Abstract
A collagen-degrading thermophile, Geobacillus collagenovorans MO-1, was found to produce two metallopeptidases that hydrolyze the synthetic substrate 4-phenylazobenzyloxycarbonyl-Pro-Leu-Gly-Pro-d-Arg (Pz-PLGPR), containing the collagen-specific sequence -Gly-Pro-X-. The peptidases, named Pz peptidases A and B, were purified to homogeneity and confirmed to hydrolyze collagen-derived oligopeptides but not collagen itself, indicating that Pz peptidases A and B contribute to collagen degradation in collaboration with a collagenolytic protease in G. collagenovorans MO-1. There were many similarities between Pz peptidases A and B in their catalytic properties; however, they had different molecular masses and shared no antigenic groups against the respective antibodies. Their primary structures clarified from the cloned genes showed lower identity (22%). From homology analysis for proteolytic enzymes in the database, the two Pz peptidases belong to the M3B family. In addition, Pz peptidases A and B shared high identities of over 70% with unassigned peptidases and oligopeptidase F-like peptidases of the M3B family, respectively. Those homologue proteins are putative in the genome database but form two distinct segments, including Pz peptidases A and B, in the phylogenic tree. Mammalian thimet oligopeptidases, which were previously thought to participate in collagen degradation and share catalytic identities with Pz peptidases, were found to have lower identities in the overall primary sequence with Pz peptidases A and B but a significant resemblance in the vicinity of the catalytic site.
Collagens are the most abundant proteins occurring in all animal and some insect cells as an extracellular matrix component (13, 23, 33). Their degradation by microorganisms has prospective possibilities leading to development with applications in the field of antibiotics against pathogenic microorganisms and drugs related to collagen breakdown. This is because collagens are crucial for various cellular events, while only a limited number of proteases present in animal cells as well as natural habitats participate in collagen degradation. The synthetic substrate 4-phenylazobenzyloxycarbonyl-Pro-Leu-Gly-Pro-d-Arg (Pz-PLGPR), which contains the collagen-specific sequence -Gly-Pro-X-, has been used as a substrate in the simple assay method for bacterial collagenases (43). Bacterial collagenases cleave this substrate at the Leu-Gly bond, and the more hydrophobic product, Pz-Pro-Leu, extracted in ethyl ether, is spectrophotometrically assayed. In spite of the fact that most bacterial collagenases hydrolyze collagens and Pz-PLGPR, there are some distinct peptidases, so called Pz peptidases, that hydrolyze Pz-PLGPR but do not act on protein substrate collagens (41). Pz peptide-hydrolyzing activity is considered to be essential for collagen biodegradation in collagen-degrading microorganisms, in collaboration with their collagenolytic proteases. Thus far, one type of Pz peptidases had been purified from Bacillus licheniformis (2), and its gene cloning was later reported (1). However, to date, little is known about the diversity of other bacterial Pz peptidases and their real nature with regard to collagen degradation and molecular functions. Although similar bacterial metallooligopeptidases acting on neuropeptides have been found to be inactive on Pz-PLGPR (10, 26-28, 38), Pz peptidases are distinct from many bacterial metallooligopeptidases in substrate specificity and must play a critical role for cell growth in a collagen-containing medium.
On the other hand, Pz peptidases have been studied independently in all forms, including those in eukaryotes, from humans to yeasts (4, 6, 9, 19, 37). Early studies on the Pz peptidases were inspired by the idea that the enzyme might participate in the physiological turnover of collagen. Mammalian Pz peptidases, which were renamed as thimet oligopeptidases (TOPs) thereafter, were recognized as different from collagenases in spite of having the same hydrolyzing site (6). TOPs are primarily cytosolic and thiol-sensitive metallopeptidases and are limited to action on oligopeptides, in particular, on the neuropeptides of 3 to 17 amino acid residues (11, 37). Judging from a large volume of accumulated data, the physiological role of TOPs is now presumed to be as neuropeptide-metabolizing enzymes, which extinguish the signaling action of neuropeptides (39). However, the enzymological characteristics are quite similar to those of bacterial Pz peptidases, and some bacterial Pz peptidases predicted from genome research have been assigned as TOPs (e.g., accession number NP_764245 in Staphylococcus epidermidis ATCC 12228). From the relevance of bradykinin to bacterial infection, the activation of the bradikinin-generating cascade is an intriguing event, and thus bradykinin breakdown, which is one of the functions of Pz peptidases, is of interest (25). It is of further importance to examine the bacterial Pz peptidases and compare them with eukaryotic TOP. We have isolated a thermophilic Bacillus sp. strain, MO-1 (later renamed as Geobacillus collagenovorans) from soil; this strain can degrade collagen in the neutral pH range (32). In the process of purifying the collagenolytic protease with a large molecular mass, strain MO-1 was found to produce the two types of Pz peptidase activities that were separated from the collagenolytic protease activity. Here, we investigated the two Pz peptidases (Pz peptidases A and B) produced by G. collagenovorans MO-1, compared them with other bacterial oligopeptidases and mammalian TOPs on the basis of enzymological and genetic data, and discussed their molecular structures and divergence.
MATERIALS AND METHODS
Bacterial strain and culture.
G. collagenovorans MO-1, which was used in this study, was described in a previous report (32). This bacterium was spore forming and gram positive, and its 16S rRNA sequence was 98% identical to Geobacillus caldoxylosilyticus DSM12041T (14). Since the DNA-DNA reassociation analysis showed 63% reassociation between strains MO-1 and DSM12041 and strain MO-1 hardly grew on several sugars (arabinose, galactose, sucrose, sorbitol, fructose, mannitol, maltose, raffinose), strain MO-1 was named Geobacillus collagenovorans. Strain MO-1 was grown in L broth (1% tryptone, 0.5% yeast extract, 0.5% NaCl, pH 7.2) at 65°C with reciprocal shaking (95 rpm). For gene cloning, the Escherichia coli DH5α strain was used. All recombinant E. coli strains were cultivated aerobically in L broth appropriately supplemented with ampicillin (50 μg/ml).
Purification of Pz peptidases A and B.
All operations were performed at 4°C unless otherwise stated.
(i) Pz-peptidase A. Step 1.
Cells harvested from the culture (total 3 liters, 8 h of cultivation) were resuspended in 300 ml of buffer I (50 mM Tris-HCl, pH 7.5) after washing with saline and then disintegrated by sonication (5-min pulse).
Step 2.
The cell extract obtained by centrifugation at 12,000 × g for 20 min was fractionated with solid (NH4)2SO4. The precipitate from 50 to 95% (NH4)2SO4 saturation [104.1 g (NH4)2SO4 was added after excluding the precipitate formed with 93.9 g (NH4)2SO4] was recovered by centrifugation, dissolved in buffer I, and dialyzed against the same buffer at 4°C.
Step 3.
The dialysate was applied to a DEAE-cellulose column (4.4 by 11.8 cm) equilibrated with buffer I and then eluted with a linear gradient from 0 to 0.5 M NaCl in buffer I (total, 1.2 liter). The active fractions (2.9 ml each) were pooled, concentrated by ultrafiltration through an Amicon PM-10 membrane (Millipore, Bedford, MA), and dialyzed against 30% (NH4)2SO4-saturated buffer I.
Step 4.
The dialysate was applied to a butyl-TOYOPEARL 650S column (7.0 by 11.0 cm) equilibrated with 30% (NH4)2SO4-saturated buffer I. Elution was performed with a linear gradient of 30 to 0% saturated (NH4)2SO4 (total, 4 liters). Two active fractions (5.0 ml each) were observed, and the major one was pooled and concentrated to about 4 ml by ultrafiltration, as indicated above.
Step 5.
The concentrate was put on a Sephadex G-200 column (1.5 by 91 cm) equilibrated with buffer I. Active fractions (1.2 ml each) were pooled and concentrated by ultrafiltration.
Step 6.
The concentrate was put on the same Sephadex G-200 column (1.5 by 91 cm) again, and the active fractions were pooled.
Step 7.
The solution was reduced in volume by ultrafiltration and put on a DEAE-Sepharose CL-6B column (1.9 by 84 cm) equilibrated with buffer I. The column was eluted with 1 liter of a linear gradient from 0 to 1.0 M NaCl. The active fractions (3.1 ml each) were pooled and concentrated by ultrafiltration.
Step 8.
The concentrate was applied to a Sephadex G-200 column as indicated above, and the active fractions (1.2 ml each) were pooled and concentrated to 1.0 ml by ultrafiltration using the Centricon YM-10 (Millipore, Bedford, MA).
(ii) Pz peptidase B. Step 1.
Cells harvested from the culture (total 3 liters, 4.5 h of cultivation) were used for purification.
Step 2.
The fractionation with ammonium sulfate was carried out as for Pz peptidase A. The precipitate from 50 to 90% (NH4)2SO4 saturation was obtained by centrifugation, dissolved in buffer I, and dialyzed against this buffer at 4°C.
Step 3.
The dialysate was applied to a DEAE-cellulose column (7.0 by 10.3 cm), equilibrated with buffer I, and then eluted with a linear gradient from 0 to 0.5 M NaCl in buffer I (total, 1 liter). The active fractions (6.1 ml each) were pooled and concentrated to 191 ml by ultrafiltration through the Amicon PM-10 membrane.
Step 4.
The concentrate was put on a DEAE-Sepharose CL-6B column (1.9 by 84 cm) equilibrated with buffer I. The column was eluted with 1 liter of a linear gradient from 0 to 1.0 M NaCl. Two peaks of active fractions (4.2 ml each) were observed, and the minor one was pooled and concentrated by ultrafiltration. The concentrate was dialyzed against a 20 mM potassium phosphate buffer (pH 7.5).
Step 5.
The dialysate was applied to a DEAE-Sepharose CL-6B column (1.9 by 84 cm) equilibrated with the same buffer. Elution was performed with a linear gradient from 0 to 0.5 M NaCl in the buffer. The active fractions (4.2 ml each) were pooled and concentrated by ultrafiltration.
Step 6.
The concentrate was put on a Sephadex G-200 column (1.5 by 91 cm) equilibrated with buffer I. Active fractions (1.2 ml each) were pooled, concentrated, and dialyzed against 30% (NH4)2SO4-saturated buffer I.
Step 7.
The dialysate was put on a butyl-TOYOPEARL 650S column (2.0 by 12.7 cm) equilibrated with the dialysis buffer. Elution was performed with a linear gradient of 30 to 0% saturated (NH4)2SO4 (total, 4 liters). Active fractions (2.1 ml each) were pooled and concentrated by ultrafiltration. The concentrate was dialyzed against 15% (NH4)2SO4-saturated buffer I.
Step 8.
The dialysate was applied to a phenyl-Sepharose CL-6B column (2.0 by 12.7 cm) equilibrated with the dialysis buffer. Elution was performed with a linear gradient of 15% (NH4)2SO4-saturated buffer I and 70% (vol/vol) ethylene glycol in buffer I. Active fractions (2.1 ml each) were pooled and dialyzed against 45% (NH4)2SO4-saturated buffer I.
Step 9.
The dialysate was applied to an ethyl-agarose column (7.0 by 11.0 cm) equilibrated with the dialysis buffer. The column was eluted with a linear gradient of 45% to 0% (NH4)2SO4-saturated buffer I. Active fractions (1.1 ml each) were pooled and concentrated by ultrafiltration.
Step 10.
The concentrate was applied to a Sephacryl S-300 column (2.0 by 95 cm) equilibrated with buffer I. The column was eluted with buffer I, and active fractions (1.75 ml each) were pooled and concentrated as a final sample.
Enzyme assay.
Pz peptidase activity was measured basically by following the method described previously (32). The reaction mixture (100 μl), which included 50 μg Pz-PLGPR peptide and appropriate enzyme (5 to 10 μl) in 50 mM HEPES-NaOH (pH 7.6 for Pz peptidase A and pH 8.4 for Pz peptidase B), was incubated at 60°C. At an appropriate interval (e.g., 0, 5, and 10 min), 100 μl 1.25% (wt/vol) citrate solution was added to the mixture to stop the reaction. Then 1.0 ml ethyl acetate was added and the mixture was vortexed for 10 s. After centrifugation at 10,000 × g for 10 min, the upper phase (ethyl acetate) was recovered, and its absorbance was determined at 320 nm. The molar extinction coefficient (ɛ = 21,000) for Pz-Pro-Leu in the phase was used for the calculation of the activity. One unit of enzyme activity was defined as the amount of enzyme that can hydrolyze 1 μmol Pz peptide in a minute. The hydrolysis of other peptides, hypertensin, bradykinin, substance P, and neurotensin, was examined in 100 μl of a reaction mixture, as for of Pz peptidase, and confirmed by high-performance liquid chromatography using a reverse-phase chromatography (WS-II5C18AR column; Wakosil, Osaka, Japan; elution, 0 to 95% acetonitrile gradient in water/0.1% [vol/vol] trifluoroacetic acid); after that, peptide hydrolysis products were analyzed by time-of-flight mass spectrometry (Applied Biosystems, Voyager DE STR-D1) to determine the cleavage sites. Peptides with the repeat tripeptide sequence of Gly-Pro-Leu (collagen-specific sequences) were synthesized using the peptide synthesizer PSSM-8 (Shimadzu, Kyoto, Japan). Protein was determined by the method of Lowry with bovine serum albumin as the standard (24). Protease activities against collagen, elastin, gelatin, keratin, and casein were determined by the modified ninhydrin method described before (12, 32). The reaction temperature was 60°C.
Enzyme inhibition by chemical agents and metal ions.
The reaction mixtures containing the inhibitors and enzyme were preincubated for 30 min, and then the substrate was added to the mixture, except for dithiothreitol.
Polyclonal antibody preparation and Western blotting analysis.
To obtain polyclonal antibodies against Pz peptidases A and B, 6-week-old female rabbits were immunized with the purified enzyme samples (100 μg per rabbit) emulsified with Titer Max Gold adjuvant (0.2 ml; CytRx Corp., Norcross, GA). The procedures for gel electrophoresis, semidry electrotransfer, blocking of membrane with skim milk, hybridization with polyclonal antibodies, washing, and visualization with diaminobenzidine followed the methods described previously (22, 29).
Protein sequencing.
The N-terminal and internal protein sequence of the purified enzyme was determined by automated Edman degradation using a type PPSQ-21 protein sequencer (Shimadzu, Kyoto Japan). For determining the internal amino acid sequence, a purified protein sample (5 to 10 μg) was mixed with a 4-μl aliquot of a 100-μg/ml solution of Staphylococcus aureus V8 protease in 30 mM Tris-HCl (pH 6.8) containing 1% (wt/vol) sodium dodecyl sulfate (SDS), 5 mM mercaptoethanol, 10% (wt/vol) sucrose, and 0.001% (wt/vol) bromophenol blue. Immediately following the mixing, the electrophoresis run was begun. Digestion of the proteins by V8 protease was allowed to proceed within the gel by briefly stopping electrophoresis for 15 min once the sample had migrated about halfway through the 3% polyacrylamide stacking gel, at which time the components appeared to be maximally concentrated. After running the sample through the 10% polyacrylamide running gel, partially digested proteins were recovered by transfer onto a polyvinylidene difluoride membrane by semidry electrotransfer and subjected to the protein sequencer.
Gene cloning and analysis.
The chromosomal DNA of G. collagenovorans MO-1 was isolated from the cells cultivated in L broth for 8 h and digested with HindIII, SphI, or EcoRI to construct libraries with the vector pUC119 and the host Escherichia coli DH5α, following the method described previously (42). Two pairs of PCR primers were designed from the N-terminal and internal amino acid sequences of Pz peptidases A (primer AF, CGCGGATCCATGAAATTYWSBGAATTTCGCTATGAA; primer AR, RTTGGATCCRTTTTCRTANACRAARTGTTGRAATTC [B = C, G, or T; R = A or G; S = C or G; W = A or T; Y = C or T]) and B (primer BF, CGCGGATCCATGGAAGCAAAACAAACNAAAAAA; primer BR, YSWGGATCCTTGCATNACTTCNGCNGCTTGNGCNAG) to amplify the DNA probe for colony hybridization. A 1.3-kb probe for Pz peptidase A and a 0.5-kb probe for Pz peptidase B were prepared by PCR (30 cycles of denaturation at 94°C for 15 s, annealing at 50°C for 30 s, and extension at 68°C for 2 min), using KOD plus DNA polymerase (TOYOBO, Osaka, Japan). The probes were labeled using the digoxigenin (DIG) labeling kit (Roche, Mannheim, Germany). Colonies carrying a hybrid plasmid containing the desired DNA fragment were screened using the DIG luminescent detection kit (Roche) according to the supplier's specifications. Both strands of the cloned fragments were sequenced using PRISM dye primer sequencing kits on a 310NT DNA sequencer (Applied Biosystems, Foster City, CA).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in the DDBJ/EMBL/GenBank nucleotide sequence databases with accession numbers AB191721 for the Pz peptidase A gene and AB191722 for the Pz peptidase B gene.
RESULTS
Purification.
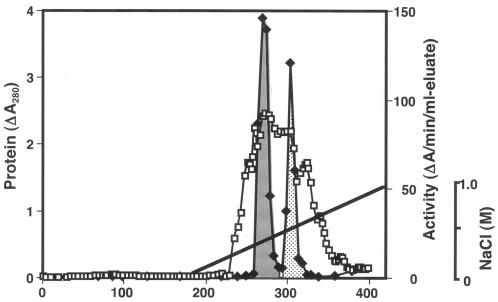
Pz peptidases A and B were purified to homogeneity from cells (Table 1). The Pz peptidase activity appeared mainly inside the cells but not in the culture supernatant in the growing phase (data not shown). At the beginning, the G. collagenovorans MO-1 strain was thought to produce one species of Pz peptidase. However, we found that Pz peptidase activity was divided into two peaks in the elution profile of the butyl-TOYOPEARL 650S column chromatography (step 4). Two active peaks were more clearly observed in the elution profile of the DEAE-Sepharose CL-6B column chromatography for Pz peptidase B purification (Fig. 1). Pz peptidases A and B were finally purified by the combination of effective procedures, even though there might be more efficient ways to isolate these peptidases. In spite of the large total activity in the cell extract, the significant loss in activity after butyl-TOYOPEARL column chromatography resulted in rather low yields for the final samples of both Pz peptidases A and B.
TABLE 1.
Summary of purification of Pz peptidases A and B
| Purification step | Total protein (mg) | Sp act (U/mg protein) | Fold | Total activity (U) | Yield (%) |
|---|---|---|---|---|---|
| Pz peptidase A | |||||
| 1. Cell-free extract | 7,370 | 0.045 | 1 | 332 | 100 |
| 2. Ammonium sulfate fractionation | 2,120 | 0.101 | 2.24 | 214 | 64.5 |
| 3. DEAE cellulose | 1,150 | 0.161 | 3.58 | 185 | 55.7 |
| 4. Butyl-TOYOPEARL 650S | 91.5 | 0.798 | 17.7 | 73.0 | 22.0 |
| 5. 1st Sephadex G-200 | 11.8 | 4.91 | 109 | 57.9 | 17.4 |
| 6. 2nd Sephadex G-200 | 4.42 | 9.76 | 217 | 43.1 | 13.0 |
| 7. DEAE Sepharose CL-6B | 1.66 | 18.4 | 409 | 30.5 | 9.2 |
| 8. 3rd Sephadex G-200 | 0.92 | 24.7 | 549 | 22.7 | 6.8 |
| Pz peptidase B | |||||
| 1. Cell-free extract | 5,890 | 0.0477 | 1 | 281 | 100 |
| 2. (NH4)2SO4 fractionation | 3,230 | 0.0845 | 1.77 | 273 | 97.2 |
| 3. DEAE cellulose | 1,530 | 0.144 | 3.01 | 220 | 78.3 |
| 4. DEAE Sepharose CL-6B | 225 | 0.267 | 5.60 | 60.0 | 21.3 |
| 5. 2nd DEAE Sepharose CL-6B | 94.9 | 0.363 | 7.09 | 34.5 | 12.3 |
| 6. Sephadex G-200 | 67.9 | 0.481 | 10.1 | 32.7 | 11.6 |
| 7. Butyl-TOYOPEARL 650S | 29.3 | 0.345 | 7.23 | 10.1 | 3.6 |
| 8. Phenyl Sepharose CL-4B | 11.2 | 0.730 | 15.3 | 8.18 | 2.9 |
| 9. Ethyl agarose | 5.93 | 1.45 | 30.4 | 8.60 | 3.1 |
| 10. Sephacryl S-300 | 2.35 | 2.32 | 48.6 | 5.45 | 1.9 |
FIG. 1.
Elution patterns of Pz peptidases A and B in DEAE-Sepharose CL-6B column chromatography for Pz peptidase B. The fractions of Pz peptidases A and B are depicted in dark gray and light gray, respectively. Protein (□) and enzyme activity (⧫) were analyzed. The gradient concentrations of NaCl for elution are indicated by solid lines.
Antigenicity, molecular mass, and amino acid sequence.
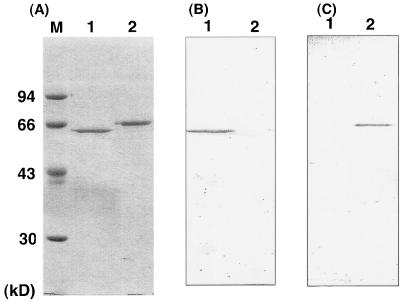
The difference between Pz peptidases A and B in antigenicity was investigated using the polyclonal antibodies specific to Pz peptidases A and B, respectively (Fig. 2). The two Pz peptidases shared no antigenic groups against the respective antibodies, indicating that they should be translated from the distinct genes in a cell.
FIG. 2.
(A) SDS-PAGE pattern for purified Pz peptidases A and B. (B, C) Western blotting analysis of purified Pz peptidases A and B. Lanes 1 and 2 always include purified Pz peptidases A and B, respectively. Lane M contains molecular mass standards. In panel B, the antibody raised against Pz peptidase A was used, while the one against Pz peptidase B was used in panel C.
The Mr of Pz peptidases A and B was estimated as 62,000 and 69,000, respectively, by SDS-polyacrylamide gel electrophoresis (PAGE). In contrast, Pz peptidase A behaved as a monomer of Mr 54,000 on gel filtration with Sephadex G-200, whereas Pz peptidase B behaved as an active Mr 250,000 globular protein with Sephacryl S-300 (data not shown). Pz peptidase B is most likely to consist of a tetramer.
The 15 N-terminal amino acid sequences of the purified enzymes were determined to be Met-Lys-Phe-Ser-Glu-Phe-Arg-Tyr-Glu-Arg-Pro-Asn-Ile-Glu-Lys for Pz peptidase A and Met-Glu-Ala-Lys-Gln-Thr-Lys-Lys-Ser-Leu-Pro-Leu-Leu-Ser-Glu for Pz peptidase B. The sequences did not meet any bacterial signal sequence for secretion. Protein fragments partially digested with V8 protease for the internal amino acid sequence were recovered as an Mr 16,000 band for Pz peptidase A and as an Mr 19,000 band for Pz peptidase B on a polyvinylidene difluoride membrane. The N-terminal sequences of these internal peptides were determined to be Phe-Gln-His-Phe-Val-Tyr-Glu-Asn-Pro-Asn-Ala-Thr-Pro-Ala-Glu for Pz peptidase A and Ala-Leu-Leu-Ala-Gln-Ala-Ala-Glu-Val-Met-Gln-Ser-Ser-Ser for Pz peptidase B.
Temperature and pH dependence of Pz peptidase activity.
Pz peptidases A and B were most active at 65°C and 70°C, respectively. At 37°C, Pz peptidases A and B showed 29% and 36% activity relative to the optimum, respectively. Both enzymes were stable up to 60°C and 70°C for 30 min at their optimum pH, respectively. A half-life of 30 min was found at 70°C for Pz peptidase A and at 75°C for Pz peptidase B. Pz peptidase B is slightly more thermophilic than Pz peptidase A. Pz peptidase A was active between pH 6.5 and 8.3, with an optimum at pH 7.6, while Pz peptidase B was active between pH 7.5 and 9.2, with an optimum at pH 8.4. The pH profile of Pz peptidase B was slightly shifted to more alkaline than that of Pz peptidase A, but the width of the active pH range was similar.
Effect of various chemical agents.
Pz peptidases A and B were 100% inhibited by EDTA and 1,10-phenanthroline, both of which are inhibitors of metalloenzymes (Table 2). The activity was not influenced by the serine protease inhibitors phenylmethylsulfonyl fluoride and diisopropyl fluorophosphate even after incubation for 30 min at room temperature. The enzymes were partly inhibited (67% for Pz peptidase A, 31% for Pz peptidase B) by the thiol reagent N-ethylmaleimide. Dithiothreitol also affected the hydrolyzing activity, but the degree of inhibition was different from that of N-ethylmaleimide. Although the metal agents CaCl2 and MgCl2 hardly affected the activity, FeCl2, ZnCl2, NiCl2, and CuCl2 completely inhibited the activities of both enzymes at 2 mM concentration. Metallocarboxypeptidases containing a zinc atom are inhibited by a millimolar excess of zinc together with other exo- and endometalloproteases, and this characteristics has been often exhibited for metallopeptidases due to a second zinc, observed to bind to the enzyme active site, establishing a distorted tetrahedrally coordinated complex which involves the third ligand glutamate residue (the general base for catalysis), a water molecule, a chloride ion, and a hydroxide ion (17). In contrast, a lower concentration of CoCl2 (0.01 mM) and MnCl2 (0.5 mM) activated Pz peptidases A and B by 50% to 200%, respectively. However, the addition of the metal agents at 2 mM resulted in the inhibition of activity for both enzymes. The observation that the two enzymes were remarkably activated by CoCl2 and MnCl2 can be attributed to the same reason in the case of thermolysin, where the electrostatic environment of thermolysin with cobalt substituted is more stable than that of the native enzyme (21). The susceptibilities of Pz peptidases A and B to various agents were quite similar but differed in extent.
TABLE 2.
Inhibitor sensitivity and metal ion dependence of Pz peptidases A and B
| Agenta | Concn (mM) | Remaining activity (%)
|
|
|---|---|---|---|
| Pz peptidase A | Pz peptidase B | ||
| Control | 100 | 100 | |
| EDTA | 2 | 0 | 0 |
| 1,10-Phenanthroline | 2 | 0 | 0 |
| PMSF | 2 | 106 | 97 |
| DFP | 2 | 122 | 81 |
| N-Ethylmaleimide | 2 | 33 | 69 |
| Dithiothreitolb | 0.1 | 88 | 85 |
| 1 | 68 | 13 | |
| CaCl2 | 2 | 86 | 119 |
| MgCl2 | 2 | 81 | 108 |
| FeCl2 | 2 | 0 | 8 |
| ZnCl2 | 2 | 0 | 0 |
| NiCl2 | 2 | 0 | 0 |
| CuCl2 | 2 | 0 | 0 |
| CoCl2 | 0.01 | 148 | 336 |
| 2 | 8 | 12 | |
| MnCl2 | 0.5 | 217 | 321 |
| 2 | 87 | 84 | |
PMSF, phenylmethylsulfonyl fluoride; DFP, diisopropyl fluorophosphate.
Dithiothreitol was preincubated with the enzymes overnight in the concentration indicated, and due to significant noise by dithiothreitol, the enzyme activity was assayed with the sample diluted 200-fold.
Substrate specificity.
Pz peptidases A and B showed no aminopeptidase activity on X-p- nitroanilide (X:Pro, Gly, Leu, Arg), succinyl-Arg-p-nitroanilide, Gly-Pro-p-nitroanilide, carboxybenzyloxy-Gly-Pro-p-nitroanilide, succinyl-Ala-Ala-Ala-p-nitroanilide, and succinyl-Ala-Ala-Pro-Phe-p-nitroanilide. In addition, the enzymes cleaved none of the protein substrates, such as collagen, gelatin, elastin, keratin, and casein. Table 3 shows the hydrolytic activity of Pz peptidases A and B on a selection of collagen-derived substrate peptides and hormone peptides. An alternative synthetic collagenase substrate, furylacryloyl-Leu-Gly-Pro-Ala (FA-LGPA), was the shortest among the peptides hydrolyzed by both enzymes (40), while neurotensin (13 amino acid residues) was the longest. The cleavage sites of the enzymes were the same for shorter peptide substrates, such as FA-LGPA and Pz-PLGPR, but different for longer hormone peptides, such as substance P and neurotensin (Table 3). Pz peptidase A had two hydrolyzing sites on substance P, whereas Pz peptidase B had one site. Although the single cleavage site of Pz peptidase B is in common with one of two sites of TOPs, two sites of Pz peptidase A were distinct from those of Pz peptidase B and TOPs (37). However, Pz peptidases A and B shared the same digestion sites for neurotensin and bradykinin with TOPs in addition to one extra cleavage site for Pz peptidase A. The cleavage pattern of these substrates by Pz peptidase B was more similar to that of TOPs than that by Pz peptidase A. Furthermore, a repeat of tripeptide Gly-Pro-Leu as a collagen-specific sequence was examined for hydrolysis (Table 3). In spite of the fact that the monomer of tripeptide [Gly-Pro-Leu] was not hydrolyzed, the dimer [Gly-Pro-Leu]2, trimer [Gly-Pro-Leu]3, and tetramer [Gly-Pro-Leu]4 peptides were hydrolyzed by both Pz peptidases between Leu-Gly, suggesting that the enzymes should have a significant role in collagen degradation. The pentamer [Gly-Pro-Leu]5 and longer peptides were too insoluble in water to perform the assay. From these results, Pz peptidases A and B are endopeptidases and fail to hydrolyze less than tetrapeptides. Although the enzymes preferentially recognize and hydrolyze the collagen-specific sequence Gly-Pro-X (X = Leu here), there are hardly strict rules to predict the bonds cleaved by Pz peptidases A and B. This is the case with TOPs (37).
TABLE 3.
Peptide bonds hydrolyzed in collagen-derived peptide substrates and hormone peptides by Pz peptidases A and B and thimet oligopeptidase
| Substratea | Peptide bond(s) hydrolyzed
|
||
|---|---|---|---|
| Pz peptidase A | Pz peptidase B | Thimet oligopeptidaseb | |
| FA-LGPA | fsFA-L↓G-P-A | FA-L↓G-P-A | No data |
| Pz-PLGPR | Pz-PL↓G-P-R | Pz-P-L↓G-P-R | Pz-P-L↓G-P-R |
| Hypertensin | N-R-V-Y-V↓H-P-F | N-R-V-Y-V↓H-P-F | No data |
| Bradykinin | R-P-P-G-F↓S-P-F-R | R-P-P-G-F↓S-P-F-R | R-P-P-G-F↓S-P-F-R |
| Substance P | R-P-K-P-Q↓Q↓F-F-G-L-M | R-P-K-P-Q-Q-F-F↓G-L-M | R-P-K-P-Q-Q-F↓F↓G-L-M |
| Neurotensin | pE-L-Y-E-N-K-P-R↓R-P↓Y-I-L | pE-L-Y-E-N-K-P-R↓R-P-Y-I-L | pE-L-Y-E-N-K-P-R↓R-P-Y-I-L |
| [GPL]1 | No digestion | No digestion | No data |
| [GPL]2 | G-P-L↓G-P-L | G-P-L↓G-P-L | No data |
| [GPL]3 | G-P-L↓G-P-L↓G-P-L | G-P-L↓G-P-L↓G-P-L | No data |
| [GPL]4 | G-P-L↓G-P-L↓G-P-L↓G-P-L | G-P-L↓G-P-L↓G-P-L↓G-P-L | No data |
| Proteins (casein, collagen, gelatin) | No digestion | No digestion | No digestion |
FA-LGPA, furylacryloyl-Leu-Gly-Pro-Ala; Pz-PLGPR, 4-phenylazobenzyloxycarbonyl-Pro-Leu-Gly-Pro-d-Arg; GPL, Gly-Pro-Leu. The concentration of the substrates in the reaction mixture was 0.2 mM, except for Pz-PLGPR (0.64 mM [50 μg/100-μl reaction mixture], as shown in Materials and Methods).
Data were derived from those described in reference 37.
Participation of Pz peptidases A and B in collagen degradation in collaboration with a collagenolytic protease.
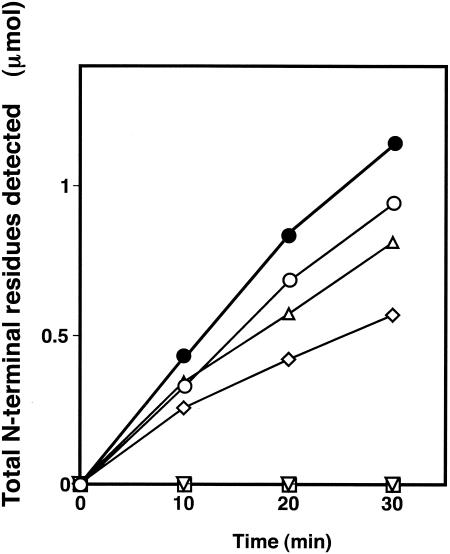
It was investigated whether Pz peptidases A and B can actually hydrolyze collagen peptides resulting from collagen degradation by a collagenolytic protease of the same strain, MO-1 (32). Figure 3 shows that the addition of purified Pz peptidases A and B to a reaction mixture containing collagen and the previously purified collagenolytic protease resulted in an accelerated degradation of collagen, which was a more efficient formation of peptides than that formed exclusively with collagen and the collagenolytic protease. Pz peptidases A and B have shown no direct degrading activity on collagen. In addition, the collagenolytic protease caused no detectable increase in the total amount of free N termini as a result of the digestion of Pz peptidases A and B in the absence of collagen (data not shown). Consequently, Pz peptidases A and B are responsible for collagen degradation with the collagenolytic protease, suggesting that the degradation by Pz peptidases A and B of collagen-derived peptides should contribute to the cell growth of G. collagenenovorans MO-1.
FIG. 3.
Participation of Pz peptidases A and B in collagen degradation. The reaction mixture, which contained 10 mg collagen (type I, bovine tendon), 1.2 μg purified collagenolytic protease from strain MO-1 (32), and purified Pz peptidase A (9.2 μg) or Pz peptidase B (30 μg) in 5 ml of a 50 mM HEPES buffer (pH 7.6 for Pz peptidase A and pH 8.4 for Pz peptidase B), was incubated with shaking at 60°C. An aliquot (300 μl) was extracted every 10 min and mixed with 0.5 M EDTA (6 μl) to stop the reaction. The increase in the number of N-terminal residues by degradation was measured following the ninhydrin method, which was described previously (12, 32). Symbols: •, Pz peptidase A, Pz peptidase B, and collagenolytic protease; ▵, Pz peptidase A and collagenolytic protease; ○, Pz peptidase B and collagenolytic protease; ◊, collagenolytic protease only; □ Pz peptidase A only; ▿, Pz peptidase B only.
Gene cloning and analysis of the nucleotide sequence.
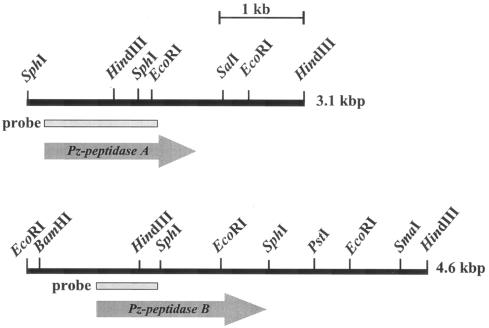
Gene cloning for Pz peptidases A and B was carried out by using the probes that were prepared by PCR with primers designed from N-terminal and internal amino acid sequences. Two independent hybrid plasmids containing the gene for Pz peptidase A were obtained from two separate libraries of the HindIII-digested and SphI-digested G. collagenovorans chromosomes, using a 1.3-kb probe. Although the DNA fragments included in those hybrid plasmids contained the partial genes for Pz peptidase A, the two fragments linked together (3.1 kb) were enough to cover the entire gene (Fig. 4). Likewise, two hybrid plasmids for the Pz peptidase B gene were screened separately from two libraries of the HindIII-digested and EcoRI-digested chromosomes with a 0.5-kb probe. The two fragments were linked together (4.6 kb) to cover the gene for Pz peptidase B (Fig. 4).
FIG. 4.
Composite showing the restriction map of the Pz peptidase A and B regions of G. collagenovorans MO-1, the probes for colony hybridization, and the deduced open reading frames.
The nucleotide sequences of the 3.1-kb DNA fragment for Pz peptidase A and of the 4.6-kb DNA fragment for Pz peptidase B were determined. Nucleotide sequences identical to the N-terminal and internal sequences of the proteins were found in both DNA fragments; they revealed an open reading frame (ORF) for Pz peptidase A of 1,692 bp, capable of coding for a protein of 564 amino acids with a putative Mr of 66,400, and an ORF for Pz peptidase B of 1,812 bp, capable of coding for a protein of 604 amino acids with a putative Mr of 70,000. These values, deduced from the nucleotide sequences of the genes for Pz peptidases A and B, are rather close to the Mrs of 62,000 and 69,000 calculated from the SDS-PAGE analysis, respectively. A potential ribosome-binding site complementary to the 3′ end of 16S rRNA was found 7 bp upstream of the start codons in both cases. In the 5′ flanking region of the genes for Pz peptidases A and B, −35 and −10 consensus sequences for the transcriptional promoter were found 53 bp (ATGATA) and 26 bp (TATGAT) upstream of the start codon for the Pz peptidase A gene and 76 bp (TTGATT) and 49 bp (TATGAA) upstream of that for the Pz peptidase B gene. There were no remarkable inverted repeats that can function for transcriptional regulation in the 5′ flanking region.
A comparison of the amino acid sequence of Pz peptidases A and B with those of other peptidases using a BLAST program equipped in the MEROPS database (http://merops.sanger.ac.uk/) revealed a high similarity to peptidases belonging to the M3 family, in particular, the M3B subfamily of proteolytic enzymes (6, 35). Pz peptidase A shows the highest identity to Bacillus cereus ATCC 10987 unassigned peptidase (protein ID AAS40381.1; identity, 71%), Bacillus anthracis strain Ames unassigned peptidase (protein ID AAP25299.1; identity, 71%), and Clostridium perfringens strain 13 unassigned peptidase (protein ID BAB82029; identity, 61%). The functions of proteins indicating such high scores in identity with Pz peptidase A are not definite yet because they are all putative peptidases derived from their genome databases. Therefore, no biochemical information on the homologous peptidases is available. On the other hand, Pz peptidase B showed the highest identity with several oligopeptidase F-like peptidases. The peptidases from B. anthracis strain Ames (protein ID AAP25167.1), B. cereus ATCC 10987 (protein ID AAS40243.1), and Bacillus subtilis 168 (protein ID CAB13011.1), which are assigned as oligopeptidase F in the genome database, showed identities of 73%, 73%, and 72% to Pz peptidase B, respectively. In addition to these putative peptidases, Pz peptidase B shared 71% identity with the Pz peptidase from B. licheniformis N22 (protein ID BAA13561.1), on which enzymological analyses had already been done (1). Although the high identity demonstrated that Pz peptidase B and B. licheniformis Pz peptidase share many similarities in function and structure, there are differences in localization, molecular weight, and thermostability (2). In contrast, much lower identity (47%) between Pz peptidase B and the most investigated oligopeptidase F, from Lactococcus lactis NCDO 763, was observed (28). The gap in identity suggests that Pz peptidase B is biochemically different from the typical oligopeptidase F. Moreover, it should be noted that there is only 22% identity between Pz peptidases A and B. Hence, Pz peptidase A is distinct from Pz peptidase B and B. licheniformis N22 Pz peptidase and is a new type of peptidase that participates in collagen degradation. In addition, TOPs having a similar function to that of Pz peptidases showed lower identity to Pz peptidases A and B. Human TOP showed 11% identity to both Pz peptidases A and B in amino acid sequence.
Phylogenetic analysis was done with respect to Pz peptidases A and B and those proteins selected from the database due to their high scores of identity in amino acid sequence (34). Pz peptidase A belongs to a group with unassigned peptidases from B. cereus ATCC 10987, B. anthracis strain Ames, and C. perfringens strain 13, which is distinct from oligopeptidase F-like proteins and Pz peptidase B (data not shown).
The consensus sequence HEXXH, characteristic of a Zn-dependent metallopeptidase, was found in positions 356 to 360 (HEAGH) for the Pz peptidase A gene and in positions 395 to 399 (HEFGH) for the Pz peptidase B gene (18). The consensus region for zinc binding is essential for catalytic activity and is observed between proteins sharing high identity. It is noteworthy that human TOP has a significant resemblance at the vicinity with those peptidases, in particular, to Pz peptidase A.
DISCUSSION
We are reporting the presence of two homologous Pz peptidases in G. collagenovorans MO-1. Two distinct peaks of Pz peptidase activities appeared during the purification process. In the early studies of collagen degradation by the thermophile G. collagenovorans MO-1, Pz peptidase activity was not expected to be separated from the collagenolytic activity (32). The presence of two Pz peptidases was interesting and raised the questions of (i) what the difference between them is and (ii) why two isozymes occur in the cell. To answer these questions, experiments related to biochemical characteristics were first carried out. As a result, the two Pz peptidases were found to resemble one another in many biochemical aspects (substrate specificity, pH, temperature dependence, thermostability, and susceptibility to agents), whereas there are definite differences in molecular mass and antigenicity. Collagen degradation by the two Pz peptidases in collaboration with the collagenolytic protease in vitro made it possible to conclude that the Pz peptidases participate in collagen degradation. The collagen degradation, as shown in Fig. 3, suggests a model in which collagens are extracellularly digested as a primary step by the collagenolytic protease of G. collagenovorans MO-1, and, subsequently, the truncated collagen fragments are further hydrolyzed intracellularly by the two Pz peptidases. This pattern of breakdown is just similar to the scheme that extracellular lactoceptins and intracellular peptidases are responsible for casein hydrolysis by lactic acid bacteria, although the protein substrates are different (7). There were only minor differences in the digestion patterns of the two peptidases for peptide substrates (Table 3), indicating that they share a similar function, i.e., collagen degradation. In some gram-positive bacteria, macromolecular proteins are degraded by extracellular proteases and then the resulting oligopeptides are transported to the inside of cells with the use of oligopeptide-binding proteins to be further hydrolyzed by various peptidases for optimum growth (16). Further studies of the genes for the two Pz peptidases support the model. The genes of two Pz peptidases contain no signal sequence for secretion, and there is no putative transcriptional element in the 5′-flanking regions of the genes. These features of their genes are in accord with the observations that two Pz peptide-hydrolyzing activities were detected from the cellular fraction but not from the extracellular one and that the Pz peptidase activities of this strain are not induced by the protein substrates, such as collagen and casein, but are constitutive (data not shown). From these facts, one reason that the two Pz peptidases occur in the cell is probably the enhanced ability to degrade collagen fragments so that they can grow in the medium, a phenomenon that is related to the second question.
From the homology analyses using their gene structures, we found that there are several homologous but putative proteins derived from the genome databases of various bacteria. Interestingly, two isozymes similarly to the pair of Pz peptidases A and B in G. collagenovorans MO-1 are also present in other bacteria: e.g., B. anthracis strain Ames and B. cereus ATCC 10987. Although both isozymes belong to the metallopeptidase family M3B in the peptidase database MEROPS, the function of one group homologous with Pz peptidase A is not well characterized; however, the other group homologous with Pz peptidase B is assigned as an oligopeptidase F-like peptidase from the sequences (http://merops.sanger.ac.uk). The identity in the amino acid sequence between these two groups is much lower, as is the case with Pz peptidases A and B (22%). In contrast, some L. lactis strains that have two homologous genes coding for oligopeptidase F share an identity as high as 80%. Both genes are in the chromosome of strain IL-1403. One is in the chromosome, and the other is in the plasmid of strain NCDO763 (31). Deletion of those genes has not proved to be lethal in the bacterium. The major roles of the oligopeptidases in L. lactis are probably necessary for optimal growth in a milk medium, protein turnover, and degradation of signal peptides, although it is still uncertain whether they have the same function. On the other hand, the fact that a gene homologous to oligopeptidase F was detected in Mycobacterium genitalium, which has one of the smallest genomes among eubacteria, suggested that the oligopeptidase F homologues could have an important role in the cells (15, 30). However, as stated above, we have proposed that the main role of the two Pz peptidases is to support the collagen degradation in G. collagenovorans MO-1. It is intriguing that B. anthracis strain Ames and B. cereus ATCC 10987, which are equipped with two Pz peptidase homologues, are both known to be pathogenic and to infect animals. Those pathogenic bacteria are presumed to adhere to the surface of the animals to degrade collagen molecules in situ and to display the pathogenicity. It is plausible that the Pz peptidase homologues are integrated into a mechanism in which pathogenic microorganisms attach to the skin of animals, decompose collagens, and, finally, display pathogenicity. G. collagenovorans MO-1, which belongs to the group of thermophilic bacilli, is phylogenetically far from B. anthracis and B. cereus strains and was thus concluded to be a nonpathogenic or less-pathogenic strain (3). From the perspective of a phylogenetic tree, the Pz peptidase A and B genes were possibly segregated as a distinct gene in the early stages and evolved independently or were acquired from some other sources.
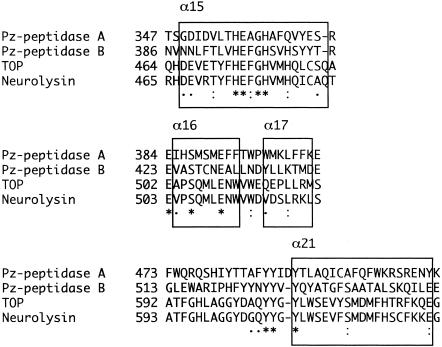
The comparison of Pz peptidases with TOPs is of great interest because the substrate specificities of Pz peptidases and TOPs are similar (6). This suggests that their hydrolytic mechanisms should be adopted in a similar manner; therefore, common structural features between them are expected. However, the identity of Pz peptidases and TOPs in overall amino acid sequence is rather low. In contrast, there is significant homology in the vicinity around the zinc-binding and substrate-binding sites of Pz peptidases and TOPs (Fig. 5). Very recently, the X-ray crystallographic analysis was completed for human TOP, based on the structural data of neurolysin, which is its most homologous protein (8, 36). The consensus zinc-binding motif, the HEXXH sequence, was observed in the region corresponding to the helix α15 of TOP, suggesting that these four enzymes function in a similar manner, adopting the equally folded structure. The glutamate residue of the HEXXH motif is thought to play an important role in the reaction mechanism, polarizing and orienting a zinc-coordinating water molecule that is the attacking nucleophile (20). The two histidines coordinate the zinc ion cofactor. The conservative glutamate residues, Glu384 for Pz peptidase A and Glu423 for Pz peptidase B, should serve as the third zinc ligands that are characteristic of the metallopeptidase M3 family. The regions following the glutamate residues are supposed to form two helices equivalent to α16 and α17 of TOP. The loop regions proceeding to the helix α21 of TOP are rich in tyrosine. Those tyrosine residues would likely interact with the bound substrate (36). Pz peptidases A and B also have tyrosines at the equivalent positions. The loop is presumed to be responsible for the differences in cleavage site specificity between TOP and neurolysin. This is the case with Pz peptidases A and B. Several glycine residues in the loops of TOP and neurolysin are demonstrated to contribute to high flexibility and mobility for the substrate entrance. However, Pz peptidases A and B have no glycine in the loop region, suggesting that the region is less flexible to fit the peptide substrates so that the substrate specificity of Pz peptidases is different from that of TOPs. Furthermore, the regulation in length of the peptides cleaved by Pz peptidases A and B is close to that of TOP. This allows us to presume that Pz peptidases A and B have a deep, narrow, and long channel containing the active site at the bottom (36), although there is no evidence of the channel for Pz peptidases A and B. The channel in TOP restricts the length of the peptides cleaved by the enzyme (36). In addition, Pz peptidase A has six Cys residues, as do TOPs, while Pz peptidase B has two. TOP is activated by treatment of dithiothreitol at a 0.1 mM concentration, and Cys residues have been reported to be concerned with the oligomerization of the molecules of the enzyme (5). Although such activation by reducing reagents was not observed for the two Pz peptidases, some Cys residues should play critical roles in the enzymatic reaction mechanism. To resolve the points stated above, a molecular comparison with TOP using the accurate and detailed structure of Pz peptidases is necessary. We are trying to crystallize the Pz peptidases overexpressed in E. coli cells.
FIG. 5.
Homology in the vicinity around the zinc-binding and substrate binding sites of Pz peptidases A and B, TOP, and neurolysin (protein ID P42676). Strictly conserved amino acid residues are indicated by asterisks, semiconservative ones are indicated by stacked dots, and less conservative ones are indicated by single dots. Boxed regions denote helices assigned in TOP.
REFERENCES
- 1.Akiyama, K., K. Mori, and R. Takata. 1999. Cloning and sequencing of the Pz-peptidase gene from Bacillus licheniformis N22. J. Biosci. Bioeng. 87:231-233. [DOI] [PubMed] [Google Scholar]
- 2.Asdornnithee, S., E. Himeji, K. Akiyama, T. Sasaki, and R. Takata. 1995. Isolation and characterization of Pz-peptidase from Bacillus licheniformis N22. J. Ferm. Bioeng. 79:200-204. [Google Scholar]
- 3.Ash, C., J. A. Farrow, S. Wallbanks, and M. D. Collins. 1991. Phylogenetic heterogeneity of the genus Bacillus revealed by comparative analysis of small-subunit-ribosomal RNA sequences. Lett. Appl. Microbiol. 13:202-206. [Google Scholar]
- 4.Barrett, A. J., M. A. Brown, P. M. Dando, C. G. Knight, N. McKie, N. D. Rawlings, and A. Serizawa. 1995. TOP and oligopeptidase M or neurolysin. Methods Enzymol. 248:529-556. [DOI] [PubMed] [Google Scholar]
- 5.Barrett, A. J., and M. A. Brown. 1990. Chicken liver Pz-peptidase, a thiol-dependent metallo-endopeptidase. Biochem. J. 271:701-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett, A. J., and J. M. Chen. 2004. TOP, p. 352-356. In A. J. Barrett, N. D. Rawlings, and J. F. Woessner (ed.), Handbook of proteolytic enzymes, 2nd ed. Elsevier, New York, N.Y.
- 7.Broadbent, J. R., M. Barnes, C. Brennand, M. Strickland, K. Houck, M. E. Johnson, and J. L. Steele. 2002. Contribution of Lactococcus lactis cell envelope proteinase specificity to peptide accumulation and bitterness in reduced-fat cheddar cheese. Appl. Environ. Microbiol. 68:1778-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, C. K., K. Madauss, W. Lian, M. R. Beck, W. D. Tolbert, and D. W. Rodgers. 2001. Structure of neurolysin reveals a deep channel that limits substrate access. Proc. Natl. Acad. Sci. USA 98:3127-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Büchler, M., U. Tisljar, and D. H. Wolf. 1994. Proteinase yscD (oligopeptidase yscD). Structure, function and relationship of the yeast enzyme with mammalian TOP (metalloendopeptidase, EP 24.15). Eur. J. Biochem. 219:627-639. [DOI] [PubMed] [Google Scholar]
- 10.Christensson, C., H. Bratt, L. J. Collins, T. Coolbear, R. Holland, M. W. Lubbers, P. W. O'Toole, and J. R. Reid. 2002. Cloning and expression of an oligopeptidase, PepO, with novel specificity from Lactobacillus rhamnosus HN001 (DR20). Appl. Environ. Microbiol. 68:254-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dando, P. M., M. A. Brown, and A. J. Barrett. 1993. Human TOP. Biochem. J. 294:451-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doi, E., D. Shibata, and T. Matoba. 1981. Modified colorimetric ninhydrin methods for peptidase assay. Anal. Biochem. 118:173-184. [DOI] [PubMed] [Google Scholar]
- 13.Engel, U., O. Pertz, C. Fauser, J. Engel, C. N. David, and T. W. Holstein. 2001. A switch in disulfide linkage during minicollagen assembly in Hydra nematocysts. EMBO J. 20:3063-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fortina, M. G., D. Mora, P. Schumann, C. Parini, P. L. Manachini, and E. Stackebrandt. 2001. Reclassification of Saccharococcus caldoxylosilyticus as Geobacillus caldoxylosilyticus (Ahmad et al. 2000) comb. nov. Int. J. Syst. Evol. Microbiol. 51:2063-2071. [DOI] [PubMed] [Google Scholar]
- 15.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, J. L. Fritchman, J. F. Weidman, K. V. Small, M. Sandusky, J. Fuhrmann, D. Nguyen, T. R. Utterback, D. M. Saudek, C. A. Phillips, J. M. Merrick, J.-F. Tomb, B. A. Dougherty, K. F. Bott, P.-C. Hu, and T. S. Lucier. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 16.Garault, P., D. Le Bars, C. Besset, and V. Monnet. 2002. Three oligopeptide-binding proteins are involved in the oligopeptide transport of Streptococcus thermophilus. J. Biol. Chem. 277:32-39. [DOI] [PubMed] [Google Scholar]
- 17.Gomez-Ortiz, M., F. X. Gomis-Ruth, R. Huber, and F. X. Aviles. 1997. Inhibition of carboxypeptidase A by excess zinc: analysis of the structural determinants by X-ray crystallography. FEBS Lett. 400:336-340. [DOI] [PubMed] [Google Scholar]
- 18.Häse, C. C., and R. A. Finkelstein. 1993. Bacterial extracellular zinc-containing metalloproteases. Microbiol. Rev. 57:823-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ibrahim-Granet, O., and C. D'Enfert. 1997. The Aspergillus fumigatus mepB gene encodes an 82 kDa intracellular metalloproteinase structurally related to mammalian TOPs. Microbiology 143:2247-2253. [DOI] [PubMed] [Google Scholar]
- 20.Kester, W., and B. W. Matthews. 1977. Crystallographic study of the binding of dipeptide inhibitors to thermolysin: implications for the mechanism of catalysis. Biochemistry 16:2506-2516. [DOI] [PubMed] [Google Scholar]
- 21.Kuzuya, K., and K. Inouye. 2001. Effects of cobalt-substitution of the active zinc ion in thermolysin on its activity and active-site microenvironment. J. Biochem. 130:783-788. [DOI] [PubMed] [Google Scholar]
- 22.Kyhse-Andersen, J. 1984. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J. Biochem. Biophys. Methods 10:203-209. [DOI] [PubMed] [Google Scholar]
- 23.Linsenmayer, T. F. 1991. Collagen, p. 7-44. In E. D. Hay (ed.), Cell biology of extracellular matrix, Plenum Press, New York, N.Y.
- 24.Lowry, O. H., N. J. Rosenbrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 25.Maeda, H., and T. Yamamoto. 1996. Pathogenic mechanisms induced by microbial proteases in microbial infections. Biol. Chem. Hoppe-Seyler 377:217-226. [DOI] [PubMed] [Google Scholar]
- 26.Makinen, K. K., and P. L. Makinen. 1987. Purification and properties of an extracellular collagenolytic protease produced by the human oral bacterium Bacillus cereus (strain Soc 67). J. Biol. Chem. 262:12488-12495. [PubMed] [Google Scholar]
- 27.Makinen, K. K., P. L. Makinen, and S. A. Syed. 1992. Purification and substrate specificity of an endopeptidase from the human oral spirochete Treponema denticola ATCC35405, active on furylacryloyl-Leu-Gly-Pro-Ala and bradykinin. J. Biol. Chem. 267:14285-14293. [PubMed] [Google Scholar]
- 28.Monnet, V., M. Nardi, A. Chopin, M. C. Chopin, and J. C. Gripon. 1994. Biochemical and genetic characterization of PepF, an oligopeptidase from Lactococcus lactis. J. Biol. Chem. 269:32070-32076. [PubMed] [Google Scholar]
- 29.Murai, A., Y. Tsujimoto, H. Matsui, and K. Watanabe. 2004. An Aneurinibacillus sp. strain AM-1 produces a proline-specific aminopeptidase useful for collagen degradation. J. Appl. Microbiol. 96:810-818. [DOI] [PubMed] [Google Scholar]
- 30.Mushegian, A. R., and E. V. Koonin. 1996. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl. Acad. Sci. USA 93:10268-10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nardi, M., P. Renault, and V. Monnet. 1997. Duplication of the pepF gene and shuffling of DNA fragments on the lactose plasmid of Lactococcus lactis. J. Bacteriol. 179:4164-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okamoto, M., Y. Yonejima, Y. Tsujimoto, Y. Suzuki, and K. Watanabe. 2001. A thermostable collagenolytic protease with a very large molecular mass produced by thermophilic Bacillus sp. strain MO-1. Appl. Microbiol. Biotechnol. 57:103-108. [DOI] [PubMed] [Google Scholar]
- 33.Özbek. S., O. Pertz, M. Schwager, A. Lustig, T. Holstein, and J. Engel. 2002. Structure/function relationships in the minicollagen of Hydra nematocysts. J. Biol. Chem. 277:49200-49204. [DOI] [PubMed] [Google Scholar]
- 34.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 35.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ray, K., C. S. Hines, J. Coll-Rodriguez, and D. W. Rodgers. 2004. Crystal structure of human TOP provides insight into substrate recognition, regulation, and localization. J. Biol. Chem. 279:20480-20489. [DOI] [PubMed] [Google Scholar]
- 37.Tisljar, U. 1993. TOP—a review of a thiol dependent metallo-endopeptidase also known as Pz-peptidase, endopeptidase 24.15 and endo-oligopeptidase. Biol. Chem. Hoppe-Seyler 374:91-100. [PubMed] [Google Scholar]
- 38.Tobiassen, R. O., T. Sørhaug, and L. Stepaniak. 1997. Characterization of an intracellular oligopeptidase from Lactobacillus paracasei. Appl. Environ. Microbiol. 63:1284-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tullai, J. W., P. M. Cummins, A. Pabon, J. L. Roberts, M. C. Lopingco, C. N. Shrimpton, A. I. Smith, J. A. Martignetti, E. S. Ferro, and M. J. Glucksman. 2000. The neuropeptide processing enzyme EC 3.4.24.15 is modulated by protein kinase A phosphorylation. J. Biol. Chem. 275:36514-36522. [DOI] [PubMed] [Google Scholar]
- 40.van Wart, H. E., and D. E. Steinbrink. 1981. A continuous spectrophotometric assay for Clostridium histolyticum collagenase. Anal. Biochem. 113:356-365. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe, K. 2004. Collagenolytic proteases from bacteria. Appl. Microbiol. Biotechnol. 63:520-526. [DOI] [PubMed] [Google Scholar]
- 42.Watanabe, K., H. Iha, A. Ohashi, and Y. Suzuki. 1989. Cloning and expression in Escherichia coli of an extremely thermostable oligo-1,6-glucosidase gene from Bacillus thermoglucosidasius. J. Bacteriol. 171:1219-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wünsch, E., and H. G. Heidrich. 1963. Zur quantitiativen Bestimmung der Kollagenase. Hoppe-Seyler's Z Physiol. Chem. 333:149-151. [DOI] [PubMed] [Google Scholar]