Abstract
Gene conversion has been defined as the nonreciprocal transfer of information between homologous sequences. Despite its broad interest for genome evolution, the occurrence of this mechanism in bacteria has been difficult to ascertain due to the possible occurrence of multiple crossover events that would mimic gene conversion. In this work, we employ a novel system, based on cointegrate formation, to isolate gene conversion events associated with crossovers in the nitrogen-fixing bacterium Rhizobium etli. In this system, selection is applied only for cointegrate formation, with gene conversions being detected as unselected events. This minimizes the likelihood of multiple crossovers. To track the extent and architecture of gene conversions, evenly spaced nucleotide changes were made in one of the nitrogenase structural genes (nifH), introducing unique sites for different restriction endonucleases. Our results show that (i) crossover events were almost invariably accompanied by a gene conversion event occurring nearby; (ii) gene conversion events ranged in size from 150 bp to 800 bp; (iii) gene conversion events displayed a strong bias, favoring the preservation of incoming sequences; (iv) even small amounts of sequence divergence had a strong effect on recombination frequency; and (v) the MutS mismatch repair system plays an important role in determining the length of gene conversion segments. A detailed analysis of the architecture of the conversion events suggests that multiple crossovers are an unlikely alternative for their generation. Our results are better explained as the product of true gene conversions occurring under the double-strand break repair model for recombination.
Gene duplication is the main mechanism that gives rise to gene families in both eukaryotic and prokaryotic genomes (28). One common observation is that members of a multigene family tend to maintain a higher degree of sequence conservation at the intraspecific level than that seen in interspecific comparisons. This is a clear indication that members of a multigene family are evolving in a concerted way, a process called concerted evolution (5). Concerted evolution of multigene families occurs not only in eukaryotic organisms but in prokaryotes as well (39). Evidence for its occurrence has been reported for the rRNA genes (16S rRNA) in Bacteria and Archaea (17), the 23S rRNA intervening sequences in Salmonella enterica serovar Typhimurium and Salmonella enterica serovar Typhi (21, 22), the genes coding for the translation factor EF-Tu (tuf) in bacteria (1, 16), the flagellin genes (fla) in Campylobacter (23), the family of outer membrane proteins (bab) in Helicobacter pylori (30), and the genes coding for the nitrogenase enzyme (nifH) in Rhizobium etli (34; E. Sepúlveda and D. Romero, unpublished data). Although several molecular processes may participate in concerted evolution of gene families, it is generally thought that gene conversion among repeated genes is responsible for this evolutionary trend. Specialized systems for gene conversion also participate in the generation of antigenic variation in several pathogenic bacteria (reviewed in reference 39).
Gene conversion is one of the possible outcomes of a recombination event and has been defined as the nonreciprocal transfer of genetic information from one DNA duplex to another. This process was initially demonstrated in ascomycete fungi (18), and the yeast Saccharomyces cerevisiae is still the preferred organism for its study due to the ability to recover all the products of a meiotic recombination event. This ability facilitates the demonstration of nonreciprocal transfer events. The need to understand gene conversion and its association with crossovers was the main motivation for the development of the Holliday model for recombination (18); the possibility for gene conversion has been retained in successive models of recombination, including the double-strand break repair model (18, 43, 44).
Gene conversion has been more difficult to study in other organisms, including bacteria. The main problem is that, since the recovery of all the products of a recombination event is not possible, the characteristic nonreciprocity of gene conversion events cannot be ensured. In fact, it has been argued that several possible examples of gene conversion in bacteria may be due to selection for rare double crossovers rather than to gene conversion (40). Despite this limitation, some groups have provided convincing evidence for gene conversion in bacteria, including Escherichia coli (13, 45) and Salmonella enterica serovar Typhimurium (1, 3, 11), using substrates harboring repeated sequences in an inverted orientation. In these reports, selection is applied for gene conversion events occurring between inverted repeats, afterwards exploring their relation with crossover. In at least one case, there is convincing albeit indirect evidence for the nonreciprocal origin of these events (3).
Our group has been studying Rhizobium etli, an α-proteobacterium that is able to form nitrogen-fixing symbiotic associations with bean plants. Besides its symbiotic capabilities, R. etli is also interesting because of the presence of reiterated gene elements (8), which can play important roles in shaping genomic structure. One important multigene family is the nifH family (encoding one component of the nitrogenase enzyme), comprised of three identical members (31, 32) located in a large plasmid (371 kb) called the symbiotic plasmid or pSym (9, 35). Homologous recombination among the members of this family promotes different genomic rearrangements in pSym, having important symbiotic consequences (36, 37, 47). Phylogenetic evidence indicates the existence of concerted evolution among members of this family (E. Sepúlveda and D. Romero, unpublished data), perhaps generated through gene conversion (34). A previous evaluation of the occurrence of gene conversion in R. etli relied on the introduction of a 28-bp insertion into one of the nifH copies, followed by its elimination by recombinational interactions with either of the other two nifH copies (34). Although products consistent with the occurrence of gene conversion were isolated, it is formally possible that at least some of these arose from repeated reciprocal exchanges rather than true gene conversion (34).
In this work, we employ a novel approach to evaluate gene conversion, using a genetic system based on the cointegration between sequences harboring planned sequence alterations, or restriction fragment length polymorphism (RFLPs). In this approach, selection is only applied for cointegration; any gene conversion arises as an unselected event. This avoids weaknesses present in previous approaches, where direct selection for gene convertants raises the possibility of multiple crossovers to explain its generation. Characterization of the gene convertants obtained through this approach allowed us to evaluate the association of recombination with gene conversion, the length of converted tracts, and the role of sequence heterology. Our results show that (i) crossover events are frequently accompanied (98%) by a gene conversion event occurring nearby; (ii) gene conversion events frequently encompass more than half of the length of this gene; (iii) gene conversion events display a strong polarity, favoring the preservation of incoming sequences; (iv) even small amounts (1.6%) of sequence divergence have a strong effect on recombination frequency; and (v) the MutS mismatch repair system plays an important role in determining the length of gene conversion segments.
MATERIALS AND METHODS
Bacterial strains and media.
Escherichia coli strains were grown in LB medium (25) at 37°C. Rhizobium etli strains were grown in PY medium (27) at 30°C. Antibiotics were added to the media when needed at the following concentrations (in micrograms per milliliter): carbenicillin, 100; chloramphenicol, 15; kanamycin, 15; nalidixic acid, 20; spectinomycin 100; and tetracycline, 10 (E. coli) or 2 (R. etli). For selection in cloning experiments, 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) was added to LB plates at 30 μg ml−1.
General DNA manipulations and mutagenesis of the nifH gene.
All DNA manipulations were done using standard procedures (38). Most of the plasmid transformations employed Escherichia coli DH5∝ as a host (10). PCRs were done in a Techgene thermocycler using Platinum Taq High Fidelity DNA polymerase (Invitrogen) for mutagenesis and conventional Taq DNA polymerase for analytical characterization. For ligations, T4 polynucleotide ligase (Amersham Biosciences) was used. Restriction enzymes were purchased from diverse companies and used according to the recommendations of the supplier. Custom oligonucleotides were synthesized at the Unidad de Síntesis de Oligonucleótidos (Instituto de Biotecnología, Universidad Nacional Autónoma de México, México).
Introduction of specific restriction sites into the nifH gene was done by a variation of published PCR mutagenesis procedures (26). Specific oligonucleotide primers (see Table 1) containing one or more modified nucleotides to introduce a restriction site at the time of polymerization were designed. Only single base changes (either transitions or transversions) were used, avoiding the introduction of stop or otherwise rare codons. To introduce mutations, two PCR products were generated, using either the Hindu/Narl primer combination (product size, 567 bp) or the Apalu/Xbal combination (size 557 bp), employing as a template DNA from R. etli CFN42. Both products were gel purified, using a GeneClean II kit (Bio 101). The purified PCR products were mixed, heat denatured (at 90°C for 2 min), and annealed (at 60°C for 1 min), taking advantage of a 200=bp overlap between both products. A mixture of the four deoxyribonucleotides as well as Platinum Taq DNA polymerase was added to the annealing mixture and incubated at 72°C for 6 min to allow the generation of a complete nifH sequence. After this, the Hindu and Xbal oligonucleotides were added to the reaction and subjected to PCR (30 cycles with denaturation at 92°C for 1 min, annealing at 56°C for 1 min, and extension at 72°C for 2 min). The PCR products were cloned into pUC19 (48) and analyzed to verify the introduction of the restriction sites in the nifH sequence. Two full-sized nifH clones were obtained, one containing the HindIII, ApaLI, and XbaI restriction sites and the other with the HindIII, NarI, and XbaI sites.
TABLE 1.
PCR primers used in this worka
| Primer | Sequenceb | Source/positionc | GenBank accession no. |
|---|---|---|---|
| Hindu | AGGAAGCTTATATGTCAGATTTGC | nifHa, (−)11-13 | M10587 |
| Bamhu | GGATCCGACCCGAAAGCC | nifHa, 115-132 | M10587 |
| Apalu | GTGCACATGACGATGTCGACT | nifHa, 344-364 | M10587 |
| NarI | GCGCCCGTTACAGATCAG | nifHa, 564-547 | M10587 |
| MluI | TCCGACGCGTACTGGATCA | nifHa, 701-683 | M10587 |
| BclI | CTTGATCATGCCGAAGTCGAG | nifHa, 831-811 | M10587 |
| XbaI | TCCTCTAGACAGCGGCAGTTAT | nifHa, 912-891 | M10587 |
| 1 | CTGAAACCCAACAAAAG | nifHa, (−)135-(−)119 | M10587 |
| 2 | GCAAGGCGATTAAGTTG | pIC20R, 385-369 | L08913 |
| 3 | AGTCGGCAAATAATGTC | ΩTc, 2543-2559 | U35135 |
| 4 | AAAACGCTGTCATTCTC | nifHa, 1033-1017 | U80928 |
All the oligonucleotides are shown in the 5′ to 3′ direction.
Nucleotides that were modified to generate the corresponding restriction site (underlined) are shown in boldface.
Positions correspond to the start codon of the indicated sequence (nifHa) or to the initial nucleotide in the reported sequence (pIC20R and ΩTc).
The whole process was repeated cyclically to introduce new mutations, using as templates the product of the preceding step and primer pairs Bamhu/XbaI (to introduce mutations in the 5′ end of the gene) or Hindu/Mlul and Hindu/Bcll (to mutagenize the 3′ end of nifH). This process generates two clones, one harboring the BamHI and ApaLI sites in the 5′ end of nifH and another with the NarI, MluI, and BclI sites in the 3′ end; both clones share a native BglII site and are flanked by HindIII and XbaI sites. The mutations were combined in a single gene by digesting the first clone with HindIII and BglII and the second with BglII and XbaI and ligating the desired fragments into pUC19. Introduction of these mutations was verified by manually sequencing both strands of the modified nifH gene by the Sanger dideoxy chain termination method, using a thermosequenase cycle sequencing kit (Amersham Biosciences). Oligonucleotides were labeled by kinasing with [γ-33P]ATP and T4 DNA kinase (Amersham Biosciences). Sequencing reactions were electrophoresed in 6% polyacrylamide-8 M urea gels. Besides the planned mutations, spontaneous sequence changes, perhaps occurring during the successive PCR steps, generated a novel MaeIII site. A map of the relevant restriction sites in the nifH gene is shown in Fig. 1.
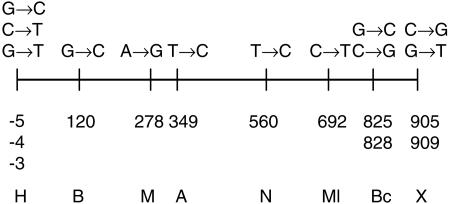
FIG. 1.
Locations of the nucleotide substitutions introduced on the nifH gene. Numbers below the horizontal line indicate the position of the base substitutions (showed above the line) with respect to the starting nucleotide of the gene. These substitutions generate unique sites for different restriction enzymes, which are indicated by a one-letter code (H, HindIII; B, BamHI; M, MaeIII; A, ApaLI; N, NarI; Ml, MluI; Bc, BclI; X, XbaI).
Construction of integrative plasmids.
For construction of integrative vectors, pIC20R (20) was modified by introduction of an RK2 oriT sequence (29, 46). This was done by PCR amplification from pEYM1 (47), using custom-made oligonucleotides that introduce a XhoI site. The PCR product was cloned into the single XhoI site in pIC20R. This was used as a vector for introduction, by HindIII-XbaI digestion and ligation, of the wild-type nifH sequence, the nifH gene harboring the eight RFLPs (Fig. 1), and other variants, such as one having the mutations HindIII, BamHI, MaeIII, and ApaLI in the 3′ end of nifH and the other with the NarI, MluI, BclI, and XbaI mutations in the 5′ zone. Finally, a 1.8-kb HindIII-HindIII tetracycline resistance cassette from pBSL193 (2) was introduced into each of these plasmids, giving rise to pMC0, pMC11, pMC32, and pMC63, respectively.
To simplify cointegrate selection in the mutS background, a kanamycin-resistant derivative from pMC11 was constructed by substitution of the HindIII-HindIII tetracycline resistance cassette with a 2.2-kb HindIII-HindIII kanamycin resistance cassette from pHP45Ω-Km (7), giving rise to pJGus28.
Molecular characterization of transconjugants.
Escherichia coli S17-1 (F− pro-82 thi-1 endA1 hsdR17 supE44 recA13, chromosomally integrated RP-4-2 [Tc::Mu, Km::Tn7]) was used as a host for conjugative transfer of integrative plasmids (42). To that end, biparental matings were set up on solid media between Escherichia coli S17-1 harboring the desired plasmid and R. etli as described previously (47); transconjugants were selected by its resistance to nalidixic acid and tetracycline. In most cases, R. etli CFNX55 (36) (harboring a large deletion that removes two of the three nifH genes in pSym) was used as a recipient. To ensure the independence of the observed events, 10 separate conjugation experiments were set up, retaining not more than five single-colony isolates from each experiment. Total DNA was isolated from each transconjugant and analyzed by PCR with specific primers (left PCR with primers 1 and 2, right PCR with primers 3 and 4, Fig. 2) to amplify both nifH products. All the PCR products were purified by using CentriSep spin columns (Applied Biosystems) before digestion with restriction enzymes. Determinations of conjugation frequency were repeated at least 10 times and are expressed as number of transconjugants per recipient cell ± standard deviation.
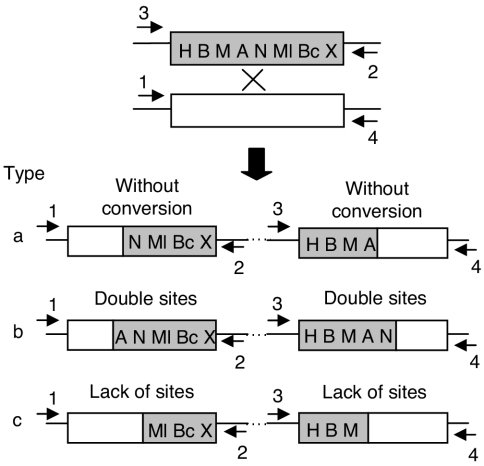
FIG. 2.
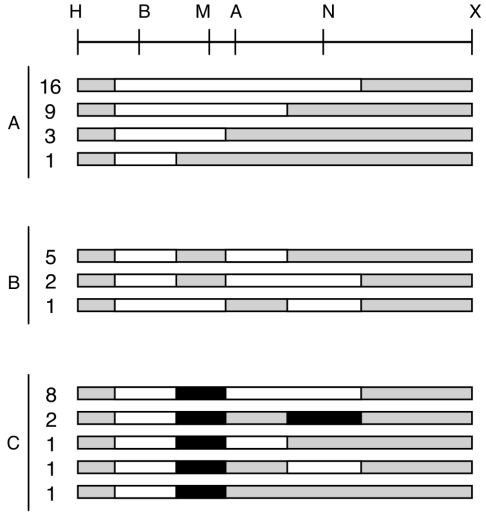
Experimental strategy to detect gene conversion associated to cointegration. A shaded rectangle represents the nifH gene harboring different RFLPs (Fig. 1); the open rectangle corresponds to the wild-type nifH gene. Cointegration between circular molecules bearing these genes (indicated by crossed lines on the top part of the figure) generates three different types of cointegrate, depending on its possible association with gene conversion. In type a, a single crossover is depicted, without gene conversion. Gain of sites on both sides of the cointegrate (type b, sites A and N) or loss of sites (type c, sites A and N) is interpreted as evidence of gene conversion. The primers used to amplify each side of the cointegrate are indicated by numbered arrowheads.
Generation of strain CFNX704.
To isolate an R. etli strain harboring the desired RFLPs on the nifH copy in pSym, plasmid pMC11 was transferred by conjugation to R. etli CFNX55, selecting integrants by their resistance to tetracycline. An integrant harboring a gene conversion event encompassing the BamHI, MaeIII, ApaLI, and NarI markers was identified by PCR. From this strain, loss of pMC11 by excision was screened by checking single-colony isolates for a tetracycline-sensitive phenotype; these were found at a frequency of 10−4. The excisant was analyzed by PCR and restriction analysis to verify the retention of the BamHI, MaeIII, ApaLI, and NarI markers. This strain, called CFNX704, was then used in crosses with the pMC0 plasmid (containing the nifH wild-type sequence). As before, we analyzed 50 transconjugants coming from 10 independent experiments.
Construction of a mutS derivative.
To evaluate the participation of the mutS repair system in gene conversion, we employed strain CFNX706, a mutS::loxPSp derivative from R. etli (J. M. Martínez-Salazar, J. Zuñiga-Castillo, and D. Romero, unpublished data). This strain harbors a loxPSp insertion in the mutS gene, interrupting codon 292. Strain CFNX706 was modified by generating a large deletion on pSym that eliminates two of the three nifH genes, using the recombination enhancement by replication system (47). In this system, activation of a supernumerary replication origin on pSym leads to the high-frequency generation of a deletion on pSym identical to the one in strain CFNX55 (47).
To apply this system, plasmid pEYM13, harboring oriV from RK2 (47) was inserted by single-crossover recombination into one of the nifH genes of strain CFNX706, selecting integrants by their resistance to kanamycin. To activate replication from the supernumerary origin, plasmid pEYM5, encoding the replication initiator protein trfA from RK2, was introduced by conjugation with several integrants; transconjugants were selected by their resistance to chloramphenicol. Over 25% of the chloramphenicol-resistant transconjugants also displayed the loss of the kanamycin resistance marker, indicating the presence of possible deletions on pSym. The presence of the desired deletion was verified by analyses of plasmid profiles (revealing a 107-kb deletion on pSym) as well as by Southern hybridization against a nifH probe (data not shown). Spontaneous loss of pEYM5 from the mutS::loxPSp derivative harboring the desired deletion was screened by checking single-colony isolates for a chloramphenicol-sensitive phenotype, giving rise to strain CFNX712. This strain was then used as a recipient in crosses with plasmid pJGus28. Fifty transconjugants, coming from 10 independent experiments, were analyzed as described before.
RESULTS
Experimental strategy.
The nifH multigene family has three completely identical members located on pSym. To evaluate the length and distribution of gene conversion tracts in this family, one of the nifH genes was modified by introducing single-base-pair changes approximately every 100 bp along the gene sequence, as described in Materials and Methods. Twelve different changes were made, generating unique recognition sites for eight different restriction enzymes (Fig. 1). Such modifications serve as convenient landmarks to evaluate the extension of gene conversion tracts (see below).
This modified nifH gene was inserted into a plasmid (pMC11) that can be transferred by conjugation from Escherichia coli to Rhizobium etli, but it is unable to replicate in this latter host. The recipient for such crosses was Rhizobium etli CFNX55 (36), a derivative that harbors only one copy of the wild-type nifH gene. Upon single-crossover recombination in this strain, three types of cointegrates are possible (Fig. 2). The first type entails cointegrate formation without associated gene conversion (Fig. 2, type a), while the other two involve cointegrate formation associated with gene conversion, favoring either incoming sequences (type b) or endogenous sequences (type c). As shown in Fig. 2, these types can be easily distinguished by looking at the distribution of restriction sites in both sides of the cointegrate. Type a recombinants have only a redistribution of restriction sites at the crossover point, while types b and c display an increase in the number of restriction sites (double gain, type b) or a reduction in restriction sites (double loss, type c). Thus, the number and position of restriction sites modified in the gene conversion events allows an evaluation of the length and position of gene conversion tracts along the nifH gene.
In this experimental approach, selection is applied only for cointegration of the plasmid; no selection whatsoever was applied for recovery of gene conversion events. This is an important difference with previous studies, because it minimizes the likelihood that gene conversion events arise through selection for rare double crossover events, which would mimic gene convertants.
Independent isolation of both sides of the cointegrate is possible through PCR amplification, using primer pairs that amplify either the left side (primers 1 and 2) or the right side (primers 3 and 4) of the cointegrate (Fig. 2). These PCR products were then subjected to restriction analysis, searching for instances in which cutting with a specific enzyme occurred in both sides or in neither side of the cointegrate; these were interpreted as examples of gene conversion.
Crossover events are strongly associated with gene conversion.
To evaluate the association of crossover formation to gene conversion, cointegrates between pMC11 (harboring eight different RFLPs) and pSym were selected, as described in Materials and Methods. In R. etli, formation of cointegrates of this kind is strictly dependent on recA (50). Interestingly, when plasmid pMC11 was used as a donor, integrants were obtained at a very low frequency (1.63 × 10−7± 0.52 × 10−7). Higher integration frequencies were seen when plasmids lacking RFLPs (pMC0, integration frequency 1.6 × 10−5± 0.05 × 10−5) or with RFLPs only on the 5′ half (pMC32, 1.18 × 10−5± 0.12 × 10−5) or on the 3′ half (pMC63, 6.6 × 10−6± 3.1 × 10−6) of the gene were used. Thus, the low integration frequency observed with pMC11 may be attributed to degree of sequence divergence (1.6%) between the recombining sequences. A similar sensitivity of recombination frequency to degree of heterology has been observed previously (33, 49).
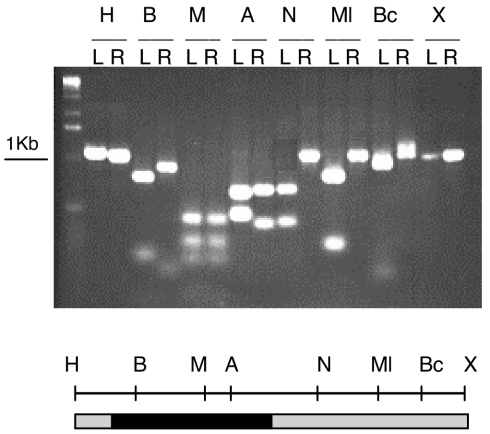
DNA was purified from 50 independent cointegrates with pMC11 and subjected to separate PCRs to amplify the left- and right-hand sides of each cointegrate, which were then subjected to restriction analysis. An example of these analyses is shown in Fig. 3. In this case, digestion with BamHI, MaeIII, and ApaLI was observed for both sides of the cointegrate, while restriction on only one side was observed for the remaining enzymes. This indicates that a continuous gene conversion event encompassing these three sites had occurred in this particular cointegrate. Since it is impossible to determine the exact endpoint of conversions occurring between two markers, the middle zone between two restriction sites was chosen to register the end of every conversion event. Thus, this particular conversion event was roughly 400 bp in size.
FIG. 3.
Detection of gene conversion in cointegrates. The left (L) and right (R) parts of a specific cointegrate were amplified by PCR (Fig. 2 and Material and Methods). These products were digested with different restriction enzymes (H, HindIII; B, BamHI; M, MaeIII; A, ApaLI; N, NarI; Ml, MluI; Bc, BclI; X, XbaI) and analyzed by agarose gel electrophoresis (top). Note that both sides of the cointegrate were cut with BamHI, MaeIII, and ApaLI, indicating a gene conversion event encompassing these markers. This is summarized in the lower part of the figure (shaded bar, regions of nifH not subjected to gene conversion; black bar, region undergoing gene conversion towards gain of the markers).
Figure 4 show the results obtained for the 50 different cointegrates analyzed. Four classes of events were observed. Classes A to C represent different kinds of gene conversion events, while class D comprises crossover events not associated with gene conversion. Interestingly, 98% of the events (49 out of 50) fall in the conversion classes (A to C), while only a single event was located in the no-conversion class (class D, 2%). Thus, crossover formation is frequently accompanied by a gene conversion event occurring nearby.
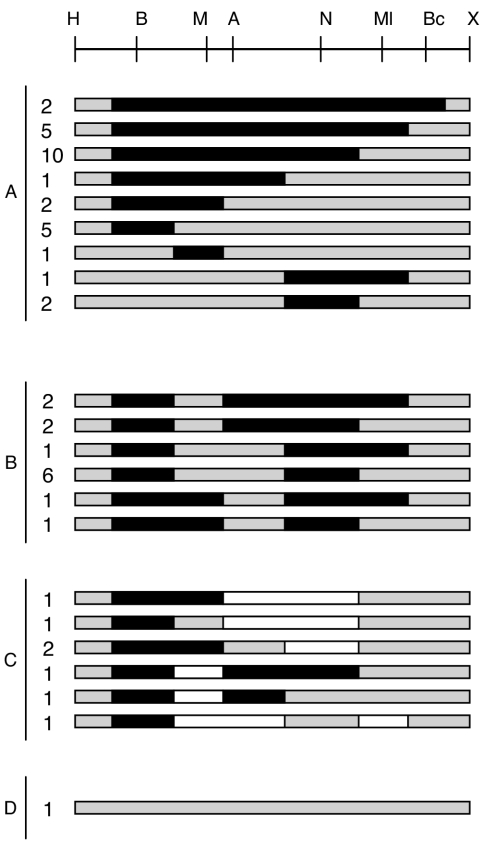
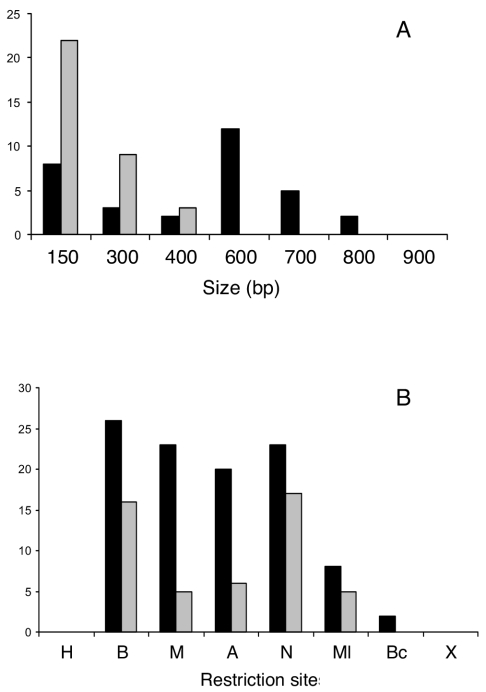
FIG. 4.
Structure of gene conversion tracts obtained upon introduction of pMC11 into R. etli CFNX55. The RFLP map of the nifH gene is shown on top as a reference. Letters at the left side represent the four cointegrate classes found (A, continuous conversion; B, discontinuous conversion; C, bipolar conversions; and D, single crossover with no evidence of gene conversion). Values indicate the number of isolates with the corresponding conversion tract. Black bars represent the extent of gene conversion tracts towards marker gain; white bars indicate gene conversion tracts showing marker loss; shaded bars are regions not subjected to gene conversion. Note that gene conversion is biased towards marker gain.
Structure of gene conversion events.
Class A events (continuous gene conversion), such as the one shown in Fig. 3, were more frequent in this sample. Fifty-eight percent of the isolates (29 out of 50) belonged to this class. As shown in Fig. 4, continuous gene conversion tracts ranged in size from 150 bp up to 800 bp; more than half of the members in this class (17 out of 29 events) displayed continuous gene conversion tracts at least 600 bp in size.
The second most frequent class corresponds to discontinuous gene conversion events (class B). Members of this class, encompassing 26% of the isolates, display two tracts of continuous gene conversion, with a marker or two between these tracts that do not display gene conversion. The most complex class corresponds to the bipolar conversions (class C), which represent 14% of the isolates characterized. Members of this class display at least two conversion tracts; these tracts are clearly discernible, because one of these displays gain of sites in the conversion tract (double gain, Fig. 2), while in the other tract the restriction sites were absent (double loss, Fig. 2). In a single isolate, the contrasting conversion tracts are contiguous, while in the rest these tracts are separated by an intervening marker that does not display conversion. Thus, class C conversions are a mixed class that contains both continuous and discontinuous events.
Length and distribution of gene conversion tracts.
To evaluate the length distribution of gene conversion tracts, continuous and discontinuous classes were analyzed separately; bipolar conversions were included in either the continuous or discontinuous class depending on architecture (Fig. 5). The size of the conversion tract was evaluated for each class; isolates harboring a continuous conversion contribute only with a single tract to the total, but isolates in classes B and C contribute with two or three tracts, depending on structure. Therefore, the data in Fig. 5 are based on the analysis of 72 conversion tracts. Gene conversion tract length for the continuous class reveals a bimodal distribution, centered at 150 bp and 600 bp. The discontinuous tracts, in contrast, show a single unimodal distribution, centered at 150 bp. This is consistent with an interpretation that continuous tracts are formed through the cooperation of two separate processes, such as gap filling and heteroduplex correction (see Discussion).
FIG. 5.
Size distribution (A) and positions (B) involved in gene conversion events. Data are derived from Fig. 4. For both panels, vertical black bars represent continuous tracts, while shaded bars correspond to discontinuous tracts. The letters in panel B represent the different RFLPs, as shown in Fig. 1.
To evaluate if all the sites are equally likely to participate in a gene conversion event, representation of each site in conversion tracts was counted separately for both continuous and discontinuous events. Figure 5 show that for continuous events the sectors containing the BamHI, MaeIII, ApaLI, and NarI markers are equally likely to participate in gene conversion. Participation of the MluI and BclI markers is somewhat reduced, while the HindIII and XbaI markers were not included in the continuous conversion events analyzed. The lack of participation of these two terminal markers may be artifactual, due to the reduced homology available at the end of the gene (Fig. 4). In contrast, for the discontinuous events, the BamHI and NarI markers are included preferentially in conversion events; markers flanking these regions, such as MaeIII, ApaLI, MluI, and BclI, were poorly represented in discontinuous conversion events. This suggests that continuous conversion tracts encompassing the BamHI or NarI marker have a significant probability of terminating in the markers surrounding them.
Biased transfer of genetic information by gene conversion.
In the system described thus far, we have taken advantage of gain or loss of markers in both sides of each cointegrate (double gain and double loss, respectively, Fig. 2) to detect a gene conversion event. In principle, both kinds of convertants should be observed in the same proportions. Interestingly, we found that conversion is strongly biased towards the double gain class. From Fig. 4, it is clear that almost 90% of the tracts observed (64 out of 72) showed gain of sites. In fact, the few tracts displaying loss of sites come exclusively from class C (bipolar) convertants.
This lack of marker parity may be explained under two contrasting hypotheses. One alternative is that all the markers employed might have an intrinsic repair preference, favoring their use as templates for heteroduplex repair over the wild-type sequence of the nifH gene. A second possibility is that the observed bias may arise as a consequence of the way in which the recombining sequences are brought together. In all our experiments, the nifH copy harboring the RFLP markers is introduced by conjugation from Escherichia coli into an R. etli strain bearing a wild-type nifH gene. In this case, the observed bias is favoring conversion towards double gain (i.e., towards the incoming sequence) rather than its restoration to a wild-type sequence. This may be explained, under the double-strand break repair model of recombination by saying that the resident copy preferentially receives a double-strand cut, thus being a receptor of information (see Discussion).
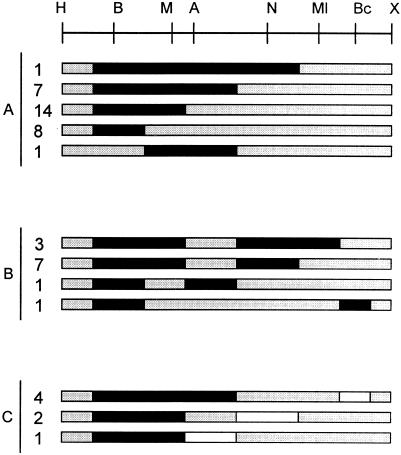
These hypotheses may be distinguished by exchanging the configuration of markers participating in conversion, putting the RFLP markers in the resident copy. If the bias is due to preferential repair, convertants should be still biased towards double gain; if the bias is due to preferential cutting of the resident sequence, the bias should be reversed towards the double loss class. To that end, the BamHI, MaeIII, ApaLI, and NarI markers were transferred to the nifH gene present in pSym of R. etli, generating strain CFNX704 (see Materials and Methods). Plasmid pMC0, harboring a wild-type nifH gene, was introduced by conjugation into strain CFNX704 to generate 50 independent cointegrates, which were screened for conversion as before. The results of this experiment are shown in Fig. 6.
FIG. 6.
Structure of gene conversion tracts obtained upon introduction of pMC0 into R. etli CFNX704. Letters at the left side represent the three cointegrate classes found (A, continuous conversion; B, discontinuous conversion; and C, bipolar conversions). Black bars represent the extent of gene conversion tracts towards marker gain; white bars indicate gene conversion tracts showing marker loss; shaded bars are regions not subjected to gene conversion. Note that gene conversion is biased towards marker loss. (H, HindIII; B, BamHI; M, MaeIII; A, ApaLI; N, NarI; X, XbaI.)
Exchanging the configuration of markers does not greatly affect the proportion of the different types of convertants; 58% of the isolates were still class A convertants (continuous gene conversion). However, there were slight differences in the proportion of classes B (discontinuous conversion, 16% versus 26%) and C (bipolar convertants, 26% versus 14%). More importantly, conversion bias is now reversed towards loss of markers. Nearly 82% of the tracts observed (68 out of 83, Fig. 6) showed double marker loss. Again, the few cases displaying double marker gain come from the bipolar class. These results indicate that, in this system, gene conversion is strongly biased towards the acquisition of markers present in the incoming sequence.
MutS mismatch repair system is an important determinant for length of gene conversion segments.
The data presented here suggest that continuous conversion segments may be formed through the cooperation of two separate processes, such as gap filling and heteroduplex correction (see Discussion). If that were the case, inactivation of the MutS system, one of the main systems for mismatch correction in bacteria (33, 49), would instigate a marked reduction in the length of gene conversion segments. To evaluate this possibility, a kanamycin-resistant derivative of pMC11 (pJGus28, containing the eight different RFLPs) was introduced by conjugation into an R. etli mutS::loxPSp derivative that harbors only one copy of the wild-type nifH gene on pSym (strain CFNX712, see Materials and Methods). As expected for knocking out one of the main barriers for recombination between divergent sequences, integrants were obtained readily in this mutant background (at a frequency of 2.8 × 10−5± 1.49 × 10−5). This frequency is 30-fold higher than the one obtained upon transfer of pJGus28 into CFNX55 (7.54 × 10−7± 3.84 × 10−7).
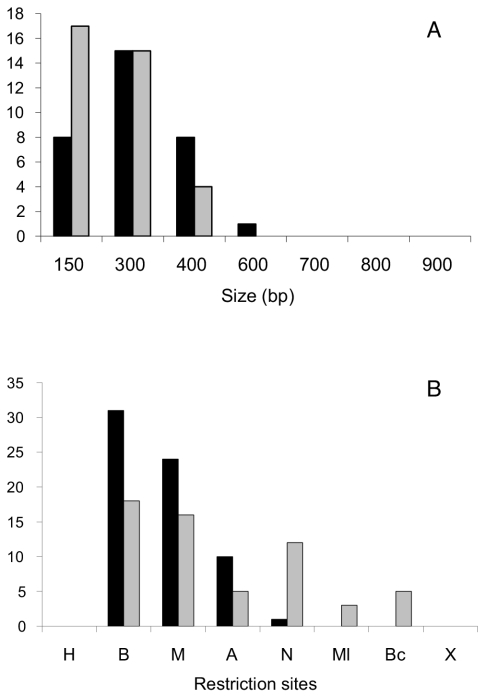
The analysis of 50 cointegrates obtained in the mutS::loxPSp derivative is shown in Fig. 7. In this mutant background, no effect was seen either on the proportion of the different conversion classes (A, 62%; B 24%; C, 14%) or on the bias towards the acquisition of markers present in the incoming sequence (62 out of 68 conversion tracts displayed double marker gain). Striking differences were detected, however, for both the length of gene conversion tracts and the sectors covered by these tracts.
FIG. 7.
Structure of gene conversion tracts obtained upon introduction of pJGus28 into R. etli CFNX712 (mutS::loxPSp). Letters at the left side represent the three cointegrate classes found (A, continuous conversion; B, discontinuous conversion; and C, bipolar conversions). Numbers indicate the amount of isolates with the corresponding conversion tract. Black bars represent the extent of gene conversion tracts towards marker gain; white bars indicate gene conversion tracts showing marker loss; shaded bars are regions not subjected to gene conversion. (H, HindIII; B, BamHI; M, MaeIII; A, ApaLI; N, NarI; Ml, MluI; Bc, BclI; X, XbaI.)
As shown in Fig. 8, both the continuous and discontinuous tracts obtained in a mutS background displayed unimodal distributions, centered at 300 bp and 150 bp, respectively. In contrast, continuous conversion tracts in the wild-type strain displayed a bimodal distribution, centered at 150 and 600 bp (Fig. 5). Moreover, while for continuous tracts in the wild-type strain, sectors encompassing the BamHI, MaeIII, ApaLI, and NarI markers are equally likely to participate in gene conversion (Fig. 5), in the mutS background the BamHI and MaeIII sectors participate preferentially (Fig. 8). Both the reduction in gene conversion tract length and the preferential use of two of the markers in the mutS background support the interpretation that the MutS mismatch repair system participates in the generation of gene conversion.
FIG. 8.
Size distribution (A) and positions (B) involved in gene conversion events in R. etli CFNX712 (mutS::loxPSp). Data are derived from Fig. 7. For both panels, vertical black bars represent the continuous tracts, while shaded bars correspond to discontinuous tracts. The letters in panel B represent the different RFLPs, as shown in Fig. 1.
DISCUSSION
The data reported here show that, when two homologous sequences recombine to form a cointegrate, the majority of the products have a structure consistent with a gene conversion event occurring nearby. These events may encompass most of the recombining sequences. Roughly one half of the events are represented by continuous gene conversion events, while the other half show discontinuous or even bipolar events. Moreover, there is a clear bias in information transfer, favoring the conversion toward the markers present in the incoming sequence.
There are two alternatives to explain these data, one based on sister exchanges and the other postulating gene conversion. As argued before (40), apparent gene conversion events may be formed through the chance formation of double crossover events between dissimilar alleles located on sister molecules; these products, upon segregation, would be scored as convertants. Thus, these products have been dubbed apparent gene convertants, to reflect the possibility that they may have formed through reciprocal events rather than the nonreciprocal events that are the hallmark of gene conversion.
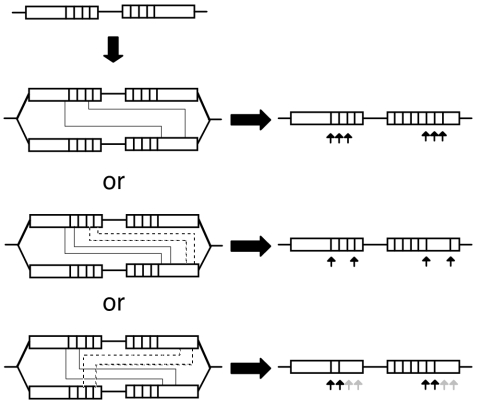
The most likely scenario for the formation of these double crossover events would be after formation of the cointegrate. In Fig. 9 we present the sequence of events required to explain our results under this hypothesis. The initial crossover event required to form the cointegrate might generate a simple redistribution of markers. Upon replication of the cointegrate structure, unequal double crossovers may generate an apparent gene conversion segment. This might be a tenable explanation for the continuous gene conversion class (class A in Fig. 4), which represents 58% of the events observed. However, more complex rationales have to be used to explain the discontinuous (class B) and bipolar (class C) events, representing 40% of the events observed here. For these, unequal double crossovers do not suffice. As shown in Fig. 9, under this hypothesis, four crossovers are needed to generate these classes. In the case of discontinuous conversions, two crossovers would be needed to generate the first conversion tract, followed by two additional crossovers farther away from the first pair. A similar situation has to be posed for most of the cases of bipolar conversions, but in this case crossovers have to involve the four copies present in the replicated structure.
FIG. 9.
Alternative model to explain gene conversion through unequal crossovers. After a cointegration event, a partially replicated molecule would generate four nifH sequences (open rectangles). Vertical lines within the open rectangles represent RFLPs. Pairs of continuous or broken lines joining the rectangles indicate the region of the unequal crossover. The events needed to generate continuous conversion tracts (top), discontinuous conversions (middle), and bipolar conversions (bottom) are shown. Arrows indicate regions transferred in an apparent gene conversion event, leading to marker gain (black) or marker loss (shaded). Note that generation of continuous conversions can be explained by two unequal crossover events, but both discontinuous and bipolar conversions require the participation of four unequal crossover events.
We consider the hypothesis of unequal multiple crossovers an unlikely alternative to explain our results. First, all the conversion events observed here were isolated in the absence of selection for the conversion event itself. The only selection applied was for the integration event; thus, selection for the conversion event cannot be invoked as the reason to observe multiple crossovers. Second, putative double crossover events (explaining class A convertants) were found at roughly the same frequency as the quadruple crossovers needed to explain convertants belonging to classes B and C; it is hard to envisage, in the absence of selection for conversion, why this has to occur at such a high frequency. Third, we have checked specifically for the occurrence of additional crossovers in our sample. In particular, we have evaluated the frequency of an additional unequal crossover leading to the formation of triplications. Its presence can be detected using PCR amplification with primers 3 and 2 (Fig. 2); only two out of 50 isolates revealed triplications (data not shown). Thus, unequal crossovers are infrequent enough to explain our results. Fourth, the unequal crossover model cannot explain the bias toward acquisition of the incoming sequence observed among the convertants. In fact, the unequal crossover model predicts that the double gain and double loss classes should appear at the same frequency. Moreover, the reversal of transfer bias observed upon exchanging the configuration of markers is also an unexpected feature under this model.
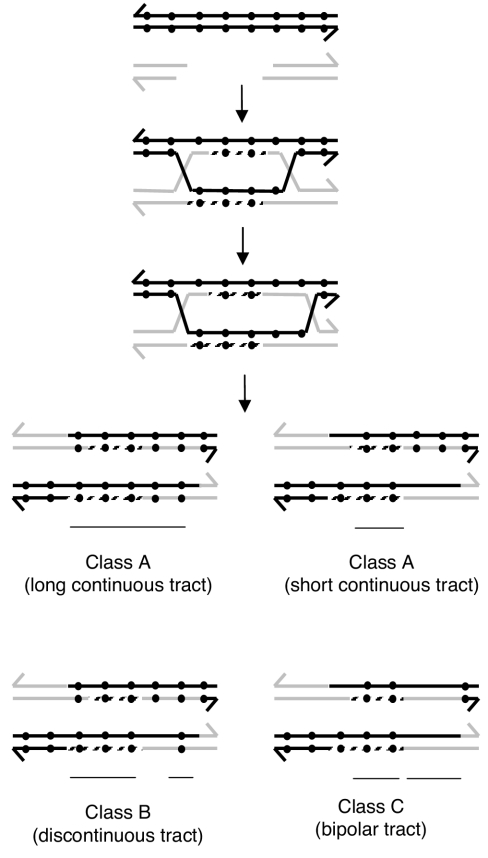
We think that our data are better explained by invoking the occurrence of gene conversion, perhaps generated under the double-strand break repair model (44). This model for recombination is now widely accepted for both prokaryotic and eukaryotic organisms (4). Variations of this model have been used to explain recombinational repair of collapsed replication forks, a rather frequent event in bacteria (15, 19, 24). As shown in Fig. 10, this model explains all our data in an economical way. In this model, a double-strand gap made on the resident nifH sequence may be repaired by the modified nifH sequence present in the incoming plasmid. The DNA synthesis associated with gap repair generates, in this case, a short conversion tract; migration of the Holliday junction generates heteroduplex DNA. If the mismatches in the heteroduplex segment are corrected using the strands containing the information for the RFLPs, a long continuous conversion tract will ensue (Fig. 10, class A). Correction favoring the strand containing the RFLP information in one heteroduplex and the wild-type information in the other would generate a short continuous tract (Fig. 10, class A). Thus, in this model, continuous tracts are formed in two ways: by gap repair and also by heteroduplex correction. The bimodal distribution observed for the size of continuous conversion tracts (Fig. 5) is consistent with this interpretation. Similar sizes for converted tracts have been observed in other systems, such as Escherichia coli (45) Salmonella enterica serovar Typhimurium (1, 11), and Acinetobacter calcoaceticus (14).
FIG. 10.
Model for gene conversion of one nifH gene to the other initiated by a double-strand break. Black dots in the black double-strand represent differences in nucleotide sequence (RFLPs). According to the double-strand break repair model, a double-strand cut on the recipient molecule is enlarged by degradation to a gap, followed by strand exchange and gap DNA resynthesis (discontinuous lines). After that, the DNA heteroduplex could appear because of the migration of the Holliday junction. Further cuts are needed to generate a cointegrate. In class A, a long continuous tract could be the result of the action of gap DNA resynthesis and the repair of both DNA heteroduplexes, favoring retention of the RFLPs. For a short continuous tract (class A), we suggest that both DNA heteroduplexes may be repaired using the wild-type sequence as a template. Thus, the short tracts may arise only by the action of gap repair. In class B, a discontinuous tract could be the result of gap resynthesis and, in some mismatches, their repair favoring the wild-type or the modified sequence. For bipolar tracts (class C), gap repair results in tracts biased toward marker gain, while mismatch repair using the wild-type sequence as a template would result in marker loss.
This model also predicts that variations in the way in which the heteroduplexes are corrected should generate additional classes. For instance, correction of an intervening marker in one heteroduplex toward the mutated sequence and to wild-type sequence in the other should generate a discontinuous conversion segment (Fig. 10, class B). Also, correction of both heteroduplexes to wild-type sequence should generate a bipolar conversion tract (Fig. 10, class C). As expected under this model, both classes were found in our data, at a cumulative frequency similar to the one for the continuous class.
The presence of continuous and discontinuous conversion segments has also been observed in other studies of gene conversion in bacteria, particularly with the tufA-tufB genes in S. enterica serovar Typhimurium (1, 11). In that case, most of the converted segments belong to the continuous class, with a minority of discontinuous events. An important difference with our work is that in the case of S. enterica serovar Typhimurium (1, 11), selection was applied for isolation of the conversion events; this would reduce the representation of the discontinuous class.
The reduction in the length of conversion tracts in the mutS background presented here lends further support to the interpretation that these tracts appear through the operation of gap filling and heteroduplex correction. These data suggest that the MutS mismatch correction system is one of the major players in mismatch correction during gene conversion. This system, however, may not be the only one to participate in heteroduplex correction. Even in its absence, classes that should be reduced in abundance, such as the discontinuous and bipolar classes, are unabated. Thus, other mismatch repair systems, such as the very short repair system, are likely to participate in heteroduplex correction. Similar conclusions were also reached for the S. enterica serovar Typhimurium tufA-tufB system (1, 11), although in that case the characterization of the conversion tracts obtained in the mutS background was not presented.
This model also explains the close association between crossover formation (a selected event) and gene conversion (an unselected event). Under the double-strand break repair model, the strong association observed here should be the result of a preference to start a crossover in regions with a gap at least 100 bp in size. This will frequently include at least one marker, thus forcing the repair of that gap and the conversion of the restriction site. Association between crossover formation and conversion has been observed for S. enterica serovar Typhimurium (1, 11).
The fact that conversion is biased towards the incoming sequence is an unexpected characteristic from our data. To our knowledge, such a strong bias has not been reported previously, with the possible exception of natural transformation in Bacillus subtilis. In this organism, a weak preference to incorporate incoming markers has been observed (12). According to the double-strand break repair model, the molecule that receives the double-strand break will be the one to receive information through gap repair (i.e., the one to be converted). To explain the observed bias, we have to postulate that the resident molecule, not the incoming molecule, is the one that frequently receives a double-strand cut. This preference should arise in different ways, including generation of a double-strand break by collapse of a replication fork in the resident molecule and through the operation of endonucleases that preferentially cut resident molecules, to mention but two.
The proposed mechanisms for the bias towards the donor sequence may also help to illuminate the way in which cointegrates are generated in this organism. It is commonly thought that during conjugation, DNA is transferred as a linear concatemeric array that provides flanking homology for the selected marker (6, 41). A double crossover on such a direct duplication substrate could give rise to an integrant. If that were the case, there must be a strong bias favoring the retention of the resident sequence, because the discontinuity on the linear array should be detected (and corrected) by the mismatch repair system. The fact that the observed bias is toward the donor sequences militates against this view. Thus, we favor an alternative view in which the donor sequence is first circularized in the recipient cell and then integrated, using existing discontinuities on the resident sequence.
Although the specific mechanism involved in the observed bias remains to be clarified, these findings suggest an easy way to introduce specific mutations into the R. etli genome using gene conversion. Moreover, if the observed bias applies to other, more natural ways of transfer, this would make R. etli a rather permissive host to incorporate variations arising in a different host. Work in progress will clarify if this is the case.
In summary, we have provided evidence consistent with the operation of gene conversion in R. etli. Since the observed conversion tracts may frequently encompass more than half of the nifH gene, this process would be a good way to explain the concerted evolution among the members of this family. Moreover, our interpretation of these data lead to the prediction that both the size of the converted segment and the classes observed should be modified in backgrounds deficient in migration of the Holliday intermediates (ruvB, recG, and radA). Experiments are under way to test these hypotheses.
Acknowledgments
We are grateful to Patricia León and Edgardo Sepúlveda for helpful discussions, Laura Cervantes and Javier Rivera for technical help and advice, Araceli Dávalos for plasmid pIC20R oriT, and Paul Gaytán and Eugenio López for oligonucleotide synthesis. We are also indebted to the anonymous reviewers for helpful comments on the manuscript.
Work in our laboratory is partially supported by grant 31753-N (CONACyT, México). G.S. was supported by scholarships from CONACyT, México, and Dirección General de Estudios de Posgrado, Universidad Nacional Autónoma de México.
REFERENCES
- 1.Abdulkarim, F., and D. Hughes. 1996. Homologous recombination between the tuf genes of Salmonella typhimurium. J. Mol. Biol. 260:506-522. [DOI] [PubMed] [Google Scholar]
- 2.Alexeyev, M. F., I. N. Shokolenko, and T. P. Croughan. 1995. Improved antibiotic-resistance gene cassettes and omega elements for Escherichia coli vector construction and in vitro deletion/insertion mutagenesis. Gene 160:63-67. [DOI] [PubMed] [Google Scholar]
- 3.Arwidsson, O., and D. Hughes. 2004. Evidence against reciprocal recombination as the basis for tuf gene conversion in Salmonella enterica serovar typhimurium. J. Mol. Biol. 338:463-467. [DOI] [PubMed] [Google Scholar]
- 4.Cromie, G. A., J. C. Connelly, and D. R. Leach. 2001. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol. Cell 8:1163-1174. [DOI] [PubMed] [Google Scholar]
- 5.Dover, G. 2002. Molecular drive. Trends Genet. 18:587-589. [DOI] [PubMed] [Google Scholar]
- 6.Erickson, M. J., and R. J. Meyer. 1993. The origin of greater-than-unit-length plasmids generated during bacterial conjugation. Mol. Microbiol. 7:289-298. [DOI] [PubMed] [Google Scholar]
- 7.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 8.Flores, M., V. González, S. Brom, E. Martínez, D. Piñero, D. Romero, G. Dávila, and R. Palacios. 1987. Reiterated DNA sequences in Rhizobium and Agrobacterium spp. J. Bacteriol. 169:5782-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.González, V., P. Bustos, M. A. Ramírez-Romero, A. Medrano-Soto, H. Salgado, I. Hernández-González, J. C. Hernández-Celis, V. Quintero, G. Moreno-Hagelsieb, L. Girard, O. Rodríguez, M. Flores, M. A. Cevallos, J. Collado-Vides, D. Romero, and G. Dávila. 2003. The mosaic structure of the symbiotic plasmid of Rhizobium etli and its relation with other symbiotic genome compartments. Genome Biol. 4:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 11.Hughes, D. 2000. Co-evolution of the tuf genes links gene conversion with the generation of chromosomal inversions. J. Mol. Biol. 297:355-364. [DOI] [PubMed] [Google Scholar]
- 12.Iglesias, A., and T. A. Trautner. 1983. Plasmid transformation in Bacillus subtilis: symmetry of gene conversion in transformation with a hybrid plasmid containing chromosomal DNA. Mol. Gen. Genet. 189:73-76. [Google Scholar]
- 13.Kobayashi, I. 1992. Mechanisms for gene conversion and homologous recombination: the double-strand break repair model and the succesive half crossing-over model. Adv. Biophys. 28:81-133. [DOI] [PubMed] [Google Scholar]
- 14.Kowalchuk, G. A., L. A. Gregg-Jolly, and L. N. Ornston. 1995. Nucleotide sequences transferred by gene conversion in the bacterium Acinetobacter calcoaceticus. Gene 153:111-115. [DOI] [PubMed] [Google Scholar]
- 15.Kuzminov, A. 2001. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc. Natl. Acad. Sci. USA 98:8241-8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lathe, W. C., III, and P. Bork. 2001. Evolution of tuf genes: ancient duplication, differential loss and gene conversion. FEBS Lett. 502:113-116. [DOI] [PubMed] [Google Scholar]
- 17.Liao, D. 2000. Gene conversion drives within genic sequences: concerted evolution of ribosomal RNA genes in bacteria and archaea. J. Mol. Evol. 51:305-317. [DOI] [PubMed] [Google Scholar]
- 18.Liu, Y., and S. C. West. 2004. Happy Hollidays: 40th anniversary of the Holliday junction. Nat. Rev. Mol. Cell. Biol. 5:937-946. [DOI] [PubMed] [Google Scholar]
- 19.Marians, K. J. 2004. Mechanisms of replication fork restart in Escherichia coli. Phil. Trans. R. Soc. Lond. B Biol. Sci. 29:71-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marsh, J. L., M. Erfle, and E. J. Wykes. 1984. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene 32:481-485. [DOI] [PubMed] [Google Scholar]
- 21.Mattatall, N. R., D. A. Daines, S. L. Liu, and K. E. Sanderson. 1996. Salmonella typhi contains identical intervening sequences in all seven rrl genes. J. Bacteriol. 178:5323-5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mattatall, N. R., and K. E. Sanderson. 1996. Salmonella typhimurium LT2 possesses three distinct 23S rRNA intervening sequences. J. Bacteriol. 178:2272-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meinersmann, R. J., and K. L. Hiett. 2000. Concerted evolution of duplicate fla genes in Campylobacter. Microbiology 146:2283-2290. [DOI] [PubMed] [Google Scholar]
- 24.Michel, B., M. J. Flores, E. Viguera, G. Grompone, M. Seigneur, and V. Bidnenko. 2001. Rescue of arrested replication forks by homologous recombination. Proc. Natl. Acad. Sci. USA 98:8181-8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Mingfu, M., and B. Robinson. 1997. Approaches to DNA mutagenesis: an overview. Anal. Biochem. 254:157-178. [DOI] [PubMed] [Google Scholar]
- 27.Noel, K. D., A. Sánchez, L. Fernández, J. Leemans, and M. A. Cevallos. 1984. Rhizobium phaseoli symbiotic mutants with transposon Tn5 insertions. J. Bacteriol. 158:148-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohno, S. 1970. Evolution by gene duplication. Springer-Verlag, Berlin, Germany.
- 29.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. A. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncP alpha plasmids. Compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 30.Pride, D. T., and M. J. Blaser. 2002. Concerted evolution between duplicated genetic elements in Helicobacter pylori. J. Mol. Biol. 316:629-642. [DOI] [PubMed] [Google Scholar]
- 31.Quinto, C., H. de la Vega, M. Flores, L. Fernández, T. Ballado, G. Soberón, and R. Palacios. 1982. Reiteration of nitrogen fixation gene sequences in Rhizobium phaseoli. Nature 299:724-726. [Google Scholar]
- 32.Quinto, C., H. de la Vega, M. Flores, J. Leemans, M. A. Cevallos, M. A. Pardo, R. Azpiroz, M. L. Girard, E. Calva, and R. Palacios. 1985. Nitrogenase reductase: a functional multigene family in Rhizobium phaseoli. Proc. Natl. Acad. Sci. USA 82:1170-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rayssiguier, C., D. Thaler, and M. Radman. 1989. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature 342:396-401. [DOI] [PubMed] [Google Scholar]
- 34.Rodríguez, C., and D. Romero. 1998. Multiple recombination events maintain sequence identity among members of the nitrogenase multigene family in Rhizobium etli. Genetics 149:785-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romero, D., and S. Brom. 2004. The symbiotic plasmids of the Rhizobiaceae, p. 271-290. In B. Funnell and G. Phillips (ed.), Plasmid biology. American Society for Microbiology, Washington, D.C.
- 36.Romero, D., S. Brom, J. Martínez-Salazar, M. L. Girard, R. Palacios, and G. Dávila. 1991. Amplification and deletion of a nod-nif region in the symbiotic plasmid of Rhizobium phaseoli. J. Bacteriol. 173:2435-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romero, D., J. Martínez-Salazar, L. Girard, S. Brom, G. Dávila, R. Palacios, M. Flores, and C. Rodríguez. 1995. Discrete amplifiable regions (amplicons) in the symbiotic plasmid of Rhizobium etli CFN42. J. Bacteriol. 177:973-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Santoyo, G., and D. Romero. 2005. Gene conversion and concerted evolution in bacterial genomes. FEMS Microbiol. Rev. 29:169-183. [DOI] [PubMed]
- 40.Segall, A. M., and J. R. Roth. 1994. Approaches to half-tetrad analysis in bacteria: recombination between repeated, inverse-order chromosomal sequences. Genetics 136:27-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silberstein, Z., and A. Cohen. 1987. Synthesis of linear multimers of oriC and pBR322 derivatives in Escherichia coli K-12: role of recombination and replication functions. J. Bacteriol. 169:3131-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon, R. 1984. High frequency mobilization of gram-negative bacterial replicons by the in vivo constructed Tn5-Mob transposon. Mol. Gen. Genet. 196:413-416. [DOI] [PubMed] [Google Scholar]
- 43.Stahl, F. 1994. The Holliday junction on its thirtieth anniversary. Genetics 138:241-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szostak, J., T. Orr-Weaver, R. Rothstein, and F. Stahl. 1983. The double-strand-break repair model for recombination. Cell 33:25-35. [DOI] [PubMed] [Google Scholar]
- 45.Takahashi, N. K., K. Kusano, T. Yokochi, Y. Kitamura, H. Yoshikura, and I. Kobayashi. 1993. Genetic analysis of double-strand break repair in Escherichia coli. J. Bacteriol. 175:5176-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas, C. M., and C. A. Smith. 1987. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu. Rev. Microbiol. 41:77-101. [DOI] [PubMed] [Google Scholar]
- 47.Valencia-Morales, E., and D. Romero. 2000. Recombination enhancement by replication (RER) in Rhizobium etli. Genetics 154:971-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 49.Zahrt, T. C., G. C. Mora, and S. Maloy. 1994. Inactivation of mismatch repair overcomes the barrier to transduction between Salmonella typhimurium and Salmonella typhi. J. Bacteriol. 176:1527-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zuñiga-Castillo, J., D. Romero, and J. M. Martínez-Salazar. 2004. The recombination genes addAB are not restricted to Gram-positive bacteria: genetic analysis of the recombination initiation enzymes RecF and AddAB in Rhizobium etli. J. Bacteriol. 186:7905-7913. [DOI] [PMC free article] [PubMed] [Google Scholar]