Abstract
KSF-1Φ, a novel filamentous phage of Vibrio cholerae, supports morphogenesis of the RS1 satellite phage by heterologous DNA packaging and facilitates horizontal gene transfer. We analyzed the genomic sequence, morphology, and receptor for KSF-1Φ infection, as well as its phylogenetic relationships with other filamentous vibriophages. While strains carrying the mshA gene encoding mannose-sensitive hemagglutinin (MSHA) type IV pilus were susceptible to KSF-1Φ infection, naturally occurring MSHA-negative strains and an mshA deletion mutant were resistant. Furthermore, d-mannose as well as a monoclonal antibody against MSHA inhibited infection of MSHA-positive strains by the phage, suggesting that MSHA is the receptor for KSF-1Φ. The phage genome comprises 7,107 nucleotides, containing 14 open reading frames, 4 of which have predicted protein products homologous to those of other filamentous phages. Although the overall genetic organization of filamentous phages appears to be preserved in KSF-1Φ, the genomic sequence of the phage does not have a high level of identity with that of other filamentous phages and reveals a highly mosaic structure. Separate phylogenetic analysis of genomic sequences encoding putative replication proteins, receptor-binding proteins, and Zot-like proteins of 10 different filamentous vibriophages showed different results, suggesting that the evolution of these phages involved extensive horizontal exchange of genetic material. Filamentous phages which use type IV pili as receptors were found to belong to different branches. While one of these branches is represented by CTXΦ, which uses the toxin-coregulated pilus as its receptor, at least four evolutionarily diverged phages share a common receptor MSHA, and most of these phages mediate horizontal gene transfer. Since MSHA is present in a wide variety of V. cholerae strains and is presumed to express in the environment, diverse filamentous phages using this receptor are likely to contribute significantly to V. cholerae evolution.
Vibrio cholerae is the host for a variety of bacteriophages (vibriophages), which include virulent phages as well as temperate phages represented by the kappa-type phages produced by most strains of the El Tor biotype (18, 36). Another group of vibriophages includes the filamentous phages, which have a single-stranded DNA (ssDNA) genome (1, 8, 23, 25, 40). Several of the V. cholerae-specific filamentous phages have been implicated in virulence gene transfer among V. cholerae strains (2, 7, 40). Filamentous phages of V. cholerae have also been found to be distinct from the well-characterized filamentous coliphages in that some of these phages can form lysogens (1, 25, 40). The most remarkable of these phages is CTXΦ (40), which exists as a prophage in toxigenic V. cholerae and encodes cholera toxin (CT).
Mechanisms associated with the induction and propagation of CTXΦ and related phages have been a major area of interest in vibriophage biology (5, 6, 9-12). In toxigenic El Tor and O139 strains of V. cholerae, CTX prophage is integrated in the bacterial genome arrayed in different tandem structures along with a related satellite phage RS1 (5, 6). The genome of RS1 contains genes encoding proteins needed for replication (RstA), integration (RstB), and regulation of gene expression (RstR and RstC) but lacks the genes encoding proteins needed for assembling and secretion of viral particles (Psh, Cep, pIIICTX, Ace, and Zot) as well as CT (5-7, 39). Thus, satellite phage RS1 can replicate autonomously but depends on its helper phage CTXΦ for assembly and secretion of RS1 viral particles (6, 12, 39). Conversely, RS1 encodes the antirepressor RstC, which is not present in CTX prophage. This protein promotes transcription of CTXΦ genome and RS1 genes by counteracting the activity of the phage repressor RstR (6). We have shown previously that production of RS1Φ particles can also occur independent of CTXΦ but using functions encoded by another putative filamentous phage, called KSF-1Φ (13). RS1Φ produced via this process is capable of infecting recipient strains by a mechanism which is independent of the CTXΦ receptor toxin-coregulated pilus (TCP). In the present study, we examined the morphology, host range, genomic sequence, and receptor for KSF-1Φ infection and analyzed phylogenetic relationships among diverse filamentous vibriophages which can mediate horizontal gene transfer.
MATERIALS AND METHODS
Bacterial strains, plasmids, and phages.
V. cholerae host strains used for phage preparations or as recipients in transduction assays were from either clinical or environmental sources. Clinical strains were from patients who attended the treatment Center of the International Centre for Diarrheal Disease Research, Bangladesh (ICDDR,B), located in Dhaka. The environmental strains were isolated from surface waters in Dhaka. Strains were stored either in lyophilized form or in sealed deep nutrient agar at room temperature. Before use, the identities of the V. cholerae strains were confirmed by biochemical reaction and serology (42), and the presence or absence of genes encoding TCP and mannose-sensitive hemagglutinin (MSHA) pilus was ascertained using DNA probes or PCR assays as described below. The genetically marked phage genome pKSF1-Km was a derivative of the replicative-form (RF) DNA of KSF1Φ, which was marked with a kanamycin resistance (Kmr) determinant as described below.
Probes and hybridization.
The KSF-1Φ genome was detected in naturally occurring V. cholerae strains by its positive hybridization with a probe derived from pKSF-1, the RF of the phage genome (13). Strand-specific oligonucleotide probes corresponding to the plus and minus strand of the KSF-1Φ genome were also used to detect the presence of the single-stranded KSF-1Φ genome. Colony blots or Southern blots were prepared using nylon filters (Hybond; Amersham Biosciences, Uppsala, Sweden) and processed by standard methods (29). The polynucleotide probes were labeled by random priming (16) using a random primers DNA labeling kit (Invitrogen Corporation, Carlsbad, CA) and [α-32P]dCTP (3,000 Ci/mmol; Amersham), and oligonucleotide probes were labeled by 3′ tailing using terminal deoxynucleotidyl transferase and [α-32P]dCTP (Amersham). Southern blots and colony blots were hybridized with the labeled probes and autoradiographed by standard methods (29).
PCR assays.
All oligonucleotides used either as probes or PCR primers were synthesized commercially by Oswel DNA Service (University of Edinburgh, Edinburgh, United Kingdom), and PCR reagents and kits were purchased from Perkin-Elmer Corp. (Norwalk, CT). Presence of tcpA and mshA genes were detected by PCR assays as described previously (26, 34). The expected sizes of the PCR amplicons were ascertained by electrophoresis with agarose gels. Identities of all PCR products were further verified using specific oligonucleotide probes.
Construction of pKSF1-Km.
The RF of KSF-1Φ genome (pKSF-1), was isolated from the native host strain 55V71 (13) by standard methods (29), and a preliminary analysis of restriction endonuclease cleavage sites within pKSF-1 was done. The pKSF-1 DNA was then marked by inserting a Kmr marker. This was done by ligating XbaI-digested linearized pKSF-1 with a Kmr determinant derived from XbaI-cleaved pCTX-Km (40). A recipient V. cholerae strain, Env-002, was electroporated with the ligated DNA, and transformants were selected on Luria agar plates containing kanamycin (50 μg/ml). Kmr colonies were analyzed for the presence of pKSF1-Km.
Preparation of phage.
Overnight cultures of V. cholerae were diluted 100-fold in fresh LB medium and grown for 6 h at 30°C with shaking. Supernatant fluids were sterilized by filtration through 0.22-μm-pore-size filters (Millipore Corporation, Bedford, MA). To confirm that the filtrates did not contain any bacterial cells, aliquots of the filtrates were streaked on Luria agar plates and incubated overnight at 37°C. For phage assays, aliquots of the sterile supernatant fluids were incubated with the recipient strains and plated on appropriate antibiotic plates as described in the following section.
For isolation and analysis of phage nucleic acids, the filtrates were mixed with one-fourth volumes of a solution containing 20% polyethylene glycol 6000 and 10% NaCl and centrifuged at 12,000 × g to precipitate the phage particles. The precipitate was dissolved in a solution containing 20 mM Tris-Cl (pH 7.5), 60 mM KCl, 10 mM MgCl, and 10 mM NaCl and digested with pancreatic DNAse I (100 units/ml) and RNase A (50 μg/ml) at 37°C for 2 h to remove possible nucleic acids carried over from lysed bacterial cells. The solution was extracted with phenol-chloroform to disrupt phage particles, and the total nucleic acids were precipitated with ethanol. The nucleic acids were analyzed by Southern hybridization using appropriate probes (13) to detect the presence of relevant phage genomes.
Transduction assays.
The susceptibilities of recipient strains to KSF-φ were assayed using the genetically marked phage KSF1-Kmφ prepared from culture supernatants of Env-002 (pKSF1-Km). Phage particles were precipitated from 50-ml aliquots of filtered sterile supernatants, and the pellet was suspended in 100 μl of TES [N -tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid] buffer (20 mM Tris-HCl [pH 7.5], 10 mM NaCl, 0.1 mM Na2-EDTA). Recipient cells were grown in Trypticase soy broth (TSB) medium (Difco Laboratories, Becton Dickinson and Company, Sparks, MD) at 37°C, precipitated by centrifugation, and washed in fresh TSB. Approximately 105 bacterial cells were mixed with 10 μl of the phage preparation in a final volume of 100 μl. Each mixture was inoculated into 5 ml of TSB and incubated for 1 h at 37°C, and aliquots of the culture were analyzed by plating on Luria agar plates containing kanamycin (50 μg/ml) and on plates without kanamycin. The ratio of Kmr transduced colonies to the total number of colonies derived from the recipient strain was calculated and expressed as the susceptibility to KSF-1Φ infection. To test the inhibition of phage infection with anti-MSHA antibody, dilutions of anti-MSHA monoclonal antibody HA17:10, provided by A.-M. Svennerholm (33), in parallel with TSB medium as a control were mixed with recipient cells and the mixtures were incubated at 37°C for 30 min prior to the addition of the phage.
Analysis of infected cells.
Representative infected colonies were grown overnight in LB containing kanamycin (50 μg/ml), and cells were precipitated by centrifugation. The supernatant fluids of the cultures were titrated for the presence of KSF1-KmΦ particles using strain Env-002 as the recipient. Total DNA or plasmids were extracted from bacterial pellets by standard methods (29) and purified using microcentrifuge filter units (Ultrafree-Probind; Sigma Chemical Company, St. Louis, MO). Integration of the phage genome into the chromosome of the recipient cells was determined by comparative Southern blot analysis of total DNA and plasmid preparations from the phage-infected and the corresponding native strains as described previously (10, 12).
MSHA testing.
Expression of MSHA was tested by slide agglutination as described previously (17) with V. cholerae grown in TSB medium (Difco) at 37°C, using chicken erythrocytes.
DNA sequencing and analysis of genomic sequence.
Nucleotide sequencing was performed with an automated DNA sequencing system (ABI Prism 310; PE Applied Biosystems, Foster City, CA) using BigDye terminator cycle sequencing ready reaction kit (PE Applied Biosystems). Initially, overlapping subclones of pKSF-1 were constructed in pUC18 and were sequenced using universal sequencing primers (Invitrogen). The nucleotide sequences of both strands of pKSF-1 were further determined by primer walking with primers derived from the preliminary sequencing of pKSF-1 subclones in pUC18. Sequences were processed using the Sequencher alignment program, Version 4.0 (Gene Codes Corporation, Inc., MI). The nucleotide sequence of KSF-1 was compared to sequences in the GenBank databases, and the protein homology search was done using the National Center for Biotechnology Information BLAST server program. Different phage gene sequences were aligned using the CLUSTAL-W multiple sequence alignment program (3), and phylogenetic analyses were conducted using Molecular Evolutionary Genetics Analysis software (MEGA version 2.1; Arizona State University, Tempe, Arizona) (27). Trees were constructed by neighbor joining using the Jukes-Cantor distance method. Bootstrap values were calculated based on 1,000 computer-generated trees.
Nucleotide sequence accession number.
The sequence of the KSF-1Φ genome has been assigned GenBank accession number AY714348.
RESULTS AND DISCUSSION
KSF-1Φ is viable and infectious.
The importance of filamentous phages in V. cholerae evolution is beginning to be understood in more detail with the discovery of new phages involved in lateral gene transfer. We previously observed the presence of extrachromosomal pKSF-1 DNA with non-O1/non-O139 strain 55V71 and a single-stranded version of pKSF-1 in phage DNA preparations from the same strain. This led us to assume that pKSF-1 is the genome of a filamentous phage. Furthermore, pKSF-1 supported morphogenesis of the V. cholerae satellite phage RS1 (13).
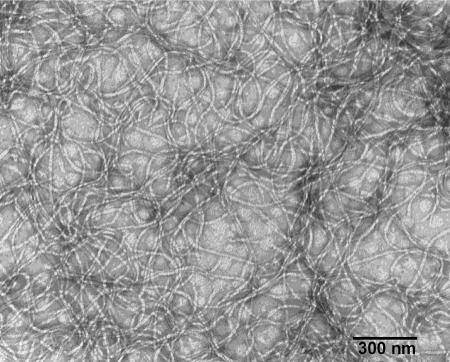
In the present study, electron microscopic examination of KSF-1Φ showed that the phage was indeed a filamentous particle (Fig. 1). Filamentous phages which belong to the genus Inovirus of the family Inoviridae are known to be long proteinaceous tubes ranging in length between 0.8 and 2 μm and containing a single-stranded circular DNA genome. The single-stranded DNA is converted to a double-stranded RF in infected cells. Consistent with this definition, the length of KSF-1Φ particle was 1.2 μm, with a width of 7 nm, and the particle contained a single-stranded DNA genome. The size of KSF-1Φ is similar to that of a previously described filamentous phage, fs2, of V. cholerae O139 (23). In this study, we further demonstrate that KSF-1 phage is able to mediate transfer of its own genome to recipient cells by the formation of infectious particles. To monitor the production of phage particles, we constructed a genetically marked derivative of the phage genome which carried a Kmr determinant. The Kmr also allowed us to conveniently monitor phage infection, since infected cells became resistant to kanamycin. We found that a variety of V. cholerae strains, including V. cholerae O1 and non-O1 strains, were infected by the phage (Table 1). The infected strains produced moderate to high titers (2.7 × 103 to 1.5 × 106) of new infectious particles. Production of the phage particles by infected cells was not associated with cell lysis, and KSF-1Φ did not form plaques on a lawn of susceptible bacteria.
FIG. 1.
Electron micrograph of KSF-1 phage particles. Phage particles were isolated from the culture supernatant of strain Env-002 infected with KSF1-KmΦ. Phage preparations from the native strain Env-002 did not show any detectable phage particles.
TABLE 1.
Susceptibility of V. cholerae strains carrying different combinations of MSHA and TCP genes to a genetically marked derivative of KSF-1φa
| Strain | Description | Presence of
|
Susceptibility to KSF1-Kmφ | |
|---|---|---|---|---|
| mshA | tcpA | |||
| AK-31047 | O1 | + | + | 5.4 × 10−3 |
| C6706 | O1 | + | + | 3.2 × 10−2 |
| KHT46 | C6706ΔmshA | − | + | 0 |
| AL-11089 | O139 | + | + | 2.3 × 10−2 |
| Env-99 | O139 | + | + | 7.5 × 10−3 |
| Env-002 | O1 | + | + | 6.3 × 10−3 |
| O395 | O1 | + | + | 2.7 × 10−5 |
| TCP-2 | O395 ΔtcpA ΔctxA | + | − | 2.1 × 10−5 |
| 55V71 | Non-O1, non-O139 | + | − | 4.2 × 10−2 |
| 761V1325 | Non-O1, non-O139 | + | − | 3.4 × 10−2 |
| 775V1346 | Non-O1, non-O139 | + | − | 6.2 × 10−2 |
| 825V1409 | Non-O1, non-O139 | − | − | 0 |
| 824V1406 | Non-O1, non-O139 | − | − | 0 |
| 828V1412 | Non-O1, non-O139 | − | − | 0 |
| 819V1399 | Non-O1, non-O139 | + | − | 5.7 × 10−2 |
| 820V1401 | Non-O1, non-O139 | + | − | 6.3 × 10−2 |
| 883V1486 | Non-O1, non-O139 | + | − | 9.9 × 10−3 |
| 886V1491 | Non-O1, non-O139 | + | − | 9.2 × 10−3 |
Presence of different genes was detected using DNA probes and PCR assays. Susceptibility to the phage was determined using a genetically marked phage (see text for details). The values are the proportions of total cells infected (number of colonies growing on kanamycin plates divided by the total number of colonies recovered). Values are averages from three independent assays.
MSHA pilus is the receptor in KSF-1Φ infection.
Filamentous phages are known to utilize pili as their receptors, and V. cholerae produces several types of pili and carries the genes for many pilin-like proteins (19). The most remarkable of these is the CTXΦ receptor TCP, which is also the major intestinal colonization factor. We previously found that RS1Φ produced by KSF-1Φ-positive strains, presumably using KSF-1Φ virion proteins, did not use the CTXΦ receptor TCP for infecting recipient cells (13). In the present study, we investigated whether a different type IV pilus, MSHA, which is produced by a wide variety of V. cholerae strains, could act as a receptor in KSF-1Φ infection. First, we identified groups of naturally occurring V. cholerae strains which were (i) negative for the mshA gene encoding MSHA and (ii) positive for the mshA gene and produced MSHA. These strains were exposed to KSF1-Kmφ, the genetically marked derivative of KSF-1Φ. While strains which were negative for mshA were resistant to the phage, the MSHA-positive strains were found to be infected (Table 1). The susceptibility varied between 2.1 × 10−5 to 6.2 × 10−2 for different strains. Classical biotype strains are mostly hemagglutinin negative. However, they carry the mshA pilin gene and express the gene, although at a very low level (24). In agreement with this previous observation, the classical biotype strain O395 and its TCP-deleted derivative TCP2 (37) were infected by KSF-1 phage at a low frequency (Table 1).
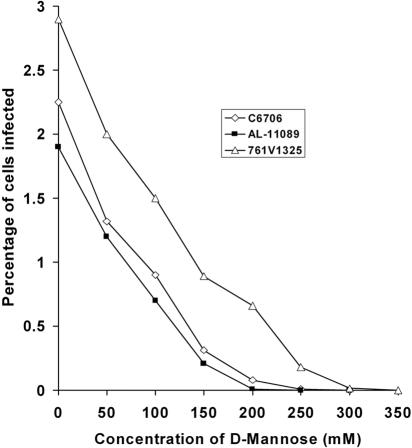
The biological activity of MSHA is known to be inhibited by d-mannose. We, therefore, presumed that if MSHA is the receptor for KSF-1 phage, the addition of d-mannose to block the receptor should render MSHA-positive strains resistant to the phage. We found that the addition of increasing concentrations of d-mannose solution to recipient cells, prior to the assay for susceptibility to KSF-1Φ, progressively inhibited infection of the MSHA-positive strains (Fig. 2). These findings indicated that MSHA pilus can act as a receptor in KSF-1Φ infection. We further tested this assumption by using an isogenic pair of strains, in one of which the mshA gene was deleted (38). While the wild-type strain was infected by KSF-1Φ at high efficiency, the mutant was completely resistant to the phage (Table 1). Finally, we used a monoclonal antibody against MSHA to block the putative receptor and test whether the antibody inhibited infection of MSHA-positive cells by KSF-1Φ. In our assay, incubation with the antibody for 30 min prior to the addition of phage particles completely inhibited infection of otherwise susceptible cells by KSF-1Φ. Taken together, these results suggested strongly that MSHA is the receptor in KSF-1Φ infection. Several other filamentous vibriophages, including fs1, fs2, 493, and VGJ, reportedly use MSHA as their receptor (2, 8, 24). Most of these phages have been either presumed or demonstrated to be involved in lateral gene transfer in V. cholerae. This study shows that KSF-1Φ, which also mediates horizontal gene transfer, constitutes another example of a filamentous phage using MSHA pili as receptors in infecting recipient strains.
FIG. 2.
Effect of d-mannose on the susceptibility of V. cholerae strains to KSF1-KmΦ. The identity (and serotype) of strains tested are C6706 (O1, El Tor), AL-11089 (O139), and 761V1325 (non-O1/non-O139).
Genomic organization of KSF-1Φ.
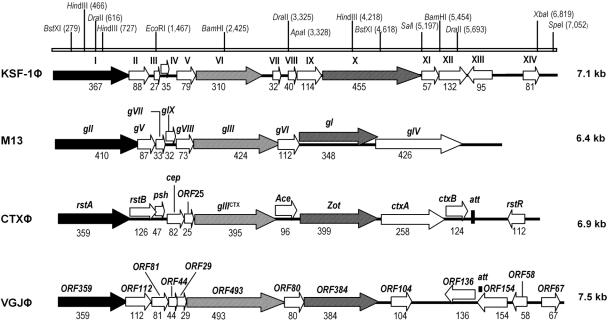
Determination of the nucleotide sequence of the KSF-1Φ genome revealed that the phage genome comprises 7,107 nucleotides and contains 14 open reading frames (ORFs; ORFI through ORFIV) as shown in Fig. 3. The ORFs were preceded by purine-rich consensus sequences (Shine-Dalgarno sequence) for potential ribosome binding sites. Of the 14 ORFs, 13 were located on one of the strands, and we assumed that this strand was the viral strand (plus strand) of the single-stranded KSF-1Φ genome. This was subsequently confirmed by hybridizing phage DNA preparations with single-strand-specific oligonucleotide probes. A homology search showed that 4 of the 14 predicted protein sequences of KSF-1Φ were homologous to previously reported phage proteins. The percent homology with the sequence of various previously reported phage proteins varied from 25% to 100%. Detailed findings of the homology search are presented in Table 2.
FIG. 3.
Genomic organization of KSF-1Φ. Linear ORF maps of M13, CTXΦ, VGJΦ, and KSF-1 phages, aligned by using the first base of the replication initiator gene. The restriction map of KSF-1Φ is shown on top. ORFs or genes are represented by arrows oriented in the direction of transcription, and filled or striped arrows represent genes included in the phylogenetic analyses.
TABLE 2.
Results of BLAST search showing reported homology of ORFs and intergenic regions of KSF-1 phage with those of other phages or bacteria
| ORF/intergenic region | Nucleotide position | Homology to gene products of other bacteriophage or bacterium
|
Percent identity | Accession number | ||
|---|---|---|---|---|---|---|
| Phage | Host bacterium | Gene product | ||||
| I | 23-283 | VGJ | V. cholerae | Rolling circle replication protein | 60 | NP835472 |
| I | 1-1101 | VSKK | V. cholerae | Putative replication protein | 98 | NP536619 |
| I | 1-1101 | VSK | V. cholerae | rep-VSK | 89 | NP542355 |
| I | 1-498 | fs1 | V. cholerae | Hypothetical protein | 99 | NP695201 |
| I | 40-1083 | CTX | V. cholerae | RstA | 38 | NP231097 |
| I | 40-1083 | Vf33, Vf12, f237, VfO3K6, VfO4K68, | V. parahaemolyticus | Putative replication protein | 38 | BAA33512, BAA33520, NP797930, NP059531, NP059541 |
| I | 40-1095 | Vibrio vulnificus | Putative replication protein | 34 | NP936848 | |
| I | 292-1101 | VGJphi | V. cholerae | Rolling circle replication protein | 100 | NP835472 |
| I | 408-491 | VSK | V. cholerae | rep-VSK | 65 | NP542355 |
| I | 469-972 | V. cholerae | Catalase | 45 | AAP84006 | |
| I | 496-1101 | fs1 | V. cholerae | Hypothetical protein | 99 | NP695202 |
| II | 1109-1327 | VGJ | V. cholerae | Putative ssDNA binding protein | 98 | NP835473 |
| II | 1109-1327 | VSKK, fs1 | V. cholerae | Hypothetical protein | 98 | NP536620NP695203 |
| II | 1136-1327 | VSK | V. cholerae | VSK-int | 92 | NP542356 |
| II | 1166-1324 | Vf33, Vf12, f237, VfO3K6, VfO4K68 | V. parahaemolyticus | Hypothetical protein | 44 | BAA33506, BAA33514, NP797931, NP059532, NP059542 |
| Intergenic | 1333-1449 | VGJ | V. cholerae | Putative ssDNA binding protein | 69 | NP835473 |
| Intergenic | 1333-1449 | fs1 | V. cholerae | Hypothetical protein | 69 | NP695203 |
| Intergenic | 1333-1425 | VSKK | V. cholerae | Hypothetical protein | 77 | NP536620 |
| Intergenic | 1333-1425 | VSK | V. cholerae | VSK-int | 77 | NP542356 |
| Intergenic | 1333-1425 | Vf33, Vf12 | V. parahaemolyticus | Hypothetical protein | 48 | BAA33506, BAA33514 |
| Intergenic | 1333-1425 | VfO3K6, FO4K68, f237 | V. parahaemolyticus | Hypothetical protein | 45 | NP059532, NP059542, NP797931 |
| IX | 3453-3782 | VfO3K6, f237, VfO4K68 | V. parahaemolyticus | Hypothetical protein | 27 | NP059536, NP797936, NP059546 |
| X | 3800-5164 | VfO3K6, VfO4K68, f237 | V. parahaemolyticus | Zot-like protein | 39 | NP059537, NP059547, NP797937 |
| X | 3800-4807 | CTX | V. cholerae | Zonula occludens toxin | 25 | AAL09680 |
| X | 3800-4762 | Pseudomonas aeruginosa | Zonula occludens toxin | 23 | ZP00138787 | |
| X | 3800-4612 | Pf1 | Pseudomonas aeruginosa | Hypothetical protein | 26 | NP039606 |
| Intergenic | 6942-7106 | VGJ | V. cholerae | Hypothetical protein | 58 | NP835484 |
| Intergenic | 6955-7032 | VGJ | V. cholerae | Hypothetical protein | 69 | NP835484 |
The peptide encoded by ORFI of KSF-1Φ is homologous to the replication proteins of previously described phages VGJ, VSK, VSKK, and fs1 (1, 8, 25). ORFI is also similar in terms of position and size to genes of previously reported filamentous phages, which mapped at the same relative position as the gII gene of M13 phage (Fig. 3). This gene encodes the pII protein, which is necessary for rolling-circle replication of the phage genome (31). The deduced amino acid sequence of the putative peptide encoded by ORFII is homologous to peptides of previously described filamentous vibriophages that mapped at the same position as the ssDNA-binding protein of Ff phages (f1, M13, and fd), and its size was similar to that of this protein (Fig. 3). We therefore assume that ORFI and ORFII of KSF-1 phage are homologues of the gII and gV genes of Ff phages and thus constitute the putative replication module of KSF-1 phage. ORFIII through ORFVII of KSF-1Φ map at the same relative position as gVII, gIX, gVIII, gIII, and gVI, respectively, of the M13 phage, although there was no significant homology of the putative peptides encoded by these ORFs with that of M13. Of these five ORFs, the first four had sizes comparable to the corresponding genes of previously described Ff phages (31), and these genes are known to encode the minor and major capsid proteins. ORFVI of KSF-1Φ is located at the same relative position as gIII of Ff phages and the gIIICTX of CTX phage and is similar in size (20). The gIII gene encodes pIII, a minor capsid protein that recognizes and interacts with the receptors and coreceptors of these phages. It thus appears that ORFIII through VII of KSF-1 phage encodes putative structural proteins of the phage, including a pIII-like protein involved in receptor binding. Protein encoded by ORFX of KSF-1Φ was found to have a modest level of homology with the Zot-like protein of Vibrio parahaemolyticus phages f237, and VfO4K68, as well as to the Zot protein of CTX phage (Table 2). The relative position and size of the zot gene is similar to the gI gene, which encodes pI in Ff phages and is needed for assembly and secretion of the viral particles (30, 35). Thus, it is likely that the product of ORFX plays a similar role in KSF-1Φ.
Additionally, some filamentous phages encode transcriptional repressors, such as RstR of CTXΦ and vpf122 of Vf33 phage, which regulate the expression of other phage genes (6). In CTXΦ and in VJGΦ, the repressor genes are transcribed in the direction opposite that of most other genes. We identified an ORF (ORFXIII) in KSF-1Φ, which also transcribes in the opposite direction, although it was not clear whether this ORF encodes a functional repressor. The functions of putative peptides encoded by ORFIX, ORFXI, ORFXII, and ORFIV are not known. The KSF-1 genome also contains an 18-bp att-like sequence, CAAGCCGATACTGCGCGA, which is similar but not identical to that of other integrative filamentous phages. It also remains to be established whether this site is involved in possible phage integration. Generally, the genome of a filamentous phage is organized in a modular structure, in which functionally related genes are grouped (21). These include the replication module containing the genes coding for rolling-circle replication protein and the ssDNA-binding protein, the structural module containing the major and minor coat protein-encoding genes, and the assembly and secretion module containing genes for morphogenesis and extrusion of the virus particles. Although the functions of all the gene products encoded by the putative ORFs of KSF-1Φ are not clearly known and, overall, KSF-1Φ appears to have a mosaic genomic structure, the genetic organization of other filamentous phages appear to be preserved in KSF-1.
Evolutionary relationships of KSF-1Φ and other filamentous vibriophages.
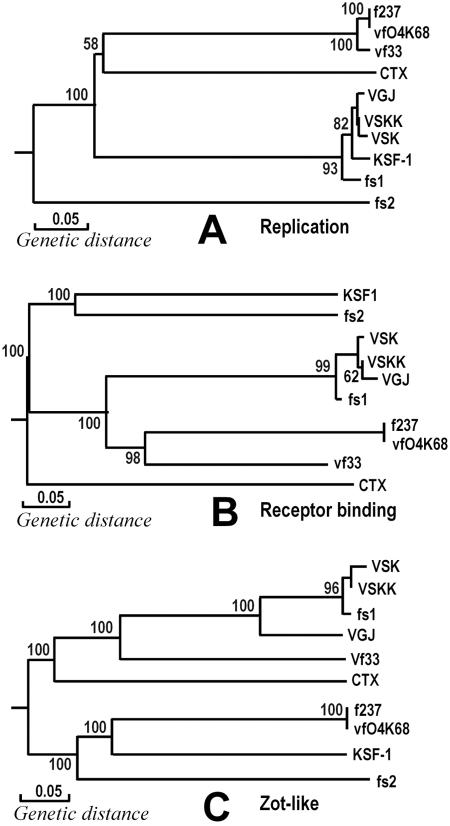
The coevolution of V. cholerae as a pathogen and genetic elements that mediate the transfer of virulence genes is becoming increasingly evident. Since the discovery of CTXΦ, a number of filamentous phages have been reported to be involved in lateral gene transfer (2, 13, 14, 32). However, the phylogenetic relationships among these filamentous phages have not been adequately explored. Construction of phylogenetic trees using different regions of the genomic sequence of KSF-1Φ and corresponding regions of previously described filamentous vibriophages allowed us to assess evolutionary relatedness and divergence between KSF-1Φ and other filamentous phages of V. cholerae as well as V. parahaemolyticus. The current trend towards using sequence data for phylogenetic analysis has raised the question of identifying the most appropriate level for analysis. Lawrence and colleagues (28) have discussed the problems associated with viral genomes having mosaic structures and that mosaicism of viral genomes undermines the validity of approaches based on comparison of whole phage genomes. It has also been suggested that phylogenetic analyses should be performed at the level of individual genes, since these represent the functional units of highly mosaic viral genomes (41). We therefore selected three essential genes or corresponding ORFs encoding putative replication proteins, pIII-like receptor-binding coat proteins, and Zot-like phage assembly proteins for phylogenetic analysis. These three ORFs in the KSF-1Φ genome all showed modest to high levels of homology with corresponding genes of one or more previously described filamentous vibriophages (Table 2). Estimates of genetic relationships based on the sequence of these genes among 10 different filamentous vibriophages are shown in Fig. 4.
FIG. 4.
Phylogenetic relationships among gene sequences of putative replication proteins (A), Zot-like proteins (B), and pIII-like receptor-binding coat proteins (C) of 10 different filamentous vibriophages. The trees were constructed from pairwise Jukes-Cantor distances by using the neighbor-joining method. Bootstrap values based on 1,000 computer-generated trees are indicated at the nodes, and only values greater than 60 are shown.
Based on the sequence of replication protein genes, the V. parahaemolyticus phages clustered together, but at least three distinct branches were observed among the V. cholerae phages (Fig. 4). These branches were represented by CTX phage, fs2 phage, and a group of five other phages, including KSF-1, VJG, VSK, VSKK, and fs1. Thus, KSF-1 appears to be closely related to VGJ, VSK, VSKK, and fs1 in its replication protein gene. However, in the phylogenetic tree based on Zot-like protein genes, KSF-1Φ appeared more closely related to phages f237 and vfO4K68 of V. parahaemolyticus than to V. cholerae-specific filamentous phages. Similarly, phage vf33 of V. parahaemolyticus belonged to a different branch, closer to a cluster V. cholerae phages. CTX phage and fs2 phage also belonged to distinct branches of this phylogenetic tree. The most notable feature of this analysis is the divergent clustering of pIII-like protein genes (Fig. 4) carried by phages sharing the same receptor. This gene encodes a minor capsid protein that recognizes and interacts with the phage receptor. Understandably, CTXΦ, which uses TCP as its receptor, belongs to a branch distinct from those of the rest of the V. cholerae phages. However, KSF-1 and another five V. cholerae phages, fs1, fs2, VGJ, VSK, and VSKK, which reportedly use MSHA as their receptors, did not cluster together but, rather, belonged to four different branches. Interestingly, phage fs2, which uses the same receptor as KSF-1, fs1, VSK, VSKK, and VGJ phages, is diverged from these phages in all three of the genomic regions examined. Overall, we found that V. cholerae phages KSF-1, fs1, VSK, VSKK, and VGJ belong to a closely related cluster in terms of their replication proteins, but KSF-1Φ is considerably diverged from this cluster in the sequence of receptor-binding proteins and Zot-like proteins. The overall sequence homology of KSF-1Φ with other filamentous phages was also not high and ranged between 34.3% for CTXΦ to 52.5% for VSK phage. These results are consistent with the mosaic genomic structure of KSF-1Φ, which seems to have evolved through extensive horizontal exchange of genetic materials among these phages and, possibly, other unidentified phages.
In contrast to the well-characterized filamentous coliphages, several of the filamentous phages of V. cholerae integrate into the host chromosome (1, 2, 40). KSF1φ genome was also occasionally found to integrate into the chromosome of susceptible V. cholerae (Fig. 5). This is consistent with the presence of genomic sequence encoding a putative protein with homology to that of an integrase protein of phage VSK described previously (25). However, the mechanisms related to this integration event are not clear from this study. As mentioned above, the KSF-1Φ genome also contains an 18-bp att-like sequence which is similar but not identical to the att site of VGJΦ and other integrative filamentous phages. Further studies are under way to characterize the chromosomal junctions and understand the mechanisms leading to KSF-1Φ integration. Phages VGJ and KSF-1 have been recently shown to participate in the transfer of CTXΦ or the RS1 satellite phage in a TCP-independent manner. Horizontal transfer of CTX and RS1 by VGJ phage involves site-specific cointegration and formation of a hybrid phage genome (2). On the other hand, transfer of RS1 by KSF-1Φ seems to occur by heterologous packaging of excised RS1 element into viral particles (13). Thus, the mechanisms involved in lateral gene transfer by these two phages appear to be different.
FIG. 5.
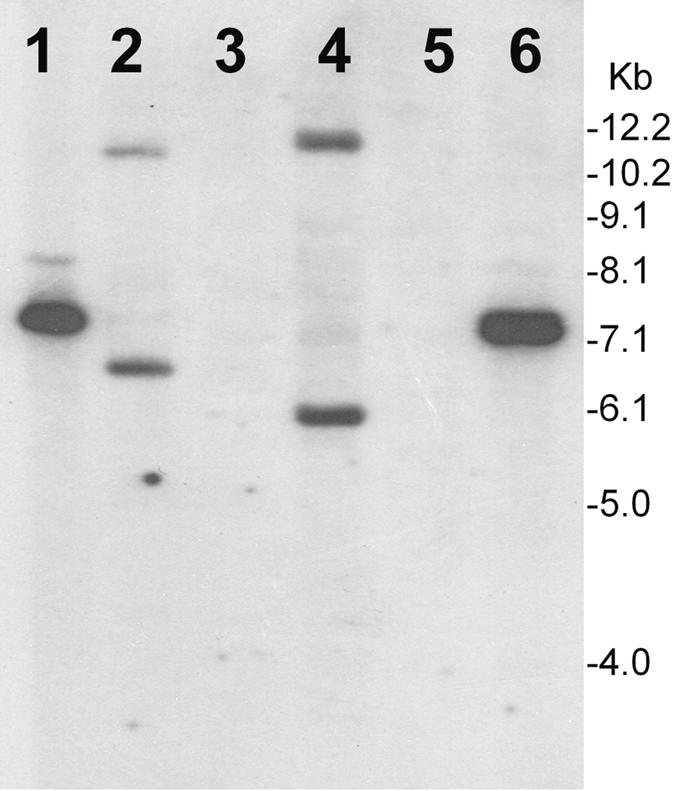
Southern hybridization analysis of total genomic DNA or plasmids isolated from V. cholerae strains infected with KSF1-KmΦ, demonstrating phage integration. Total DNA or plasmids were digested with XbaI and hybridized with the pKSF-1 probe. Lanes 1 and 6, pKSF linearized with XbaI; lanes 2 and 4, total DNA from infected strains N16961 and SA406, respectively; lanes 3 and 5, plasmid preparations of strains N16961 and SA406, respectively. Numbers indicating molecular sizes of bands correspond to a 1-kb DNA ladder (Invitrogen). The absence of bands in the lanes containing plasmid preparations and the positive hybridization of restricted chromosomal DNA of the transductants suggest integration of the KSF1-KmΦ genome into the chromosome of recipient strains.
The CT-converting phage CTXΦ is also known to carry a phage repressor gene, rstR, which encodes the RstR protein that provides immunity to superinfection by the same phage. We did not detect an ORF with a predicted protein product homologous to RstR in KSF-1, and strains carrying KSF-1Φ were susceptible to superinfection by the same phage. For example, strain 55V71, which carried a native KSF-1Φ, was also susceptible to superinfection by the genetically marked KSF1-Km phage (Table 1). Interestingly, however, the large intergenic region between ORFXIII and ORFXIV of the KSF-1Φ genome contained a 27-bp sequence (bases 6098 to 6124) which was identical to part of the nucleotide sequence of rstR gene, suggesting that KSF-1Φ might have been derived from an ancestral phage which had functional RstR.
Significance of MSHA and filamentous phages in lateral gene transfer.
Whereas CTXΦ uses the TCP pili for infecting recipient cells, at least four other phages, including fs1, fs2, VGJ, and KSF, use the MSHA pili as their receptor. It is interesting that fs1, fs2, KSF-1, and VGJ have considerable divergence in the sequences of their putative receptor-binding proteins (Fig. 4). Thus, MSHA pili can act as a receptor of genetically diverse filamentous phages, and several of these phages have been shown to mediate horizontal gene transfer. In previous studies, the mshA gene encoding MSHA was found to be present in a wide variety of V. cholerae strains (15, 34). Hence, this phage receptor is apparently ubiquitous among V. cholerae strains, and filamentous phages mediating gene transfer could spread rapidly among these strains.
Phage VGJ, which also uses MSHA as its receptor, has been reported to facilitate the transfer of CTX phage as well as RS1 satellite phage genome by forming hybrid genomes (9). In our study, however, RS1 was transferred through alternative packaging of circularized single-stranded RS1 instead of the KSF-1 genome. Particularly because of their flexible capsid structure, filamentous phages may be uniquely suited to mobilizing genes and larger elements. Hence, one or more of these vibriophages are also likely to package possible other DNA elements carrying the appropriate signals, and thus we speculate that the range of DNA transfer mediated by filamentous phages may be considerably more than currently appreciated. In this regard, it is also remarkable that a gene (orf8) associated with a filamentous vibriophage has been linked exclusively to strains associated with the “pandemic” O3:K6 clone of Vibrio parahaemolyticus (22, 32).
Little is known about the expression of TCP in the aquatic habitats of V. cholerae, but it is reasonable to assume that it would be low compared to that in the intraintestinal environment. On the other hand, MSHA pili have been shown to be involved in adherence of V. cholerae to zooplankton (4). Therefore, MSHA is likely to be expressed more in the natural aquatic habitat of V. cholerae.
The consequences of acquisition of genes by horizontal transfer is only beginning to be understood in molecular detail. The acquisition of virulence genes and pathogenicity islands contributes to the improved evolutionary fitness of V. cholerae as a pathogen and this, in turn, may promote its more efficient seeding of the environment and establishment of endemicity. In the present study, we show that phages involved in horizontal gene transfer may be diverse in terms of their evolutionary lineage and mechanism of gene transfer but can share common receptors. Moreover, lack of homoimmunity for such phages as KSF-1Φ can allow superinfection of the same host by more than one KSF-1-derived phage particle carrying different DNA elements, which could accumulate and result in quantum leaps in the evolution of V. cholerae. Clearly, further studies are required to understand how vibriophages interact with V. cholerae to promote this organism's acquisition of the critical genes which alter its virulence or adaptation to its environmental niche.
Acknowledgments
This research was funded in part by NIH Research Grant GM068851 and by Ellison Medical Foundation Grant ID-T-0007-01, under subagreements between the Harvard Medical School and the International Centre for Diarrheal Disease Research, Bangladesh (ICDDR,B). The ICDDR,B is supported by countries and agencies which share its concern for the health problems of developing countries. Current donors providing unrestricted support include the aid agencies of the governments of Australia, Bangladesh, Belgium, Canada, Japan, Kingdom of Saudi Arabia, The Netherlands, Sweden, Sri Lanka, Switzerland, and the United States of America.
We thank A.-M. Svennerholm, University of Göteborg, Sweden, for the monoclonal antibody against MSHA pilus and Afjal Hossain for secretarial assistance.
REFERENCES
- 1.Campos, J., E. Martinez, E. Suzarte, B. L. Rodríguez, K. Marrero, Y. Silva, T. Ledón, R. del Sol, R. Fando. 2003. VGJφ, a novel filamentous phage of Vibrio cholerae, integrates into the same chromosomal site as CTXφ. J. Bacteriol. 185:5685-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campos, J., E. Martínez, K. Marrero, Y. Silva, B. L. Rodríguez, E. Suzarte, T. Ledón, and R. Fando. 2003. Novel type of specialized transduction for CTXφ or its satellite phage RS1 mediated by filamentous phage VGJφ in Vibrio cholerae. J. Bacteriol. 185:7231-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. H. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiavelli, D. A., J. W. Marsh, and R. K. Taylor. 2001. The mannose sensitive hemagglutinin of Vibrio cholerae promotes adherence to zooplankton. Appl. Environ. Microbiol. 67:3220-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis, B. M., and M. K. Waldor. 2000. CTXΦ contains a hybrid genome derived from tandemly integrated elements. Proc. Natl. Acad. Sci. USA 97:8572-8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis, B. M., H. H. Kimsey, A. V. Kane, and M. K. Waldor. 2002. A satellite phage-encoded antirepressor induces repressor aggregation and cholera toxin gene transfer. EMBO J. 21:4240-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis, B. M., and M. K. Waldor. 2003. Filamentous phages linked to virulence of Vibrio cholerae. Curr. Opin. Microbiol. 6:35-42. [DOI] [PubMed] [Google Scholar]
- 8.Ehara, M., S. Shimodori, F. Kojima, Y. Ichinose, T. Hirayama, M. J. Albert, K. Supawat, Y. Honma, M. Iwanaga, and K. Amako. 1997. Characterization of filamentous phages of Vibrio cholerae O139 and O1. FEMS Microbiol. Lett. 154:293-301. [DOI] [PubMed] [Google Scholar]
- 9.Faruque, S. M., Asadulghani, A. R. M. A. Alim, M. J. Albert, K. M. N. Islam, and J. J. Mekalanos. 1998. Induction of the lysogenic phage encoding cholera toxin in naturally occurring strains of toxigenic Vibrio cholerae O1 and O139. Infect. Immun. 66:3752-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faruque, S. M., Asadulghani, M. N. Saha, A. R. M. A. Alim, M. J. Albert, K. M. N. Islam, and J. J. Mekalanos. 1998. Analysis of clinical and environmental strains of nontoxigenic Vibrio cholerae for susceptibility to CTXΦ: molecular basis for the origination of new strains with epidemic potential. Infect. Immun. 66:5819-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faruque, S. M., M. M. Rahman, Asadulghani, K. M. N. Islam, and J. J. Mekalanos. 1999. Lysogenic conversion of environmental Vibrio mimicus strains by CTXΦ. Infect. Immun. 67:5723-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faruque, S. M., Asadulghani, M. Kamruzzaman, R. K. Nandi, A. N. Ghosh, G. B. Nair, J. J. Mekalanos, and D. A. Sack. 2002. RS1 element of Vibrio cholerae can propagate horizontally as a filamentous phage exploiting the morphogenesis genes of CTXΦ. Infect. Immun. 70:163-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faruque, S. M., M. Kamruzzaman, Asadulghani, D. A. Sack, J. J. Mekalanos, and G. B. Nair. 2003. CTX phage-independent production of RS1 satellite phage by Vibrio cholerae. Proc. Natl. Acad. Sci. USA 100:1280-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faruque, S. M., and J. J. Mekalanos. 2003. Pathogenicity islands and phages in V. cholerae evolution. Trends Microbiol. 11:505-510. [DOI] [PubMed] [Google Scholar]
- 15.Faruque, S. M., N. Chowdhury, M. Kamruzzaman, M. Dziejman, M. H. Rahman, D. A. Sack, G. B. Nair, and J. J. Mekalanos. 2004. Genetic diversity and virulence potential of environmental Vibrio cholerae population in a cholera-endemic area. Proc. Natl. Acad. Sci. USA 101:2123-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feinberg, A., and B. Volgelstein. 1984. A technique for radio labelling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 137:266-267. [DOI] [PubMed] [Google Scholar]
- 17.Finkelstein, R. A., and S. Mukerjee. 1963. Hemagglutination: a rapid method for differentiating Vibrio cholerae and El Tor vibrios. Proc. Soc. Exp. Biol. Med. 112:355-359. [Google Scholar]
- 18.Guidolin, A., and P. A. Manning. 1987. Genetics of Vibrio cholerae and its bacteriophages. Microbiol. Rev. 51:285-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishman, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Ventor, and C. M. Frasier. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heilpern, A. J., and M. K. Waldor. 2003. pIIICTX, a predicted CTXφ minor coat protein, can expand the host range of coliphage fd to include Vibrio cholerae. J. Bacteriol. 185:1037-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill, D. F., N. J. Short, N. R. Perharm, and G. B. Petersen. 1991. DNA sequence of the filamentous bacteriophage Pf1. J. Mol. Biol. 218:349-364. [DOI] [PubMed] [Google Scholar]
- 22.Iida, T., A. Hattori, K. Tagomori, H. Nasu, R. Naim, and T. Honda. 2001. Filamentous phage associated with recent pandemic strains of Vibrio parahaemolyticus. Emerg. Infect. Dis. 7:477-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikema, M., and Y. Honma. 1998. A novel filamentous phage, fs-2, of Vibrio cholerae O139. Microbiology 144:1901-1906. [DOI] [PubMed] [Google Scholar]
- 24.Jouravleva, E. A., G. A. McDonald, J. W. Marsh, R. K. Taylor, M. Boesman-Finkelstein, and R. A. Finkelstein. 1998. The Vibrio cholerae mannose-sensitive hemagglutinin is the receptor for a filamentous bacteriophage from V. cholerae O139. Infect. Immun. 66:2535-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kar, S., R. K. Ghosh, A. N. Ghosh, and A. Ghosh. 1996. Integration of the DNA of a novel filamentous bacteriophage VSK from Vibrio cholerae 0139 into the host chromosomal DNA. FEMS Microbiol. Lett. 145:17-22. [DOI] [PubMed] [Google Scholar]
- 26.Keasler, S. P., and R. H. Hall. 1993. Detection and biotyping Vibrio cholerae O1 with multiplex polymerase chain reaction. Lancet 341:1661. [DOI] [PubMed] [Google Scholar]
- 27.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 28.Lawrence, J. G., G. F. Hatful, and R. W. Hendrix. 2002. Imbroglios of viral taxonomy: genetic exchange and failings of phenetic approaches. J. Bacteriol. 184:4891-4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Marvin, D. A. 1998. Filamentous phage structure, infection and assembly. Curr. Opin. Struct. Biol. 8:150-158. [DOI] [PubMed] [Google Scholar]
- 31.Model, P., and M. Russel. 1988. Filamentous bacteriophage, p. 375-456. In R. Calendar (ed.), The bacteriophages, vol. 2. Plenum Publishing Corporation, New York, N.Y. [Google Scholar]
- 32.Nasu, H., T. Iida, T. Sugahara, Y. Yamaichi, K. Park, K. Yokohama, K. Makino, H. Shinagawa, and Y. Honda. 2000. A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J. Clin. Microbiol. 38:2156-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osek, J., A.-M. Svennerholm, and J. Holmgren. 1992. Protection against Vibrio cholerae El Tor infection by specific antibodies against mannose-binding hemagglutinin pili. Infect. Immun. 60:4961-4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rivera, I. N. G., J. Chun, A. Huq, R. B. Sack, and R. R. Colwell. 2001. Genotypes associated with virulence in environmental isolates of Vibrio cholerae. Appl. Environ. Microbiol. 67:2421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russel, M., N. A. Linderoth, and A. Sali. 1997. Filamentous phage assembly: variation on a protein export theme. Gene 192:23-32. [DOI] [PubMed] [Google Scholar]
- 36.Takeya, K. 1974. Lysogeny in El Tor vibrios and prophage typing, p. 74-83. In D. Barua and W. Burrows (ed.), Cholera. The W. B. Saunders Co., Philadelphia, Pa.
- 37.Taylor, R., C. Shaw, K. Peterson, P. Spears, and J. Mekalanos. 1988. Safe, live Vibrio cholerae vaccines? Vaccine 6:151-154. [DOI] [PubMed] [Google Scholar]
- 38.Thelin, K. H., and R. K. Taylor. 1996. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect. Immun. 64:2853-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waldor, M. K., E. J. Rubin, D. N. Gregory, H. H. Kimsey, and J. J. Makalanos. 1997. Regulation, replication and integration functions of the Vibrio cholerae CTXφ are encoded by region RS2. Mol. Microbiol. 24:917-926. [DOI] [PubMed] [Google Scholar]
- 40.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous bacteriophage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 41.Wang, P. W., L. Chu, and D. S. Guttman. 2004. Complete sequence and evolutionary genomic analysis of the Pseudomonas aeruginosa transposable bacteriophage D3112. J. Bacteriol. 186:400-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.World Health Organization. 1974. World Health Organization guidelines for the laboratory diagnosis of cholera. Bacterial Disease Unit, World Health Organization, Geneva, Switzerland.