Abstract
A phosphoserine-containing peptide was identified from tryptic digests from Sulfolobus solfataricus P1 by liquid chromatography-tandem mass spectrometry. Its amino acid sequence closely matched that bracketing Ser-309 in the predicted protein product of open reading frame sso0207, a putative phosphohexomutase, in the genome of S. solfataricus P2. Open reading frame sso0207 was cloned, and its protein product expressed in Escherichia coli. The recombinant protein proved capable of interconverting mannose 1-phosphate and mannose 6-phosphate, as well as glucose 1-phosphate and glucose 6-phosphate, in vitro. It displayed no catalytic activity toward glucosamine 6-phosphate or N-acetylglucosamine 6-phosphate. Models constructed using the X-ray crystal structure of a homologous phosphohexomutase from Pseudomonas aeruginosa predicted that Ser-309 of the archaeal protein lies within the substrate binding site. The presence of a phosphoryl group at this location would be expected to electrostatically interfere with the binding of negatively charged phosphohexose substrates, thus attenuating the catalytic efficiency of the enzyme. Using site-directed mutagenesis, Ser-309 was substituted by aspartic acid to mimic the presence of a phosphoryl group. The Vmax of the mutationally altered protein was only 4% that of the unmodified form. Substitution of Ser-309 with larger, but uncharged, amino acids, including threonine, also decreased catalytic efficiency, but to a lesser extent—three- to fivefold. We therefore predict that phosphorylation of the enzyme in vivo serves to regulate its catalytic activity.
In recent years a significant body of genomic and biochemical evidence has accumulated indicating that archaeal proteins are the targets of covalent modification by the phosphorylation and dephosphorylation of the hydroxyl amino acids serine, threonine, and tyrosine (reviewed in reference 23). However, only a small handful of archaeal phosphoproteins have been identified to date (2, 4, 6, 11, 19), and information on the functional consequences of these phosphorylation events remains lacking. Among those members of the Archaea whose genome sequences have been determined, the extreme acidothermophile Sulfolobus solfataricus P2 contains the largest number of open reading frames (ORFs) encoding potential protein-serine/threonine/tyrosine kinases (23); the protein products of at least two exhibit protein-serine/threonine kinase activity in vitro (29, 30). As S. solfataricus represents a potentially rich source of information on protein phosphorylation in the Archaea, we incubated soluble protein extracts with trypsin and searched for peptides containing covalently bound phosphoryl groups by mass spectrometry. Herein we report the first results of this exploration, evidence that Ser-309 is the site of a phosphorylation event in the protein product of ORF sso0207, a putative phosphohexomutase, that inhibits its catalytic activity.
MATERIALS AND METHODS
Materials.
High-pressure liquid chromatography (HPLC)-grade methanol and enzymes used for the assay of phosphohexomutase activity were from Sigma-Aldrich (St. Louis, MO). HPLC-grade acetonitrile was from Fisher Scientific (Pittsburgh, PA). Chelating Sepharose Fast-Flow was from Amersham Biosciences (Piscataway, NJ). BamHI was from New England Biolabs (Beverly, MA). HindIII and sequencing-grade trypsin were from Promega (Madison, WI). Protease inhibitor cocktail (minus EDTA) was from Boehringer-Mannheim (Indianapolis, IN). The phoshopeptide isolation kit was from Pierce Biotechnology, Inc. (Rockford, IL). Pfu Turbo DNA polymerase, BL21-CodonPlus(DE3)-RIL cells, and a Quik-Change II site-directed mutagenesis kit were from Stratagene (LaJolla, CA). QIAquick PCR purification and QIAprep spin miniprep kits were from QIAGEN (Valencia, CA). Expression vector pET-29b was from Novagen (San Diego, CA). Escherichia coli TOP10 cells and all oligonucleotides used in this study (Table 1) were from Invitrogen (Carlsbad, CA). Genomic DNA from Sulfolobus solfataricus P2 was from the American Type Culture Collection (ATCC) (Manassas, VA). General laboratory reagents were from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA).
TABLE 1.
Oligonucleotides used in this study
| Primer | Primer sequence (5′ to 3′)a | Purpose of mutation(s) |
|---|---|---|
| PMM PET FOR | catttatagGaTCCtatgggtaagttgtttggaactg | BamHI |
| PMM PET REV | cctataAGCTTtactagttcactcactaacttttcc | HindIII |
| PS309N FOR | gtggactaaggtaggtaAcatagatattgcgc | S309N |
| PS309N REV | gcgcaatatctatgTtacctaccttagtccac | S309N |
| PS309A FOR | gtggactaaggtaggtGCcatagatattgcgc | S309A |
| PS309A REV | gcgcaatatctatgGCacctaccttagtccac | S309A |
| PMMS2Q FOR | gtggactaaggtaggtCAGatagatattgcgc | S309Q |
| PMMS2Q REV | gcgcaatatctatCTGacctaccttagtccac | S309Q |
| PMMS2V FOR | gtggactaaggtaggtGTGatagatattgcgc | S309V |
| PMMS2V REV | gcgcaatatctatCACacctaccttagtccac | S309V |
| PMMS2T FOR | gtggactaaggtaggtaCcatagatattgcgc | S309T |
| PMMS2T REV | gcgcaatatctatgGtacctaccttagtccac | S309T |
| PMMS2D FOR | gtggactaaggtaggtGAcatagatattgcgc | S309D |
| PMMS2D REV | gcgcaatatctatgTCacctaccttagtccac | S309D |
Uppercase letters indicate base substitutions to create restriction sites or mutations.
Standard procedures.
Protein concentrations were determined by the method of Bradford (1) using premixed reagent and a standardized solution of bovine serum albumin, both from Pierce Biotechnology, Inc. (Rockford, IL). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed as described by Laemmli (26). Gels were stained with Coomassie blue as described by Fairbanks et al. (8).
Phosphopeptide isolation and identification.
Sulfolobus solfataricus P1 (ATCC 35091) was grown at 75°C with vigorous aeration in 50 ml of ATCC medium 1304 with the concentration of yeast extract increased to 2 g/liter as described previously (30). Upon reaching stationary phase, cells were harvested by centrifugation at 4,000 × g for 30 min at 4°C. Cell pellets were stored at −20°C until needed.
Cell pellets from two 50-ml cultures were thawed on ice and then resuspended with stirring in a total of 5 ml of 50 mM morpholinepropanesulfonic acid (MOPS), pH 7.0, containing 2 mM EDTA, 2 mM NaF, 0.2 mM sodium pyrophosphate, and 0.2 mM sodium orthovanadate. Cells were lysed by sonic disruption, six periods of 30 s duration each, using a Branson Sonic Power Co. (Plainview, NY) sonifier cell disruptor (model W185) set at full power. The lysate was cooled on ice for 2 min between each round of sonic disruption. Cell debris was removed by centrifugation at 4,000 × g for 30 min at 4°C.
The supernatant liquid was transferred to a prechilled tube, and 5 volumes of ice-cold methanol were added. The solution was mixed well and incubated at −20°C for 1 h. The precipitated protein was collected by centrifugation at 4,000 × g for 30 min at 4°C. The protein pellet was rinsed twice with 25 ml ice-cold methanol and then air dried. The dried protein pellet was dissolved in 3 ml of freshly prepared 100 mM ammonium bicarbonate. The protein concentration of the resuspended pellet was 5.7 mg/ml. The suspension was divided into 150-μl portions (850 μg each) and stored at −80°C until needed.
Freshly prepared 100 mM ammonium bicarbonate was added to an aliquot of the protein suspension to a final volume of 500 μl. Next, 20 μg of sequencing-grade trypsin was added. The mixture was incubated at 37°C overnight. Insoluble material was removed by centrifugation, and the supernatant liquid was dried under vacuum. Methanolic hydrochloride, 2 M, was prepared from methanol and acetyl chloride as described previously (10). An 800-μl aliquot of a freshly prepared solution was added to the dried pellet, and the mixture was incubated at room temperature for 2 h to methylate free carboxyl groups. The mixture was dried under vacuum. A 250-μl volume of 0.1% (vol/vol) acetic acid containing 10% (vol/vol) acetonitrile was added, and the mixture agitated vigorously on a Vortex mixer. Insoluble material was pelleted by centrifugation, and the supernatant applied to a gallium spin column from a phosphopeptide isolation kit (36). The column was then washed and developed according to the manufacturer's instructions. Those fractions eluted with freshly prepared 100 mM ammonium bicarbonate (3 × 25 μl) were pooled, and 75 μl of 5% (vol/vol) acetic acid was added. Liquid chromatography-tandem mass spectrometry analysis (LC/MS/MS) of this material then was performed as described by Potters et al. (37).
Peak list files generated by the Xcalibur software were analyzed using Matrix Science's (Boston, MA) Mascot MS/MS ions search. Selected search parameters indicated analysis using an ion trap mass spectrometer equipped with an electrospray ionization source, trypsin digestion with up to two possible missed cleavages, an average peptide mass tolerance of ±2 Da, and a fragment ion mass tolerance of ±1.2 Da. The variable modification parameters selected included methyl esterification of C-terminal and side-chain carboxylates, phosphorylation of threonine, serine, and tyrosine residues, and oxidation of methionine. Search results were then verified manually.
Cloning of sso0207 and expression and purification of its recombinant protein product, rSsoPHM.
Open reading frame sso0207 was amplified by PCR using 550 ng of S. solfataricus P2 genomic DNA as a template, primers PMM PET FOR and PMM PET REV (Table 1), and 2.5 units Pfu Turbo DNA polymerase according to the manufacturer's recommendations, except that MgCl2 was added to a concentration of 2.5 mM. The primers introduced a BamHI site at the 5′ end and a HindIII site at the 3′ end of the PCR product to facilitate cloning. The resulting PCR product was purified using a QIAquick PCR purification kit according to the manufacturer's instructions. The purified PCR product was cloned into the expression vector pET-29b, and the resulting plasmid used to transform chemically competent E. coli TOP10 cells. DNA sequencing was performed to verify the presence of the insert and the fidelity of PCR amplification. Site-directed mutagenesis was performed using the Quik-Change II site-directed mutagenesis kit and the primers listed in Table 1 according to the manufacturer's protocol, except that MgCl2 was added to the PCRs to a concentration of 2.5 mM. DNA sequencing was used to verify the fidelity of the altered open reading frame.
The gene product of sso0207, SsoPHM, and mutagenically altered versions thereof were produced as recombinant fusion proteins. E. coli BL21-CodonPlus(DE3)-RIL cells were transformed with the appropriate plasmid DNA (approximately 50 ng) and cultured in 5 ml of LB medium supplemented with 100 μg/ml kanamycin and 34 μg/ml chloramphenicol. The culture was incubated overnight with shaking at 37°C and then used to inoculate 250 ml of LB supplemented with 100 μg/ml kanamycin, 34 μg/ml chloramphenicol, and 4 mM l-arginine. Following 2 h of incubation at 37°C with shaking, isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 0.8 mM and the culture was incubated for an additional 4 h. The cells were then harvested by centrifugation and stored at −80°C until use.
Cell pellets were thawed on ice and then resuspended by stirring in 5 ml of 50 mM MOPS, pH 7.0, containing 150 mM NaCl, 20 mM imidazole, 10 mM β-mercaptoethanol, 250 μg/ml lysozyme, and protease inhibitor cocktail (without EDTA) as suggested by the manufacturer. Cell suspensions were sonicated on ice for five rounds of 30 s each as described above. The suspension was cooled on ice for 1 minute between each period of sonic disruption. Cell debris was removed by centrifugation. The supernatant was transferred to a prechilled tube and further clarified by centrifugation at 10,000 × g for 20 min at 4°C. The supernatant liquid was incubated in a 65°C water bath for 20 min and cooled, and precipitated proteins removed by centrifugation at 4,000 × g for 20 min at 4°C.
The supernatant liquid was applied to a 1-ml column of chelating Sepharose Fast-Flow that had been charged with NiCl2 as described by the manufacturer and equilibrated in 50 mM MOPS, pH 7.0, containing 150 mM NaCl, 20 mM imidazole, and 10 mM β-mercaptoethanol. The column was extensively washed with 25 ml of 50 mM MOPS, pH 7.0, containing 150 mM NaCl, 20 mM imidazole, and 10 mM β-mercaptoethanol. rSsoPHM was then eluted with 5 ml of 50 mM MOPS, pH 7.0, containing 150 mM NaCl, 500 mM imidazole, and 10 mM β-mercaptoethanol. The elution solution was then brought to 95% saturation by the slow addition of 3.25 g ammonium sulfate with constant stirring. The solution was incubated on ice for 20 min. The mixture was centrifuged at 10,000 × g for 20 min at 4°C. Protein pellets were dissolved in 0.5 to 1 ml of 50 mM morpholineethanesulfonic acid (MES), pH 6.5, containing 10 mM MnCl2, 2 mM dithiothreitol, and 15% (vol/vol) glycerol. The solution was divided into aliquots and stored at −80°C until use. A 250-ml culture typically yielded 2 to 5 mg of rSsoPHM.
Assays of phosphohexomutase activity.
Phosphomannomutase and phosphoglucomutase activities in the 1-P to 6-P direction were measured using coupling enzymes to generate NADPH from the hexose 6-phosphates formed (35). The conversion of α-d-glucose 1-phosphate to d-glucose 6-phosphate was determined by incubating rSsoPHM, 1 to 5 μg, at 65°C in 1.0 ml of 25 mM MES, pH 6.5, containing 5 mM MnCl2, 10 μM glucose 1,6-bisphosphate, and 0.5 mM α-d-glucose 1-phosphate. The mixture was cooled, and the quantity of d-glucose 6-phosphate produced was determined by adding 0.25 mM NADP+ and 10 μg/ml yeast glucose 6-phosphate dehydrogenase and then incubating at 25°C until the absorbance at 340 nm was constant. The conversion of α-d-mannose 1-phosphate to d-mannose 6-phosphate was monitored using similar procedures except that (i) α-d-mannose 1-phosphate was substituted for α-d-glucose 1-phosphate and (ii) phosphomannose isomerase, 3.5 μg/ml, and phosphoglucose isomerase, 10 μg/ml, were present during the second incubation step of the coupled assay (35).
The conversion of hexose 6-phosphates to hexose 1-phosphates was measured using a modification of the method of Naught and Tipton (33). rSsoPHM, 1 to 5 μg, was incubated for 1 h at 65°C in 100 μl of 25 mM MES, pH 6.5, containing 5 mM MnCl2, 50 μM glucose 1,6-bisphosphate, and the appropriate hexose 6-phosphate at a final concentration of 0.5 mM. Following incubation, a 30-μl portion was removed and added to an equal volume of 2 N HCl. The mixture was heated at 100°C for 5 min, during which time the phosphoester bonds of the 1-phospho, but not the 6-phospho, sugars were hydrolyzed (33). The quantity of free phosphate liberated was then measured using Malachite green (27).
Computer modeling of SsoPHM and mutagenically altered variants.
Three independent models, SsoPHM itself, and variants in which Ser-309 had been replaced with aspartic acid or glutamine were constructed using the program MODELLER 6.0 (31, 40) and the crystal structure of phosphomannomutase/phosphoglucomutase from Pseudomonas aeruginosa (PDB [Protein Data Bank] code 1K35) as the template (39). Pair-wise alignments were obtained using ClustalW (14) and then visualized using the BOXSHADE program (43) at the Biology Workbench website. MODELLER yielded five PDB files. The best-scoring model for each model was chosen and subjected to energy minimizations. A fourth model, in which Ser-309 was phosphorylated in silico, was constructed from the model of SsoPHM using the SYBYL v. 6.7 molecular modeling package (Tripos Inc., St. Louis, MO). The model was first minimized using the steepest descent method for 100 steps and then using the conjugate gradient method for 300 steps with a nonbonded cutoff of 10.0 Å and a dielectric constant of 10. After initial minimization, the phosphorylated model was created by addition of a phosphate to Ser-309. The phosphorylated model was subjected to a second minimization, as described above. All figures were prepared using visual molecular dynamics (15).
RESULTS AND DISCUSSION
Identification of a phosphorylated peptide from an S. solfataricus P1 protein digest.
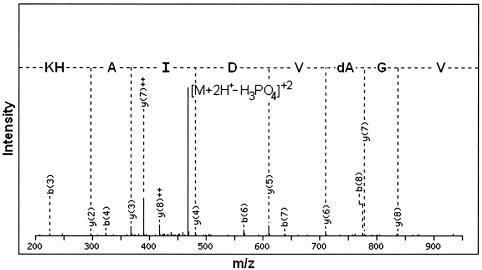
A soluble extract from S. solfataricus P1 was probed for the presence of phosphoproteins using mass spectrometry. Briefly, the component proteins within the extract were cleaved into peptides using trypsin and applied to a gallium spin column to enrich for any phosphorylated species present (36). Methyl ester derivatization of carboxylate moieties was used to reduce nonspecific binding. The adherent fraction was then analyzed by liquid chromatography-tandem mass spectrometry using a Thermo LCQ Deca XP ion trap mass spectrometer. Figure 1 shows the MS/MS fragmentation pattern of a peptide with a +2 charge and an m/z of 517.54. The most abundant fragment [(M + 2H+-H3PO4)+2] was separated from the +2-charged precursor ion by Δm/z = 49, equivalent to the mass signature characteristic for the neutral loss of phosphoric acid, 98 Da. Peptide fragments, particularly those belonging to the -y ion series (labeled in Fig. 1), provided data on the amino acid sequence of the peptide. Note that due to the loss of phosphoric acid the serine is converted to dehydroalanine and the aspartic acid residue and carboxyl terminus of the peptide have been derivatized to methyl esters.
FIG. 1.
Identification of a phosphopeptide from Sulfolobus solfataricus P1. Shown is the MS/MS fragmentation of the [M + 2H+]2+ ion of the phosphopeptide VGpSVDIAHK (m/z = 517.54). The prominent ion corresponding to the neutral loss of phosphoric acid (Δm/z = 49 for the +2 ion) is labeled ([M + 2H+-H3PO4]2+), as are the fragments corresponding to the -y ion series, which yielded amino acid sequence data. Note that because of the loss of phosphoric acid, serine 309 is converted to dehydroalanine (dA) and that the aspartic acid residue and the carboxy terminus of the peptide were derivatized during work up to the corresponding methyl esters.
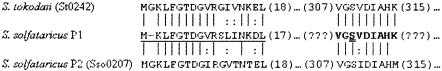
The sequence of the phosphopeptide (VGpSVDIAHK) closely matched a predicted tryptic peptide for the protein encoded by ORF st0242 in the genome of Sulfolobus tokodaii (22), a predicted α-phosphohexomutase (Fig. 2). A BLAST search identified an ORF, sso0207, in the genome of S. solfataricus P2 (42) encoding a protein with 71% identity and 82% similarity to St0242. Further inspection revealed that the N-terminal sequence of Sso0207 closely matched that of a polypeptide from S. solfataricus P1 that behaved like a phosphohexomutase in vitro (44).
FIG. 2.
The phosphopeptide from Sulfolobus solfataricus P1 is derived from a potential phosphohexomutase. Shown is a comparison of the selected segments of the DNA-deduced amino acid sequences of the predicted α-phosphohexomutases from S. tokodaii (St0242 [22]) and S. solfataricus P2 (Sso0207 [42]), with the phosphopeptide identified by mass spectrometry in this study isolated from a protein digest of S. solfataricus P1 (bold) as well as the N-terminal sequence of a phosphohexomutase previously identified from S. solfataricus P1 (44) (underlined). Amino acid identities between Sso0207 and the other proteins are indicated by vertical lines. Conservative differences are indicated by a colon. The phosphorylated serine residue is denoted by a double underline.
rSsoPHM exhibits α-phosphohexomutase activity.
ORF sso0207 was cloned, and its protein product overexpressed as a “histidine-tagged” recombinant fusion protein, rSsoPHM, in the E. coli strain BL21-Codon Plus(DE3)-RIL. This particular strain of E. coli was found to be essential for consistent overexpression of the protein, presumably because sso0207 utilizes an abundance of codons that are considered rare in E. coli (i.e., AGG, AGA, AUA, and CUA [http://www.kazusa.or.jp/codon/]). rSsoPHM was purified by metal-affinity chromatography to greater than 95% homogeneity as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown).
rSsoPHM catalyzed the conversion of α-d-mannose 1-phosphate and α-d-glucose 1-phosphate to the corresponding 6-phosphohexoses with comparable efficiency: 2.5 and 2.6 nmol/min/mg, respectively, at a substrate concentration of 0.5 mM. This observation was verified by steady-state kinetic analysis of the conversion of mannose and glucose 6-phosphates to the corresponding 1-phosphohexoses. No activity was detected toward glucosamine 6-phosphate or N-acetylglucosamine 6-phosphate, however (data not shown). rSsoPHM required a divalent metal, preferentially Mn2+ or Mg2+, for maximal activity, as had been reported for previously characterized members of this enzyme family (21, 33, 41).
In the phosphohexomutases, catalysis proceeds via the formation of a phosphoenzyme intermediate (41). Briefly, the enzyme must first be primed by phosphorylation of a catalytic serine residue using either a diphosphohexose or ATP as phosphodonor (21). The primed enzyme then transfers its phosphoryl group to the 6- or 1-hydroxyl of a phosphohexose substrate to transiently form a 1,6-bisphosphohexose. Reciprocal transfer of the 1- or 6-phosphoryl group, respectively, to the catalytic serine regenerates the primed enzyme and the 6- or 1-phosphohexose product. Attempts to prime rSsoPHM with either ATP or glucose 1,6-bisphosphate had no noticeable effect on catalytic activity, while incubation with [γ-32P]ATP resulted in little phosphate incorporation. Since we had observed previously that the endogenous SsoPHM in soluble extracts of S. solfataricus P1 could be primed on the catalytic serine by incubating with [γ-32P]ATP (44), we suspect that the recombinantly expressed protein, rSsoPHM, emerged from the E. coli host with its active-site serine residue already phosphorylated (data not shown). An examination of the literature revealed that many of the phosphohexomutases produced by recombinant expression in E. coli emerged with a portion of their active-site serine residues in a phosphorylated state (20, 45).
It is unclear whether either glucose or mannose phosphate represents the cellular substrate(s) for SsoPHM. The Km values for rSsoPHM toward glucose 6-phosphate and mannose 6-phosphate were in the low millimolar range, significantly higher than that expected for a physiological substrate. Published Km values for established phosphogluco- and phosphomannomutases typically fall in the mid-micromolar range (20, 32, 34), while those for other, physiologically irrelevant substrates generally fall in the millimolar range (3, 20, 45). On the other hand, S. solfataricus contains glycogen (24), and its genome encodes both a deduced glycogen synthase (sso0987) and a deduced UDP-glucose pyrophosphorylase (sso0813) (42). Since the protein encoded by sso0207 is the only identifiable phosphohexomutase in the S. solfataricus P2 genome, SsoPHM provides the only recognizable avenue for interconverting glucose 6-phosphate and glucose 1-phosphate during glycogen synthesis and breakdown.
Probing the potential role of the phosphoryl group on Ser-309.
The phosphorylated serine identified by mass spectrometry, which corresponded to Ser-309 in the protein encoded by sso0207, was well separated from the predicted catalytic serine, Ser-97. Moreover, while mutagenic alteration of Ser-97 to alanine yielded a protein with little or no catalytic activity, substitution of Ser-309 by alanine resulted in only a small diminution in Vmax (Table 2). Given that the phosphorylation of Ser-309 was not integrally involved in catalysis, we asked whether it might play a regulatory role.
TABLE 2.
Kinetic values for rSsoPHM and mutagenically altered variants thereofa
| Variant | Km [G-6-P], mMa | Vmax (μmol G-1-P/min/mg) | % of WTb |
|---|---|---|---|
| rSsoPHM | 13 | 0.45 | 100 |
| S309A | 9 | 0.26 | 58 |
| S309T | 8 | 0.16 | 36 |
| S309V | 12 | 0.16 | 36 |
| S309N | 8 | 0.12 | 27 |
| S309Q | 6 | 0.10 | 22 |
| S309D | 2 | 0.02 | 4 |
Shown are the Michaelis constants toward α-d-glucose 6-phosphate [G-6-P] for rSsoPHM and mutagenically altered variants in which Ser-309 has been replaced by the indicated residues.
WT, wild type.
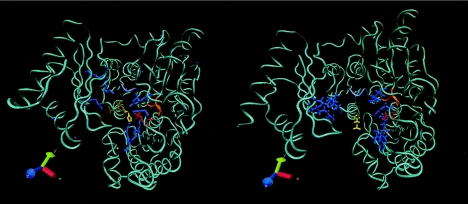
A model for SsoPHM was generated using the crystal structure of AlgC, a homologous α-phosphomannomutase from P. aeruginosa (39), as a scaffold. The amino acid sequences of AlgC and SsoPHM share 27% identity and 43% similarity. The left panel of Fig. 3 shows the crystal structure of AlgC complexed with α-d-glucose 1-phosphate (38). Basic residues that bind the 1- and 6-phosphate groups on substrates and the diphosphohexose intermediate are shown in blue. The active-site serine is shown in red, and the metal-binding loop in orange. A loop purported to play a role in substrate discrimination is shown in purple. The side chain of the histidine residue that aligns with Ser-309 in SsoPHM is shown in yellow. The right panel in Fig. 3 shows a model of the phosphorylated form of SsoPHM, with the side chain of Ser-309, including the phosphoryl group, shown in yellow (root mean square deviation, 0.86 Å). The presence of this relatively bulky and negatively charged phosphoryl group near the floor of the active site would be predicted to impair binding of anionic phosphohexoses in a manner analogous to that by which phosphorylation electrosterically blocks substrate binding by isocitrate dehydrogenase in enteric bacteria (7, 16).
FIG. 3.
The phosphorylated serine in SsoPHM is located within the substrate binding site. (Left) Crystal structure of AlgC, an α-phosphomannomutase from Pseudomonas aeruginosa that is homologous to SsoPHM, complexed with the substrate α-d-glucose 1-phosphate (38). (Right) Model of the phosphorylated form of SsoPHM generated using AlgC as a scaffold. The basic residues that form the two phosphate binding pockets are shown in blue, the active-site serine is shown in red, the metal-binding loop is shown in orange, the loop responsible for sugar-specificity is in purple, and phospho-Ser-309 of SsoPHM and the corresponding residue, His-308, of AlgC are shown in yellow. The amino acid sequence of SsoPHM is 27% identical and 43% similar to that of AlgC. The root mean square deviation value for the model was 0.86 Å.
Attempts to verify the above prediction by phosphorylating rSsoPHM on Ser-309 in vitro were thwarted by the inability of the protein kinases on hand, the recombinant products of open reading frames sso0433 and sso0469 encoded by the genome of S. solfataricus, to modify the enzyme (temperature = 65°C). We therefore attempted to mimic the phosphorylation of Ser-309 by replacing it with a series of alternative amino acids via site-directed mutagenesis. Table 2 illustrates the effects these amino acid substitutions exerted on the catalytic activity of rSsoPHM. Introduction of an aspartic acid residue in place of a phosphoacceptor serine, threonine, or tyrosine has often been observed to mimic the effect of phosphorylation on a protein's structure and activity (see references 5, 9, 13, 18, and 28 for examples). In rSsoPHM, the replacement of Ser-309 with aspartic acid yielded a variant of the enzyme exhibiting less than 5% of the maximal activity of the wild-type enzyme. Replacement of Ser-309 by alanine, threonine, valine, asparagine, or glutamine also yielded enzymes whose Vmax decreased progressively with the increasing size of the mutagenically introduced side chain. However, in no case did the introduction of a neutral amino acid approach the dramatic attenuation of activity attained using the ionizable side chain of aspartic acid.
While the presence of a serine residue at the position corresponding to residue 309 in SsoPHM is not highly conserved among the phosphohexomutases, a subgroup has been identified in which aspartic acid is (41). Intriguingly, open reading frames encoding deduced representatives of this subgroup have been identified in the archaeons Pyrococcus horikoshii and Thermococcus kodakarensis. For these proteins, the presence of an aspartic acid residue at this location is thought to render them specific for amino sugars. However, when the mutationally altered form of rSsoPHM in which Ser-309 was substituted by aspartic acid was challenged with glucosamine 6-phosphate, no activity was detected (data not shown).
Physiological implications of regulation of Sso0207 by phosphorylation.
The α-phosphohexomutases play important and diverse roles in carbohydrate metabolism (41) and therefore represent likely targets for regulation by posttranslational modification. Examination of the projected structure of SsoPHM and the behavior of mutationally altered forms strongly suggest that phosphorylation of Ser-309 inhibits the enzyme's catalytic activity. Recent studies on phosphoglucomutases in higher organisms have revealed two examples of regulation by covalent phosphorylation. The activity of human phosphoglucomutase 1 can be stimulated twofold by phosphorylation of Thr-466 using p21-activated protein kinase 1 (12). This activation appears to be physiologically relevant, as the phosphoglucomutase activity in cultured cells increased in parallel with the level of p21-activated protein kinase 1 activity. Similarly, overexpression of a constitutively active protein-tyrosine kinase in Drosophila melanogaster led to an increase in the phosphoglucomutase activity in cell extracts and greater immunoreactivity of the enzyme toward antibodies against phosphotyrosine (17).
The site of phosphorylation on phosphoglucomutase 1, Thr-466, lies well outside the conserved catalytic core domain, while the site of phosphorylation in the Drosophila protein remains to be determined. However, mass spectrometric analyses of a pair of phosphoglucomutase isozymes from Paramecium tetraurelia revealed that these proteins were phosphorylated at multiple serine and threonine residues in vivo (25). Intriguingly, two of these, Thr-280 and Thr-373, are located at the entrance to and inside the catalytic cleft of these proteins. Unfortunately, the effects of these phosphorylation events on catalytic activity were not reported. However, it is tempting to speculate that both S. solfataricus and P. tetraurelia employ protein phosphorylation to modulate the activity of phosphohexomutases in a similar manner. Further work—in particular the identification of the protein kinase that phosphorylates Ser-309 in SsoPHM—is needed, however, to definitively address this hypothesis.
Summary.
We have determined that a phosphohexomutase from the archaeon S. solfataricus is modified by covalent modification in vivo. The phosphoacceptor serine, Ser-309, maps near the bottom of the active-site cleft. Introduction of a phosphate group at this location would be expected to inhibit phosphohexomutase activity. Studies of mutagenically altered forms of rSsoPHM supported this prediction. Specifically, introduction of an aspartic acid residue in place of the phosphoacceptor serine yielded an enzyme whose catalytic efficiency was markedly attenuated. To the best of our knowledge, this represents the first instance in which the phosphorylation of an archaeal protein on a hydroxyl amino acid has been linked to an alteration in the functional properties of the affected protein.
Acknowledgments
This work was supported by grants MCB-0315122 from the National Science Foundation (P.J.K.), GM55067 from the National Institutes of Health (P.J.K.), N00014-01-1-0852 from the Multidisciplinary University Research Initiative (MURI) program of the Department of Defense (R.F.H.), and N00173-02-1-G016 from the Defense Sciences Office of DARPA (R.F.H.) and by the Thomas F. and Kate Miller Jeffress Memorial Trust (T.J.L.).
We thank David Bevan for expert assistance with our molecular modeling studies.
REFERENCES
- 1.Bradford, M. M. 1976. A rapid and simple method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 2.Cardona, S., F. Remonsellez, N. Guiliani, and C. A. Jerez. 2001. The glycogen-bound polyphosphate kinase from Sulfolobus acidocaldarius is actually a glycogen synthase. Appl. Environ. Microbiol. 67:4773-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng, P.-W., and D. M. Carlson. 1979. Mechanism of phosphoacetylglucosamine mutase. J. Biol. Chem. 254:8353-8357. [PubMed] [Google Scholar]
- 4.Condo, I., D. Ruggero, R. Reinhardt, and P. Londel. 1998. A novel aminopeptidase associated with the 60 kDa chaperonin in the thermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 29:775-785. [DOI] [PubMed] [Google Scholar]
- 5.Cuevas, B. D., Y. Lu, M. Mao, J. Zhang, R. LaPushin, K. Siminovitch, and G. B. Mills. 2001. Tyrosine phosphorylation of p85 relieves its inhibitory activity on phosphatidylinositol 3-kinase. J. Biol. Chem. 276:27455-27461. [DOI] [PubMed] [Google Scholar]
- 6.Daas, P. J. H., R. W. Wassenaar, P. Willemsen, R. J. Theunissen, J. T. Keltjens, C. van der Drift, and G. D. Vogels. 1996. Purification and properties of an enzyme involved in the ATP-dependent activation of the methanol:2-mercaptoethanesulfonic acid methyltransferase reaction in Methanosarcina barkeri. J. Biol. Chem. 271:22339-22345. [DOI] [PubMed] [Google Scholar]
- 7.Dean, A. M., M. H. I. Lee, and D. E. Koshland, Jr. 1989. Phosphorylation inactivates Escherichia coli isocitrate dehydrogenase by preventing isocitrate binding. J. Biol. Chem. 264:20482-20486. [PubMed] [Google Scholar]
- 8.Fairbanks, G., T. L. Steck, and D. F. H. Wallace. 1971. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry 10:2606-2617. [DOI] [PubMed] [Google Scholar]
- 9.Ferrari, G., R. Rossi, D. Arioso, A. Vindigni, G. Biamonti, and A. Montecucco. 2003. Cell cycle-dependent phosphorylation of human DNA ligase I at the cyclin-dependent kinase sites. J. Biol. Chem. 278:37761-37767. [DOI] [PubMed] [Google Scholar]
- 10.Ficarro, S. B., M. L. McCleland, P. T. Stukenberg, D. J. Burke, M. M. Ross, J. Shabanowitz, D. F. Hunt, and F. M. White. 2002. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20:301-305. [DOI] [PubMed] [Google Scholar]
- 11.Grabowski, B., and Z. Kelman. 2001. Autophosphorylation of archaeal Cdc6 homologs is regulated by DNA. J. Bacteriol. 183:5459-5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gururaj, A., C. J. Barnes, R. K. Vadlamudi, and R. Kumar. 2004. Regulation of phosphoglucomutase 1 phosphorylation and activity by a signaling kinase. Oncogene 23:8118-8127. [DOI] [PubMed] [Google Scholar]
- 13.Haase, C., J. T. Steiler, T. Arendt, and M. Holzer. 2004. Pseudophosphorylation of tau protein alters its ability for self-aggregation. J. Neurochem. 88:1509-1520. [DOI] [PubMed] [Google Scholar]
- 14.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 15.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33-38. [DOI] [PubMed] [Google Scholar]
- 16.Hurley, J. H., A. M. Dean, P. E. Thorsness, D. E. Koshland, Jr., and R. M. Stroud. 1990. Regulation of isocitrate dehydrogenase by phosphorylation involves no long-range conformational change in the enzyme. J. Biol. Chem. 265:3599-3602. [DOI] [PubMed] [Google Scholar]
- 17.Inoue, H., S. Kondo, Y. Hinohara, N. Juni, and D. Yamamoto. 2003. Enhanced phoshorylation and enzymatic activity of phosphoglucomutase by the Btk29A tyrosine kinase in Drosophila. Arch. Biochem. Biophys. 413:207-212. [DOI] [PubMed] [Google Scholar]
- 18.Iyer, D., N. Belaguli, M. Fluck, B. G. Rowan, L. Wei, N. L. Weigel, F. W. Booth, H. F. Epstein, R. J. Schwartz, and A. Balasubramanyan. 2003. Novel phosphorylation target in the serum response factor MADS box regulates α-actin transcription. Biochemistry 42:7477-7486. [DOI] [PubMed] [Google Scholar]
- 19.Jeon, S.-J., S. Fujiwara, M. Takagi, T. Tanaka, and T. Imanaka. 2002. Tk-PTP, a protein tyrosine/serine phosphatase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1: enzymatic characteristics and identification of its substrate proteins. Biochem. Biophys. Res. Commun. 295:508-514. [DOI] [PubMed] [Google Scholar]
- 20.Jolly, L., P. Ferrari, J. van Heijenoort, F. Fassy, and D. Mengin-Lecreulx. 1999. Reaction mechanism of phosphoglucosamine mutase from Escherichia coli. Eur. J. Biochem. 262:202-210. [DOI] [PubMed] [Google Scholar]
- 21.Jolly, L., F. Pompeo, J. van Heijenoort, F. Fassy, and D. Mengin-Lecreulx. 2000. Autophosphorylation of a phosphoglucosamine mutase from Escherichia coli. J. Bacteriol. 182:1280-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawarabayasi, Y., Y. Hino, H. Horikawa, K. Jin-no, M. Takahashi, M. Sekine, S. Baba, A. Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Kato, T. Yoshizawa, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, S. Masuda, M. Yanagii, M. Nishimura, A. Yamagishi, T. Oshima, and H. Kikuchi. 2001. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain 7. DNA Res. 8:123-140. [DOI] [PubMed] [Google Scholar]
- 23.Kennelly, P. J. 2003. Archaeal protein kinases and protein phosphatases—insights from genomics and biochemistry. Biochem. J. 370:373-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konig, H., R. Skorko, W. Zillig, and W.-F. Reiter. 1982. Glycogen in thermoacidophilic archaebacteria of the genera Sulfolobus, Thermoproteus, Desulfurococcus, and Thermococcus. Arch. Microbiol. 132:297-303. [Google Scholar]
- 25.Kussmann, M., K. Hauser, R. Kissmehl, J. Breed, H. Plattner, and P. Roepstorff. 1999. Comparison of the in vivo and in vitro phosphorylation of exocytosis-sensitive protein PP63/perfusin by differential MALDI mass spectrometric peptide mapping. Biochemistry 38:7780-7790. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Lanzetta, P. A., L. J. Alvarez, P. S. Reinach, and O. A. Candia. 1979. An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 100:95-97. [DOI] [PubMed] [Google Scholar]
- 28.Lee, H., N. Rezai-Zadeh, and E. Seto. 2004. Negative regulation of histone deacetylase 8 activity by cyclic AMP-dependent protein kinase A. Mol. Cell. Biol. 24:765-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lower, B. H., M. B. Potters, and P. J. Kennelly. 2004. A phosphoprotein from the archaeon Sulfolobus solfataricus with protein-serine kinase activity. J. Bacteriol. 186:463-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lower, B. H., and P. J. Kennelly. 2003. Open reading frame sso2387 from the archaeon Sulfolobus solfataricus encodes a polypeptide with protein-serine kinase activity. J. Bacteriol. 185:3436-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin-Renom, M. A., A. C. Stuart, A. Fiser, R. Sanchez, F. Melo, and A. Sali. 2000. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29:291-295. [DOI] [PubMed] [Google Scholar]
- 32.Mio, T., T. Yamada-Okabe, M. Arisawa, and H. Yamada-Okabe. 2000. Functional cloning and mutational analysis of the human cDNA for phosphoacetylglucosamine mutase: identification of the amino acid residues essential for catalysis. Biochim. Biophys. Acta 1492:369-376. [DOI] [PubMed] [Google Scholar]
- 33.Naught, L. E., and P. A. Tipton. 2001. Kinetic mechanism and pH dependence of the kinetic parameters of Pseudomonas aeruginosa phosphomannomutase/phosphoglucomutase. Arch. Biochem. Biophys. 396:111-118. [DOI] [PubMed] [Google Scholar]
- 34.Naught, L. E., C. Regni, L. J. Beamer, and P. A. Tipton. 2003. Roles of active site residues in Pseudomonas aeruginosa phosphomannomutase/phosphoglucomutase. Biochemistry 42:9946-9951. [DOI] [PubMed] [Google Scholar]
- 35.Pirard, M., Y. Achouri, J.-F. Collet, E. Schollen, G. Matthijs, and E. van Schaftingen. 1999. Kinetic properties and tissue distribution of mammalian phosphomannomutase enzymes. Biochem. J. 339:201-207. [PMC free article] [PubMed] [Google Scholar]
- 36.Posewitz, M. C., and P. Tempst. 1999. Immobilized gallium(III) affinity chromatography of phosphopeptides. Anal. Biochem. 71:2883-2892. [DOI] [PubMed] [Google Scholar]
- 37.Potters, M. B., B. T. Solow, K. M. Bischoff, D. E. Graham, B. H. Lower, R. Helm, and P. J. Kennelly. 2003. Phosphoprotein with phosphoglycerate mutase activity from the archaeon Sulfolobus solfataricus. J. Bacteriol 185:2112-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Regni, C., L. Naught, P. A. Tipton, and L. J. Beamer. 2004. Structural basis of diverse substrate recognition by the enzyme PMM/PGM from P. aeruginosa. Structure 12:55-63. [DOI] [PubMed] [Google Scholar]
- 39.Regni, C., P. A. Tipton, and L. J. Beamer. 2002. Crystal structure of PMM/PGM: an enzyme in the biosynthetic pathway of P. aeruginosa virulence factors. Structure 10:269-279. [DOI] [PubMed] [Google Scholar]
- 40.Sali, A., and T. L. Blundell. 1993. Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 41.Shackelford, G. S., C. A. Regni, and L. A. Beamer. 2004. Evolutionary trace analysis of the α-D-phosphohexomutase family. Protein Sci. 13:2130-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C. Chan- Weiher, I. G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. Gordon, I. Heikamp-de Jong, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, R. F., B. A. Wiese, M. K. Wojzynski, D. B. Davidson, and K. C. Worley. 1996. BCM search launcher—an integrated interface to molecular biology data base search and analysis services available on the World Wide Web. Genome Res. 6:454-462. [DOI] [PubMed] [Google Scholar]
- 44.Solow, B., K. M. Bischoff, M. J. Zylka, and P. J. Kennelly. 1998. Archaeal phosphoproteins. Identification of a hexosephosphate mutase and the α-subunit of succinyl-CoA synthetase in the extreme acidothermophile Sulfolobus solfataricus. Protein Sci. 7:105-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tavares, I. M., L. Jolly, F. Pompeo, J. H. Leitao, A. H. Fialho, I. Sa-Correria, and D. Mengin-Lecreulx. 2000. Identification of the Pseudomonas aeruginosa glmM gene, encoding a phosphoglucosamine mutase. J. Bacteriol. 182:4453-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]