Abstract
Target site selection of transposable elements is usually not random but involves some specificity for a DNA sequence or a DNA binding host factor. We have investigated the target site selection of the long terminal repeat-containing retrotransposon Tf1 from the fission yeast Schizosaccharomyces pombe. By monitoring induced transposition events we found that Tf1 integration sites were distributed throughout the genome. Mapping these insertions revealed that Tf1 did not integrate into open reading frames, but occurred preferentially in longer intergenic regions with integration biased towards a region 100–420 bp upstream of the translation start site. Northern blot analysis showed that transcription of genes adjacent to Tf1 insertions was not significantly changed.
INTRODUCTION
Transposable elements are discrete DNA segments that can translocate between non-homologous insertion sites. Although they usually insert into many different sites, most elements display some degree of target selectivity. This serves to facilitate element propagation and to avoid insertions into essential genes of the host. A target site may be selected through direct interactions between the transposable element and the target DNA or through its association with accessory proteins that may be element- and/or host-encoded (reviewed in 1).
Well-studied examples of transposable element targeting include Drosophila P elements and the Saccharomyces cerevisiae long terminal repeat (LTR)-containing retrotransposons Ty1–Ty5. P elements have a preference for 5′ untranslated regions and for regions 100–200 bp upstream of the transcription start site. This region-specific integration can result in changes in the expression level of adjacent genes and has been exploited for genetic screens (for review see 2,3). In the case of Ty1–Ty4, >90% of native insertions in the genome are found in upstream regions of genes transcribed by RNA polymerase III (Pol III), such as tRNA genes (4). Ty1 integrates 70–700 bp upstream of the transcription start site of these genes and it has been demonstrated that this is due to interactions between the retrotransposon and a component of the RNA Pol III transcriptional apparatus and/or through its recognition of chromatin structures associated with RNA Pol III transcription (5–9). Furthermore, Ty1 integration also occurs within pre-existing LTRs (10), as well as upstream or within a variety of Pol II-transcribed genes which can result in alterations in the expression levels of these genes (11–13). For Ty3, target specificity appears to depend on the ability of the Ty3 integration machinery to compete with Pol III for interactions with TFIIIB and TFIIIC transcription factors (14,15). Ty3 inserts only at Pol III-transcribed genes and integrates in a site-specific manner within a few base pairs of the transcription start site. Ty5 typically integrates near telomeres or the silent mating loci, HML and HMR (16,17), and it is thought that integration complexes recognise chromatin or other DNA-bound proteins (18).
The LTR-retrotransposon Tf1 isolated from the fission yeast Schizosaccharomyces pombe (19) contains a single open reading frame (ORF) (20) encoding Gag, protease, reverse transcriptase and integrase proteins (19,21). Tf1 integrates into genomic DNA due to transposition events, and it has been shown that this transposition activity is independent of temperature (21). Like Ty3, Tf1 is a member of the gypsy family of transposable elements (19), but it remains to be determined whether it also has a preference for Pol III promoters. Here we report the analysis of the target site specificity of Tf1. By examining the genomic distribution of induced transposition events we found that Tf1 is preferentially targeted to intergenic regions close to the 5′ end of ORFs with no preference for pol III promoters.
MATERIALS AND METHODS
Media, strains and plasmids
Strains were grown in complete media (YES) and in minimal media (EMM) (22). Amino acids and pyrimidines were supplemented at a final concentration of 225 µg/ml. Vitamin B1 (thiamine, 15 µM) was added to EMM to repress the nmt1 promoter. Phloxin B (Sigma) was added at 5 mg/l to facilitate screening for cdc25 revertants. 5-Fluoroorotic acid (5-FOA) (Milford Laboratories) plates were made by adding 1 mg/ml of 5-FOA to EMM. YES + 5-FOA + G418 plates contained 1 mg/ml of 5-FOA and 0.5 mg/ml (corrected for purity) of Geneticin (Gibco). The following yeast strains were used: ura4-D18 leu1-32 h- (strain 513) and cdc25-22 ura4-297 leu1-32 h- (strain 2586). The plasmid pHL414 (Fig. 1A) was described earlier (20).
Figure 1.
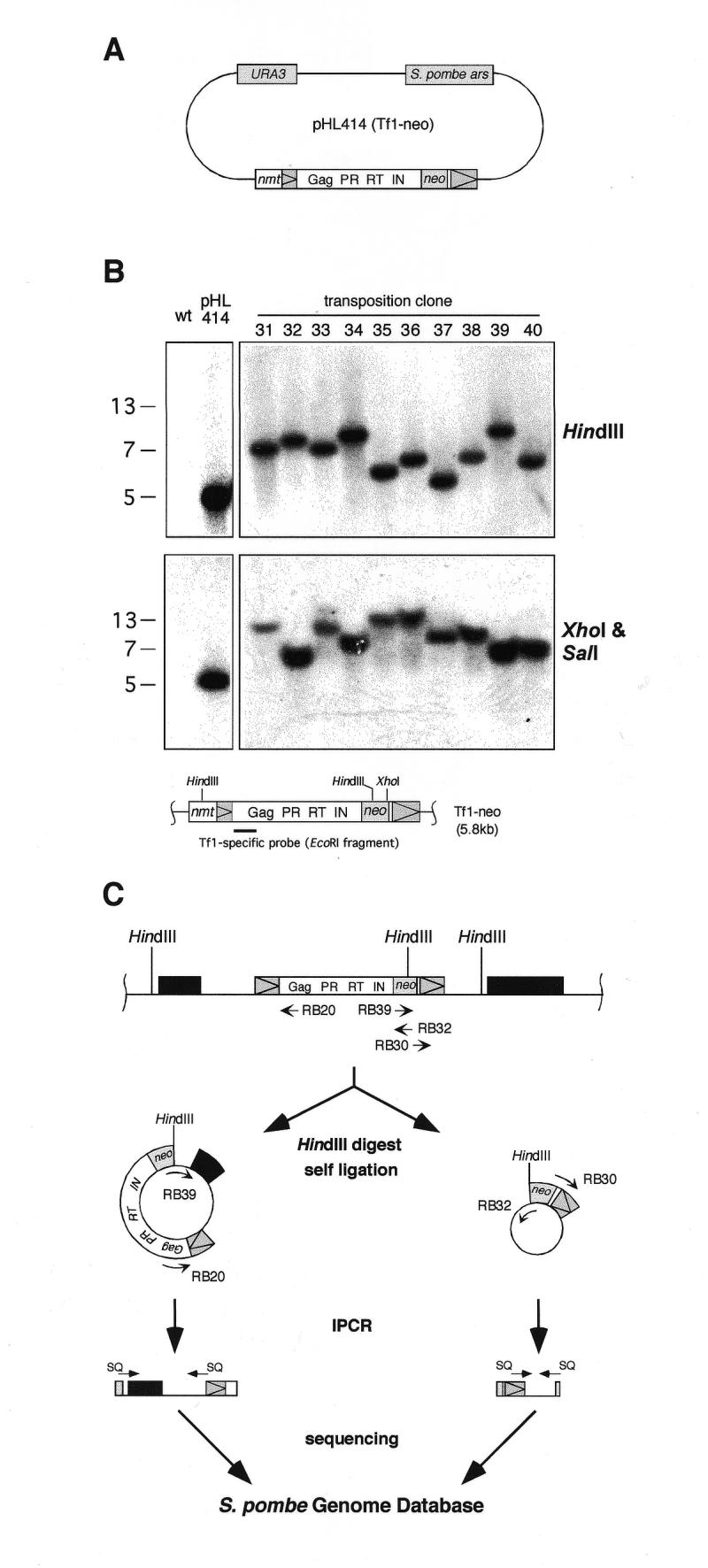
Determination of genomic Tf1-neo insertions. (A) The transposition assay plasmid pHL414 contained the URA3 gene of S.cerevisiae and the Tf1-neo element fused to an inducible nmt promoter. The neo gene inserted in Tf1 confers G418 resistance. The large white box represents the single full-length ORF of Tf1. The positions of the protein domains Gag, protease (PR), reverse transcriptase (RT) and integrase (IN) are indicated. Triangles within grey boxes represent LTRs. The plasmid contains an autonomous replication sequence of S.pombe (ars). (B) Southern blot analysis of Tf1 transposition clones. DNA from G418-resistant clones was cut with restriction enzymes as indicated, blotted, and hybridised to a Tf1-specific probe shown in the schematic presentation of the nmt–Tf1-neo fusion construct. As controls, DNA from an isogenic strain without Tf1 insertion (wt) and from the same strain carrying the pHL414 plasmid (pHL414) in the presence of thiamine were used. Of 40 analysed transposition clones, 10 representative clones are shown here. Molecular weights are shown to the left of the gel. (C) The genomic locations of induced Tf1-neo insertion events were determined using IPCR and the S.pombe genome database. IPCR primers (see Materials and Methods; RB30, RB32: primer set 1; RB20, RB39: primer set 2) and sequencing primers (SQ) are shown as arrows. HindIII restriction sites are indicated, and genes are represented as black boxes.
Tf1 transposition assay
The transposition assay was described previously (20). Strain 513 carrying the pHL414 plasmid was first streaked out to single colonies on EMM + leucine + thiamine plates. Individual colonies were picked and used to inoculate the growth of patches on EMM + leucine plates. After 2 days at 32°C, the patches were used to inoculate a culture of EMM + leucine at an optical density at 595 nm (OD595) of 0.1. These cultures were grown for another 2 days at 32°C before plating on EMM + 5-FOA + uracil + leucine at a density of 500 cells per plate. After 3 days at 32°C, 5-FOA-resistant colonies were replica plated onto YES + 5-FOA + G418 plates and incubated for a further 2 days to obtain colonies that had undergone a transposition event. From each of four independent transposition assays 10 G418-resistent colonies were picked and analysed by Southern blotting. Small and large colonies were selected in order to take into account that Tf1 integration could have affected the growth rate of some clones.
Southern blotting
Southern blots (23) of genomic DNA of putative transposition clones were hybridised to a 32P-labelled EcoRI fragment (19) of Tf1 (Fig. 1B). DNA extracted from strain 513 did not hybridise to this probe because this strain was isogenic to strain 972, which does not contain a Tf1 copy (19). The URA3 probe used was a 1040 bp HinfI fragment of the URA3 gene (24). A 673 bp probe of the neo gene was obtained by PCR (Expand High Fidelity PCR system, Boehringer Mannheim) using the pFA6-kanMX6 plasmid as template (25).
Determination of Tf1 insertion sites
To isolate regions flanking the insertion sites of the retrotransposon, 2 µg of DNA from clones shown by Southern blotting to have undergone a transposition event were digested with HindIII which cuts within the neo gene of Tf1-neo. DNA was diluted to 1 ng/µl, T4 Ligase (NEB) was added at 1 U/µl and the mix was incubated at 16°C overnight. After phenol–chloroform extraction and ethanol precipitation, 100 ng of self-ligated DNA was used as template for inverse PCR (IPCR) (Expand High Fidelity PCR system, Boehringer Mannheim). Self-ligated fragments were amplified during 35 cycles in a 50 µl reaction using either of two inverse primer sets (Fig. 1C). Primer set 1 (annealing at 55°C, 30 s) was RB30 (5′-GGAGTACGGATAAAATGCTTGATGG-3′) and RB32 (5′-CGGTTTGGTTGATGCGAGTG-3′) corresponding to sequences in the neo gene downstream of the HindIII site. Primer set 2 (annealing at 53°C, 30 s) was RB20 (5′-TGTCCCAGTAAGAATCACCTCTCTG-3′) and RB39 (5′-AGGAGAAAACTCACCGAGGCAG-3′) corresponding to a sequence in the unique EcoRI fragment of Tf1 and to a sequence in the neo gene upstream of the HindIII site in Tf1-neo, respectively. The extension reaction was carried out at 68°C with extension times chosen according to the size of the fragments, as predicted by Southern blot analysis. PCR products were gel-purified (Qiagen) and sequenced using Big Dye Terminator cycle sequencing (Perkin-Elmer). Oligonucleotides derived from the LTR and the neo gene were used as primers. From the sequencing data, the Tf1 genomic insertion site was determined using the S.pombe genome database of the Sanger Centre (http://www.sanger.ac.uk).
Screen for the disruption of wee1 in a cdc25 temperature-sensitive strain
Strain 2586 was transformed with plasmid pHL414 and transformants were streaked out to single colonies on EMM + leucine + thiamine plates at 25°C. An individual colony was inoculated into 10 ml EMM + leucine + thiamine and grown at 25°C overnight. After washing the cells twice with water, 100 ml of EMM + leucine was inoculated at a cell density of OD595 = 0.1 and incubated at the permissive temperature of 25°C for 24 h. Cells were then plated on EMM + leucine + uracil + 5-FOA + Phloxin B at a density of 107 cells per plate and incubated at the restrictive temperature of 36°C for 4 days. 5-FOA resistant colonies were replica plated onto YES + 5-FOA + G418 and incubated for 2 days at 36°C. G418-resistant revertants were examined for the wee1 phenotype by light microscopy and then analysed for disruption of the wee1 gene by Southern blotting. Genomic DNA from revertants was digested with AlwNI, blotted and hybridised to Tf1-specific and wee1-specific probes (2086 bp KpnI–BglII fragment of the wee1 ORF, see Fig. 4).
Figure 4.
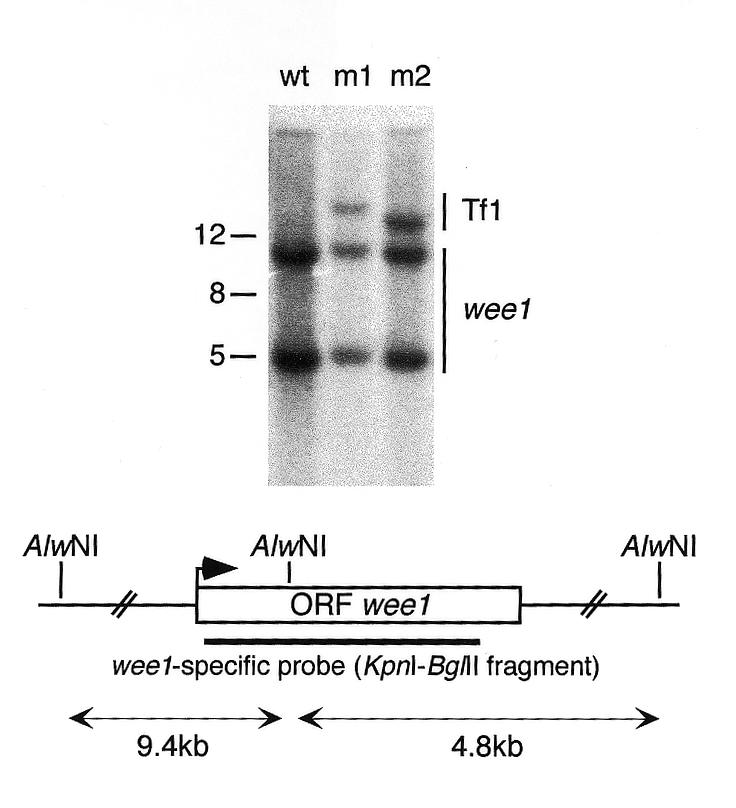
Screen for Tf1 insertion into the wee1 gene. The pHL414 plasmid was transformed into a cdc25 temperature-sensitive strain and transposition was induced at the permissive temperature. G418-resistant revertants which grew at the restrictive temperature were analysed for the disruption of the wee1 gene. Genomic DNA from such clones (m1, m2) was digested with AlwNI and analysed by Southern blotting using wee1- and Tf1-specific (see Fig. 1B) probes. The blot shows that the Tf1 insertions were not on the same genomic fragment as the wee1 gene. As control, DNA from a strain without Tf1 insertion was included (wt). Molecular weights are shown to the left of the gel. The wee1 ORF is presented as a box with an arrow indicating the translation start site, and the AlwNI restriction sites in the vicinity of the wee1 gene are indicated.
RNA preparation and northern blot analysis
Northern blots of RNA (26) were hybridised to 32P-labelled probes prepared by random oligo priming using a Prime-It kit (Stratagene). As a loading control, blots were either stained with methylene blue for the 1.8 and 3.5 kb rRNAs, or hybridised to a probe of the his3 ORF (DraI–EcoRV fragment). Probes for the following (predicted) ORFs were obtained by PCR using genomic DNA from strain 513 as template: ubc4, SPBC119.01, SPAC22G7.09, SPAC22G7.10, SPBC649.05, uvi15, SPCC1223.10, ptc2, SPCC777.07, SPCC777.06, SPCC1672.01, SPCC962.06, SPAC4F8.13, SPAC4F8.12, SPCC14G10.05, SPCC14G10.04. The size of the probes ranged from 230 to 1038 bp, and they did not contain intronic sequences.
RESULTS AND DISCUSSION
In order to investigate target site preference for Tf1 LTR retrotransposon insertion we used the plasmid pHL414 kindly provided by H. L. Levin (20), which contained Tf1 under control of the nmt1 promoter (Fig. 1A). The transposon contained the neo gene allowing insertional events to be selected on the basis of stable G418 resistance, and the plasmid contained the S.cerevisiae URA3 gene which can complement S.pombe ura4 mutations. The plasmid was transformed into ura4– cells, and transposition was induced by depleting thiamine from the medium in order to activate the nmt1 promoter. Next, cells were plated on 5-FOA to select for uracil auxotrophs to ensure that the original plasmid was lost from the cells, and colonies were selected for G418 resistance ensuring that Tf1 was still present in the cells. Usually, G418 resistance can only occur if a Tf1 transposition event from the plasmid to the chromosome has taken place.
The majority of Tf1 insertions were at different locations in the genome
To investigate the distribution of Tf1 insertions within the genome we analysed 40 putative transposition events by Southern blotting (Fig. 1B). Of these, 34 (85%) showed unique restriction patterns indicating that insertions occurred at sites distributed throughout the genome. The remaining six insertions (15%) consisted of three pairs with each pair having an identical restriction pattern which differed from the other two pairs. The two insertions of each of these three pairs could have occurred on the same genomic fragment. However, they could also have coincidentally occurred on different fragments of similar size as was the case with clones 31 and 33, which showed a similar restriction pattern but mapped to different locations on chromosome I (Table 1). Probing the blots with a URA3-specific probe gave no signal showing that the pHL414 plasmid was not present in the clones, either as free plasmid or integrated in the genome (data not shown). We also confirmed the integrity of the transposon by reprobing the blots for the neo gene (data not shown). These results indicate that Tf1 insertion occurs at sites distributed throughout the genome.
Table 1. Location of 27 Tf1 insertion sites.
| Insertion clone | Chromosome | 5′-Tf1a to ORF | Name ORF | 3′-Tf1a to ORF | Name ORF | Sizea intergenic region | Orientationb gene Tf1 gene |
|---|---|---|---|---|---|---|---|
| 1 | 3 | 1093 | SPCC794.10 | 394 | SPCC794.09 | 1487 | ← ← → |
| 2 | 2 | 980 | SPBC119.01 | 258 | ubc4 | 1238 | → → → |
| 5 | 2 | 3582 | SPBC18H10.19 | 346 | SPBC18H10.18 | 3928 | ← ← → |
| 8 | 1 | 141 | SPAC23G3.12 | 1458 | SPAC23G3.13 | 1599 | ← → ← |
| 9 | 1 | 273 | SPAC22G7.09 | 221 | SPAC22G7.10 | 494 | ← → → |
| 12 | 3 | 2089 | SPCC188.09 | 1124 | SPCC188.10 | 3213 | ← → ← |
| 13 | 2 | 534 | SPBC649.05 | 789 | uvi15 | 1323 | → ← → |
| 14 | 2 | 3476 | SPBP4H10.15 | 421 | SPBP4H10.14 | 3897 | ← ← → |
| 15 | 3 | 1008 | SPCC1223.10 | 900 | ptc2 | 1908 | ← → → |
| 16 | 2 | 1007 | SPBC14F5.03 | 571 | pgk1 | 1578 | ← → ← |
| 17 | 1 | 101 | SPAC11H11.05 | 681 | SPAC11H11.06 | 782 | ← → → |
| 20 | 2 | 874 | SPBC29A3.01 | 535 | SPBP23A10.16 | 1409 | ← → ← |
| 21 | 3 | 1771 | SPCC777.07 | 307 | SPCC777.06 | 2078 | ← ← → |
| 24 | 1 | 238 | ypt3 | 1718 | SPAC18G6.02 | 1956 | ← ← → |
| 25 | 3 | 1576 | SPCC1672.01 | 287 | SPCC962.06 | 1863 | ← ← → |
| 26 | 2 | 308 | his5 | 978 | SPBC21H7.08 | 1286 | ← → → |
| 27 | 1 | 191 | SPAC20G8.09 | 135 | SPAC20G8.10 | 326 | ← → ← |
| 28 | 1 | 541 | SPAC4F8.13 | 211 | SPAC4F8.12 | 752 | ← ← ← |
| 29 | 2 | 261 | SPBC26H8.12 | 2535 | SPBC26H8.11 | 2796 | ← ← → |
| 30 | 3 | 176 | SPCC70.04 | 4493 | SPCC70.05 | 4669 | ← → ← |
| 31 | 1 | 2194 | SPAC1639.02 | 898 | SPAC1639.01 | 30921 | ← ← ← |
| 32 | 1 | 272 | ckb1 | 600 | SPAC1851.02 | 872 | → ← → |
| 33 | 1 | 2427 | SPAC29B12.08 | 1012 | SPAC29B12.07 | 3439 | → ← → |
| 37 | 3 | 1514 | SPCC14G10.04 | 169 | SPCC14G10.05 | 1683 | → → → |
| 38 | 1 | 330 | SPAC2G11.09 | 576 | SPAC2G11.08 | 906 | ← ← → |
| 39 | 3 | 144 | SPCC777.05 | 469 | SPCC777.04 | 613 | → ← → |
| 40 | 3 | 4296 | SPCC1884.01 | 770 | SPCC1884.02 | 5066 | → → → |
aDistance in base pairs to the closest ORF (5′ or 3′ end).
bArrows indicate 5′ to 3′ orientation; the arrows representing Tf1 are printed in bold and are shown between the arrows representing the two genes adjacent to each insertion site.
Tf1 inserted preferentially in a 320 bp band at the 5′ end of ORFs
Next, we used IPCR and the S.pombe genome database to determine the genomic location of 27 Tf1 insertion sites (Fig. 1C). The mapped Tf1 copies were flanked by 5 bp duplications of the insertion site indicating that they were the result of a true transposition event and not due to recombination (21).
As shown in Table 1, all insertions occurred in intergenic regions. Our analysis of the available S.pombe genome (J.Sgouros, personal communication, based on sequence information made available from the S.pombe Genome Project EU Consortium) suggested that 62% of the genome consists of coding sequences and from comparison with S.cerevisiae we predict that ~83% of these would be non-essential (27). Thus, we would expect that 52% of the transposition clones would have an insertion in a non-essential ORF. Given the lack of any such event, we can conclude that Tf1 integration was not random, but was significantly biased towards intergenic regions.
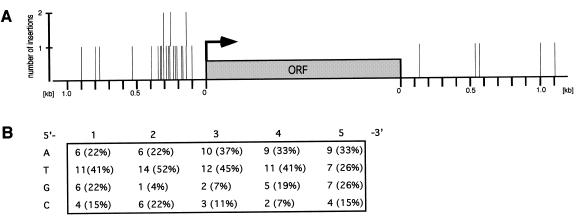
To investigate whether the insertion of Tf1 within intergenic regions was random, we analysed the distribution of insertion sites with respect to the 5′ or 3′ region of the nearest gene. In order to do so, we divided the targeted intergenic regions in half and allocated the insertions to the 5′ or 3′ region of the closest ORF. On this basis, 10 of 15 insertions between tandem ORFs and all 12 Tf1 insertions between divergent ORFs mapped closer to the 5′ than to the 3′ end of an ORF. Correspondingly, Figure 2A shows 22 Tf1 insertions in the 5′ region and five insertions in the 3′ region of an representative ORF. The 5′ region of Tf1-targeted genes had a mean size of 980 bp as determined by calculating the mean size of Tf1-targeted intergenic regions flanked by ORFs in divergent orientation (1.96 ± 1.1 kb, n = 12). Thus, if transposition into the 5′ region of genes was completely random, 33% of the insertions would have been found in a region of 320 bp. However, our analysis revealed that 82% (18/22) of the insertions were in a band 100–420 nt upstream of the translation start site of an ORF. This significant bias of Tf1 integration towards a 320 bp band close to the 5′ end of ORFs is further supported by the lack of any insertion between convergent ORFs. Taking into account the frequency and the average size of the different intergenic regions, we would have expected to find 10–11 Tf1 insertions in intergenic regions flanked by divergent ORFs (27%, 1.34 ± 1.1 kb), 12–13 insertions in regions flanked by tandem ORFs (45%, 0.97 ± 0.8 kb) and four to five insertions in regions between convergent ORFs (28%, 0.53 ± 0.5 kb). We found 12 insertions between divergent ORFs and 15 insertions between tandem ORFs, but no insertion between convergent ORFs (Table 1). This suggests that Tf1 avoids convergent intergenic regions entirely, presumably because of its preference for 5′ ends of ORFs.
Figure 2.
Analysis of Tf1 integration sites within intergenic regions. (A) The location of all mapped Tf1 insertion sites with regard to the closest ORF (5′ or 3′ end) is shown. Bars indicate single Tf1 insertions. Long bars are used to represent two Tf1 insertions which occurred at about the same distance to the closest ORF (clone 8, 141 bp and clone 39, 144 bp; clone 2, 258 bp and clone 29, 261 bp; clone 21, 307 bp and clone 26, 308 bp). The arrow indicates the 5′ end of an ORF (grey box). (B) Composition of 5 bp Tf1 target site duplication. The total and average base usage at the 5 bp duplicated target site is indicated. The total number of analysed target sites was 27 and the orientation of the target site is 5′ to 3′ and parallel to the Tf1 element.
If Tf1 simply had a preference for 5′ ends of ORFs we would have expected that the mean size of Tf1-targeted intergenic regions would have been equivalent to the mean size of the corresponding regions in the genome. However, we found that the mean size of Tf1-targeted intergenic regions flanked by divergent (1.95 ± 1.1 kb, n = 12) and tandem (2.06 ± 1.5 kb, n = 15) ORFs was larger than the mean size of these regions in the genome (1.34 ± 1.1 kb for divergent ORFs; 0.97 ± 0.8 kb for tandem ORFs). This difference was highly significant for regions flanked by tandem ORFs (P < 0.01, two-tailed paired t-test), and showed a good significance for regions flanked by divergent ORFs (P = 0.06). Thus, our analysis suggests that Tf1 integrated preferentially into particular large intergenic regions, and that this was not directly related to its preference for a region at the 5′ ends of ORFs.
We examined next whether Tf1 inserted at 5′ ends of ORFs by targeting directly or indirectly a particular DNA sequence. The retroelement did not show a preference for AT-rich regions as has been reported for Ty1 elements (28). We determined the AT content of 64 ± 1% for the S.pombe genome by analysing 10 randomly picked cosmids. Regions comprising 500, 100 and 20 bp on either side of the insertion sites had an average AT content of 67 ± 2%, 68 ± 5% and 67 ± 9%, respectively. Analysis of the duplicated Tf1 insertion site (21) revealed no consensus sequence, but did suggest that the four bases were not used equally. The second position was found to be enriched for T and depleted for G, the third position was depleted for G and C and the fourth position was depleted for C (Fig. 2B). Further, the target site showed an elevated AT content at the second (74%), third (84%) and fourth (74%) position when compared to the AT content of the S.pombe genome. To consider the possibility that Tf1 is targeted by a host DNA binding factor we searched for potential binding sites 500 bp upstream and downstream of the different insertion sites using the MACAW software as well as multiple alignment and PROFILE searches, but could not find any consensus sequence.
It has been shown previously that Ty1 elements insert with high frequency within pre-existing LTRs (10). In S.pombe Tf1 solo LTRs are distributed throughout the genome and occur at a frequency of 1.5 LTRs every 27 intergenic region, as determined by our genome analysis. The Tf1 insertions in clones 5, 15 and 40 were at a distance of 27 bp, 473 bp and 2.6 kb to a LTR, respectively, and thereby closer to a LTR than to at least one of the two adjacent ORFs. In clone 12, Tf1 inserted even within a LTR. Although the significance of these findings is difficult to evaluate it remains a possibility that Tf1 has a preference for LTRs, either by being targeted directly to LTR sequences or by a chromatin feature which targeted ancient retroelements to these regions.
For budding yeast Ty1 and Ty3 elements, a preference for 5′ ends of tRNA genes has been reported previously (6,7). However, we did not observe this preference for Tf1. Furthermore, our analysis revealed that Tf1 integration was not biased towards a particular chromosome or a region on a chromosome (Table 1), with the exception of two Tf1 insertion events which occurred within a distance of 3 kb of each other (clones 21 and 39). In this region there was no evidence for a local recombination event or the presence of particular sequences, like LTRs or rRNA and tRNA genes.
Tf1 integration had no significant effect on transcription of adjacent genes
It has been reported that Ty1 inserts into 5′ untranslated regions and can activate, inactivate or confer constitutive expression or mating-type regulation on neighbouring transcriptional units (13). P elements can mutate genes in a similar way by preferentially inserting into the 5′ untranslated region as well as into a region 100–200 bp upstream of the transcription start site (2). To analyse the effects of Tf1 insertion on the transcription of adjacent genes, we examined eight transposition clones by northern blot analysis (Table 2). As shown in Figure 3, the changes in the transcription level of the examined genes were <2-fold and the size of the transcripts remained unchanged when compared to a control. These results suggest that, unlike Ty1 and P elements, Tf1 does not preferentially target 5′-UTRs and that the retrotransposon can insert close to ORFs without interfering with transcription. Since very little is known about the size of promoter regions in S.pombe we speculate that these might be small in some genes and remain unaffected by an upstream Tf1 insertion. Alternatively, Tf1 might have affected the function of some transcription regulation elements upon insertion, but the retroelement controls the expression of adjacent genes with its own LTR-located promoter and enhancer elements, as it has been described for Ty1 elements (29,30).
Table 2. Transposition clones used in northern blot analysis.
| Clone | Closest genesa | Encoded protein | Distanceb |
|---|---|---|---|
| 2 | 5′-ubc4 | ubiquitin-conjugating enzyme | 258 |
| 3′-SPBC119.01 | putative proteosome subunit | 980 | |
| 9 | 5′-SPAC22G7.09 | similar to nucleoporin nup49/nsp49 | 273 |
| 5′-SPAC22G7.10 | hypothetical protein | 221 | |
| 13 | 5′-SPBC649.05 | hypothetical protein | 534 |
| 3′-uvi15 | UV-induced protein uvi15 | 789 | |
| 15 | 5′-SPCC1223.10 | hypothetical protein | 1008 |
| 5′-ptc2 | protein phospatase 2C homolog 2 | 900 | |
| 21 | 5′-SPCC777.07 | similar to alpha-1,2 mannosyltransferase ktr1 | 1771 |
| 5′-SPCC777.06 | similar to metal dependent hydrolase | 307 | |
| 25 | 5′-SPCC1672.01 | similar to histidinol-phosphatase | 1576 |
| 5′-SPCC962.06 | similar to mammalian splicing factor SF1 | 287 | |
| 28 | 5′-SPAC4F8.12 | similar to splicing factor prp8 | 211 |
| 3′-SPAC4F8.13 | similar to rasgap-related protein | 541 | |
| 37 | 5′-SPCC14G10.05 | similar to phospholipase | 169 |
| 3′-SPCC14G10.04 | similar to extensin-like protein precursor | 1514 |
a5′ and 3′ indicate that Tf1 inserted close to the 5′- or 3′-end of the ORF, respectively.
bIn bp to the closest ORF.
Figure 3.
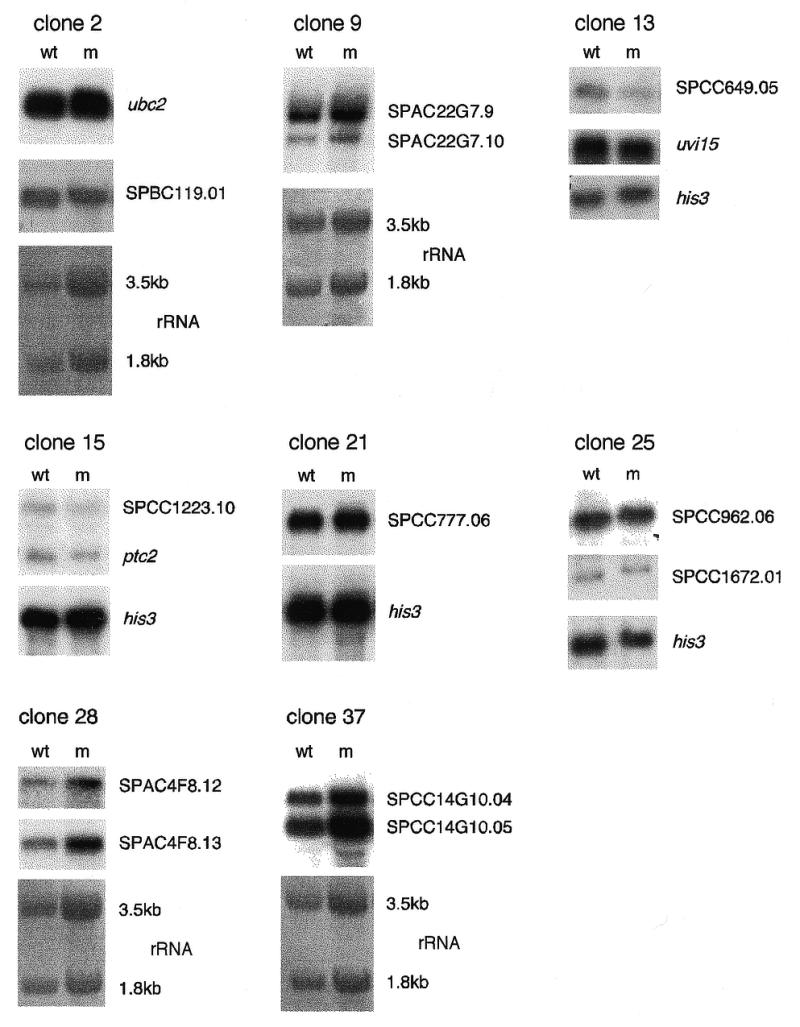
Northern blot analysis of genes adjacent to Tf1 insertions. The transcription level and the size of the transcript of the two genes adjacent to a Tf1 insertion were analysed by northern blot. The names of the gene transcripts detected from transposition (m) and control clones (wt) are shown to the right of the gels. No transcript was detected for SPCC777.07 (clone 21). As a loading control, rRNA was stained with methylene blue for the 3.5 and 1.8 kb band, or the his3 transcript level was monitored.
Tf1 did not disrupt the wee1 ORF in a screen for cdc25 revertants
To investigate the frequency of Tf1 insertion into a particular gene we screened for transposon induced revertants of a temperature-sensitive cdc25-22 mutant because generally such revertants are due to inactivation of the wee1 ORF (31). We induced transposition from the pHL414 plasmid in a cdc25 temperature-sensitive strain at the permissive temperature, followed by selection of cells able to grow at the restrictive temperature on 5-FOA and G418. We screened >8 × 106 cells carrying a Tf1 insertion and would have expected to obtain approximately 1700 Tf1 integrations in the wee1 ORF if transposition were completely random. However, we obtained only two transposition clones which grew at 36°C and their revertant phenotype was not due to a Tf1 insertion within or close to the wee1 gene as shown by Southern blotting (Fig. 4). This result is consistent with our previous findings that Tf1 targets 5′ ends of ORFs without affecting the expression of the encoded protein. In preliminary experiments we used a modified transposition protocol and screened for transposition clones auxotrophic for glutamate, adenine, histidine or lysine. No auxotrophic mutants with a transposon insertion were found.
Physiological stress did not alter Tf1 transposition
In order to see if we could increase or randomise transposition, we induced transposition while subjecting S.pombe cells to various treatments which were reported to increase transposition of Ty1 elements (32–34). We investigated transposition during (i) damaging DNA with 20 µg/ml bleomycin or 4.2 mM 4-nitroquinoline-1-oxide, (ii) osmotic stress with 100 mM β-mercaptoethanol or 10% dimethyl sulphoxide, (iii) changing the chromatin structure by depolymerising microtubules with 100 µg/ml thiabendazole or 25 µg/ml methyl-β-cyclodextrin, (iv) meiosis, (v) nitrogen starvation, and (vi) in block and release assays using hydroxyurea and temperature shifts. However, none of these treatments resulted in significant changes in the quantity or quality of Tf1 transposition (data not shown) as monitored by determining the transposition efficiency (21) and by using the screen for cdc25 revertants, respectively.
Tf1 might target 5′ ends of ORFs by interacting with a host factor
We have shown that Tf1 does not insert into ORFs and that Tf1 insertion occurred preferentially in large intergenic regions, with a bias towards a 320 bp region located 100 bp upstream of the translation start site. Northern blot analysis of genes adjacent to Tf1 insertions did not reveal any significant changes in the transcription level or size of the corresponding transcript. Consistent with these results, we propose that Tf1 is targeted region-specific close to the 5′ ends of ORFs by a DNA binding host factor. This could be a component associated with chromatin structure because it has been shown previously that Tf1 transposition is decreased in the presence of the histone deacytelase inhibitor Trichostatin A (35). Furthermore, the retrotransposon encodes a chromo motif (chromatin organisation modifier) (36) which could mediate interactions with chromatin-associated factors (37). Alternatively, or in addition, a transcription-related factor could target Tf1 to 5′ ends of ORFs, as it has been described for Ty1 elements (5,7–9,38). In the case of Tf1, however, this factor is more likely to be associated with the Pol II transcription complex because Tf1 insertion is not biased towards Pol III-transcribed genes. However, we failed to detect any DNA binding factor sites although these may have been too degenerate or short to be detected.
As the S.pombe genome is densely packed with ORFs, the size of intergenic regions is limited (see earlier), and so larger intergenic regions may contain a higher number of target sequences for factors involved in the regulation of chromatin structure and/or transcription. Therefore, Tf1 might preferably insert into large intergenic regions because the host factor(s) targeting the retroelement is more likely to be present in these regions.
The isolation of Tf1 insertions in our study was based on the expression of the neo gene. Thus, it is possible that some other Tf1 targets were not detected because they are in silent regions of the genome, such as the centromere, the telomeres or the mating type loci (39–41). Analysis of unselected transposition events will therefore be an important task for future studies.
Application of Tf1 as tool for molecular biology
Transposons have proven to be very helpful molecular tools. They have been used as insertional mutagens to tag genes (42), and to generate transcriptional and translational fusions (43). Our analysis has shown that Tf1 is not suitable for insertional mutagenesis; however, altering the target site specificity of Tf1 through random mutagenesis might turn it into a useful tool for this application (44). In this respect it would also be important to know whether target site specificity of Tf1 is changed in mutant strains with an altered chromatin structure, as is the case for some Ty elements (30,45–47). Similarly, treating cells with Trichostatin A, which results in a reduced Tf1 transposition (35), could alter target site preference. In addition to providing information about transposition and target site selectivity, the Tf1 system could provide a useful tool with which to probe and dissect chromatin structure in vivo as already suggested for retrotransposons (1,7,47).
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to H. L. Levin for kindly providing the pHL414 plasmid and to John Sgouros and the Sanger Centre for provding information on the S.pombe genome. We thank David Ish-Horowicz, Joyce Müller, Heidi Browing, Damian Brunner and all other members of the cell cycle laboratory for help and comments on the manuscript. This work was funded by ICRF. R.B. was also supported by a fellowship of Boehringer Ingelheim Fonds.
REFERENCES
- 1.Craig N.L. (1997) Annu. Rev. Biochem., 66, 437–474. [DOI] [PubMed] [Google Scholar]
- 2.Spradling A.C., Stern,D.M., Kiss,I., Roote,J., Laverty,T. and Rubin,G.M. (1995) Proc. Natl Acad. Sci. USA, 92, 10824–10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engels W.R. (1996) Curr. Top. Microbiol. Immunol., 204, 103–123. [DOI] [PubMed] [Google Scholar]
- 4.Kim J.M., Vanguri,S., Boeke,J.D., Gabriel,A. and Voytas,D.F. (1998) Genome Res., 8, 464–478. [DOI] [PubMed] [Google Scholar]
- 5.Bryk M., Banerjee,M., Murphy,M., Knudsen,K.E., Garfinkel,D.J. and Curcio,M.J. (1997) Genes Dev., 11, 255–269. [DOI] [PubMed] [Google Scholar]
- 6.Chalker D.L. and Sandmeyer,S.B. (1992) Genes Dev., 6, 117–128. [DOI] [PubMed] [Google Scholar]
- 7.Devine S.E. and Boeke,J.D. (1996) Genes Dev., 10, 620–633. [DOI] [PubMed] [Google Scholar]
- 8.Smith J.S. and Boeke,J.D. (1997) Genes Dev., 11, 241–254. [DOI] [PubMed] [Google Scholar]
- 9.Weinstock K.G., Mastrangelo,M.F., Burkett,T.J., Garfinkel,D.J. and Strathern,J.N. (1990) Mol. Cell. Biol., 10, 2882–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji H., Moore,D.P., Blomberg,M.A., Braiterman,L.T., Voytas,D.F., Natsoulis,G. and Boeke,J.D. (1993) Cell, 73, 1007–1018. [DOI] [PubMed] [Google Scholar]
- 11.Natsoulis G., Thomas,W., Roghmann,M.C., Winston,F. and Boeke,J.D. (1989) Genetics, 123, 269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilke C.M., Heidler,S.H., Brown,N. and Liebman,S.W. (1989) Genetics, 123, 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boeke J.D. and Sandmeyer,S.B. (1990) In Broach,J.R., Jones,E.W. and Pringle,J. (eds), Molecular and Cellular Biology of the Yeast Saccharochomyces cerevisiae: Genome Dynamics, Protein Synthesis, and Energetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 14.Kirchner J., Connolly,C.M. and Sandmeyer,S.B. (1995) Science, 267, 1488–1491. [DOI] [PubMed] [Google Scholar]
- 15.Connolly C.M. and Sandmeyer,S.B. (1997) FEBS Lett., 405, 305–311. [DOI] [PubMed] [Google Scholar]
- 16.Zou S., Kim,J.M. and Voytas,D.F. (1996) Nucleic Acids Res., 24, 4825–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou S., Ke,N., Kim,J.M. and Voytas,D.F. (1996) Genes Dev., 10, 634–645. [DOI] [PubMed] [Google Scholar]
- 18.Zou S. and Voytas,D.F. (1997) Proc. Natl Acad. Sci. USA, 94, 7412–7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levin H.L., Weaver,D.C. and Boeke,J.D. (1990) Mol. Cell. Biol., 10, 6791–6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levin H.L., Weaver,D.C. and Boeke,J.D. (1993) EMBO J., 12, 4885–4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levin H.L. and Boeke,J.D. (1992) EMBO J., 11, 1145–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreno S., Klar,A. and Nurse,P. (1991) Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- 23.Southern E.M. (1992) Biotechnology, 24, 122–139. [PubMed] [Google Scholar]
- 24.Bach M.L., Lacroute,F. and Botstein,D. (1979) Proc. Natl Acad. Sci. USA, 76, 386–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wach A., Brachat,A., Pohlmann,R. and Philippsen,P. (1994) Yeast, 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- 26.Baum B., Wuarin,J. and Nurse,P. (1997) EMBO J., 16, 4676–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winzeler E.A., Shoemaker,D.D., Astromoff,A., Liang,H., Anderson,K., Andre,B., Bangham,R., Benito,R., Boeke,J.D., Bussey,H. et al. (1999) Science, 285, 901–906. [DOI] [PubMed] [Google Scholar]
- 28.Oyen T.B. and Gabrielsen,O.S. (1983) FEBS Lett., 161, 201–206. [DOI] [PubMed] [Google Scholar]
- 29.Garfinkel D.J., Mastrangelo,M.F., Sanders,N.J., Shafer,B.K. and Strathern,J.N. (1988) Genetics, 120, 95–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian Z., Huang,H., Hong,J.Y., Burck,C.L., Johnston,S.D., Berman,J., Carol,A. and Liebman,S.W. (1998) Mol. Cell. Biol., 18, 4783–4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fantes P.A. (1981) J. Bacteriol., 146, 746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rolfe M., Spanos,A. and Banks,G. (1986) Nature, 319, 339–340. [Google Scholar]
- 33.Bradshaw V.A. and McEntee,K. (1989) Mol. Gen. Genet., 218, 465–474. [DOI] [PubMed] [Google Scholar]
- 34.Ribeiro-dos-Santos G., Schenberg,A.C., Gardner,D.C. and Oliver,S.G. (1997) Mol. Gen. Genet., 254, 555–561. [DOI] [PubMed] [Google Scholar]
- 35.Dang V.D., Benedik,M.J., Ekwall,K., Choi,J., Allshire,R.C. and Levin,H.L. (1999) Mol. Cell. Biol., 19, 2351–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malik H.S. and Eickbush,T.H. (1999) J. Virol., 73, 5186–5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paro R. and Hogness,D.S. (1991) Proc. Natl Acad. Sci. USA, 88, 263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boeke J.D. and Devine,S.E. (1998) Cell, 93, 1087–1089. [DOI] [PubMed] [Google Scholar]
- 39.Allshire R.C., Javerzat,J.P., Redhead,N.J. and Cranston,G. (1994) Cell, 76, 157–169. [DOI] [PubMed] [Google Scholar]
- 40.Nimmo E.R., Cranston,G. and Allshire,R.C. (1994) EMBO J., 13, 3801–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klar A.J. (1992) Trends Genet., 8, 208–213. [DOI] [PubMed] [Google Scholar]
- 42.Kleckner N., Roth,J. and Botstein,D. (1977) J. Mol. Biol., 116, 125–159. [DOI] [PubMed] [Google Scholar]
- 43.Casadaban M.J. and Cohen,S.N. (1979) Proc. Natl Acad. Sci. USA, 76, 4530–4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bender J. and Kleckner,N. (1992) EMBO J., 11, 741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Devine S.E. and Boeke,J.D. (1994) Nucleic Acids Res., 22, 3765–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rinckel L.A. and Garfinkel,D.J. (1996) Genetics, 142, 761–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu Y., Zou,S., Wright,D.A. and Voytas,D.F. (1999) Genes Dev., 13, 2738–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]