Microbiol. 6:7-14. [DOI] [PubMed] [Google Scholar]
- 59.Petnicki-Ocwieja, T., D. J. Schneider, V. C. Tam, S. T. Chancey, L. Shan, Y. Jamir, L. M. Schechter, M. D. Janes, C. R. Buell, X. Tang, A. Collmer, and J. R. Alfano. 2002. Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 99:7652-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petnicki-Ocwieja, T., K. van Dijk, and J. R. Alfano. 2005. The hrpK operon of Pseudomonas syringae pv. tomato DC3000 encodes two proteins secreted by the type III (Hrp) protein secretion system: HopB1 and HrpK, a putative type III translocator. J. Bacteriol. 187:649-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 62.Schechter, L. M., K. A. Roberts, Y. Jamir, J. R. Alfano, and A. Collmer. 2004. Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J. Bacteriol. 186:543-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schesser, K., E. Frithz-Lindsten, and H. Wolf-Watz. 1996. Delineation and mutational analysis of the Yersinia pseudotuberculosis YopE domains which mediate translocation across bacterial and eukaryotic cellular membranes. J. Bacteriol. 178:7227-7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shan, L., H.-S. Oh, J. Chen, M. Guo, J. Zhou, J. R. Alfano, A. Collmer, X. Jia, and X. Tang. 2004. The HopPtoF locus of Pseudomonas syringae pv. tomato DC3000 encodes a type III chaperone and a cognate effector. Mol. Plant-Microbe Interact. 17:447-455. [DOI] [PubMed] [Google Scholar]
- 65.Singer, A. U., D. Desveaux, L. Betts, J. H. Chang, Z. Nimchuk, S. R. Grant, J. L. Dangl, and J. Sondek. 2004. Crystal structures of the type III effector protein AvrPphF and its chaperone reveal residues required for plant pathogenesis. Structure 12:1669-1681. [DOI] [PubMed] [Google Scholar]
- 66.Sory, M.-P., A. Boland, I. Lambermont, and G. R. Cornelis. 1995. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc. Natl. Acad. Sci. USA 92:11998-12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sory, M. P., and G. R. Cornelis. 1994. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14:583-594. [DOI] [PubMed] [Google Scholar]
- 68.Stebbins, C. E., and J. E. Galán. 2001. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature 414:77-81. [DOI] [PubMed] [Google Scholar]
- 69.Thomas, N. A., and B. B. Finlay. 2003. Establishing order for type III secretion substrates—a hierarchical process. Trends Microbiol. 11:398-403. [DOI] [PubMed] [Google Scholar]
- 70.van Dijk, K., V. C. Tam, A. R. Records, T. Petnicki-Ocwieja, and J. R. Alfano. 2002. The ShcA protein is a molecular chaperone that assists in the secretion of the HopPsyA effector from the type III (Hrp) protein secretion system of Pseudomonas syringae. Mol. Microbiol. 44:1469-1481. [DOI] [PubMed] [Google Scholar]
- 71.Wehling, M. D., M. Guo, Z. Q. Fu, and J. R. Alfano. 2004. The Pseudomonas syringae HopPtoV protein is secreted in culture and translocated into plant cells via the type III protein secretion system in a manner dependent on the ShcV type III chaperone. J. Bacteriol. 186:3621-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wulff-Strobel, C. R., A. W. Williams, and S. C. Straley. 2002. LcrQ and SycH function together at the Ysc type III secretion system in Yersinia pestis to impose a hierarchy of secretion. Mol Microbiol. 43:411-423. [DOI] [PubMed] [Google Scholar]
- 73.Xiao, Y., S. Heu, J. Yi, Y. Lu, and S. W. Hutcheson. 1994. Identification of a putative alternate sigma factor and characterization of a multicomponent regulatory cascade controlling the expression of Pseudomonas syringae pv. syringae Pss61 hrp and hrmA genes. J. Bacteriol. 176:1025-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yip, C. K., B. B. Finlay, and N. C. Strynadka. 2005. Structural characterization of a type III secretion system filament protein in complex with its chaperone. Nat. Struct. Mol. Biol. 12:75-81. [DOI] [PubMed] [Google Scholar]
- 75.Zwiesler-Vollick, J., A. E. Plovanich-Jones, K. Nomura, S. Bandyopadhyay, V. Joardar, B. N. Kunkel, and S. Y. He. 2002. Identification of novel hrp-regulated genes through functional genomic analysis of the Pseudomonas syringae pv. tomato DC3000 genome. Mol. Microbiol. 45:1207-1218. [DOI] [PubMed] [Google Scholar]