Abstract
The nar operon, coding for the respiratory nitrate reductase of Thermus thermophilus (NRT), encodes a di-heme b-type (NarJ) and a di-heme c-type (NarC) cytochrome. The role of both cytochromes and that of a putative chaperone (NarJ) in the synthesis and maturation of NRT was studied. Mutants of T. thermophilus lacking either NarI or NarC synthesized a soluble form of NarG, suggesting that a putative NarCI complex constitutes the attachment site for the enzyme. Interestingly, the NarG protein synthesized by both mutants was inactive in nitrate reduction and misfolded, showing that membrane attachment was required for enzyme maturation. Consistent with its putative role as a specific chaperone, inactive and misfolded NarG was synthesized by narJ mutants, but in contrast to its Escherichia coli homologue, NarJ was also required for the attachment of the thermophilic enzyme to the membrane. A bacterial two-hybrid system was used to demonstrate the putative interactions between the NRT proteins suggested by the analysis of the mutants. Strong interactions were detected between NarC and NarI and between NarG and NarJ. Weaker interaction signals were detected between NarI, but not NarC, and both NarG and NarH. These results lead us to conclude that the NRT is a heterotetrameric (NarC/NarI/NarG/NarH) enzyme, and we propose a model for its synthesis and maturation that is distinct from that of E. coli. In the synthesis of NRT, a NarCI membrane complex and a soluble NarGJH complex are synthesized in a first step. In a second step, both complexes interact at the cytoplasmic face of the membrane, where the enzyme is subsequently activated with the concomitant conformational change and release of the NarJ chaperone from the mature enzyme.
Membrane-bound nitrate reductases are heterotrimeric enzymes that catalyze the reduction of nitrate, coupled to the generation of a proton-motive force across the cytoplasmic membrane during anaerobic respiration. In the nitrate reductase A (NRA) of Escherichia coli, different primary dehydrogenases transfer electrons to quinones, which function as electron donors to NarI, a di-heme b-type cytochrome with quinol dehydrogenase activity. From the outward-facing acceptor site of NarI, electrons are channeled to the inner face of the membrane, where they are transferred to NarH, a protein with four iron-sulfur clusters. Electrons are then transferred to an iron-sulfur cluster of NarG (23), finally to reach its Mo-bis molybdopterin guanine dinucleotide cofactor (MGD), where nitrate reduction takes place (1, 2, 31).
The NarG and NarH subunits form a complex that interacts with the NarI subunit at the inner face of the cytoplasmic membrane. The three-dimensional structure of NRA shows the existence of interactions between a C-terminal β-strand of the NarI subunit and two β-hairpins, one located at the N terminus of NarG and the other at an internal region of NarH (2). Therefore, attachment of the NarGH complex to the membrane depends on interactions between NarI and both components of the NarGH complex. Despite such interactions, a fraction of the NarGH active complex can be irreversibly detached from the membrane after cell disruption (17). This soluble NarGH complex can still reduce nitrate in the presence of artificial electron donors, such as reduced viologens, which donate the electrons at a currently unknown point of this complex, probably the MGD cofactor (5). Moreover, Escherichia coli mutants that do not synthesize the NarI subunit produce a soluble and active NarGH complex, thus showing that its attachment to the membrane is not required for the synthesis of an active enzyme (3).
In all the organisms so far analyzed, the three structural subunits of the enzyme are expressed from a single operon, which also encodes a fourth protein, named NarJ. In E. coli, NarJ is required for the production of an active NarGH complex because of its role as a specific chaperone required for MGD incorporation into NarG (3, 4). It has been proposed for E. coli that such a maturation step takes place in the cytoplasm, before the attachment of the active NarGH complex to the NarI membrane subunit (4, 27).
In addition to NarGHJI homologues, three additional proteins are encoded by the nar operon of Thermus thermophilus. Two of these genes are located downstream of narI and code for nitrate/nitrite transporters (NarK and NarT). Although the presence of at least one of them is required for anaerobic growth, their absence does not affect the synthesis of an active NRT (22). The first gene of the operon is also specific to T. thermophilus and codes for a membrane-bound di-heme c-type cytochrome (NarC). We have shown that in the absence of NarC, the NarG protein remains soluble and inactive in nitrate reduction (29). Consequently, we proposed that NarC was required for the attachment of the NarGH complex to the membrane and that this binding was necessary for a subsequent “maturation” step. Bearing in mind that biochemical (9), genetic (3), and structural (2) evidence demonstrated that the attachment site for this respiratory enzyme in other bacteria was the NarI protein, we wondered what the actual role of NarI was in the synthesis and maturation of the thermophilic nitrate reductase.
In this article we investigate the role of the cytochrome b (NarI) on the attachment to the membrane and activation of the NRT and its relationship with the periplasmic cytochrome c (NarC). Our data demonstrate that NarI and NarC interact and that both are required for NarJ-dependent activation of the NarGH complex, which, unlike the NRA of E. coli, occurs after its binding to the membrane.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
Table 1 lists the bacterial strains and plasmids used in this work. The E. coli strains were grown in Luria broth (LB) at 37 or 30°C. T. thermophilus was grown in TB medium (20), either aerobically with shaking (150 rpm) or anaerobically in TB with KNO3 (40 mM) (7). Induction of the NRT synthesis was carried out under microaerobic conditions, as described previously (7). The minimal M162 medium was used to isolate ΔleuB derivatives of T. thermophilus (8).
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Description or use | Reference or source |
|---|---|---|
| E. coli DH5α | supE44 ΔlacU169 (Φ80 lacZΔM15) hsdR17 recA endA1 gyrA96 thi-1 relA1 | 10 |
| E. coli BTH101 | F′ cya-99 araD139 galE15 galK16 rpsL1(Strr) hsdR2 mcrA1 mcrB1 | 12 |
| T. thermophilus HB8 | Wild-type strain | 19; ATCC 27634 |
| T. thermophilus HB8narC | HB8 narC::kat | 29 |
| T. thermophilus HB8narI | HB8 narI::kat | This work |
| T. thermophilus HB8narJ | HB8 narJ::kat | This work |
| T. thermophilus HB27nar | T. thermophilus HB27::nar | 21 |
| T. thermophilus L | T. thermophilus HB27::nar ΔleuB | This work |
| T. thermophilus LnarC | T. thermophilus HB27::nar ΔleuB narC::kat | This work |
| pUC118/119 | E. coli cloning plasmids; Ampr | 28 |
| pUP3B | pUC119 derivative; clone carrying the whole nrc operon; Ampr | 7 |
| pKT1 | E. coli plasmid carrying the kanamycin thermostable resistance gene (kat); Ampr, Kanr | 15 |
| pTH3ΔEcoRV | Plasmid for the isolation of ΔleuB mutants | 26 |
| pIW | Suicidal vector; selection for leucine prototrophy | 26 |
| pIWnarC | Insertion of the wild-type narC gene into the leuB locus | This work |
| pT18 | pUC19 derivative expressing T18 fragment of CyaA; Ampr | 12 |
| pT25 | pACYC184 derivative expressing T25 fragment of Cya, Cmr | 12 |
| pT18zip | pT18, leucine zipper fused to T18 fragment | 12 |
| pT25zip | pT25, leucine zipper fused to T25 fragment | 12 |
| pT25gdh | pT25 derivative, T25-Gdh fusion protein, Cmr | 7 |
| pT18gdh | pT18 derivative, Gdh-T18 fusion protein, Ampr | 7 |
| pT25narJ | pT25 derivative, T25-NarJ fusion protein, Cmr | 7 |
| pT18narJ | pT18 derivative, NarJ-T18 fusion protein, Ampr | 7 |
| pT25narG | pT25 derivative, T25-NarG fusion protein, Cmr | 7 |
| pT18narG | pT18 derivative, NarG-T18 fusion protein, Ampr | 7 |
| pT25narH | pT25 derivative, T25-NarH fusion protein, Cmr | This work |
| pT18narI | pT18 derivative, NarI-T18 fusion protein, Ampr | This work |
| pT25narC-ΔPs | pT25 derivative T25-NarCΔPs fusion protein, Cmr | This work |
| pT18narC-ΔPs | pT18 derivative, NarCΔPs-T18 fusion protein, Ampr | This work |
| pT25narC-Cd | pT25 derivative, T25-NarCCd fusion protein, Cmr | This work |
| pT18narC-Cd | pT18 derivative, NarCCd-T18 fusion protein, Ampr | This work |
Transformation was carried out by natural competence in T. thermophilus (13) and by conventional methods in E. coli (10). Selection of the transformed cells was done on TB or LB plates with kanamycin (30 mg/liter), ampicillin (100 mg/liter), and/or chloramphenicol (20 mg/liter).
Nucleic acid and protein analysis.
Standard methods were used for DNA purification, restriction analysis, PCR, and DNA sequencing. Topology predictions were carried out online using the TMHMM program available at http://www.cbs.dtu.dk/services/TMHMM-1.0/ (25). For cell fractionation, samples from induced cultures of T. thermophilus were centrifuged, and the cells were resuspended in 1/10 volume of TS buffer (50 mM Tris-HCl, 50 mM NaCl, pH 7.5) containing a protease inhibitor cocktail (C-Complete Mini; Roche) at the concentration recommended by the manufacturer. After cell disruption by sonication (Braun Labsonic; 1 min in 0.5 s pulses, maximal power), the samples were centrifuged to discard unbroken cells (5,000 × g, 5 min), and soluble and particulate fractions were separated by ultracentrifugation (150,000 × g, 15 min at 4°C). Sensitivity to trypsin was assayed in 80-μl cell fraction samples containing 2 mg/ml of protein. Two microliters of trypsin solution (50 mg/ml in HCl 1 mM) were added to the samples, which were then incubated at room temperature for the indicated times. Digestion was stopped by boiling in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer, and the proteins were separated by SDS-PAGE (14). NarG and NarJ were identified by Western blot with specific rabbit antisera (29) and detected by chemiluminescence with the ECL detection kit (Amersham).
Nitrate reductase activity.
The activity of the nitrate reductase from T. thermophilus was measured at 80°C (20), with reduced methyl-viologen (MVr) as electron donor and potassium nitrate (40 mM) as electron acceptor (24). One enzyme unit was defined as the amount required to produce 1 nmol of nitrite per min. For practical reasons, the total units were normalized to 1 ml of a culture at an optical density at 550 nm (OD550) of 1 (∼109 cells).
Isolation of insertion mutants.
The isolation of the narC::kat mutant has already been described (29). In this mutant, the kat cassette interrupted narC at nucleotide position 125 from its ATG translation start codon. This kat gene codes for a thermostable resistance to kanamycin expressed from a constitutive promoter and does not have any transcriptional terminator to allow the expression of downstream genes (15). Therefore, in all the mutants used in this work the kat gene was inserted in the same transcription sense as the nar operon.
For the isolation of narI and narJ mutants, the kat gene was inserted into a PstI restriction site for narI and a SacI site for narJ in such a way that the coding sequences were interrupted at positions 522 and 324 relative to their ATG start codon, respectively. Upstream and downstream DNA sequences were added in both cases to provide appropriate arms for homologous recombination. Transformation and mutant selection were carried out as described previously (29). Mutants were checked by PCR and Southern blot.
Complementation of the narC::kat mutants.
Complementation of the narC::kat mutant was carried out by insertion into its chromosome of a wild-type narC gene expressed under the control of the narp promoter. Selection was based on the complementation of a ΔleuB mutant (26) constructed in T. thermophilus HB27::nar. This strain is a facultative anaerobic derivative of the high transformation efficiency HB27 strain, to which the cluster for anaerobic respiration of the HB8 strain was transferred (21). Isolation of the ΔleuB derivative was carried out by transformation with plasmid pTH3ΔEcoRV (26) followed by negative selection with ampicillin (6) of leucine auxotrophs in M162 medium. A narC::kat derivative of this ΔleuB mutant was isolated as above to render the host strain used for complementation.
For complementation, the narC gene, expressed from the narp promoter, was inserted into the suicidal vector pIW, which contains a wild-type leuB gene (26), obtaining the pIWnarC plasmid. After transformation of the ΔleuBnarC host strain with the latter, leucine prototrophs were selected on M162 medium with kanamycin. The simultaneous presence of the narC::kat mutation and the wild-type gene was assessed by PCR with the appropriate primers (Table 2).
TABLE 2.
Oligonucleotides used
| Name | Sequence | Purpose |
|---|---|---|
| H1 | 5′-CTGGGTACCTCAGGACGGGAAGGCCCTCTACGG-3′ | pT18narC-ΔSP |
| H2 | 5′-ATTCGATATCCTCCTAAGCTGGGCGCGGATC-3′ | pT18narC-ΔSP/Cd |
| H4 | 5′-CTGGGTACCTGTCCTCTGGCAGCTTCGGCCGAG-3′ | pT18narC-Cd |
| H7 | 5′-AAAACTGCAGGGATGAAGGTTAGAGCCCACATGTCC-3′ | pT25narH |
| H8 | 5′-TTTAGGTACCTCGCCCCGCAAGGGGGGCTGGATG-3′ | pT25narH |
| H17 | 5′-AAAACTGCAGGGCAGGACGGGAAGGCCCTCTACGG-3′ | pT25narC-ΔSP |
| H18 | 5′-ATTCGATATCCCTCCTAAGCTGGGCGCGGATC-3′ | pT25narC-ΔSP/Cd |
| H20 | 5′-AAAACTGCAGGGGTCCTCTGGCAGCTTCGGCCGAG-3′ | pT25narC-Cd |
| H24 | 5′-ACGCGTCGACGATGAAGGTTAGAGCCCACAT-3′ | pT18narH |
| H25 | 5′-ATTCGATATCTCGCCCCGCAAGGGGGGCTGGA-3′ | pT18narH |
| H26 | 5′-ACGCGTCGACGATGAAGTGGAACGCCGCGCTCTTC-3′ | pT18narI |
| H27 | 5′-ATTCGATATCAGCTGACCTCGCCAGTTGCG-3′ | pT18narI |
| o27-35 | 5′-CGCAGTACCAGTCGTAG-3′ | Presence of narC::kat |
| okat2 | 5′-GAAACTTCTGGAATCGC-3′ | Presence of narC::kat |
| okat3 | 5′-GGAACGAATATTGGATA-3′ | Presence of kat |
| okat4 | 5′-AGAAATTCTCTAGCGAT-3′ | Presence of kat |
Bacterial two-hybrid assays.
A bacterial two-hybrid system based on the functional reconstitution of the Bordetella pertussis adenylate cyclase from its T18 and T25 domains was used to determine the putative interactions between the nar gene products (11, 12). For this, the coding sequences of the nar genes were cloned into plasmids pT18 or pT25 in order to be expressed as N- or C-terminal domains fused to the T18 or T25 domains of the adenylate cyclase, respectively (Table 1). Fusions containing deletion derivatives of NarC expressing the whole protein without its signal peptide (ΔSP, from amino acid positions 20 to 262) or its cytoplasmic domain (Cd, the C-terminal 20 amino acids) were also expressed by both plasmids. Constructions were carried out by adding appropriate enzyme restriction sites during amplification (Table 2).
The homo-oligomeric glutamate dehydrogenase, Gdh, from T. thermophilus HB8 was used as a control for positive interactions between thermophilic proteins. Negative controls were also carried out by assaying our fusion proteins against fusions expressing a leucine-zipper domain (12). Interactions were assayed through the β-galactosidase activity (12) of E. coli BTH101 cells cotransformed with pT18 and pT25 derivatives.
RESULTS
NarC and NarI are required for membrane attachment and maturation of the NRT.
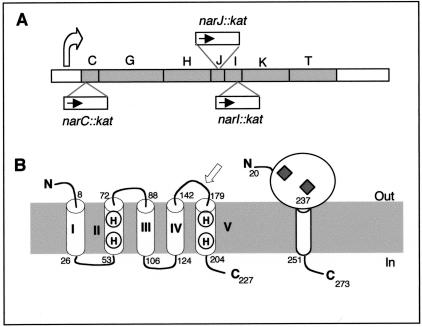
Figure 1A shows a scheme of the narCGHJIKT operon of T. thermophilus, in which the approximate insertion points and the transcription sense of the kat gene in the three nar::kat mutants used in this work are indicated.
FIG. 1.
The nar::kat mutants. A) Scheme showing the structure of the nar operon, the approximate location of the inserted kat gene inside narC, narI, and narJ, and the narp promoter location (curved arrow). B) Proposed topology for the NarI and NarC proteins. Amino acid positions that define the five membrane-spanning helices (I to V) in NarI and NarC are labeled. Conserved histidines involved in heme B coordination are indicated on helices II and V. The white arrow indicates the position where the NarI protein is interrupted in the narI::kat mutant. Diamonds indicate the presence of heme C groups in NarC. The cytoplasmic (in) and periplasmic (out) faces of the membrane are indicated.
Computer predictions about the NarC amino acid sequence (accession number Q934F6 in the Translated European Molecular Biology Laboratory TrEMBL database) suggest a periplasmic location with a C-terminal anchor region consisting of a transmembrane helix followed by a positively charged cytoplasmic domain (Fig. 1B). Sequence comparisons also show the presence of two heme C binding motifs. Consistent with this, a specific staining procedure showed the presence of a cytochrome c of the size expected for NarC in membrane fractions of the wild-type strain, which was absent from the narC::kat mutant (29).
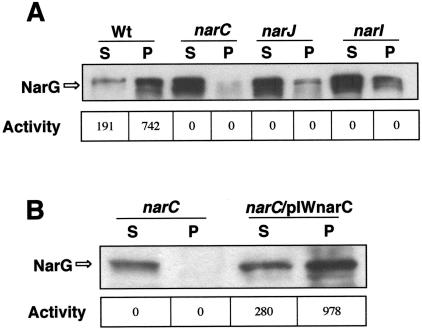
As expected from our previous work (29), the narC::kat mutant synthesized a soluble form of NarG (accession number O06459), the large subunit of the NRT (Fig. 2A). This soluble NarG was not able to reduce nitrate with MVr as the electron donor. To exclude the possibility that such a phenotype could be the consequence of a putative polar effect of the kat gene on the expression of genes located downstream of narC in the operon, we inserted a wild-type narC gene under the control of the narp promoter into the leuB locus of the chromosome (Materials and Methods). As shown in Fig. 2B, a normal membrane location and activity of the enzyme was recovered in the complemented strain. Therefore, the absence of NarC was responsible for the phenotype observed in the narC::kat mutant.
FIG. 2.
Phenotype of nar::kat mutants. A) Western blot for detecting NarG in soluble (S) and particulate (P) fractions of nitrate/anoxia-induced cultures of the wild-type strain (Wt) and its three nar::kat derivatives (narC, narJ, narI). B) The presence of NarG is analyzed in the soluble (S) and particulate (P) fractions from a narC::kat mutant (narC) and from a derived strain complemented with a wild-type narC gene (narC/pIWnarC) by transformation with plasmid pIWnarC. The enzyme activities (nmol of nitrite/min and OD550 unit) from each fraction are indicated.
NarI (accession no. O06462 in TrEMBL) is predicted to have five membrane-spanning α-helices (I to V), its N- and C-termini facing the periplasm and the cytoplasm, respectively (Fig. 1B). As in its E. coli counterpart, four histidine residues are located in α-helices II and V, which coordinate two heme B groups in the mesophilic protein, implying a similar role in T. thermophilus. In the narI mutant, the kat gene was inserted close to the 3′ end of the gene (arrow in Fig. 1B). Therefore, the fifth transmembrane helix (V), which carries two of these histidine residues required for heme B coupling, was deleted in this mutant.
The expected inability of the narI::kat mutant to couple the electron flow to the reduction of nitrate was confirmed by the absence of anaerobic growth with nitrate (not shown). When analyzed in more detail, the narI::kat mutants showed a phenotype almost identical to that of narC::kat mutants, in the sense that a large amount of NarG was found in the cytoplasmic fraction and that the enzyme was completely inactive in nitrate reduction with MVr as the electron donor (Fig. 2A).
Interaction between NarC and NarI.
The results above showed that NarC and NarI were required for the attachment of NarG to the membrane (Fig. 2A). Therefore, it is reasonable to speculate that a membrane complex between both cytochromes could constitute the attachment site for the catalytic subunit of the enzyme. If this were true, some interactions between them should exist.
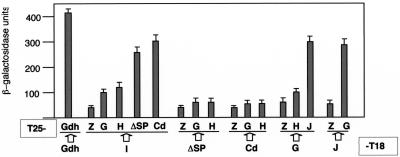
To test this hypothesis, we used a bacterial two-hybrid system, which has proved to be effective for detecting interactions between components of the MGD biosynthesis pathway (18), between membrane components of the type IV secretion systems of the conjugative apparatus of E. coli (16) and also between membrane and soluble subunits of a new type of respiratory NADH that constitutes the natural electron donor for the NRT (7). The results are shown in Fig. 3. As may be seen, significant β-galactosidase activities (∼300 units) were detected when the interaction between NarG and NarJ was assayed, independently of their location as N- or C-terminal domains of the fusion proteins. This was consistent with the role of NarJ as a NarG-specific chaperone proposed for its homologue in E. coli (4). The values for the interaction detected between NarG and NarJ were only ∼25% lower than those detected between our positive control, a homotrimeric glutamate dehydrogenase (Gdh) that is synthesized as enzymatically active forms both as C- and N-terminal fusion proteins from these plasmids (7).
FIG. 3.
Two-hybrid assays. Bacterial two-hybrid assays were developed, and the mean values of the β-galactosidase activity corresponding to six samples in two independent experiments were represented. The whole coding regions of narI (I), narG (G), narH (H), and narJ(J) and deletion derivatives of narC coding for a signal peptide-defective protein (ΔPs) or for its cytoplasmic C-terminal (Cd) domain were cloned into plasmid pT18 and/or pT25 to generate the appropriate X-T18 and T25-X protein fusions, as indicated. Pairs of pT18 and pT25 derivatives were assayed for β-galactosidase expression in E. coli BTH101 cells. Controls for negative (Z) and positive (Gdh/Gdh) interactions were used.
The values obtained for the interaction between NarI and NarC derivatives lacking its signal peptide (ΔSP from amino acid position 20 to 262) to prevent its secretion were similar to that obtained for the interaction between NarJ and NarG. Moreover, protein fusions carrying only the cytoplasmic (Cd, the C-terminal 20 amino acids) domain of NarC, rendered even higher β-galactosidase signals with NarI. On the other hand, lower-but-significantly-higher-than-background β-galactosidase activities were detected in NarI than with both the NarG and NarH proteins. By contrast, interactions were not detected when the NarC derivatives were assayed with these proteins.
Thus, our data show that NarC interacts with NarI, essentially through its cytoplasmic domain, and suggest that the major contact of a NarCI complex with NarGH most probably takes place through NarI, as happens in the NRA of E. coli (2).
NarG is misfolded in narI::kat and in narC::kat mutants.
When MVr was used as a direct electron donor, nitrite production was catalyzed by both the soluble (∼20% of total activity) and the particulate (∼80% of total activity) fractions of the wild-type strain (Fig. 2). However, neither the soluble nor the particulate fractions of the narC::kat and the narI::kat mutants were active in nitrate reduction (Fig. 2). As MVr donates the electrons directly to the NarGH complex (5), these data suggest that NarC and NarI are both involved in a maturation step of the enzyme.
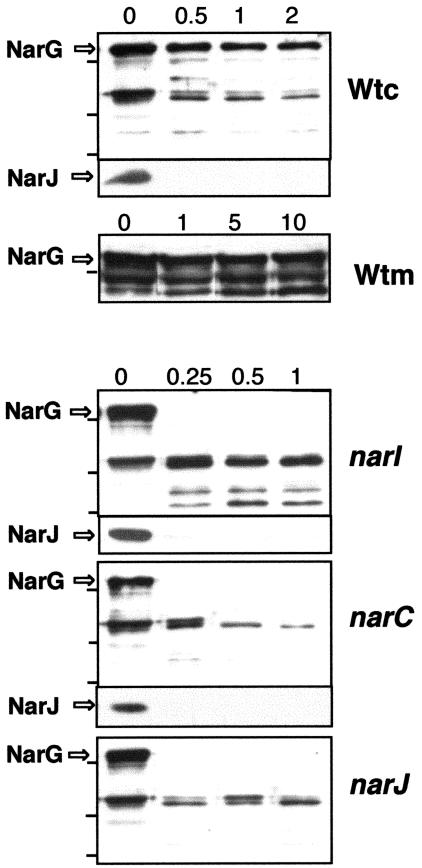
To check whether such absence of activity was related to enzyme folding, we compared the sensitivity of the NarG protein to trypsin in the soluble fraction of each mutant with that of the soluble fraction of the wild-type strain. As shown in Fig. 4, whereas the NarG protein from either of the nar::kat mutants was digested in less than 15 seconds by the protease, undigested NarG was detected in the wild-type strain even after 2 min of treatment. As an internal control, the NarJ protein was detected by Western blot in the same samples, where it showed identical protease sensitivity in the mutant and in the wild-type strains.
FIG. 4.
Sensitivity of NarG to trypsin in narI, narC, and narJ mutants. The sensitivities to trypsin of NarG and NarJ from the soluble fraction of the wild-type strain (Wtc) and its three nar::kat derivatives were compared. The sensitivity of NarG was also assayed in membrane fractions of the wild type (Wtm). The samples were incubated with the protease for the indicated times (min).
It was of particular note that the sensitivity of NarG to the protease in the soluble fraction of the narJ::kat mutant was similar to that of the narC::kat or the narI::kat mutants. Therefore, the NarC and NarI proteins are as important for the maturation of the NarG protein as its specific chaperone NarJ.
NarJ is also required for membrane attachment of NarG.
As shown above, the T. thermophilus narJ::kat mutant synthesized an inactive and misfolded form of NarG (Fig. 4). However, whereas most of this inactive NarG protein remains attached to the membrane in narJ mutants of E. coli (3), most of the NarG protein appeared in the soluble fraction in the narJ::kat mutant of T. thermophilus (Fig. 2). Thus, in the thermophile, NarJ is also required for the attachment of the enzyme to the membrane NarCI complex.
DISCUSSION
The ability of T. thermophilus to grow anaerobically depends on the presence of a conjugative element, which encodes a complete respiratory electron transport chain consisting of a terminal reductase (NRT), and a primary electron donor, a new type of NADH dehydrogenase (NRC). Both enzyme complexes are expressed under the control of transcription factors that are also encoded by this “respiratory island” (7, 20, 21).
The structure of the E. coli NRA has been resolved at a resolution of 1.9 Angstrom (2). The enzyme consists of three subunits, containing eight redox centers that conduct the electrons from the outer face of the membrane through two heme B groups of NarI, four iron-sulfur clusters of NarH, to reach another iron-sulfur cluster of NarG, and finally its MGD cofactor, where nitrate reduction takes place. The amino acid residues that participate in the coordination of these eight redox centers are conserved in the sequence of the corresponding thermophilic homologues from the NRT, supporting the idea that the electron transfer pathway and, therefore, their spatial location in the structure of the enzyme are probably similar.
Similarly, the high degree of sequence identity between NarG (50%) and NarH (52.3%) of the NRT and their NRA counterparts (20) is consistent with the hypothesis of the conservation in the thermophilic enzyme of the extended interactions observed between their mesophilic homologues. In fact, the long α-helix that in the E. coli NarG (Asn28-Gln40) is engulfed by NarH, is also well conserved in the thermophilic NarG protein, implying the existence of a similar interaction between the thermophilic proteins. Despite this, we could not detect any significant interactions between NarG and NarH (Fig. 3), and, in parallel experiments, between their mesophilic homologues NarG and NarH (data not shown). Since extensive interactions between these proteins are evident from their structure (2), our data suggest the requirement of an additional component, probably the NarJ protein, to allow the detection of effective interactions between them. In support of this, the presence of the three proteins, NarG, NarH, and NarJ, of NRA is required to obtain positive interactions with the proteins of the MGD biosynthesis pathway in this two-hybrid assay (27).
In contrast to the great similarity of NarG and NarH to their E. coli counterparts, the NarI protein shows a lower degree of identity (28%) with the E. coli NarI. In fact, the C-terminal cytoplasmic domain, which is involved in important electrostatic and hydrogen-bonded interactions with the NarGH complex, is longer and more hydrophobic in the thermophilic NarI protein, suggesting that there are differences in the way in which the NarGH complex attaches to the membrane in the NRT. Concomitantly, the thermophilic NarG protein presents interesting sequence differences with the E. coli NarG near to its N terminus, which should affect the way in which it binds to NarI: the N-terminal α-helix (Ser1-Arg6), involved in membrane anchoring, and the immediate β-hairpin, which forms a twisted β-sheet with the C-terminal β-strand of NarI, are absent from the thermophilic NarG protein. Therefore, the existence of a different set of interactions between NarI and the NarGH complex in the nitrate reductase of T. thermophilus is supported at the sequence level.
In this context, our data strongly suggest that NarC constitutes an additional subunit of the NRT. Computer predictions of the topology of this protein suggest that a single transmembrane α-helix keeps the protein anchored to the membrane, with a small (20 amino acids) and positively charged (6 residues of Arg) C-terminal domain protruding into the cytoplasm. Our two-hybrid assays indicate that this small domain is responsible for its interaction with NarI (Fig. 3) but does not rule out the possible existence of interactions between this protein and either NarG, NarH, or even NarJ, whose presence is required for binding the enzyme to the membrane. Overall, and although putative conformational effects on NarI require consideration, the most likely explanation of the solubilization of NarG in the absence of NarC is the existence of some kind of interaction between them.
In addition to being required for membrane attachment, NarC contains two heme C-binding sequences, implying that two more redox centers are present in the NRT than in its mesophilic counterparts. Based on the aforementioned topology, and on the inhibition of electron transport toward the NRT by inhibitors of the quinone cycle (7), we propose that one of these heme C groups of NarC constitutes the first electron acceptor from the reduced menaquinone-8, the major quinone present in the membranes of anaerobically grown cells of T. thermophilus. Two unpublished results support this hypothesis. First, electron transport towards the NRT is sensitive to antimycin A, an inhibitor that acts between cytochromes b and c1 in mitochondrial respiration. Second, reduced TMPD (tetramethyl-para-phenyl-diamine), which donates the electrons preferentially to the cytochrome c in mitochondria (between complexes III and IV) (30), also functions as an electron donor for the NRT in a rotenone-insensitive manner.
In addition to the heterotetrameric nature of NRT, there are also important differences between the maturation pathways of the mesophilic and the thermophilic enzymes. First, a deficiency in either NarI or NarC results in an inactive enzyme, whereas in E. coli an active enzyme is synthesized even in the absence NarI (3, 5). Therefore, binding to the membrane is required for the activation of the enzyme in T. thermophilus but not in E. coli. Second, whereas a deficiency in NarJ results in a membrane-bound apo-nitrate reductase in E. coli, in T. thermophilus the absence of NarJ results in soluble apo-nitrate reductase. Consequently, NarJ is also required for the binding of the enzyme to the membrane in T. thermophilus.
All these data lead us to propose the model of enzyme maturation shown in Fig. 5. In this model, the synthesis of the NRT starts in two parallel cytoplasmic and membrane-bound pathways. We propose that a NarCI membrane complex is formed, as suggested by the two-hybrid assays and by the effect of the inactivation of any of them in the solubility of NarG. Thus, it can be inferred from our data that the cytoplasmic domain of NarC interacts with one or more of the cytoplasmic domains of NarI to form the binding site for the cytoplasmic complex.
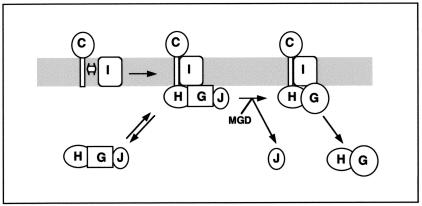
FIG. 5.
Model for the synthesis of nitrate reductase in T. thermophilus. A membrane NarCI and a soluble NarGHJ complex are initially formed. In the soluble complex, the NarG protein is inactive (squares). After attachment to the membrane, an inactive pentameric apo-nitrate reductase is formed. The enzyme is subsequently activated through the insertion of the Mo-bis-MGD cofactor (MGD) and the concomitant exit of the NarJ chaperone to render an active, heterotetrameric enzyme (circled NarG). Eventually, the active NarGH complex irreversibly separates from the membrane, giving rise to soluble forms that are active in nitrate reduction with artificial electron donors but not functional in nitrate respiration.
The cytoplasmic pathway is more similar to that proposed for E. coli. It has been recently shown in this organism that a tripartite NarJ-NarG-NarH complex is required for the interaction with enzymes of the MGD synthesis pathway (27). Interactions between NarG and NarJ were detected in the T. thermophilus model, and there is no doubt that extensive interactions between NarG and NarH are required for the activity of the enzyme. The presence of a significant fraction (50 to 60%) of protease-sensitive apo-nitrate reductase in the cytoplasm of wild-type cells of T. thermophilus during the synthesis of the NRT supports the existence of such a soluble and inactive NarGHJ complex.
In a subsequent step, this tripartite complex binds to the membrane, probably in a reversible manner. The requirement for NarJ in this complex was deduced from the solubilization of NarG in narJ::kat mutants and also from the presence of minor amounts of NarJ in membrane fractions (not shown). Moreover, the small amount of protease-sensitive NarG in the membrane (<10% [Fig. 4]) reflects the presence of such a membrane-bound complex before activation.
Once the five-member complex is bound to the cytoplasmic face of the membrane, activation takes place through the catalyzed insertion of the MGD cofactor and the concomitant conformational change, which produces a protease-resistant enzyme, and the exit of NarJ from the complex. In the E. coli system, this step requires interaction with proteins of the MGD biosynthesis pathway and takes place in the cytoplasm (27). Consequently, the participation of additional unknown factors in this activation process is a likely requirement in T. thermophilus. In this context, the soluble activity found after cell fractionation is explained as the result of some kind of turnover process that is not yet understood, as revealed by the increase in the amount of soluble activity after inhibition of the protein synthesis (not shown). Therefore, the soluble fraction of a wild-type cell growing anaerobically contains a mixture of “new” inactive and sensitive-to-protease apo-nitrate reductase and “old” but still active enzyme that has been separated from the membrane, probably irreversibly, as happens with its E. coli counterpart (9).
Acknowledgments
This work has been supported by project BIO2004-02671 from the “Ministerio de Educación y Ciencia”. An institutional grant from the Fundación Ramón Areces to CBMSO is acknowledged. O. Zafra and F. Cava held fellowships from the Comunidad de Madrid and the “Ministerio de Educación y Ciencia,” respectively.
We greatly appreciate the kind provision of the bacterial two-hybrid system by D. Ladant (Pasteur Institute, Paris, France) and L. Selig (Hybrigenics S.A., Paris, France). We are also grateful to M. Tamakoshi for sending us the leuB-based complementation system.
REFERENCES
- 1.Berks, B. C., S. J. Ferguson, J. W. Moir, and D. J. Richardson. 1995. Enzymes and associated electron transport systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim. Biophys. Acta 1232:97-173. [DOI] [PubMed] [Google Scholar]
- 2.Bertero, M. G., R. A. Rothery, M. Palak, C. Hou, D. Lim, F. Blasco, J. H. Weiner, and N. C. Strynadka. 2003. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat. Struct. Biol. 10:681-687. [DOI] [PubMed] [Google Scholar]
- 3.Blasco, F., J. Pommier, V. Augier, M. Chippaux, and G. Giordano. 1992. Involvement of the narJ and narW gene product in the formation of active nitrate reductase in Escherichia coli. Mol. Microbiol. 6:221-230. [DOI] [PubMed] [Google Scholar]
- 4.Blasco, F., J.-P. d. Santos, A. Magalon, C. Frixon, B. Guigliarelli, C.-L. Santini, and G. Giordano. 1998. NarJ is a specific chaperone required for molybdenum cofactor assembly in nitrate reductase A of Escherichia coli. Mol. Microbiol. 28:435-447. [DOI] [PubMed] [Google Scholar]
- 5.Buc, J., C. L. Santini, F. Blasco, R. Giordani, M. L. Cárdenas, M. Chippaux, A. Cornish-Bowden, and G. Giordano. 1995. Kinetic studies of a soluble alpha beta complex of nitrate reductase A from Escherichia coli. Use of various alpha beta mutants with altered beta subunits. Eur. J. Biochem. 234:766-772. [DOI] [PubMed] [Google Scholar]
- 6.Castán, P., M. A. de Pedro, C. Risco, C. Vallés, L. A. Fernández, H. Schwarz, and J. Berenguer. 2001. Multiple regulatory mechanisms act on the 5′ untranslated region of the S-layer gene from Thermus thermophilus HB8. J. Bacteriol. 183:1491-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cava, F., O. Zafra, A. Magalon, F. Blasco, and J. Berenguer. 2004. A new type of NADH dehydrogenase specific for nitrate respiration in the extreme thermophile Thermus thermophilus. J. Biol. Chem. 279:45369-45378. [DOI] [PubMed] [Google Scholar]
- 8.Degryse, E., N. Glansdorff, and A. Piérard. 1978. A comparative analysis of extreme thermophilic bacteria belonging to the genus Thermus. Arch. Microbiol. 117:189-196. [DOI] [PubMed] [Google Scholar]
- 9.Demoss, J. A., T. Y. Fan, and R. H. Scott. 1981. Characterization of subunit structural alterations which occur during purification of nitrate reductase from Escherichia coli. Arch. Biochem. Biophys. 206:54-64. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan, D. 1983. Studies on transformation of E. coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 11.Karimova, G., J. Pidoux, A. Ullmann, and D. Ladant. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. USA 95:5752-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karimova, G., A. Ullmann, and D. Ladant. 2000. A bacterial two-hybrid system that exploits a cAMP signaling cascade in Escherichia coli. Methods Enzymol. 328:59-73. [DOI] [PubMed] [Google Scholar]
- 13.Koyama, Y., T. Hoshino, N. Tomizuka, and K. Furukawa. 1986. Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J. Bacteriol. 166:338-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laemmli, U., and M. Favre. 1973. Maturation of the head of bacteriophage T4.I; DNA packaging events. J. Mol. Biol. 80:575-599. [DOI] [PubMed] [Google Scholar]
- 15.Lasa, I., J. R. Castón, L. A. Fernandez-Herrero, M. A. Pedro, and J. Berenguer. 1992. Insertional mutagenesis in the extreme thermophilic eubacteria Thermus thermophilus. Mol. Microbiol. 11:1555-1564. [DOI] [PubMed] [Google Scholar]
- 16.Llosa, M., S. Zunzunegui, and F. de la Cruz. 2003. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc. Natl. Acad. Sci. USA 100:10465-10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacGregor, C. H. 1976. Biosynthesis of membrane-bound nitrate reductase in Escherichia coli: evidence for a soluble precursor. J. Bacteriol. 126:122-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magalon, A., C. Frixon, J. Pommier, G. Giordano, and F. Blasco. 2002. In vivo interactions between gene products involved in the final stages of molybdenum cofactor biosynthesis in Escherichia coli. J. Biol. Chem. 277:48199-48204. [DOI] [PubMed] [Google Scholar]
- 19.Oshima, M., and K. Imahori. 1974. Description of Thermus thermophilus (Yoshida and Oshima) comb. nov., a nonsporulating thermophilic bacterium from a Japanese hot spa. Int. J. Syst. Bacteriol. 24:102-112. [Google Scholar]
- 20.Ramírez-Arcos, S., L. A. Fernández-Herrero, and J. Berenguer. 1998. A thermophilic nitrate reductase is responsible for the strain specific anaerobic growth of Thermus thermophilus HB8. Biochem. Biophys. Acta 1396:215-227. [DOI] [PubMed] [Google Scholar]
- 21.Ramírez-Arcos, S., L. A. Fernández-Herrero, I. Marín, and J. Berenguer. 1998. Anaerobic growth, a property horizontally transferred by an Hfr-like mechanism among extreme thermophiles. J. Bacteriol. 180:3137-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramírez-Arcos, S., R. Moreno, O. Zafra, P. Castán, C. Vallés, and J. Berenguer. 2000. Two nitrate/nitrite transporters are encoded within the mobilizable plasmid for nitrate respiration of Thermus thermophilus HB8. J. Bacteriol. 182:2179-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothery, R. A., M. G. Bertero, R. Cammack, M. Palak, F. Blasco, N. C. Strynadka, and J. H. Weiner. 2004. The catalytic subunit of Escherichia coli nitrate reductase A contains a novel [4Fe-4S] cluster with a high-spin ground state. Biochemistry 43:5324-5333. [DOI] [PubMed] [Google Scholar]
- 24.Snell, F. D., and C. T. Snell. 1949. Colorimetric methods of analysis. Van Nostrand, New York, N.Y.
- 25.Sonnhammer, E., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences, p. 175-182. Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology (ISMB98). [PubMed]
- 26.Tamakoshi, M., M. Uchida, K. Tanabe, S. Fukuyama, A. Yamagishi, and T. Oshima. 1997. A new Thermus-Escherichia coli shuttle integration vector system. J. Bacteriol. 179:4811-4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vergnes, A., K. Gouffi-Belhabich, F. Blasco, G. Giordano, and A. Magalon. 2004. Involvement of the molybdenum cofactor biosynthetic machinery in the maturation of the Escherichia coli nitrate reductase A. J. Biol. Chem. 279:41398-41403. [DOI] [PubMed] [Google Scholar]
- 28.Vieira, J., and J. Messing. 1987. Production of single-stranded plasmid DNA. Methods Enzymol. 153:3-11. [DOI] [PubMed] [Google Scholar]
- 29.Zafra, O., S. Ramírez-Arcos, P. Castán, R. Moreno, F. Cava, C. Vallés, E. Caro, and J. Berenguer. 2002. A cytochrome c encoded by the nar operon is required for the synthesis of active respiratory nitrate reductase in Thermus thermophilus. FEBS Lett. 523:99-102. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, Z., L. Huang, V. M. Shulmeister, Y. I. Chi, K. K. Kim, L. W. Hung, A. R. Crofts, E. A. Berry, and S. H. Kim. 1998. Electron transfer by domain movement in cytochrome bc1. Nature 392:677-684. [DOI] [PubMed] [Google Scholar]
- 31.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]