Abstract
In Escherichia coli, three mechanisms have been proposed to maintain proper regulation of replication so that initiation occurs once, and only once, per cell cycle. First, newly formed origins are inactivated by sequestration; second, the initiator, DnaA, is inactivated by the Hda protein at active replication forks; and third, the level of free DnaA protein is reduced by replication of the datA site. The datA site titrates unusually large amounts of DnaA and it has been reported that reinitiation, and thus asynchrony of replication, occurs in cells lacking this site. Here, we show that reinitiation in ΔdatA cells does not occur during exponential growth and that an apparent asynchrony phenotype results from the occurrence of rifampin-resistant initiations. This shows that the datA site is not required to prevent reinitiation and limit initiation of replication to once per generation. The datA site may, however, play a role in timing of initiation relative to cell growth. Inactivation of active ATP-DnaA by the Hda protein and the sliding clamp of the polymerase was found to be required to prevent reinitiation and asynchrony of replication.
Chromosome replication of Escherichia coli is initiated at a single origin, oriC, by the initiator protein, DnaA (28). In rapidly growing cells, rounds of replication overlap and two, four, or even eight origins coexist and are initiated simultaneously only once per cell cycle (45). The new origins are immediately inactivated by sequestration. Sequestration relies on binding of the SeqA protein to the newly replicated, hemimethylated origins. Subsequent methylation by Dam methylase ends the sequestration (12, 35, 50). Sequestration is one of the processes necessary to limit initiation of replication to once and only once per cell cycle. In addition to the sequestration, a reduction of the initiation potential (i.e., sufficient amounts of active DnaA) must be achieved during the sequestration period, such that when origins are released from sequestration they cannot be reinitiated (46), and initiation cannot occur again until one generation later, when enough initiation potential again has accumulated. In this way the E. coli cell does not need to keep track of how many chromosomal origins have been initiated every generation; it is sufficient to first initiate all and then sequester all origins. This explains why E. coli can harbor large numbers of minichromosomes (oriC plasmids) without problems of incompatibility. The minichromosomal origins are initiated in synchrony with the chromosomal origins (30, 31) and sequestered in the same way.
The reduction in initiation potential during sequestration can be achieved in two ways, (i) by reducing the availability of DnaA and (ii) by reducing the activity of DnaA. The availability of DnaA at oriC is affected by binding sites (DnaA boxes) distributed around the E. coli chromosome (19). One site in particular, datA, with five DnaA boxes, titrates unusually large amounts of DnaA protein in vivo (24) and is the main contributor to the DnaA titration mechanism (41). Inactivation of DnaA occurs by the RIDA mechanism (regulatory inactivation of DnaA) which is dependent on the DnaN sliding clamp of DNA polymerase III and the Hda protein which together hydrolyze the active ATP form of DnaA to the inactive ADP form (22, 23, 51).
In the present work we investigated whether both the titration of DnaA by datA and the inactivation of DnaA by RIDA are necessary mechanisms for taking down the initiation potential during sequestration and found that the latter is but the former is not.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids. All bacteria used were E. coli K-12 and are listed in Table 1. The dnaA46 allele linked to tna::Tn10 was transduced into W3110 and RSD448, and Δhda::Tet (11) was transduced into W3110. Cells were grown in AB minimal medium (13) supplemented with 10 μg/ml of thiamine, 0.2% glucose, 0.5% Casamino Acids (ABTG-CAA) at 37°C, or as otherwise indicated. For the measurement of origin incompatibility, cells were grown in Luria-Bertani medium. Kanamycin (25 μg/ml), chloramphenicol (25 μg/ml), and tetracycline (10 μg/ml) were added when required for selection. Plasmids pFH871 (3), pALO22 (14, 34), and pALO1 (32, 34) were described previously.
TABLE 1.
Bacterial strains
Flow cytometry.
DNA distributions and cell mass analysis by flow cytometry were done as described previously (40), with the exception that 300 μg/ml of rifampin and 10 μg/ml of cephalexin were used to block initiation of DNA replication (45) and cell division (8), respectively. The drugs were added at optical density at 450 nm of 0.15. Different concentrations (35, 75, 150, 300, 450, 600, 800, and 1,200 μg/ml) of rifampin were also used to see the effect of high concentrations of rifampin on rifampin-resistant initiation.
Cell cycle analysis.
The cell cycle was analyzed essentially as previously described (48). The chromosome of strain W3110 contains an inversion which leads to a displacement of oriC (26). On this chromosome the distance that one fork must travel from oriC to terC is about 13% longer than that of the other fork. In the simulation of the C period of W3110 it was assumed that the rate of DNA synthesis towards the end of the C period was reduced to half when only one fork was under way. Attempts at simulating the C period of strain W3110 have been made before but were unsuccessful, possibly because the chromosomal inversion was not taken into account (1). The individual values of C and D periods were obtained by iterative comparison until a best fit of different theoretical exponential DNA histograms to the experimental ones was found, as described elsewhere (39).
Southern hybridization.
Southern hybridization was performed as described previously (34) to measure the relative gene dosage of different markers (argE, oriC, trpS, and cpdB) to the terminus region (yddB). Chromosomal DNA extracted from exponentially growing cells (optical density at 450 nm of 0.3) was double digested with EcoRI and HindIII prior to gel electrophoresis. In this digestion, the probe of argE, oriC, trpS, cpdB, and terminus region (yddB) are hybridized on fragments of 12, 2.1, 6.1, 7.6, and 4.7 kb, respectively. A rifampin runout sample was used to normalize the band intensity. Probe fragments were made by PCR amplification of MG1655 genomic DNA using the following primers: 5′-GCGATTGCCTATTGTTTCCTCC and 5′-GGTCGTCTTCTTTTTCATCAGTCG (argE), 5′-AAGAATGGCTGGGATCGTGG and 5′-ATCGAGGTTACTGCGGATCA (oriC), 5′-TCATCACCACCAATCACCACG and 5′-GAAAATGGCTGTAAAGCGGGTC (trpS), 5′-CCACCTGATAAATCGCAAACCC and 5′-AACGCTTGGCAACCACGAG (cpdB), and 5′-AGGCTGCTATCCAGTTTTTCGTC and 5′-GTTTCGCATCAACCCTACTTCG (yddB). Southern blotting carried out to determine the incompatibility of chromosomal and minichromosomal origins was done as previously described (34, 46).
RESULTS
Cells lacking the Hda protein show origin incompatibility but cells lacking the datA site do not.
Mutants that have lost the ability to sequester oriC (e.g., dam and seqA mutants) cannot harbor autonomously replicating minichromosomes (34, 46). The reason for this is presumably that new, unsequestered origins are free to compete for initiation factors, allowing some of them to reinitiate at the expense of some of the other origins that will remain “uninitiated.” The cell has thus lost the ability to initiate every origin once, and only once. In this situation, one way to maintain both chromosomes and minichromosomes is to integrate them on the same replicon. This explains why the minichromosomes are integrated at the chromosomal origin when grown with selection for the minichromosomal marker (46).
Also, cells with intact sequestration experience origin incompatibility if the initiation potential is not reduced sufficiently to hinder reinitiation when sequestration is over (46). For instance, the presence of a sevenfold excess of DnaA protein leads to a permanently high initiation potential and to the loss of free minichromosomes by integration.
A common feature of strains that show origin incompatibility is the inability to ensure that each origin is initiated once, and only once, every generation (46). Thus, we wished to use origin compatibility as a tool to investigate whether titration of DnaA, or inactivation of DnaA, or both mechanisms, contribute to the reduction of initiation potential during sequestration. Since both ΔdatA and Δhda mutants have been reported to replicate asynchronously (and therefore presumably reinitiate replication) (11, 25), we expected that both mechanisms would be required to reduce the initiation potential sufficiently to prevent reinitiation.
Wild-type, ΔdatA, Δhda, and Δdam cells were transformed with a minichromosome, and 8 or 16 transformants of each were grown in the presence of tetracycline for 50 to 100 generations. All Δdam transformants harbored chromosomes with minichromosomes integrated at the chromosomal origin (Table 2), in accordance with previous reports (34, 46). Most of the Δhda transformants also harbored chromosomes with minichromosomes integrated at the chromosomal origin, indicating that reinitiation occurred in these cells. The result indicates that the Hda protein is required to limit replication to only once per generation.
TABLE 2.
Minichromosomes replicate autonomously in cells without datA but not in cells lacking the Hda protein
| Recipient strain | Relevant genotype | No. of transformants/totala |
|---|---|---|
| MG1655 | Wild type | 8/8 |
| RSD448 | ΔdatA | 8/8 |
| KS0003 | Δdam | 0/8b |
| JE202 | Δhda | 2/16c |
Transformants were examined by Southern blotting (see Materials and Methods) after more than 100 generations. Data are shown as the number of transformants harboring free minichromosomes relative to the total number tested.
Transformation was performed with unmethylated pALO22. All eight transformants had pALO22 integrated at the chromosomal origin.
Transformation was performed with pALO1(Kanr) instead of pALO22(Tetr) (because JE202 harbours tet as part of the hda deletion) and examined after 50 generations. Fourteen transformants had pALO1 integrated at the chromosomal origin.
None of the ΔdatA or wild-type transformants contained chromosomes with integrated minichromosomes; the minichromosomes were all free, even after more than 100 generations of growth (Table 2). This result suggests that there is no excess initiation potential after the sequestration period is over in the ΔdatA cells and that titration of DnaA by datA is not a mechanism that contributes to limiting replication to only once per generation.
The ΔdatA cell cycle has a wild-type replication pattern.
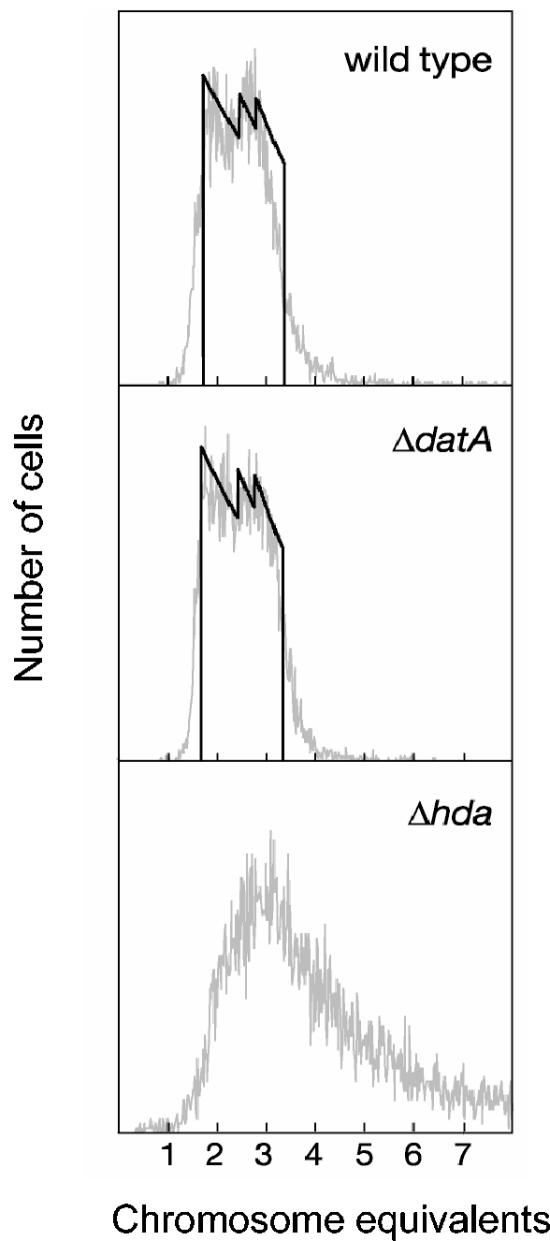
The results of the origin compatibility experiment above indicated that reinitiation does not occur in ΔdatA cells and conflicts with the results of Kitagawa et al. (25). We therefore analyzed DNA replication patterns by two additional methods: flow cytometry and marker frequency analysis. The DNA distributions of wild-type and ΔdatA cells, which grew with about the same doubling times (Table 3), were quite similar (Fig. 1), indicating that excess replication did not occur in the ΔdatA cells during exponential growth. An analysis of the replication patterns was made by running simulations of theoretical replication patterns and determining the replication (C) and postreplication (D) periods that generated the best fit to the experimental histograms (39, 48). In W3110 wild-type cells, the C period was found to be about 66 min and the D period about 18 min (Fig. 1). The unusually long C period may in part be due to a chromosomal inversion in strain W3110 that causes the forks to travel unequal distances from origin to terminus (see Materials and Methods). In the W3110ΔdatA cells, the cell cycle parameters were similar to those of the wild-type cells with a C period of about 68 min and a D period of about 18 min (Fig. 1). These results show that the replication pattern of W3110ΔdatA is essentially the same as that of W3110 and that no significant reinitiation occurs in the absence of datA. Analysis of other wild-type strains (MG1655 and AB1157) and their ΔdatA derivatives was also performed and gave essentially the same results (data not shown).
TABLE 3.
Rifampin-resistant initiation of replication in ΔdatA but not in Δhda cells
| Strain | Doubling time (min) | Average DNA content per cella
|
||
|---|---|---|---|---|
| Exponential | Rifampin treated | Rifampin/ exponential | ||
| W3110 | 41 (±0.3) | 1.00 (±0.01) | 1.52 (±0.01) | 1.52 |
| W3110 ΔdatA | 42 (±0.6) | 0.94 (±0.05) | 2.03 (±0.07) | 2.15 |
| W3110 Δhda | 72 (±0.3) | 1.98 (±0.03) | 3.16 (±0.06) | 1.59 |
Three independent experiments were performed. The values are relative to the exponentially growing wild-type cells.
FIG. 1.
The ΔdatA cells replicate with a wild-type pattern whereas the Δhda cells overreplicate. Experimentally obtained DNA histograms of 10,000 cells (thin grey lines) and computer simulated histograms (thick black lines) representing DNA distributions of idealized cell cultures where all cells have exactly the same C and D periods (i.e., with no cell-to-cell variability) are shown. The simulations giving the best fit to the experimental data had C and D periods of 68 and 18 min and 66 and 18 min for the ΔdatA mutant and its parent wild type (W3110), respectively. A computer simulated DNA distribution of W3110 Δhda cells was not made due to asynchrony of initiation.
The DNA distribution of the W3110Δhda cells showed that these cells contained more DNA than the wild type and that the DNA contents were more heterogeneous (Fig. 1 and Table 3), indicating that reinitiation and overreplication occurs when the Hda protein is missing. The result is in accordance with previous findings (11).
We also analyzed the abundance of oriC relative to a marker near the terminus (yddB) in W3110 and W3110ΔdatA cells and found the ratios to be 2.06 and 2.15, respectively (Fig. 2). The values are similar and indicate that no, or very little, reinitiation occurs in the absence of datA. In contrast, the origin-to-terminus ratios reported by Kitagawa et al. were 1.5 and 2.0, respectively, for W3110 and W3110ΔdatA (25). Because of this discrepancy we extended the marker frequency analysis to also include three additional markers 5 to 20 min away from oriC (Fig. 2A). The frequencies of these markers relative to the terminus were similar in the ΔdatA and wild-type strains (Fig. 2B). We do not know the reason for the discrepancy but conclude that the replication patterns of the two strains are essentially the same and that reinitiation does not occur in the absence of datA.
FIG. 2.
The ΔdatA cells have essentially the same origin-to-terminus ratio as wild-type cells. (A) Chromosome map of wild-type strain W3110, which has an inversion relative to other E. coli K-12 strains between rrnD and rrnE (black line). The longer (dark grey) and shorter (light grey) arms are indicated. (B) Relative frequencies of all markers to the terminus (yddB) were obtained by Southern blotting and plotted as a function of the distance from oriC. The values are the average of nine samples for wild type and six samples for the mutant.
Rifampin-resistant rounds of replication occur in ΔdatA cells.
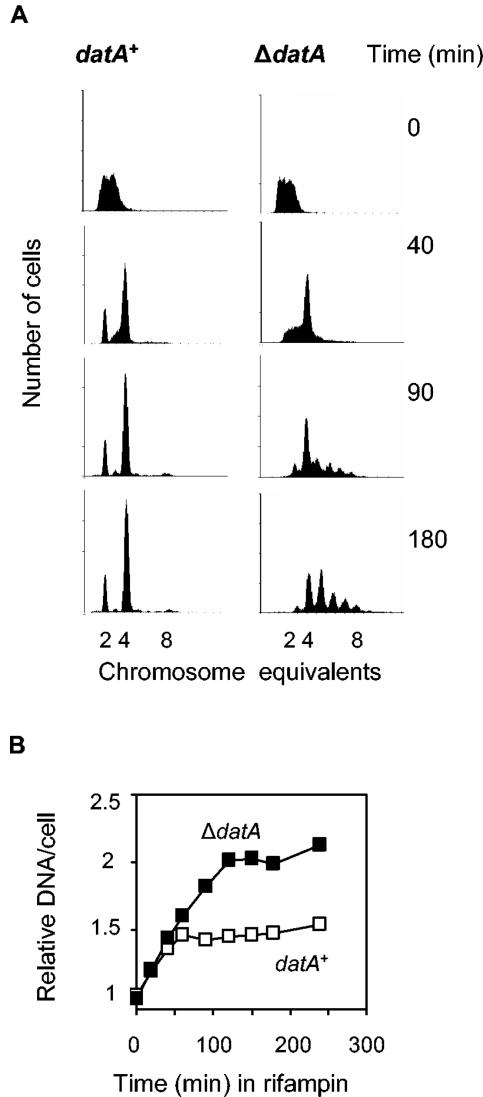
The above results contrast with the fact that the asynchrony of replication phenotype has been found in ΔdatA cells after treatment with rifampin and cephalexin and subsequent runout of replication (25). Here, a similar replication runout experiment was performed and the same result, i.e., an asynchrony phenotype, was found (Fig. 3A). The DNA histograms show that most of the wild-type cells contained four chromosomes after runout of replication, whereas the ΔdatA cells contained mostly four, five, six, or seven chromosomes (Fig. 3A).
FIG. 3.
Rifampin-resistant initiation of chromosome replication in ΔdatA cells. (A) Wild-type (W3110) and ΔdatA mutant cells were grown exponentially, and rifampin (300 μg/ml) and cephalexin (10 μg/ml) were added at 0 min. Samples taken at the indicated times were analyzed by flow cytometry. (B) Average DNA contents in wild-type and ΔdatA cells, calculated from flow cytometry histograms (40), were plotted as a function of time after addition of rifampin and cephalexin. Values are relative to the wild type at 0 min after addition of the drugs.
Several examples of the asynchrony phenotype have been reported previously (45), and the mechanisms behind this phenotype seem to be of four types: (i) compromised initiation of replication so that origins are not initiated simultaneously [dnaA(Ts)] (49), (ii) initiation more than once per generation (reinitiation) (ΔseqA [10, 35] and oriCm3 [6]), (iii) degradation of some of the newly synthesized DNA strands (recA) (44), and (iv) initiation of rifampin-resistant rounds of replication during runout (ihf) (52). Cells that show the asynchrony phenotype due to one of the latter two mechanisms do not necessarily have an altered timing of initiation and show a lower and a higher increase, respectively, in cellular DNA contents than expected after runout.
The average DNA content per wild-type cell stopped increasing at about 60 min after rifampin and cephalexin were added (Fig. 3B) and was then about 50% higher than that of the exponentially growing cells (Fig. 3B and Table 3). This is the expected DNA increase when all forks have reached the terminus. In the ΔdatA cells, the average DNA content continued to increase for about 120 min after drug treatment, ending at a value twofold higher than that of the exponentially growing cells (Fig. 3B and Table 3). This indicates that more forks than those that were under way at the time of drug addition contributed to the average DNA content at 120 min and suggests that rifampin-resistant initiation occurs in ΔdatA cells. This result explains the apparent replication asynchrony of ΔdatA cells. Rifampin-resistant replication in the absence of the datA site was also found using other strains (MG1655 or AB1157) (data not shown).
Overproduction of wild-type DnaA protein has been shown to lead to excess initiation of replication, with most of the extra forks aborted shortly after initiation (5, 43, 47). The rifampin-resistant rounds of replication may have been initiated after rifampin was added but may alternatively represent forks that were already present but that were aborted during exponential growth. The marker frequency analysis of regions 5 to 20 min away from oriC (Fig. 2) shows that the former is true. The ratios of these markers to the terminus marker (yddB) were, in the ΔdatA cells, gradually reduced as the distance from oriC was increased, in a fashion similar to that in the wild type (Fig. 2). Thus, there was no evidence that forks were aborted in the ΔdatA cells. This confirms that the extra rounds of replication found to occur after rifampin addition (Fig. 3) did not represent completion of replication by forks already present during exponential growth but resulted from initiations that occurred after rifampin was added.
Rifampin-resistant initiation of replication is dependent on DnaA functions not present in the DnaA46 protein and is suppressed by increasing the level of rifampin.
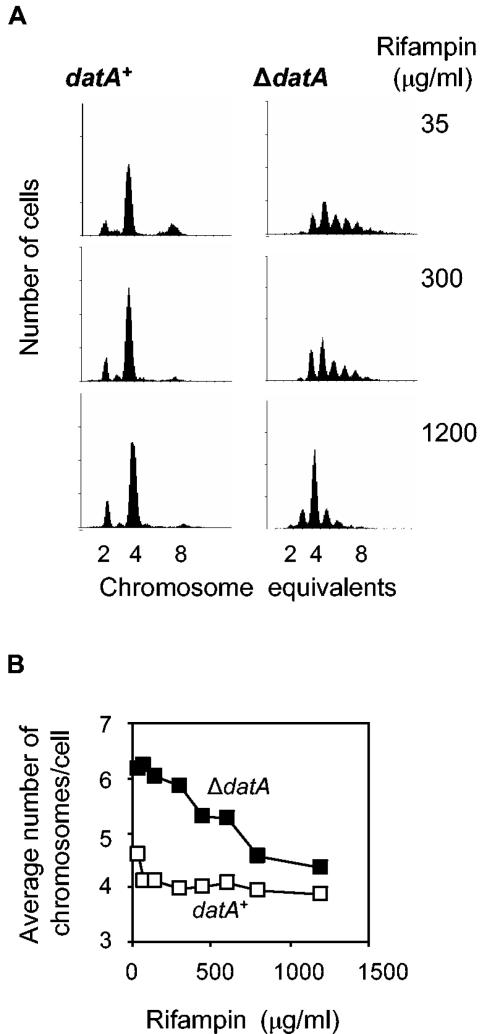
After replication runout in the presence of a fourfold increased rifampin concentration (1,200 μg/ml), the DNA content of ΔdatA cells was only slightly higher than that of the wild type (Fig. 4). This shows that the extra replication is inhibited by higher amounts of rifampin. The histogram of the ΔdatA cells incubated with the high amount of rifampin still showed 3- and 5-chromosome cells (Fig. 4A), probably because not all the rifampin-resistant initiations were suppressed.
FIG. 4.
A high concentration of rifampin suppresses the rifampin-resistant replication in ΔdatA cells. (A) Exponentially growing W3110 wild-type and ΔdatA cells were treated for four to five generations with the indicated concentrations of rifampin and 10 μg/ml of cephalexin and analyzed by flow cytometry. (B) Average numbers of chromosomes per cell calculated from the DNA histograms were plotted as a function of the concentrations of rifampin.
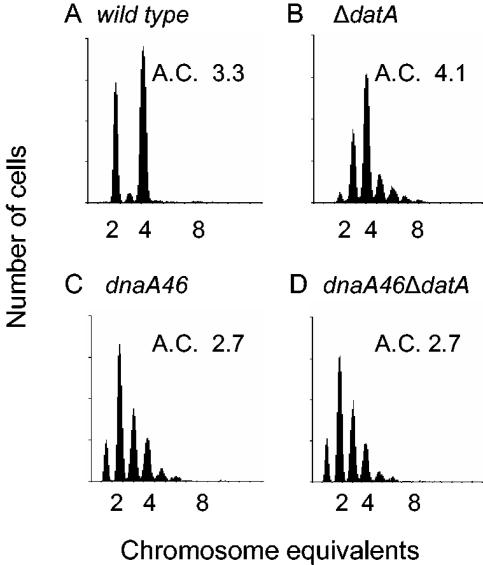
We also investigated whether the rifampin-resistant rounds of replication in the ΔdatA strain were dependent on wild-type DnaA protein, by comparing rifampin replication runout histograms of dnaA46 ΔdatA and dnaA46 cells grown at the permissive temperature (Fig. 5). Both strains showed the asynchrony of initiation characteristic of dnaA46 cells (49). The histograms were almost identical, and the average chromosome content per cell was 2.7 for both strains (Fig. 5C and D). This shows that rifampin-resistant replication did not occur in dnaA46 ΔdatA cells and indicates that it is not stable DNA replication (27) but originates in oriC and is prevented by the dnaA46 mutations.
FIG. 5.
Rifampin-resistant initiation in ΔdatA cells did not occur with DnaA46 mutant protein. Wild-type strain W3110 (A) and ΔdatA (B), dnaA46 (C), and dnaA46 ΔdatA (D) mutants were grown exponentially at 30°C, treated with rifampin (300 μg/ml) and cephalexin for four to five generations, and analyzed by flow cytometry. The average number of chromosomes per cell was calculated and shown as the A.C. number.
Reinitiation does occur in cells lacking the ability to inactivate ATP-DnaA.
Reinitiation and asynchrony of DNA replication has also been reported to occur in Δhda cells lacking the RIDA mechanism (11). To investigate whether rifampin-resistant initiation occurs also in Δhda mutants, we investigated the DNA distribution and the increase in cellular DNA content after rifampin and cephalexin treatment. The DNA histogram of exponentially growing Δhda cells showed, as mentioned above, a wider distribution and a twice as high average value compared to that of the parent wild-type cells (Fig. 1 and Table 3). The DNA increase during replication runout, however, was only slightly higher than in the wild type (Table 3), showing that no new replication forks were generated after rifampin was added. These results show that the asynchrony and overreplication phenotypes of Δhda cells represent bona fide reinitiation and confirm that the mechanism of DnaA inactivation by RIDA is required to prevent rereplication (11).
ΔdatA cells are smaller than wild-type cells.
The results reported above show that the datA site does not have a role in preventing reinitiation of replication, i.e., in ensuring that initiation does not occur again in the same cell cycle. It is, however, possible that the datA site is important for other aspects of timing of replication. The other main aspect of timing is ensuring that the (synchronous) initiations occur at the correct point and not too early or too late. To address this issue, we investigated the mass of ΔdatA cells to see if the time of initiation were changed relative to mass increase. The protein content, as well as the DNA content, of each cell was measured by flow cytometry. The protein content may be taken as a measure of the cell mass (9). The mass of ΔdatA cells was found to be 10 to 20% lower than the mass of wild-type cells (Table 4). This means that both the mass at initiation and the mass at division were lower in the ΔdatA cells compared to wild type. Thus, these two parameters had changed in the same direction after the datA site was deleted, indicating that the controls affecting initiation mass and division mass react in similar ways to deletion of the datA site. This result suggests that the presence of the datA site (and presumably the availability of DnaA protein) affects both timing of initiation and timing of division relative to mass increase. Similar results were found using AB medium with glucose or glycerol and using strain AB1157 (data not shown).
TABLE 4.
Cell cycle parameters in wild-type and ΔdatA cells
| Strain | Doubling time (min) | Average DNA content per cella | Average cell massa | Average DNA per massa |
|---|---|---|---|---|
| W3110 | 41 (±0.2) | 1.00 (±0.03) | 1.00 (±0.04) | 1.00 |
| W3110 ΔdatA | 42 (±0.4) | 0.94 (±0.05) | 0.82 (±0.04) | 1.15 |
Seven experiments with a pair of wild-type and ΔdatA cultures were performed. The values are relative to those of the wild-type cells of each individual experiment.
DISCUSSION
Inactivation of DnaA, but not titration of DnaA by the datA site, is required to restrict initiation of replication to once per generation. We have found that the apparent reinitiation in datA-less cells (25) does not occur in exponentially growing cells but is due to rifampin-resistant initiation of replication. Our results are supported by an independent study by Camara and coworkers (J. Camara, A. Breier, N. Cozzarelli, and E. Crooke, personal communication). In this study, a lack of increase in origin dosage in exponentially growing ΔdatA cells compared to wild-type cells was found by chromosome microarray experiments. We conclude that titration of DnaA protein by the datA site is not necessary in order to prevent reinitiation in E. coli.
The results obtained here (Tables 2 and 3) and earlier (11, 35, 46) using sequestration-deficient cells and Δhda cells show that the mechanisms of sequestration and RIDA are both required to inhibit more than one initiation event per origin per generation. We therefore also conclude that the “once-per-cell-cycle” regulation of initiation in E. coli seems to work by specifically inactivating the new origins by sequestration and then bringing down the initiation potential by inactivating DnaA as soon as new forks are under way.
The availability of DnaA protein may affect timing of initiation and division relative to mass growth.
Although the datA site does not seem to be required for preventing rereplication, it does seem to affect another aspect of timing. We find here that cells lacking datA initiate replication at a lower mass than wild-type cells. The regulatory machinery responsible for coordinating initiation of replication with cell growth thus responds as if the cells were larger, when the datA site is lacking. The regulatory machinery responsible for coordinating cell division with growth responds in the same way. How (or whether) the DnaA protein is involved in this regulation is not clear because we do not know whether the effect is caused by lack of titration of DnaA or by lack of some other unknown function of datA. It has been suggested that the amount of DnaA protein per origin is one of the parameters that determine the cell mass at which replication initiates (19). It has also been found that the DnaA concentrations in different E. coli K-12 strains grown under different conditions are similar (18), indicating that production of DnaA follows the bulk of other protein synthesis. The production and availability of DnaA protein could thus provide a link between initiation of replication and mass growth. This notion is supported by the present results provided they represent lack of titration of DnaA.
Since the cell mass at division also was reduced, the DnaA protein may also link cell division to mass growth. In addition to being an initiator protein, DnaA is a gene regulatory protein (38). It is thus possible that the availability of DnaA protein affects the expression of regulatory elements for cell division. In accordance with this, cells growing with a fourfold excess of datA (provided on a miniR1 plasmid) were found to exhibit a delayed cell division (40).
The fact that a change in timing of initiation is observed when datA is deleted demonstrates that small changes in timing can be made in E. coli without overreplication or disturbance of synchrony. It also suggests that titration of DnaA by the datA site plays a role in ensuring that initiation occurs at the correct time point relative to mass increase.
Loss of the datA site causes extra initiation in the presence of rifampin.
ΔdatA cells were capable of initiating rounds of replication after rifampin was added, indicating that an excess initiation potential was present that could be released. These initiations did not occur in synchrony at all origins present in a cell. Rifampin-resistant initiation of replication has also been found to occur in ihf mutants (52). The IHF protein is an important component of the initiation complex at oriC (15, 20, 42). Cells lacking the IHF protein have problems with initiation of replication and seem to require larger than normal amounts of DnaA protein to accomplish the initiation event (52). Thus, a common feature of the ΔdatA and ihf mutants may be that they contain an increased availability of active DnaA protein.
To investigate whether an elevated level of DnaA protein causes rifampin-resistant initiations, we investigated strain MG1655/pFH871 in which DnaA protein is expressed at an eightfold higher level than in the wild type (40). In agreement with earlier studies (4, 33, 47), we found increased DNA and origin contents (about 20% and 50%, respectively) and a reduced rate of replication fork movement (data not shown). In addition, 10 to 15% of the origins initiated during rifampin treatment (data not shown), supporting the notion that a surplus of DnaA allows initiation to occur in the presence of rifampin. Thus, it is reasonable to assume that the extra initiations released in the presence of rifampin in ΔdatA cells is caused by a surplus of free, active DnaA protein. It is not clear, however, why initiation of some but not all origins was allowed. It is also not clear what factor holds back initiation in the presence of a surplus of DnaA in the absence of rifampin.
Rifampin inhibits transcription by RNA polymerase which is required for initiation of replication (29, 36). It might thus appear that initiation had become independent of transcription in or near oriC. However, increasing the concentration of rifampin fourfold strongly reduced the amount of rifampin-resistant initiation of replication (Fig. 3). This result indicates that in datA-less cells, the RNA polymerase at the origin is less susceptible to rifampin than in wild-type cells. Since mass growth was inhibited with similar kinetics in ΔdatA and wild-type cells (data not shown), it is reasonable to assume that uptake of rifampin was not affected by the datA deletion.
As mentioned above, the DnaA protein acts as a gene regulatory protein affecting transcription from many promoters, two of them (the mioC and gid promoters) near oriC (37). It is possible that DnaA protein interacts with the RNA polymerases at DnaA-stimulated promoters in such a way that binding of rifampin is inhibited. If so, the excess of free DnaA presumably found in ΔdatA cells would increase the resistance to rifampin. Different dnaA(Ts) mutants are suppressed by different mutations in the beta subunit of RNA polymerase (2, 7). Many of these RNA polymerase mutants are also rifampin-resistant, suggesting that the sites of DnaA and rifampin interaction may be near each other. Rifampin-resistant initiation has also been found in some dnaA(Ts) mutants (dnaA606, dnaA601, and dnaA5) but not others (dnaA167 and dnaA46), upon shift-down to permissive temperature after prolonged incubation at nonpermissive temperature (17). It is possible that the former mutants retain the capacity of DnaA to “protect” RNA polymerase from rifampin, while the latter ones do not. The latter observation is in accordance with the lack of rifampin-resistant initiation found here in the dnaA46 ΔdatA strain.
Acknowledgments
We are very grateful to Mali Strand Ellefsen and the Department of Biophysics Flow Cytometry Core Facility for excellent technical assistance in performance of flow cytometry and Anne Wahl for her great help in the laboratory work. We also thank J. E. Camara, A. M. Breier, N. R. Cozzarelli, and E. Crooke for sharing results prior to publication and Erik Boye, Trond Bach, and Ingvild Flåtten for critical reading of the manuscript.
F. M. was supported by the Secretaria de Estado de Educacion y Universidades (Spain) y el Fondo Social Europeo. This work was supported by the European Commission FP5, the Norwegian Research Council, and the Norwegian Cancer Society.
REFERENCES
- 1.Allman, R., T. Schjerven, and E. Boye. 1991. Cell cycle parameters of Escherichia coli K-12. J. Bacteriol. 173:7970-7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atlung, T. 1984. Allele-specific suppression of dnaA(Ts) mutations by rpoB mutations in Escherichia coli. Mol. Gen. Genet. 197:125-128. [DOI] [PubMed] [Google Scholar]
- 3.Atlung, T., E. S. Clausen, and F. G. Hansen. 1985. Autoregulation of the dnaA gene of Escherichia coli K12. Mol. Gen. Genet. 200:442-450. [DOI] [PubMed] [Google Scholar]
- 4.Atlung, T., and F. G. Hansen. 1993. Three distinct chromosome replication states are induced by increasing concentrations of DnaA protein in Escherichia coli. J. Bacteriol. 175:6537-6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atlung, T., A. Løbner-Olesen, and F. G. Hansen. 1987. Overproduction of DnaA protein stimulates initiation of chromosome and minichromosome replication in Escherichia coli. Mol. Gen. Genet. 206:51-59. [DOI] [PubMed] [Google Scholar]
- 6.Bach, T., and K. Skarstad. 2004. Re-replication from non-sequesterable origins generates three-nucleoid cells which divide asymmetrically. Mol. Microbiol. 51:1589-1600. [DOI] [PubMed] [Google Scholar]
- 7.Bagdasarian, M. M., M. Izakowska, and M. Bagdasarian. 1977. Suppression of the DnaA phenotype by mutations in the rpoB cistron of ribonucleic acid polymerase in Salmonella typhimurium and Escherichia coli. J. Bacteriol. 130:577-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boye, E., and A. Løbner-Olesen. 1991. Bacterial growth control studied by flow cytometry. Res. Microbiol. 142:131-135. [DOI] [PubMed] [Google Scholar]
- 9.Boye, E., H. B. Steen, and K. Skarstad. 1983. Flow cytometry of bacteria: a promising tool in experimental and clinical microbiology. J. Gen. Microbiol. 129:973-980. [DOI] [PubMed] [Google Scholar]
- 10.Boye, E., T. Stokke, N. Kleckner, and K. Skarstad. 1996. Coordinating DNA replication initiation with cell growth: differential roles for DnaA and SeqA proteins. Proc. Natl. Acad. Sci. USA 93:12206-12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camara, J. E., K. Skarstad, and E. Crooke. 2003. Controlled initiation of chromosomal replication in Escherichia coli requires functional Hda protein. J. Bacteriol. 185:3244-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell, J. L., and N. Kleckner. 1990. E. coli oriC and the dnaA gene promoter are sequestered from Dam methyltransferase following the passage of the chromosomal replication fork. Cell 62:967-979. [DOI] [PubMed] [Google Scholar]
- 13.Clark, D. J., and O. Maaløe. 1967. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 23:99-112.
- 14.Gerdes, K., F. W. Bech, S. T. Jorgensen, A. Løbner-Olesen, P. B. Rasmussen, T. Atlung, L. Boe, O. Karlstrom, S. Molin, and K. von Meyenburg. 1986. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 5:2023-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimwade, J. E., V. T. Ryan, and A. C. Leonard. 2000. IHF redistributes bound initiator protein, DnaA, on supercoiled oriC of Escherichia coli. Mol. Microbiol. 35:835-844. [DOI] [PubMed] [Google Scholar]
- 16.Guyer, M. S., R. R. Reed, J. A. Steitz, and K. B. Low. 1981. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb. Symp. Quant. Biol. 45:135-140. [DOI] [PubMed] [Google Scholar]
- 17.Hansen, F. G. 1995. Reinitiation kinetics in eight dnaA(Ts) mutants of Escherichia coli: rifampicin-resistant initiation of chromosome replication. Mol. Microbiol. 15:133-140. [DOI] [PubMed] [Google Scholar]
- 18.Hansen, F. G., T. Atlung, R. E. Braun, A. Wright, P. Hughes, and M. Kohiyama. 1991. Initiator (DnaA) protein concentration as a function of growth rate in Escherichia coli and Salmonella typhimurium. J. Bacteriol. 173:5194-5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen, F. G., B. B. Christensen, and T. Atlung. 1991. The initiator titration model: computer simulation of chromosome and minichromosome control. Res. Microbiol. 142:161-167. [DOI] [PubMed] [Google Scholar]
- 20.Hwang, D. S., and A. Kornberg. 1992. Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J. Biol. Chem. 267:23083-23086. [PubMed] [Google Scholar]
- 21.Jensen, K. F. 1993. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 175:3401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katayama, T., T. Kubota, K. Kurokawa, E. Crooke, and K. Sekimizu. 1998. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell 94:61-71. [DOI] [PubMed] [Google Scholar]
- 23.Kato, J., and T. Katayama. 2001. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 20:4253-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitagawa, R., H. Mitsuki, T. Okazaki, and T. Ogawa. 1996. A novel DnaA protein-binding site at 94.7 min on the Escherichia coli chromosome. Mol. Microbiol. 19:1137-1147. [DOI] [PubMed] [Google Scholar]
- 25.Kitagawa, R., T. Ozaki, S. Moriya, and T. Ogawa. 1998. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 12:3032-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knott, V., D. J. Blake, and G. G. Brownlee. 1989. Completion of the detailed restriction map of the E. coli genome by the isolation of overlapping cosmid clones. Nucleic Acids Res. 17:5901-5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kogoma, T., and K. von Meyenburg. 1983. The origin of replication, oriC, and the dnaA protein are dispensable in stable DNA replication (sdrA) mutants of Escherichia coli K-12. EMBO J. 2:463-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kornberg, A., and T. A. Baker. 1992. DNA replication, 2nd ed. Freeman, New York, N.Y.
- 29.Lark, K. G. 1972. Evidence for the direct involvement of RNA in the initiation of DNA replication in Escherichia coli 15T. J. Mol. Biol. 64:47-60. [DOI] [PubMed] [Google Scholar]
- 30.Leonard, A. C., and C. E. Helmstetter. 1986. Cell cycle-specific replication of Escherichia coli minichromosomes. Proc. Natl. Acad. Sci. USA 83:5101-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Løbner-Olesen, A. 1999. Distribution of minichromosomes in individual Escherichia coli cells: implications for replication control. EMBO J. 18:1712-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Løbner-Olesen, A., T. Atlung, and K. V. Rasmussen. 1987. Stability and replication control of Escherichia coli minichromosomes. J. Bacteriol. 169:2835-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Løbner-Olesen, A., K. Skarstad, F. G. Hansen, K. von Meyenburg, and E. Boye. 1989. The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell 57:881-889. [DOI] [PubMed] [Google Scholar]
- 34.Løbner-Olesen, A., and U. von Freiesleben. 1996. Chromosomal replication incompatibility in Dam methyltransferase deficient Escherichia coli cells. EMBO J. 15:5999-6008. [PMC free article] [PubMed] [Google Scholar]
- 35.Lu, M., J. L. Campbell, E. Boye, and N. Kleckner. 1994. SeqA: a negative modulator of replication initiation in E. coli. Cell 77:413-426. [DOI] [PubMed]
- 36.Messer, W. 1972. Initiation of deoxyribonucleic acid replication in Escherichia coli B/r: chronology of events and transcriptional control of initiation. J. Bacteriol. 112:7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Messer, W., and C. Weigel. 1997. DnaA initiator—also a transcription factor. Mol. Microbiol. 24:1-6. [DOI] [PubMed] [Google Scholar]
- 38.Messer, W., and C. Weigel. 2003. DnaA as a transcription regulator. Methods Enzymol. 370:338-349. [DOI] [PubMed] [Google Scholar]
- 39.Molina, F., and K. Skarstad. 2004. Replication fork and SeqA focus distributions in Escherichia coli suggest a replication hyperstructure dependent on nucleotide metabolism. Mol. Microbiol. 52:1597-1612. [DOI] [PubMed] [Google Scholar]
- 40.Morigen, A. Løbner-Olesen, and K. Skarstad. 2003. Titration of the Escherichia coli DnaA protein to excess datA sites causes destabilization of replication forks, delayed replication initiation and delayed cell division. Mol. Microbiol. 50:349-362. [DOI] [PubMed] [Google Scholar]
- 41.Ogawa, T., Y. Yamada, T. Kuroda, T. Kishi, and S. Moriya. 2002. The datA locus predominantly contributes to the initiator titration mechanism in the control of replication initiation in Escherichia coli. Mol. Microbiol. 44:1367-1375. [DOI] [PubMed] [Google Scholar]
- 42.Polaczek, P. 1990. Bending of the origin of replication of E. coli by binding of IHF at a specific site. New Biol. 2:265-271. [PubMed] [Google Scholar]
- 43.Simmons, L. A., A. M. Breier, N. R. Cozzarelli, and J. M. Kaguni. 2004. Hyperinitiation of DNA replication in Escherichia coli leads to replication fork collapse and inviability. Mol. Microbiol. 51:349-358. [DOI] [PubMed] [Google Scholar]
- 44.Skarstad, K., and E. Boye. 1993. Degradation of individual chromosomes in recA mutants of Escherichia coli. J. Bacteriol. 175:5505-5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skarstad, K., E. Boye, and H. B. Steen. 1986. Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J. 5:1711-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skarstad, K., and A. Løbner-Olesen. 2003. Stable co-existence of separate replicons in Escherichia coli is dependent on once-per-cell-cycle initiation. EMBO J. 22:140-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skarstad, K., A. Løbner-Olesen, T. Atlung, K. von Meyenburg, and E. Boye. 1989. Initiation of DNA replication in Escherichia coli after overproduction of the DnaA protein. Mol. Gen. Genet. 218:50-56. [DOI] [PubMed] [Google Scholar]
- 48.Skarstad, K., H. B. Steen, and E. Boye. 1985. Escherichia coli DNA distributions measured by flow cytometry and compared with theoretical computer simulations. J. Bacteriol. 163:661-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skarstad, K., K. von Meyenburg, F. G. Hansen, and E. Boye. 1988. Coordination of chromosome replication initiation in Escherichia coli: effects of different dnaA alleles. J. Bacteriol. 170:852-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slater, S., S. Wold, M. Lu, E. Boye, K. Skarstad, and N. Kleckner. 1995. E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell 82:927-936. [DOI] [PubMed] [Google Scholar]
- 51.Su'etsugu, M., M. Takata, T. Kubota, Y. Matsuda, and T. Katayama. 2004. Molecular mechanism of DNA replication-coupled inactivation of the initiator protein in Escherichia coli: interaction of DnaA with the sliding clamp-loaded DNA and the sliding clamp-Hda complex. Genes Cells 9:509-522. [DOI] [PubMed] [Google Scholar]
- 52.von Freiesleben, U., K. V. Rasmussen, T. Atlung, and F. G. Hansen. 2000. Rifampicin-resistant initiation of chromosome replication from oriC in ihf mutants. Mol. Microbiol. 37:1087-1093. [DOI] [PubMed] [Google Scholar]