Abstract
When starved, Myxococcus xanthus cells send signals to each other that coordinate their movements, gene expression, and differentiation. C-signaling requires cell-cell contact, and increasing contact brought about by cell alignment in aggregates is thought to increase C-signaling, which induces expression of many genes, causing rod-shaped cells to differentiate into spherical spores. C-signaling involves the product of the csgA gene. A csgA mutant fails to express many genes that are normally induced after about 6 h into the developmental process. One such gene was identified by insertion of Tn5 lac at site Ω4406 in the M. xanthus chromosome. Tn5 lac fused transcription of lacZ to the upstream Ω4406 promoter. In this study, the Ω4406 promoter region was identified by analyzing mRNA and by testing different upstream DNA segments for the ability to drive developmental lacZ expression in M. xanthus. The 5′ end of Ω4406 mRNA mapped to approximately 1.3 kb upstream of the Tn5 lac insertion. A 1.0-kb DNA segment from 0.8 to 1.8 kb upstream of the Tn5 lac insertion, when fused to lacZ and integrated at a phage attachment site in the M. xanthus chromosome, showed a similar pattern of developmental expression as Tn5 lac Ω4406. The DNA sequence upstream of the putative transcriptional start site was strikingly similar to promoter regions of other C-signal-dependent genes. Developmental lacZ expression from the 1.0-kb segment was abolished in a csgA mutant but was restored upon codevelopment of the csgA mutant with wild-type cells, which supply C-signal, demonstrating that the Ω4406 promoter responds to extracellular C-signaling. Interestingly, the 0.8-kb DNA segment immediately upstream of Tn5 lac Ω4406 inhibited expression of a downstream lacZ reporter in transcriptional fusions integrated at a phage attachment site in the chromosome but not at the normal Ω4406 location. To our knowledge, this is the first example in M. xanthus of a chromosomal position-dependent effect on gene expression attributable to a DNA segment outside the promoter region.
Myxococcus xanthus is a gram-negative, primarily soil-dwelling bacterium (6). It moves by gliding over solid surfaces. Swarms of these bacteria secrete enzymes that lyse prey bacteria and degrade macromolecules to provide nutrients. When nutrients become scarce, cells alter their behavior, piling on top of one another to form mounds. Within the mounds, cells differentiate from metabolically active rods to dormant spheres, resulting in mature, spore-filled fruiting bodies. Outside the fruiting bodies, cells differentiate into another type called peripheral rods (38, 39). Spores are resistant to environmental insults and allow M. xanthus to survive in unfavorable conditions. When nutrients again become available, spores in the fruiting body germinate to produce a swarm of rod-shaped cells capable of growth.
The developmental process of M. xanthus has long been studied as a model to understand how cells interact with each other (reviewed in references 19, 21, 48, and 50). These studies are of broad significance because many, if not most, bacteria exist in microbial communities called biofilms, and within biofilms cells send signals to each other and respond by changing the expression of certain genes (reviewed in reference 41). Likewise, M. xanthus cells send signals to each other during development that coordinate changes in gene expression, which cause cells to alter their movement, metabolism, and shape.
M. xanthus mutants defective in signaling were first described more than 25 years ago (14), but much remains to be learned about the molecular mechanisms of signaling and response. Among five classes of signaling mutants identified so far (5, 14), the signaling mechanism is well understood in two cases, A- and C-signaling. A-signaling involves the production of extracellular proteases early in development, which release peptides and amino acids that allow cells to assess the population density (reviewed in reference 21). Only if the number of cells per unit area is sufficient will development proceed. C-signaling involves the product of the csgA gene, a 25-kDa protein that appears to be associated with the outer membrane, where it is thought to be cleaved to a 17-kDa form that serves as the C-signal (25, 34). Transmission of the C-signal requires cell movement (23), possibly to allow cells to make end-to-end contacts (44). Upon transmission, C-signal modifies the motility behavior of the recipient cell (reviewed in references 19 and 50). At the population level, a low level of C-signaling causes rippling (accumulation of cells in parallel ridges that demonstrate movement in time-lapse microscopy), a higher level is needed for mound formation, and extensive cell-cell contacts within mounds, resulting in very high C-signaling, is believed to trigger sporulation (24, 31). Hence, C-signaling appears to coordinate cell behaviors during the middle to late stages of development.
Given the importance of A- and C-signaling for morphological development to proceed, it is not surprising that many developmentally regulated genes depend on these signaling interactions for expression. Many of the developmentally regulated genes that have been studied were identified by transposition of Tn5 lac into the M. xanthus chromosome (28). Tn5 lac can generate a transcriptional fusion between an M. xanthus promoter and the Escherichia coli lacZ gene (26). Among 21 such fusions to developmentally regulated genes, 18 failed to be expressed in asg mutants defective in A-signaling (29). The results suggested that A-signaling is required in the first few hours of development for the expression of most developmental genes. The response to A-signaling involves the SasS-SasR-SasN histidine kinase-response regulator-negative regulator three-component signal transduction system (59, 60). By measuring expression of Tn5 lac fusions in a csgA mutant, it was revealed that C-signaling begins to be required at about 6 h into development (27). Genes induced after 6 h exhibited either reduced expression in a csgA mutant, or no expression. This partial or absolute dependence on csgA for expression appears to reflect dependence on extracellular C-signaling, because in every case tested, expression in the csgA mutant could be restored by codevelopment with wild-type cells, which provide C-signal in the mixture (4, 10, 11, 27). Only one component of the C-signal transduction pathway is known, FruA (8, 40). This protein is similar to response regulators of two-component signal transduction systems. It has a critical aspartate residue that is thought to be phosphorylated in response to C-signal, although the putative kinase has not been identified. The direct targets of FruA regulation are also unknown.
Just as rippling, mound formation, and sporulation require different levels of C-signaling, the expression of different developmental genes requires distinct threshold levels of C-signal (24, 31). How the C-signal transduction pathway establishes such thresholds to ensure proper temporal and spatial regulation of C-signal-dependent genes is an important question.
To investigate the regulation of C-signal-dependent genes, the promoter regions of several have been identified and characterized. Mutational analyses of the Ω4403 promoter region (56), which exhibits absolute dependence on C-signaling (11, 27), and of the Ω4400 and Ω4499 promoter regions (63, 64), which exhibit partial dependence (4, 10, 27), have defined critical cis-acting DNA elements. One motif found in all three promoter regions is a C box (consensus sequence CAYYCCY, in which Y means C or T) preceded 6 to 8 bp upstream by a 5-bp element (consensus sequence GAACA) (56, 63, 64). Although these sequences are crucial for the activity of all three promoters, the pattern of effects on expression of single base pair mutations in these sequences is different for each of the three promoters, indicating that the motif functions differently in each case. It has been proposed that a family of related transcription factors might recognize the C box and 5-bp element (and in some cases the sequence in between), each binding to a specific promoter region and activating transcription (63, 64).
Here, we report the identification of the Ω4406 promoter region. Expression of the Ω4406 promoter depends absolutely on C-signaling, like that of the Ω4403 promoter, and both are expressed with similar timing during development. Interestingly, expression of both depends strongly on the location in the M. xanthus chromosome. We show that a DNA segment located 500 to 1,300 bp downstream of the Ω4406 promoter inhibits expression of a lacZ reporter when a transcriptional fusion is integrated into the M. xanthus chromosome by site-specific recombination at a phage attachment site but not when the fusion is present at the native Ω4406 location. We believe this is the first case in M. xanthus of a chromosomal position-dependent effect on gene expression attributable to a DNA segment outside the promoter region. We discuss possible mechanisms of the position effect, and we discuss the sequence of the Ω4406 promoter region in comparison with other C-signal-dependent promoter regions.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strains and plasmids used in this work are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | φ80 lacZΔM15 ΔlacU169 recA1 endA1 hsdR17 supE44 thi-1 gyrA relA1 | 16 |
| JM83 | ara Δlac-pro strA thi φ80 lacZΔM15 | 36 |
| M. xanthus | ||
| DK1622 | Wild type | 20 |
| DK4294 | Tn5 lac (Kmr) Ω4406 | 28 |
| DK4506 | Tn5 lac (Kmr) Ω4506 | 28 |
| MMF107, -108, -109 | attB::pREG4406 | This work |
| MJL100 | Tn5 lac (Tcr) Ω4406 | This work |
| MJL123-1, -5, -7, -8, -9 | Tn5 lac (Tcr) Ω4406 Ω4406::pJL23 | This work |
| MPV4406-3.2 | attB::pPV4406-3.2 | This work |
| MPV4406-2.4 | attB::pPV04406-2.4 | This work |
| MPV4406-1.8 | attB::pPV04406-1.8 | This work |
| MPV4406-1.0 | attB::pPV04406-1.0 | This work |
| MPV1.8-1175 | Ω4406::pPV1.8-1175 | This work |
| MPV2.4-1175 | Ω4406::pPV2.4-1175 | This work |
| MPV4406-2.4SC | attB::pPV04406-2.4SC | This work |
| MPV0.74kbins | Tn5 lac (Tcr) Ω4406 orf2::pPV0.74kbins | This work |
| LS203 | csgA653 | 49 |
| MPV1.8C | csgA653 attB::pPV04406-1.8 | This work |
| MPV1.0C | csgA653 attB::pPV04406-1.0 | This work |
| Plasmids | ||
| pGEM-7Zf | Aprlacα | Promega |
| pGEM4406 | Apr (pGEM-7Zf)a; 12-kb XhoI fragment from DK4294 | This work |
| pREG1666/pREG1727 | Apr Kmr P1-inc attP ′lacZ | 11 |
| pREG4406 | Apr Kmr (pREG1666); 3.2-kb XhoI-BamHI fragment from pGEM4406 | This work |
| pJL30 | Apr (pGEM-7Zf); 3.2-kb XhoI-BamHI fragment from pGEM4406 | This work |
| pREG429 | Apr Kmr P1-inc | 13 |
| pJL23 | Apr Kmr (pREG429); 3.2-kb ClaI-BamHI fragment from pJL30 | This work |
| pUC19 | Aprlacα | 61 |
| pJL40 | Apr (pUC19); 1.0-kb SalI-SalI fragment from pJL30 | This work |
| pBR322 | Apr Tcr | 3 |
| pKV322m | Apr Tcr (pBR322); a C to T mutation eliminated the BamHI site in pBR322 | This work |
| pKV322mlacZ | Apr Tcr (pKV322m); 3′ end of lacZ preceded by a unique EcoRI site | This work |
| pPV12K | Apr Tcr (pKV322mlacZ); 7.6-kb EcoRI fragment containing attP ′lacZ from pREG1727 | This work |
| pPV4406-3.2 | Apr Tcr (pPV12K); 3.2-kb XhoI-BamHI fragment from pJL30 | This work |
| pCR2.1-TOPO | Apr Kmrlacα | Invitrogen |
| pPV4406-2.4 | Apr Kmr (pCR2.1-TOPO); 2.4-kb Ω4406 upstream DNA segment generated by PCR | This work |
| pPV04406-2.4 | Apr Tcr (pPV12K); 2.4-kb XhoI-BamHI fragment from pPV4406-2.4 | This work |
| pPV4406-1.8 | Apr Kmr (pCR2.1-TOPO); 1.8-kb Ω4406 upstream DNA segment generated by PCR | This work |
| pPV04406-1.8 | Apr Tcr (pPV12K); 1.8-kb XhoI-BamHI fragment from pPV4406-1.8 | This work |
| pPV4406-1.0 | Apr Kmr (pCR2.1-TOPO); 1.0-kb Ω4406 upstream DNA segment generated by PCR | This work |
| pPV04406-1.0 | Apr Tcr (pPV12K); 1.0-kb XhoI-BamHI fragment from pPV4406-1.0 | This work |
| pREG1175 | Apr Kmr P1-inc ′lacZ | 12 |
| pPV1.8-1175 | Apr Kmr (pREG1175); 1.8-kb XhoI-BamHI fragment from pPV04406-1.8 | This work |
| pPV2.4-1175 | Apr Kmr (pREG1175); 2.4-kb XhoI-BamHI fragment from pPV4406-2.4 | This work |
| pPV4406-2.4SC | Apr Kmr (pPV4406-2.4); a GTG to TGA mutation changed the putative start codon of orf2 to a stop codon | This work |
| pPV04406-2.4SC | Apr Tcr (pPV12K); 2.4-kb XhoI-BamHI fragment from pPV4406-2.4SC | This work |
| pPV0.74kbins | Apr Kmr (pCR2.1-TOPO); 747-bp segment of orf2 generated by PCR | This work |
The vector is indicated in parentheses.
Growth and development.
E. coli cells were grown at 37°C in Luria-Bertani medium (45) containing 50 μg of ampicillin, 25 μg of kanamycin, or 10 μg of tetracycline per ml, as necessary. M. xanthus was grown at 32°C in CTT medium (18) in liquid culture or on agar (1.5%) plates with 40 μg of kanamycin or 12.5 μg of oxytetracycline per ml when required. Fruiting body development was performed on TPM (10 mM Tris-HCl, pH 8.0, 1 mM KH2PO4, 8 mM MgSO4 [final pH, 7.6]) agar (1.5%) plates as described previously (28).
Molecular cloning.
Recombinant DNA work was performed using standard techniques (45). Plasmid DNA was prepared from E. coli DH5α or JM83.
To clone the DNA upstream of Tn5 lac Ω4406, chromosomal DNA was prepared (30) from M. xanthus DK4294 and digested with XhoI, the fragments were ligated to XhoI-digested pGEM-7Zf, and the mixture was transformed into E. coli DH5α, selecting for both ampicillin resistance (Apr) of the vector and kanamycin resistance (Kmr) of the desired insert. One transformant with a plasmid bearing an insert of the expected size was named pGEM4406. A 3.2-kb XhoI-BamHI restriction fragment from pGEM4406, including M. xanthus DNA upstream of Ω4406, and the left end of Tn5 lac, was gel purified and ligated to pREG1666 to construct pREG4406. In this and other subcloning steps described in Table 1, vectors were digested with the same restriction enzymes used to produce the fragments, unless noted otherwise below.
To construct pJL30, the 3.2-kb XhoI-BamHI restriction fragment from pGEM4406 described above was ligated to HindIII-BamHI-digested pGEM-7Zf, the XhoI and HindIII ends were filled in with the Klenow fragment of DNA polymerase I, and the blunt ends were ligated.
To construct pPV12K, the BamHI site in the tetracycline resistance (Tcr) gene of pBR322 was changed from GGATCC to GGATTC by site-directed mutagenesis using the QuikChange kit (Stratagene), resulting in pKV322m, which lacks a BamHI site, as verified by DNA sequencing, but still confers Tcr in E. coli. This plasmid was digested with EcoRI and ligated to a short, synthetic DNA segment made by annealing LK559 (5′-AATTCCAGCTGAGCGCCGGTCGCTACCATTACCAGTTGGTCTGGTGTCAAAAATAA-3′) and LK560 (5′-AATTTTATTTTTGACACCAGACCAACTGGTAATGGTAGCGACCGGCGCTCAGCTGG-3′). The oligonucleotides were treated with T4 polynuceotide kinase and then denatured and annealed in buffer (10 mM Tris-HCl, pH 7.9, 50 mM NaCl, 10 mM MgCl2, and 1 mM dithiothreitol) by heating in a boiling water bath for 5 min and cooling at room temperature for 3 to 5 h, respectively. The resulting DNA segment was expected to have four-base overhangs (underlined) compatible with DNA ends generated by digestion with EcoRI. The segment includes 16 codons downstream of the EcoRI site in the E. coli lacZ gene and a stop codon (boldface). A plasmid, pKV322mlacZ, was identified by diagnostic PCR, in which the orientation of the 3′ end of lacZ was the same as the Tcr gene. DNA sequencing confirmed this orientation, as well as the sequence of the insert and the presence of an EcoRI site preceding the 3′ end of lacZ. This EcoRI site is unique in pKV322mlacZ because only LK559 (not LK560) was designed to regenerate an EcoRI site upon ligation. Plasmid pKV322mlacZ was digested with EcoRI, treated with calf intestinal phosphatase, gel purified, and ligated to a 7.6-kb fragment containing a segment from myxophage Mx8 (attP) that promotes site-specific recombination with the Mx8 attB site in the M. xanthus chromosome, followed by a multiple cloning site and a ′trpA lacZ′ fusion segment for generating transcriptional fusions to lacZ. The 7.6-kb fragment had been gel purified after digestion of pREG1727 with EcoRI, so it was expected to be missing the 3′ end of lacZ. A plasmid, pPV12K, in which the 7.6-kb fragment had inserted in the correct orientation to produce a full-length lacZ gene was identified by restriction digestion, and DNA sequencing confirmed that the 3′ end of lacZ was intact.
Segments of Ω4406 upstream DNA were amplified by PCR using pJL30 as template and the following primers: LK805 (5′-GCCTCGAGGAAATCCTCCAGGTGGCTC-3′) and LK757 (5′-GCGGATCCTTCCGAGAATCCTCGTCTTGC-3′) (restriction sites are underlined) to produce the 2.4-kb segment, LK756 (5′-GCCTCGAGGTCGACTCTGGCGAGCTGC-3′) and LK801 (5′-TCACACAGGAAACAGCTATGAC-3′, which is complementary to vector DNA near its junction with M. xanthus DNA in pJL30, so that a BamHI site at the junction is present in the PCR product) to produce the 1.8-kb segment, and LK756 (see above) and LK757 (5′-GCGGATCCTTCCGAGAATCCTCGTCTTGC-3′) to produce the 1.0-kb segment. These segments were cloned into pCR2.1-TOPO. The inserts were sequenced, and the sequences matched sequences present in the Cereon Microbial Sequence Database, which have been transferred to The Institute for Genomic Research and are available at http://www.tigr.org/tdb/mdb/mdbinprogress.html. Each insert was subcloned into pPV12K. The 2.4-kb insert in pPV4406-2.4 was also subjected to site-directed mutagenesis using the QuikChange kit (Stratagene) to mutate GTG to TGA, changing the putative start codon of orf2 to a stop codon in pPV4406-2.4SC.
To construct pPV0.74kbins, a 747-bp segment spanning codons 114 to 361 of predicted orf2 was amplified by PCR using pPV4406-2.4 as template and primers LK1058 (5′-CCCTCGAGCGTGCCCGTGGTCCCCG-3′) and LK1059 (5′-CCGGATCCCCGGAGTTCAACCCGGAG-3′) (restriction sites are underlined). The PCR product was gel purified, cloned into pCR2.1-TOPO, and verified by DNA sequencing.
Construction of M. xanthus strains.
Strains containing pREG1666 or pPV12K derivatives integrated at Mx8 attB were constructed by P1 specialized transduction from the rec+ E. coli strain JM83 (13) or by electroporation (22) of the wild-type M. xanthus strain DK1622 or the csgA mutant strain LS203. Transductants and transformants were selected on CTT-kanamycin or CTT-oxytetracycline plates. In the case of pREG4406, transductants containing a single copy of the plasmid integrated at Mx8 attB were identified by Southern blot hybridization. In agreement with previous experience in our laboratory (4, 10, 11, 17), the majority of transductants had a single copy of the plasmid integrated at Mx8 attB. Therefore, in the case of other plasmids expected to integrate at Mx8 attB, we employed a simple screen to eliminate colonies with unusual developmental lacZ expression. At least 10 transformants were transferred to TPM agar plates containing 40 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml. Rare transformants with unusual expression of lacZ were discarded, and of the remaining candidates, three independent isolates of each mutant construct were chosen for development. In all cases, the three transformants gave similar results when developmental β-galactosidase activity was measured as described previously (28).
Strain MJL100 was constructed by transducing bacteriophage P1::Tn5 lac (Tcr) (a gift from R. Gill) into DK4294 with selection for oxytetracycline resistance. Screening for kanamycin-sensitive transductants identified MJL100 in which the Kmr gene was replaced by the Tcr gene, as verified by Southern blot analysis (4).
MJL123-1, -5, -7, -8, and -9 are Kmr Tcr strains resulting from P1 specialized transduction of pJL23 from E. coli JM83 into M. xanthus MJL100, in which the plasmid integrated by homologous recombination (single crossover), as verified by Southern blot analysis.
MPV1.8-1175 and MPV2.4-1175 strains were constructed by electroporation of wild-type DK1622 with pPV1.8-1175 and pPV2.4-1175, respectively, selecting for Kmr. Transformants in which pPV2.4-1175 had integrated by homologous recombination (single crossover) were identified by diagnostic PCR using chromosomal DNA prepared as described previously (17) as template and primers LK1150 (5′-CGCGCCCCCGTATCCTC-3′) and LK764 (5′-CGGGCCATCCGCCAGTGG-3′). Transformants in which pPV1.8-1175 had integrated by homologous recombination were also identified by diagnostic PCR, but the chromosomal DNA template was prepared by a new method, and the primers were LK1304 (5′-GGTCCCCCACATGGACAAG-3′) and LK764 (see above). The new method of DNA preparation involved transferring part of a colony into 20 to 50 μl of 10 mM Tris-HCl, pH 8.5, buffer and mixing to disperse cells; then 20 μl of cell suspension was mixed with 20 μl of Lys-N-GO reagent (Pierce Biotechnology), the mixture was incubated at 95°C for 5 min, and 5 μl was added to a 45 μl PCR mixture.
MPV0.74kbins strains were constructed by electroporation of MJL100 with pPV0.74kbins, selecting for Kmr. Transformants in which the plasmid had integrated by homologous recombination were identified by diagnostic PCR using as template chromosomal DNA prepared by the new method described above and primers LK1088 (5′-GCCTCGAGGGCTACTCCGGCATCCGC-3′) and M13F (5′-TGTAAAACGACGGCCAGT-3′).
DNA sequencing.
Plasmid pJL40 was used as a template in sequencing reactions performed by the method of Sanger et al. (46) with a Sequenase kit (United States Biochemical). Ambiguities arising from premature termination were resolved using the protocol of Fawcett and Bartlett (9). 7-Deaza-dGTP reaction mixtures were used to resolve regions of compression. In this way, we determined the sequence of the 1.0-kb M. xanthus DNA segment in pJL40. All subsequent DNA sequencing was performed at the Michigan State University Genomics Technology Support Facility.
To determine the nature of fusions between M. xanthus DNA and the ′trp-lacZ segment in different vectors, we sequenced the junction between the multiple cloning site and the ′trp-lacZ segment of pREG1727. Based on the description of its construction (11), the unique BamHI site in the multiple cloning site of pREG1727 was expected to be joined to ′trpB, but we found that it was instead joined to ′trpA, with the junction sequence being 5′-GGATCCGGGCAT-3′, in which the BamHI site is underlined. We inferred that BamHI star activity had cleaved at the 5′-GATC-3′ sequence that begins at codon 27 of trpA (62), either during construction of pREG1727 or during construction of pREG1175 (12), from which the ′trp-lacZ segment in pREG1727 was derived. Sequencing of pREG1175 revealed that it has the same BamHI-′trpA junction as pREG1727. Therefore, the loss of the 3′ end of trpB and the 5′ end of trpA occurred during construction of pREG1175. Since pPV12K was derived from pREG1727, it also has the same BamHI-′trpA junction. If DNA is inserted into the BamHI site of pREG1175, pREG1727, or pPV12K and the DNA contains a promoter and part of a coding region, an in-frame translational fusion of the coding region to trpA can result. This might improve expression of the downstream lacZ gene relative to out-of-frame fusions, for which translation stops shortly after entering trpA, possibly creating a polar effect on transcription of lacZ (33).
RNA analysis.
RNA was prepared as described previously (11). Northern blot hybridization analysis was performed as described previously (15).
To perform the S1 nuclease protection experiment, a 1.0-kb SalI-SalI fragment of Ω4406 upstream DNA was gel purified, phosphatase treated, and 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase. The denatured probe was hybridized to 50 μg of RNA, and the DNA-RNA hybrid was digested with 50 U of S1 nuclease as described previously (11) to map the 5′ end of the Ω4406-associated transcript.
Primer extension analysis was performed as described previously (11) with the oligonucleotide 5′-CAGCGGTCAGTTCTGAATGAACATCGTGAC-3′.
RESULTS
Cloning DNA upstream of Ω4406 and testing it for promoter activity.
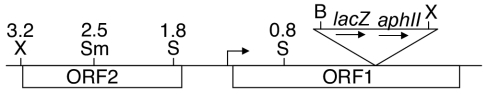
To clone the DNA upstream of the developmentally regulated Tn5 lac insertion Ω4406, we digested chromosomal DNA from M. xanthus DK4294 with XhoI restriction enzyme. Based on previous Southern blot hybridization results (28), the digestion was expected to yield a 12-kb fragment spanning from a XhoI restriction site located approximately 3 kb upstream of the Ω4406 insertion to a XhoI site in Tn5 lac (26) approximately 9 kb from its left end (Fig. 1). This fragment includes the aphII gene encoding aminoglycoside phosphotransferase, which confers Kmr when cloned in E. coli. The 12-kb fragment was cloned as described in Materials and Methods. Restriction mapping of the resulting plasmid, pGEM4406 (Table 1), showed the patterns expected on the basis of restriction sites in DNA upstream of Ω4406 (28) and restriction sites in Tn5 lac and the vector. Figure 1 shows the restriction map of DNA upstream of Ω4406.
FIG. 1.
Physical map of the Ω4406 insertion region. The diagram depicts the restriction sites in Tn5 lac (shown as a triangle that is not drawn to scale with the rest of the map) and the upstream M. xanthus chromosome that were used in this study. X, XhoI; Sm, SmaI; S, SalI; B, BamHI. Also shown are a putative transcriptional start site (right-angle arrow) and two ORFs (both oriented left to right) deduced from the DNA sequence. The numbers above the restriction sites indicate the distance in kilobases from the Tn5 lac Ω4406 insertion.
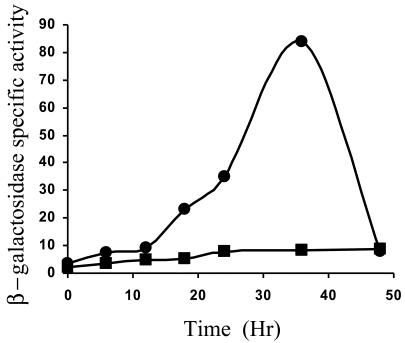
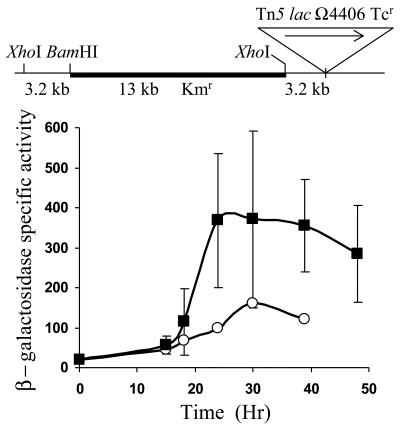
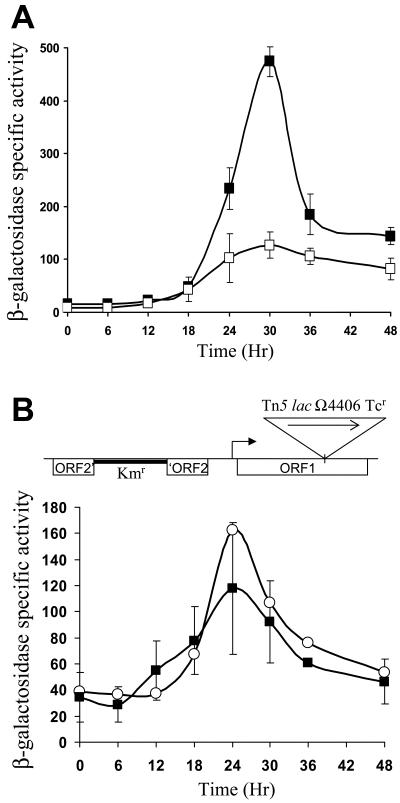
To test Ω4406 upstream DNA for promoter activity, the XhoI-BamHI restriction fragment from pGEM4406, which contains 3.2 kb of M. xanthus DNA and 53 bp of the left end of Tn5 lac (Fig. 1), was subcloned into XhoI-BamHI-digested pREG1666 to construct pREG4406 (Table 1). This plasmid contains Ω4406 upstream DNA fused to a promoterless lacZ gene. The plasmid was transduced from E. coli JM83 into the wild-type M. xanthus strain DK1622 using bacteriophage P1 specialized transduction (13). Due to the presence of the attP segment from myxophage Mx8, pREG4406 integrated efficiently into the M. xanthus chromosome at attB (51, 52). Transductants containing a single copy of pREG4406 integrated at Mx8 attB were identified by Southern blot hybridization (data not shown). Three of these transductants were assayed for β-galactosidase activity during development. Figure 2 shows that the 3.2-kb Ω4406 upstream DNA segment failed to direct developmental lacZ expression. This suggested that the Ω4406 promoter might not be present in the 3.2-kb upstream segment. Alternatively, the lacZ fusion might not be expressed when it is located at the Mx8 attB site in the M. xanthus chromosome. Reduced expression from this location has been reported previously for transcriptional lacZ fusions to developmentally regulated promoters (11, 32).
FIG. 2.
Developmental expression of Tn5 lac Ω4406 compared with the 3.2-kb Ω4406 upstream segment fused to lacZ. Developmental β-galactosidase specific activity was measured as described previously (28) for Tn5 lac-containing strain DK4294 (•) and for M. xanthus DK1622 containing a single copy of the 3.2-kb Ω4406 upstream DNA fused to promoterless lacZ within pREG1666 and integrated at Mx8 attB (▪; points are the average for three independent transductants). β-galactosidase specific activity is expressed in nanomoles of o-nitrophenyl phosphate per minute per milligram of protein.
Analysis of Ω4406 mRNA.
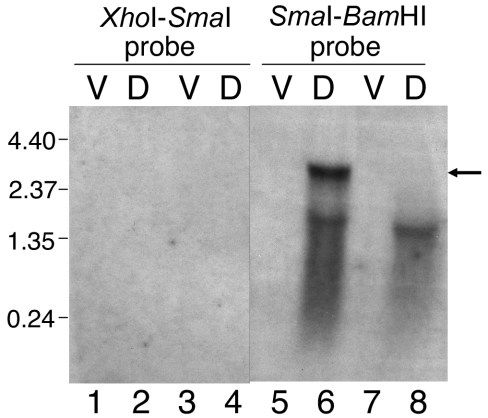
To begin to map the 5′ end of the mRNA responsible for developmental expression of Tn5 lac Ω4406, different segments of the cloned 3.2-kb Ω4406 upstream DNA were used as probes in Northern blot hybridization experiments. The 2.5-kb SmaI-BamHI fragment immediately upstream of Ω4406 (Fig. 1) detected a 3.0-kb transcript in RNA prepared from 24-h developing cells of M. xanthus DK4506 (Fig. 3, lane 6) but not in RNA prepared from vegetatively grown cells of this strain (Fig. 3, lane 5), which contains Tn5 lac inserted at a different site (Ω4506) in the chromosome. Below the 3.0-kb transcript is a smear that includes a faint but distinct signal at approximately 1.9 kb (Fig. 3, lane 6), which may reflect breakdown products of the 3.0-kb species (see below). The development-specific 3.0-kb transcript was not detected in RNA prepared from M. xanthus DK4294 cells bearing Tn5 lac Ω4406 (Fig. 3, lane 8), as expected because the fusion to lacZ would make the mRNA much longer. However, a longer development-specific transcript was not observed. Apparently, the fusion mRNA was too unstable and/or too heterogeneous in size to be detected. A faint smear with a distinct signal at approximately 1.6 kb was observed (Fig. 3, lane 8), perhaps due to breakdown products from the 5′ end of the fusion mRNA (see below).
FIG. 3.
Northern blot hybridization analysis of transcripts from the Ω4406 upstream region. RNA prepared from M. xanthus DK4506 (containing Tn5 lac Ω4506 and no insertion in the Ω4406 region) (lanes 1, 2, 5, and 6) or DK4294 (containing Tn5 lac Ω4406) (lanes 3, 4, 7, and 8) cells grown vegetatively (lanes V) or cells that had undergone 24 h of development (lanes D) was subjected to gel electrophoresis and blotting as described in Materials and Methods. Blots were hybridized with a radioactively labeled XhoI-SmaI or SmaI-BamHI fragment of Ω4406 upstream DNA (Fig. 1) as indicated at the top of the figure. The arrow indicates the 3.0-kb transcript, and the numbers to the left indicate the length (in kb) of RNA molecular size standards.
The 0.7-kb XhoI-SmaI fragment from farther upstream of Ω4406 (Fig. 1) failed to detect any transcripts (Fig. 3, lanes 1 to 4). The failure to observe the development-specific 3.0-kb transcript in RNA from DK4506 suggests that the Ω4406 mRNA 5′ end lies between the SmaI site and the site of Tn5 lac Ω4406 insertion (Fig. 1). The failure to observe the 1.9- and 1.6-kb transcripts in developmental RNA from DK4506 and DK4294, respectively, suggests that these transcripts are not derived from the upstream region between the XhoI and SmaI sites (Fig. 1).
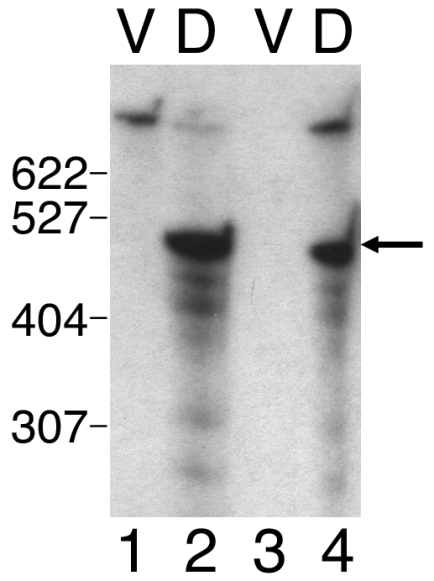
Additional probes were prepared from the 2.5-kb SmaI-BamHI segment (Fig. 1) in an attempt to further define the location of the Ω4406 mRNA 5′ end using Northern blot hybridization experiments. Both a 1.0-kb SalI-SalI fragment and a 0.8-kb SalI-BamHI fragment produced similar results (data not shown) as observed for the 2.5-kb SmaI-BamHI fragment (Fig. 3, lanes 5 to 8), except the signals observed with the 1.0-kb SalI-SalI probe were weaker than those observed with the 0.8-kb SalI-BamHI probe. These results suggested that the Ω4406 mRNA 5′ end might be located between the two SalI sites located approximately 1.8 and 0.8 kb upstream of Ω4406 (Fig. 1). To test this idea, the 1.0-kb SalI-SalI fragment was end labeled and used as a probe in a low-resolution S1 nuclease mapping experiment. Figure 4 shows that the probe yielded a major protected species of approximately 500 bases in length and several minor abundance species when hybridized to RNA prepared from DK4506 (lane 2) or DK4294 (lane 4) cells that had undergone 24 h of development but not when hybridized to RNA from vegetatively grown cells (lanes 1 and 3). The 500-base protected fragment suggests that the Ω4406 mRNA 5′ end lies about 1.3 kb upstream of the site of Tn5 lac Ω4406 insertion (Fig. 1). Since this protected fragment was observed with developmental RNA from DK4294, which contains Tn5 lac Ω4406, it supports the idea that the 1.6-kb transcript detected by Northern blot hybridization (Fig. 3, lane 8) is a breakdown product from the 5′ end of the fusion mRNA, especially since samples of the same RNA preparation were used in both experiments.
FIG. 4.
Localization of an mRNA 5′ end within the Ω4406 upstream region. RNA prepared from M. xanthus DK4506 (containing Tn5 lac Ω4506 and no insertion in the Ω4406 region) (lanes 1 and 2) or DK4294 (containing Tn5 lac Ω4406) (lanes 3 and 4) cells grown vegetatively (lanes V) or cells that had undergone 24 h of development (lanes D) was hybridized to a 1.0-kb SalI-SalI Ω4406 upstream DNA fragment (Fig. 1) that had been end labeled. The mixture was digested with S1 nuclease and analyzed by gel electrophoresis and autoradiography. The arrow indicates the major protected DNA species, and the numbers to the left indicate the length (in bases) of some of the end-labeled, MspI-digested pBR322 molecular size standard fragments. The species near the top is undigested 1.0-kb SalI-SalI probe.
DNA sequence of the Ω4406 region.
The nucleotide sequence of both strands of the 1.0-kb SalI-SalI fragment (Fig. 1) that had been used as the probe in the S1 nuclease mapping experiment (Fig. 4) was determined. A search of M. xanthus sequences in the Cereon Microbial Genome Database indicated that Ω4406 is inserted in a contig that may contain two complete open reading frames (ORFs) (Fig. 1) and two partial ORFs interrupted by the ends of the contig (data not shown). Only the complete ORFs are discussed here because both partial ORFs are oriented in the opposite direction as the complete ORFs and, therefore, cannot be coexpressed with ORF1, into which Tn5 lac Ω4406 had inserted (Fig. 1). ORF1 does not exhibit the strong bias toward usage of guanine or cytosine at the third codon position typical of M. xanthus genes. Also, the G+C content of the 2.8-kb DNA segment spanning from the end of ORF2 (Fig. 1) to the end of the partial ORF downstream of ORF1 is 59%, which is considerably lower than the 68% G+C content of the partial genome sequence. These observations may suggest that this DNA segment is a relatively recent addition to the M. xanthus genome in evolutionary time. The lack of third codon position GC bias makes the assignment of the ORF1 translational start codon less certain than for typical M. xanthus ORFs, but we predict that ORF1 begins with ATG located 448 bp upstream of the SalI site in orf1 (Fig. 1). This ATG is preceded 5 bp upstream by the sequence AGGAG, which might serve as a ribosomal binding site. If our prediction is correct, ORF1 would encode a 692-amino-acid protein, based on sequence data in the Cereon Microbial Genome Database. A BLAST search (1) with ORF1 revealed 43% similarity over a 542-amino-acid portion to a Streptomyces avermitilis hypothetical protein and 40% similarity over a 313-amino-acid portion to a Bradyrhizobium japonicum protein of unknown function.
Upstream of ORF1 is another ORF (ORF2) oriented in the same direction, but ORF2 is unlikely to be cotranscribed with ORF1 for several reasons. First, the 3.0-kb transcript detected by Northern blot hybridization with the SmaI-BamHI probe (Fig. 3, lane 6) is not long enough to express both ORF2 and ORF1. Second, the XhoI-SmaI probe, which includes the 5′ half of ORF2, did not detect the 3.0-kb transcript or any other transcripts (Fig. 3, lanes 1 to 4). Third, the 5′ end of Ω4406 mRNA appears to be located a short distance upstream of ORF1 (Fig. 4). Fourth, an inverted repeat sequence followed by TTTTT is located 40 bp downstream of the ORF2 translational stop codon. This sequence might serve as a terminator of transcription proceeding through ORF2. However, the Northern blot hybridization experiment with the XhoI-SmaI probe provided no evidence that ORF2 is transcribed under the conditions of growth and development we tested (Fig. 3, lanes 1 to 4).
Unlike ORF1, ORF2 does exhibit a strong bias toward usage of guanine or cytosine at the third codon position, which is typical for M. xanthus genes. ORF2 is predicted to begin with a GTG translational start codon, which is preceded 8 bp upstream by the sequence GAGGAAA, a potential ribosomal binding site. A BLAST search (1) with the 488-amino-acid ORF2 sequence revealed high similarity to histidine ammonia-lyase (histidase) of diverse organisms. This enzyme is the first in the histidine catabolic pathway and its synthesis is typically regulated in response to carbon and nitrogen availability (35), perhaps accounting for our inability to detect an orf2 transcript by Northern blot hybridization (Fig. 3, lanes 1 to 4).
Primer extension analysis.
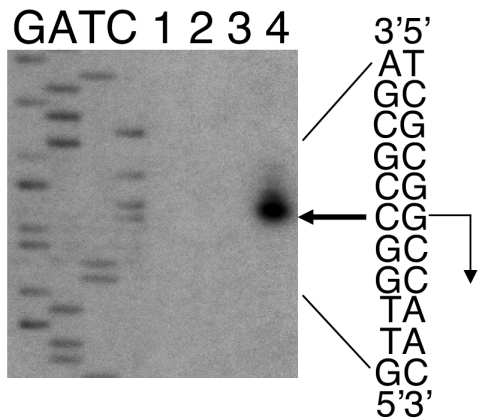
The DNA sequence of the 1.0-kb SalI-SalI fragment (Fig. 1) facilitated the design of a primer for precise localization of the Ω4406 mRNA 5′ end. When primer extension analysis was performed using RNA from M. xanthus wild-type DK1622 cells that had undergone 24 h of development, the major product localized the mRNA 5′ end to a guanine nucleotide located 55 bp upstream of the primer (Fig. 5). Minor abundance extension products that are slightly longer suggest either heterogeneity in the mRNA 5′ end or the difficulty of reverse transcriptase reaching the 5′ end. The major extension product indicates that the mRNA 5′ end is 485 bp upstream of the SalI site in orf1 (Fig. 1), which is in good agreement with the results of the S1 nuclease protection experiment (Fig. 4). No extension product was observed when the primer was hybridized to RNA prepared from vegetatively grown cells (Fig. 5, lanes 2 and 3). We conclude that the Ω4406 mRNA 5′ end is located approximately 37 bp upstream of the predicted ORF1 translational start codon (Fig. 1). From this 5′ endpoint, the 3.0-kb transcript detected by Northern blot hybridization (Fig. 3, lane 6) is predicted to extend nearly 1 kb beyond the end of ORF1 into the partial ORF located downstream, which is in the opposite orientation as ORF1.
FIG. 5.
Primer extension analysis of Ω4406 mRNA. RNA was isolated from wild-type DK1622 cells grown vegetatively (lane 2, 30 μg; lane 3, 60 μg) or cells that had undergone 24 h of development (lane 4, 30 μg) and used for primer extension analysis. Lane 1 shows a control reaction to which no RNA was added. The same primer was used to sequence Ω4406 upstream DNA. A portion of the DNA sequence is indicated at the right. The bold arrow indicates the extension product that was observed with RNA isolated from 24-h developing cells but not with RNA from vegetatively grown cells. The inferred transcriptional start site is indicated by a right-angle arrow.
Effect of chromosomal position on expression.
Our results suggest that a development-specific transcript might initiate about 1.3 kb upstream of Tn5 lac Ω4406 (Fig. 3 to 5), yet the 3.2-kb upstream segment failed to direct developmental lacZ expression when integrated at the Mx8 phage attachment site (attB) in the M. xanthus chromosome (Fig. 2). Certain developmentally regulated lacZ fusions have been reported previously to exhibit less expression upon integration at Mx8 attB than at their native location in the chromosome (11, 32). To determine whether the Ω4406 promoter is present on the 3.2-kb segment and expression from the lacZ fusion is subject to such a position effect, an M. xanthus strain was constructed to test expression of the lacZ fusion at the native chromosomal site. A plasmid (pJL23) with 3.2 kb of Ω4406 upstream DNA but devoid of Mx8 attB and lacZ was transduced into M. xanthus MJL100 containing Tn5 lac Ω4406-Tcr (in which the Kmr gene was replaced with a Tcr gene to allow selection for the incoming plasmid). A single homologous recombination event resulted in the structure depicted at the top of Fig. 6 in which the downstream copy of the 3.2-kb segment is fused to Tn5 lac Ω4406-Tcr and is separated from the DNA normally present farther upstream by the plasmid vector and the other copy of the 3.2-kb segment. The graph in Fig. 6 shows that these transductants exhibited developmental lacZ expression that was, on average, higher than observed for MJL100 containing Tn5 lac Ω4406-Tcr. The timing of lacZ expression during development was similar for the transductants and for MJL100 containing Tn5 lac Ω4406-Tcr. Taken together with our analysis of Ω4406 mRNA, these results strongly suggest that the promoter responsible for Tn5 lac Ω4406 expression is located about 1.3 kb upstream and that developmental expression of lacZ is much higher at the native location in the chromosome than at Mx8 attB.
FIG. 6.
Developmental lacZ expression from fusions at the native Ω4406 position in the chromosome. The arrangement of M. xanthus DNA (thin line) and plasmid vector DNA (thick line) is shown at the top. The triangle shows the position of Tn5 lac Ω4406-Tcr. The graph shows the average β-galactosidase specific activity of five independently isolated transductants with this structure (▪; error bars show 1 standard deviation of the data). As a control, developmental β-galactosidase specific activity was measured for Tn5 lac Ω4406-Tcr-containing strain MJL100 (○). β-galactosidase specific activity is expressed in nanomoles of o-nitrophenyl phosphate per minute per milligram of protein.
Expression at Mx8 attB with a different lacZ fusion vector.
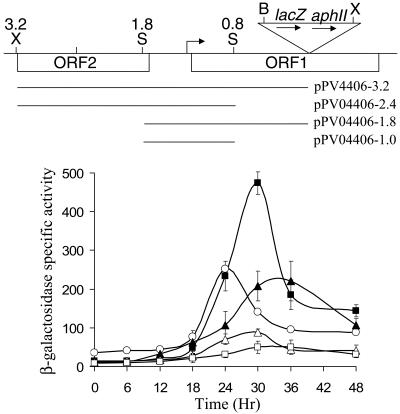
We noted that Tn5 lac Ω4406-Tcr (Fig. 6) exhibited about twofold higher developmental lacZ expression than Tn5 lac Ω4406-Kmr (Fig. 2). Although we do not understand the reason for this difference, we constructed a plasmid (pPV12K) with a Tcr gene rather than a Kmr gene located downstream of lacZ, reasoning that it might result in higher lacZ expression, as observed for the Tn5 lac fusions. Plasmid pPV12K allows fusions to be made to lacZ and integration at Mx8 attB, just like the plasmids with the Kmr gene (pREG1666 and pREG1727) that we have used previously (4, 10, 11, 17, 32, 56, 63, 64), including in the experiment shown in Fig. 2. The 3.2-kb Ω4406 upstream DNA segment was subcloned into pPV12K in the correct orientation to fuse the putative Ω4406 promoter to lacZ, and the plasmid was introduced into M. xanthus to allow integration at Mx8 attB. The transformants exhibited developmental lacZ expression, which reached a maximum of about 50 units (Fig. 7). This level of expression was well above that observed for the 3.2-kb segment in pREG1666 (Fig. 2) but only about 20% of that observed for Tn5 lac Ω4406-Tcr in the same experiment (Fig. 7). Nevertheless, the result supports the idea that the 3.2-kb segment contains the Ω4406 promoter.
FIG. 7.
Developmental expression from different segments of Ω4406 upstream DNA fused to lacZ and integrated at Mx8 attB. The top part is the physical map of the M. xanthus chromosome in the vicinity of Tn5 lac Ω4406 from Fig. 1. Underneath the map, the segments tested for promoter activity are indicated. The graph shows the average β-galactosidase specific activity of three independently isolated transformants (Materials and Methods) with the 3.2-kb (□), 2.4-kb (▪), 1.8-kb (▵), or 1.0-kb (▴) segment fused to lacZ in pPV12K. Error bars show 1 standard deviation of the data. As a control, developmental β-galactosidase specific activity was measured for Tn5 lac Ω4406-Tcr-containing strain MJL100 (○). β-galactosidase specific activity is expressed in nanomoles of o-nitrophenyl phosphate per minute per milligram of protein.
Expression at Mx8 attB of different Ω4406 upstream DNA segments.
To further localize the Ω4406 promoter region, we tested smaller segments of Ω4406 upstream DNA in pPV12K after integration at Mx8 attB. Surprisingly, when a 2.4-kb segment lacking the 0.8-kb segment immediately upstream of Ω4406 was tested, developmental lacZ expression reached about 450 units, which is ninefold higher than observed for the 3.2-kb segment and nearly twice that observed for Tn5 lac Ω4406-Tcr (Fig. 7). The 0.8-kb segment immediately upstream of Ω4406 appeared to strongly inhibit developmental lacZ expression. In qualitative agreement with this observation, a 1.0-kb segment with the same downstream endpoint as the 2.4-kb segment resulted in threefold higher developmental lacZ expression than a 1.8-kb segment with the same downstream endpoint as the 3.2-kb segment (Fig. 7). We conclude that the Ω4406 promoter is located in the 1.0-kb segment from 0.8 to 1.8 kb upstream of Ω4406, in agreement with our analysis of Ω4406 mRNA (Fig. 3 to 5), and that the 0.8-kb segment immediately upstream of Ω4406 inhibits developmental expression of a downstream lacZ reporter in pPV12K integrated at Mx8 attB in the M. xanthus chromosome.
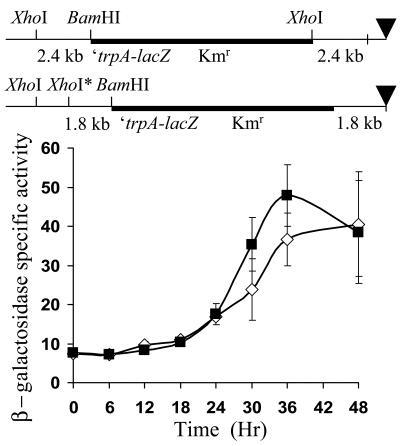
A trivial explanation of the inhibition observed for the 0.8-kb segment would be that it results in a less optimal fusion to the downstream lacZ reporter. The downstream end of the 0.8-kb segment includes 53 bp from the left end of Tn5 lac, which has translation stop codons in all three reading frames (28). This could conceivably cause a polar effect on expression of the downstream lacZ gene in the fusions generated with the 3.2- and 1.8-kb segments. On the other hand, our sequence analysis of pREG1727, from which pPV12K was derived, indicated that the fusions generated with the 2.4- and 1.0-kb segments fuse orf1 in frame with trpA (Materials and Methods), so that translation is expected to terminate at the end of trpA, near the ribosome binding site of the downstream lacZ gene. To test whether such a fusion, when integrated at the native location in the M. xanthus chromosome, would result in a higher level of developmental lacZ expression, the 2.4-kb segment was subcloned into pREG1175, fusing orf1 in frame with trpA, just as in pREG1727 (Materials and Methods). For comparison, the 1.8-kb segment, with translation stop codons in all three reading frames downstream of orf1, was also subcloned into pREG1175. Since pREG1175 lacks the Mx8 phage attachment site (attP), it does not integrate at Mx8 attB. Rather, transformants were identified in which homologous recombination occurred between the 2.4- or 1.8-kb segment of M. xanthus DNA in the plasmid and the corresponding segment of the chromosome, generating the structures depicted at the top of Fig. 8. The transformants expressed lacZ at a similar level during development (Fig. 8). These results do not support the idea that the 2.4-kb segment simply creates a better fusion for expression of lacZ than the 1.8-kb segment, as an explanation for the results shown in Fig. 7. The 0.8-kb segment immediately upstream of Ω4406 does not appear to inhibit developmental lacZ expression from fusions recombined at the native site in the chromosome (Fig. 6 and 8), in contrast to the effect of the 0.8-kb segment on expression of fusions at Mx8 attB (Fig. 2 and 7). We conclude that inhibition by the 0.8-kb segment of developmental expression from the Ω4406 promoter fused to a downstream lacZ reporter depends on chromosomal position (see Discussion).
FIG. 8.
Developmental expression from lacZ integrated by homologous recombination near the Ω4406 insertion site in the M. xanthus chromosome. The arrangement of M. xanthus DNA (thin line) and plasmid vector DNA (thick line) after recombination of pREG1175 derivatives bearing the 2.4-kb (top) or 1.8-kb (bottom) segment (Fig. 7 shows the position of these segments relative to the site of Tn5 lac Ω4406 insertion). The triangle indicates the site of the Ω4406 insertion as a reference point (Tn5 lac Ω4406 is not present). The starred XhoI site is not normally present in the chromosome at the indicated position but was designed into the primer used to synthesize the 1.8-kb segment. The graph shows the average β-galactosidase specific activity of three independently isolated transformants with the structure at the top (▪) or bottom (⋄) (Materials and Methods). β-galactosidase specific activity is expressed in nanomoles of o-nitrophenyl phosphate per minute per milligram of protein. Error bars show 1 standard deviation of the data.
Effect of orf2 on expression of orf1.
Since the 0.8-kb segment immediately upstream of Ω4406 did not appear to inhibit developmental lacZ expression from the fusion generated by insertion of Tn5 lac, it was puzzling that expression from Tn5 lac Ω4406-Tcr was about half of that from the 2.4-kb segment in pPV12K integrated at Mx8 attB (Fig. 7). On the other hand, the 1.0-kb segment in pPV12K integrated at Mx8 attB expressed lacZ during development at a comparable level as Tn5 lac Ω4406-Tcr (Fig. 7). One difference between the 1.0- and 2.4-kb segments is that orf2 appears to be intact in the 2.4-kb segment. Based on our predicted translation start codon for orf2, the upstream end of the 2.4-kb segment is just upstream of the putative ribosome binding site for orf2. Therefore, it was possible that expression of orf2 from the plasmid integrated at Mx8 attB was increasing expression of orf1 and the downstream lacZ reporter. To test this possibility, we used site-directed mutagenesis to replace the translation start codon of orf2 with a stop codon in the 2.4-kb segment. This segment was subcloned into pPV12K, the plasmid was transformed into wild-type M. xanthus DK1622, and developmental lacZ expression was measured from three transformants expected to have the plasmid integrated at Mx8 attB (Fig. 9A). Expression was reduced, reaching a maximum of only about 120 units, compared with about 450 units for the wild-type 2.4-kb segment. This result suggests that expression of orf2 from the 2.4-kb segment in pPV12K integrated at Mx8 attB boosts the level of developmental lacZ expression.
FIG. 9.
Effect of orf2 on developmental expression of orf1-lacZ fusions. (A) Developmental expression from the wild-type 2.4-kb Ω4406 upstream DNA segment (▪) or its derivative with a stop codon mutation in orf2 (□), fused to lacZ in pPV12K and integrated at Mx8 attB. Average β-galactosidase specific activity is shown for three independently isolated transformants (Materials and Methods). (B) Developmental lacZ expression from Tn5 lac Ω4406-Tcr (inserted in orf1) in M. xanthus with orf2 disrupted by plasmid integration. The arrangement of M. xanthus DNA (thin line) and plasmid vector DNA (thick line) is shown at the top. The triangle shows the position of Tn5 lac Ω4406-Tcr. The graph shows the average β-galactosidase specific activity of three independently isolated transformants with this structure (▪) (Materials and Methods). As a control, developmental β-galactosidase specific activity was measured for Tn5 lac Ω4406-Tcr-containing strain MJL100 (○). In both panels, β-galactosidase specific activity is expressed in nanomoles of o-nitrophenyl phosphate per minute per milligram of protein, and error bars show 1 standard deviation of the data.
Does expression of orf2 boost expression of orf1 at the native location in the M. xanthus chromosome? To address this question, we disrupted orf2 by plasmid integration in the chromosome of M. xanthus containing Tn5 lac Ω4406-Tcr. An internal segment of the predicted 488-codon orf2, from codon 114 to 361, was amplified by PCR and cloned into a plasmid with a Kmr gene, and the plasmid (pPV0.74kbins) was transformed into M. xanthus MJL100 (Materials and Methods). Three transformants in which the plasmid integrated by homologous recombination, resulting in the structure depicted at the top of Fig. 9B, were identified by diagnostic PCR, and developmental lacZ expression was measured. The graph in Fig. 9B shows that the plasmid insertion in orf2 had very little effect on lacZ expression from Tn5 lac Ω4406-Tcr during development. This result suggests that the boost in orf1 expression caused by orf2 expression from plasmids integrated at Mx8 attB is artifactual. Indeed, orf2 might not be expressed at its native site in the chromosome during development, since we failed to detect mRNA with a probe spanning the 5′ half of orf2 by Northern blot hybridization of RNA from 24-h developing cells (Fig. 3, lanes 2 and 4).
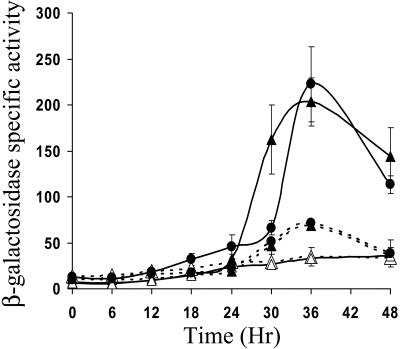
Effect of C-signaling on expression from the Ω4406 promoter.
Since orf2 had little effect on expression from the Ω4406 promoter at its native location in the chromosome (Fig. 9B), we focused on the 1.0- and 1.8-kb segments, which contain the Ω4406 promoter, but not intact orf2 (Fig. 7), to determine whether developmental lacZ expression from plasmids integrated at Mx8 attB depends on C-signaling. Previously, it was shown that developmental expression from Tn5 lac Ω4406 is abolished in csgA mutant cells that fail to make C-signal (27). Figure 10 shows that developmental lacZ expression from plasmids containing the 1.0- or 1.8-kb segment was also abolished in csgA mutant cells. When these csgA mutants were mixed with an equal number of wild-type M. xanthus DK1622 to supply the extracellular C-signal during codevelopment, expression from both the 1.0- and 1.8-kb segments was restored to the level observed for those segments in the wild-type background (Fig. 10). These results demonstrate that the Ω4406 promoter is regulated by extracellular C-signaling both in the presence and in the absence of the 0.8-kb segment immediately upstream of Ω4406, which inhibits developmental lacZ expression from the pPV12K derivative containing the 1.8-kb segment integrated at Mx8 attB of the csgA mutant.
FIG. 10.
C-signal dependence of developmental lacZ expression from the Ω4406 promoter. The average β-galactosidase specific activity of three independently isolated transformants (Materials and Methods) with the 1.0-kb (solid lines) or 1.8-kb (dashed lines) Ω4406 upstream DNA segment fused to lacZ in pPV12K and integrated at Mx8 attB of wild-type DK1622 (•) or csgA mutant LS203 in the absence (▵) or presence (▴) of an equal number of wild-type DK1622 cells (lacking lacZ but capable of C-signaling). β-galactosidase specific activity is expressed in nanomoles of o-nitrophenyl phosphate per minute per milligram of protein, and error bars show 1 standard deviation of the data.
DISCUSSION
We have localized the Ω4406 promoter region to a DNA segment located 0.8 to 1.8 kb upstream of Tn5 lac insertion Ω4406. The 5′ end of a 3.0-kb development-specific mRNA transcript mapped to the center of this 1.0-kb DNA segment, suggesting that the Ω4406 promoter is about 1.3 kb upstream of Tn5 lac Ω4406. Sequencing of the 1.0-kb DNA segment and primer extension analysis of the mRNA (Fig. 5) precisely localized the 5′ end. Mutational analysis has confirmed that the 5′ end reflects the transcriptional start site of the Ω4406 promoter (55). However, we discovered that expression of a lacZ reporter fused transcriptionally to the Ω4406 promoter depends strongly on the position of the fusion in the M. xanthus chromosome. Expression from a 3.2-kb segment of DNA upstream of Tn5 lac Ω4406 was normal at the native location in the chromosome (Fig. 6), but undetectable at a phage attachment site, Mx8 attB (Fig. 2). By constructing a new lacZ fusion vector with a different antibiotic resistance gene (Tcr), we were able to detect developmental lacZ expression from a fusion to the 3.2-kb segment integrated at Mx8 attB, but a much higher level of expression was observed when the 0.8-kb segment immediately upstream of Ω4406 was deleted (Fig. 7). The inhibitory effect of the 0.8-kb segment on developmental lacZ expression from fusions integrated at Mx8 attB has also been demonstrated with a vector (pREG1727) bearing a Kmr gene (55). In contrast, the 0.8-kb segment does not inhibit developmental expression of lacZ from fusions integrated by homologous recombination at the native Ω4406 site (Fig. 6 and 8). Taken together, our results indicate that the 0.8-kb segment mediates a chromosomal position-dependent effect on expression of a downstream lacZ reporter.
Reduced expression from transcriptional lacZ fusions integrated at Mx8 attB, relative to identical fusions located at their native location in the M. xanthus chromosome, has been reported previously (11, 32), but this is the first case in which the effect has been shown to be attributable to a DNA segment outside the promoter region. Li and Shimkets (32) found that 11 kb of DNA upstream of Tn5 lac Ω4435 was insufficient for full developmental lacZ expression when the fusion was integrated at Mx8 attB. Since expression from the fusion at Mx8 attB was about half that for Tn5 lac Ω4435 and since the strain with the fusion at Mx8 attB has a second copy of the Ω4435 promoter region, it is possible that titration of a limiting transcription factor, rather than a chromosomal position effect, accounts for the difference in expression. On the other hand, Fisseha et al. (11) observed fourfold lower developmental expression of fusions between Ω4403 upstream DNA (up to 17 kb) and lacZ when located at Mx8 attB, compared with a fusion generated at the native chromosomal site by insertion of Tn5 lac. Also, integration of a plasmid 17 kb upstream of Tn5 lac Ω4403 Tcr only slightly reduced (less than twofold) developmental lacZ expression. This strain had two copies of the 17-kb segment, separated by vector sequences, analogous to the structure depicted at the top of Fig. 6. Hence, titration of a limiting transcription factor by a second copy of the Ω4403 promoter region could only account for a slight reduction in expression, not the fourfold effect seen for fusions integrated at Mx8 attB. Therefore, it was speculated that expression of lacZ fused to the Ω4403 promoter might be partially silenced at Mx8 attB by a localized effect on chromatin structure, similar to that observed for yeast (Saccharomyces cerevisiae) mating type switching, the telomere position effect, and rRNA gene silencing (reviewed in reference 43). While these mechanisms involve histone hypoacetylation and M. xanthus does not have histones, similar mechanisms in bacteria may involve histone-like proteins, orthologs of the yeast Sir2p histone deacetylase, and proteins that bind to partitioning sequences (42). It was noted that other fusions that depend less strongly on C-signaling escaped inhibition at Mx8 attB, so it was further speculated that C-signaling leads to an altered chromosomal state of DNA integrated at Mx8 attB, partially inhibiting expression of genes that depend strongly on C-signaling (11). Our results contradict this latter speculation. Expression of lacZ fused to the 1.0-kb segment containing the Ω4406 promoter, when integrated at Mx8 attB, depends absolutely on C-signaling (Fig. 10), yet expression in wild-type cells is comparable to that observed for Tn5 lac Ω4406 (Fig. 7). Clearly, the Ω4406 promoter escapes inhibition at Mx8 attB when the 0.8-kb segment immediately upstream of Ω4406 is not present. It remains possible that a localized effect on chromatin structure explains chromosomal position-dependent expression of lacZ fusions to the Ω4403 promoter (11). Perhaps the Ω4403 promoter region is permissive for silencing, while the 1.0-kb Ω4406 promoter region is resistant to silencing. In the context of this model, perhaps the 0.8-kb segment immediately upstream of Ω4406 contains one or more sequences that facilitate attainment of a chromatin structure that impedes initiation or elongation of transcription from the Ω4406 promoter when it is located at Mx8 attB. Alternatively, one or more sequences within the 0.8-kb segment might cause termination of transcription or instability of the mRNA; however, these explanations would require that termination factors or RNases, respectively, be present at higher concentrations in the vicinity of Mx8 attB than Ω4406, in order to account for the dependence on chromosomal position. It will be interesting to determine whether the inhibitory effect of the 0.8-kb segment can be localized to a smaller segment, whether it depends on orientation and distance relative to the upstream Ω4406 promoter and the downstream lacZ reporter, and whether it would affect a different promoter.
Our primary interest in the Ω4406 promoter region was to understand its dependence on csgA for developmental expression. It was shown previously that expression from Tn5 lac Ω4406 is abolished in csgA mutant cells under conditions of development (27). Here, we showed not only that expression from the 1.0-kb segment containing the Ω4406 promoter is abolished in csgA mutant cells but also that expression is restored upon codevelopment of the csgA mutant cells with an equal number of wild-type cells, which provide C-signal (Fig. 10). This demonstrates that developmental lacZ expression from the Ω4406 promoter depends on extracellular C-signaling.
Inspection of the Ω4406 promoter region reveals striking similarity to other C-signal-dependent promoter regions (Fig. 11). The −72 to −32 bp segment of the Ω4406 promoter region is 78% identical to the −67 to −31 bp segment of the Ω4403 promoter region, if two small gaps are allowed in the latter sequence. Like the Ω4406 promoter, developmental lacZ expression from the Ω4403 promoter depends absolutely on C-signaling (11, 27). The region of similarity spans two DNA elements shown by mutational analysis to be important for Ω4403 promoter activity, a C box (consensus sequence CAYYCCY, in which Y means C or T), CATCCCT, centered at −49 bp, and a 5-bp element (consensus sequence GAACA), GACCG, centered at −61 bp (56). In the alignment shown in Fig. 11, the Ω4406 sequence diverges from the Ω4403 C-box sequence, and from the C-box consensus sequence, at the last three positions. Single base pair transversion mutations at two of these positions (Fig. 11, boldface residues) nearly abolished Ω4403 promoter activity (56). It will be interesting to see if single base pair changes in the CATCGTG sequence centered at −52 bp in the Ω4406 promoter region have a dramatic effect on promoter activity. The sequence downstream of the C box is also important for Ω4403 promoter activity (56, 57), and the Ω4406 sequence is similar in this region, making it a logical target for mutational analysis. Upstream, the Ω4406 sequence has two potential 5-bp elements, GAACC, centered at −62 bp, and GGACA, centered at −67 bp. It also has a sequence, GGCGTTTCA, centered at −86 bp (not shown in Fig. 11), that resembles a 10-bp element, GGCATGTTCA, centered at −74.5 bp in the Ω4403 promoter region, which is essential for promoter activity (56).
FIG. 11.
Alignments of the Ω4406 promoter region with the promoter regions of other C-signal-dependent M. xanthus genes. The program BESTFIT in the Wisconsin GCG sequence analysis software was used to detect the best alignment between the Ω4406 promoter region (−100 to −1) and the Ω4403 (11), Ω4499 (10), or Ω4400 (4) promoter region (−100 to −1). Nucleotides in bold caused a greater than twofold change in developmental promoter activity when mutated singly or in combination with a few adjacent nucleotides (56, 63, 64). The 7-bp C-box sequences and the 5-bp elements are underlined.
Impressive similarity between the Ω4499 and Ω4406 promoter regions also begins at about −70 bp, but it extends farther downstream than in the case of the Ω4403 promoter region (Fig. 11). Three gaps are present in the alignment, so blocks of identical sequence are present at similar but not identical positions in the Ω4499 and Ω4406 promoter regions. The two gaps in the Ω4406 sequence overlap with C boxes centered at −55 (CATTCCC) and −33 bp (CATTCCT) in the Ω4499 promoter region, which have been shown by mutational analysis to be important for promoter activity (64). Each C box in the Ω4499 promoter region is preceded by a 5-bp element (GGACA and GAACT centered at −69 and −46 bp, respectively) that is also important for promoter activity (64). The Ω4406 promoter region has an identical 5-bp sequence, GGACA, centered at −67 bp, as the 5-bp element centered at −69 bp in the Ω4499 promoter region (Fig. 11). A multiple base pair change of CCGG to AATT at −63 to −60 bp reduced Ω4499 promoter activity to 5% of the wild-type level (64), and the Ω4406 sequence matches this sequence at three positions in the alignment (Fig. 11). Downstream, the two promoter regions show two identical 8-bp sequence blocks (CGTGCGCC and TCCCATGC); however, only a few mutations have been made in this part of the Ω4499 promoter region (64). Expression from the Ω4499 promoter occurs earlier during development and does not depend as strongly on C-signaling, in comparison to the Ω4406 promoter (10, 27) (Fig. 10). We hope to understand these differences by using the sequence analysis to guide mutational studies of the Ω4406 promoter region and identify the proteins that regulate transcription.
Like the Ω4499 promoter, the Ω4400 promoter drives expression earlier, and it does not depend as strongly on C-signaling as expression from the Ω4406 promoter (4, 27) (Fig. 10). Nevertheless, the −68 to −53 bp segment of the Ω4400 promoter region is 75% identical to the −69 to −54 bp segment of the Ω4406 promoter region (Fig. 11). This segment of the Ω4400 promoter region includes a 5-bp element, GAACA, centered at −61 bp, and downstream sequence, both of which are important for developmental promoter activity, based on mutational analysis (63). The segment also includes upstream sequence, which did not appear to be important for Ω4400 promoter activity, in contrast to the corresponding sequence in the Ω4499 promoter region. We conclude that the Ω4406 promoter region exhibits striking blocks of similarity to sequences that have been shown to be important for activity of other C-signal-dependent promoters.
Another noteworthy feature of the Ω4406 promoter region sequence is a 39-bp segment with an imperfect inverted repeat (−101 TCATTCTGTGCGGCGTTTCAGGGAAACCGCACGGACAGA −63). The downstream end of the segment includes one of the potential 5-bp elements, GGACA, centered at −67 bp, and part of the other, GAACC, centered at −62 bp, in a region with similarity to other C-signal-dependent promoter regions (Fig. 11). The palindrome could be a binding site for a dimeric DNA-binding protein that regulates transcription from the Ω4406 promoter.
Tn5 lac Ω4406 is inserted near the middle of orf1 (Fig. 1), yet it causes no detectable defect in aggregation or sporulation (28). This is not unusual. Many developmentally regulated genes play a subtle role not evident under laboratory conditions or are functionally redundant with other genes (7, 28). The predicted amino acid sequence of ORF1 provides no clue about its function since it shows significant similarity only to proteins of unknown function. Upstream of orf1 and in the same orientation is orf2, predicted to encode histidase. We did not detect expression of orf2 during growth or at 24 h of development, using Northern blot analysis (Fig. 3). The gene might be expressed under different growth conditions or at a different time during development, as it is typically regulated in response to carbon and nitrogen availability in other organisms (35). If so, transcription of orf2 is unlikely to proceed downstream through orf1. First, there appears to be a transcription terminator located 40 bp downstream of the ORF2 translational stop codon. Second, disruption of orf2 by plasmid integration had little effect on expression of orf1 (Fig. 9B). On the other hand, expression of orf2 from a plasmid integrated at Mx8 attB, presumably due to readthrough transcription from a promoter in the plasmid or in the M. xanthus chromosome near Mx8 attB, appeared to boost the level of developmental expression from a lacZ reporter fused to orf1 (Fig. 7). The finding that a nonsense mutation in the putative start codon of orf2 eliminated the increased expression of the orf1-lacZ fusion (Fig. 9A) suggests that ORF2 protein, when produced aberrantly, might somehow boost expression of the downstream lacZ reporter. Alternatively, the readthrough transcription presumed to express orf2 from a plasmid integrated at Mx8 attB might not terminate downstream of orf2, boosting expression of the downstream lacZ reporter, and the nonsense mutation might cause a polar effect. In any case, orf2 expression does not appear to boost orf1 expression at the normal location in the chromosome, since orf2 disruption had little effect on orf1-lacZ expression (Fig. 9B).
In summary, we have identified the Ω4406 regulatory region and demonstrated that it responds to extracelluar C-signaling. The Ω4406 promoter region has blocks of similarity to sequences that have been shown to be important for activity of other C-signal-dependent promoters. Mutational analysis can be used to test whether these sequences define DNA elements important for Ω4406 promoter activity. Identification of proteins that directly regulate Ω4406 transcription is an important future goal. We also discovered a 0.8-kb segment located 500 to 1,300 bp downstream of the Ω4406 promoter that mediates chromosomal position-dependent inhibition of a downstream lacZ reporter. This may provide an avenue to understand position effects on gene expression in bacteria, which have frequently been described (2, 37, 54) but seldom understood, with the exceptions that proximity to the chromosomal origin of replication can increase expression during growth due to increased gene copy number (47) and proximity to sequences involved in chromosomal partitioning can determine whether expression occurs in spoIIIE mutants of Bacillus subtilis that fail to translocate 70% of one copy of the chromosome into the forespore during endosporulation (53, 58). Since neither of these mechanisms would explain the position effect mediated by the 0.8-kb segment uncovered in this study, perhaps it facilitates attainment of a chromatin structure that impedes expression, as has been proposed for gene silencing near partitioning sequences in E. coli (42).
Acknowledgments
We thank K. Viswanathan for constructing the pKV plasmids listed in Table 1. We are grateful to the Monsanto Company and The Institute for Genomic Research for providing access to their sequence databases.
This research was supported by NIH grant GM47293, by NSF grants MCB-0090478 and MCB-0416456, and by the Michigan Agricultural Experiment Station.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Beckwith, J. R., E. R. Signer, and W. Epstein. 1966. Transposition of the Lac region of E. coli. Cold Spring Harbor Symp. Quant. Biol. 31:393-401. [DOI] [PubMed] [Google Scholar]
- 3.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 4.Brandner, J. P., and L. Kroos. 1998. Identification of the Ω4400 regulatory region, a developmental promoter of Myxococcus xanthus. J. Bacteriol. 180:1995-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Downard, J., S. V. Ramaswamy, and K. Kil. 1993. Identification of esg, a genetic locus involved in cell-cell signaling during Myxococcus xanthus development. J. Bacteriol. 175:7762-7770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dworkin, M., and D. Kaiser (ed). 1993. Myxobacteria II. American Society for Microbiology, Washington, D.C.
- 7.Eichenberger, P., S. T. Jensen, E. M. Conlon, C. van Ooij, J. Silvaggi, J. E. Gonzalez-Pastor, M. Fujita, S. Ben-Yehuda, P. Stragier, J. S. Liu, and R. Losick. 2003. The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 327:945-972. [DOI] [PubMed] [Google Scholar]
- 8.Ellehauge, E., M. Norregaard-Madsen, and L. Sogaard-Andersen. 1998. The FruA signal transduction protein provides a checkpoint for the temporal co-ordination of intercellular signals in Myxococcus xanthus development. Mol. Microbiol. 30:807-817. [DOI] [PubMed] [Google Scholar]
- 9.Fawcett, T., and S. Bartlett. 1990. An effective method for eliminating “artifact banding” when sequencing double-stranded DNA templates. BioTechniques 9:46-48. [PubMed] [Google Scholar]
- 10.Fisseha, M., D. Biran, and L. Kroos. 1999. Identification of the Ω4499 regulatory region controlling developmental expression of a Myxococcus xanthus cytochrome P-450 system. J. Bacteriol. 181:5467-5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisseha, M., M. Gloudemans, R. Gill, and L. Kroos. 1996. Characterization of the regulatory region of a cell interaction-dependent gene in Myxococcus xanthus. J. Bacteriol. 178:2539-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gill, R. E., and M. C. Bornemann. 1988. Identification and characterization of the Myxococcus xanthus bsgA gene product. J. Bacteriol. 170:5289-5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gill, R. E., M. G. Cull, and S. Fly. 1988. Genetic identification and cloning of a gene required for developmental cell interactions in Myxococcus xanthus. J. Bacteriol. 170:5279-5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagen, D. C., A. P. Bretscher, and D. Kaiser. 1978. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev. Biol. 64:284-296. [DOI] [PubMed] [Google Scholar]
- 15.Halberg, R., and L. Kroos. 1992. Fate of the SpoIIID switch protein during Bacillus subtilis sporulation depends on the mother-cell sigma factor, σK. J. Mol. Biol. 228:840-849. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 17.Hao, T., D. Biran, G. J. Velicer, and L. Kroos. 2002. Identification of the Ω4514 regulatory region, a developmental promoter of Myxococcus xanthus that is transcribed in vitro by the major vegetative RNA polymerase. J. Bacteriol. 184:3348-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodgkin, J., and D. Kaiser. 1977. Cell-to-cell stimulation of motility in nonmotile mutants of Myxococcus. Proc. Natl. Acad. Sci. USA 74:2938-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaiser, D. 2003. Coupling cell movement to multicellular development in myxobacteria. Nat. Rev. Microbiol. 1:45-54. [DOI] [PubMed] [Google Scholar]
- 20.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan, H. 2003. Multicellular development and gliding motility in Myxococcus xanthus. Curr. Opin. Microbiol. 6:572-577. [DOI] [PubMed] [Google Scholar]
- 22.Kashefi, K., and P. Hartzell. 1995. Genetic supression and phenotypic masking of a Myxococcus xanthux frzF− defect. Mol. Microbiol. 15:483-494. [DOI] [PubMed] [Google Scholar]
- 23.Kim, S. K., and D. Kaiser. 1990. Cell motility is required for the transmission of C-factor, an intercellular signal that coordinates fruiting body morphogenesis of Myxococcus xanthus. Genes Dev. 4:896-905. [DOI] [PubMed] [Google Scholar]
- 24.Kim, S. K., and D. Kaiser. 1991. C-factor has distinct aggregation and sporulation thresholds during Myxococcus development. J. Bacteriol. 173:1722-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, S. K., and D. Kaiser. 1990. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell 61:19-26. [DOI] [PubMed] [Google Scholar]
- 26.Kroos, L., and D. Kaiser. 1984. Construction of Tn5 lac, a transposon that fuses lacZ expression to exogenous promoters, and its introduction into Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 81:5816-5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kroos, L., and D. Kaiser. 1987. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1:840-854. [DOI] [PubMed] [Google Scholar]
- 28.Kroos, L., A. Kuspa, and D. Kaiser. 1986. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev. Biol. 117:252-266. [DOI] [PubMed] [Google Scholar]
- 29.Kuspa, A., L. Kroos, and D. Kaiser. 1986. Intercellular signaling is required for developmental gene expression in Myxococcus xanthus. Dev. Biol. 117:267-276. [DOI] [PubMed] [Google Scholar]
- 30.Laue, B. E., and R. Gill. 1994. Use of a phase variation-specific promoter of Myxococcus xanthus in a strategy for isolating a phase-locked mutant. J. Bacteriol. 176:5341-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, S.-F., B. Lee, and L. J. Shimkets. 1992. csgA expression entrains Myxococcus xanthus development. Genes Dev. 6:401-410. [DOI] [PubMed] [Google Scholar]
- 32.Li, S.-F., and L. J. Shimkets. 1988. Site-specific integration and expression of a developmental promoter in Myxococcus xanthus. J. Bacteriol. 170:5552-5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linn, T., and G. Ralling. 1985. A versatile multiple- and single-copy vector system for the in vitro construction of transcriptional fusions to lacZ. Plasmid 14:134-142. [DOI] [PubMed] [Google Scholar]
- 34.Lobedanz, S., and L. Sogaard-Andersen. 2003. Identification of the C-signal, a contact-dependent morphogen coordinating multiple developmental responses in Myxococcus xanthus. Genes Dev. 17:2151-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magasanik, B., and F. C. Neidhardt. 1987. Regulation of carbon and nitrogen utilization. .In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Esherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 36.Messing, J. 1979. A multipurpose cloning system based on the single-stranded DNA bacteriophage M13. Recombinant DNA bulletin, publication no. 71-99, p. 43-48. National Institutes of Health, Bethesda, Md.
- 37.Nandi, S., D. Maiti, A. Saha, and R. K. Bhadra. 2003. Genesis of variants of Vibrio cholerae O1 biotype El Tor: role of the CTXphi array and its position in the genome. Microbiology 149:89-97. [DOI] [PubMed] [Google Scholar]
- 38.O'Connor, K. A., and D. R. Zusman. 1991. Behavior of peripheral rods and their role in the life cycle of Myxococcus xanthus. J. Bacteriol. 173:3342-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Connor, K. A., and D. R. Zusman. 1991. Development in Myxococcus xanthus involves differentiation into two cell types, peripheral rods and spores. J. Bacteriol. 173:3318-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogawa, M., S. Fujitani, X. Mao, S. Inouye, and T. Komano. 1996. FruA, a putative transcription factor essential for the development of Myxococcus xanthus. Mol. Microbiol. 22:757-767. [DOI] [PubMed] [Google Scholar]
- 41.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 42.Rine, J. 1999. On the mechanism of silencing in Escherichia coli. Proc. Natl. Acad. Sci. USA 96:8309-8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72:481-516. [DOI] [PubMed] [Google Scholar]
- 44.Sager, B., and D. Kaiser. 1994. Intercellular C-signaling and the traveling waves of Myxococcus. Genes Dev. 8:2793-2804. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmid, M. B., and J. R. Roth. 1987. Gene location affects expression level in Salmonella typhimurium. J. Bacteriol. 169:2872-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimkets, L. J. 1999. Intercellular signaling during fruiting-body development of Myxococcus xanthus. Annu. Rev. Microbiol. 53:525-549. [DOI] [PubMed] [Google Scholar]
- 49.Shimkets, L. J., and S. J. Asher. 1988. Use of recombination techniques to examine the structure of the csg locus of Myxococcus xanthus. Mol. Gen. Genet. 211:63-71. [DOI] [PubMed] [Google Scholar]
- 50.Sogaard-Andersen, L., M. Overgaard, S. Lobedanz, E. Ellehauge, L. Jelsbak, and A. A. Rasmussen. 2003. Coupling gene expression and multicellular morphogenesis during fruiting body formation in Myxococcus xanthus. Mol. Microbiol. 48:1-8. [DOI] [PubMed] [Google Scholar]
- 51.Stellwag, E., J. M. Fink, and J. Zissler. 1985. Physical characterization of the genome of the Myxococcus xanthus bacteriophage MX-8. Mol. Gen. Genet. 199:123-132. [DOI] [PubMed] [Google Scholar]
- 52.Stephens, K., and D. Kaiser. 1987. Genetics of gliding motility in Myxococcus xanthus: molecular cloning of the mgl locus. Mol. Gen. Genet. 207:256-266. [Google Scholar]
- 53.Sun, D., P. Fajardo-Cavazos, M. D. Sussman, F. Tovar-Rojo, R. M. Cabrera-Martinez, and P. Setlow. 1991. Effect of chromosome location of Bacillus subtilis forespore genes on their spo gene dependence and transcription by EσF: identification of features of good σF-dependent promoters. J. Bacteriol. 173:7867-7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson, A., and M. J. Gasson. 2001. Location effects of a reporter gene on expression levels and on native protein synthesis in Lactococcus lactis and Saccharomyces cerevisiae. Appl. Environ. Microbiol. 67:3434-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Viswanathan, K., P. Viswanathan, and L. Kroos. Unpublished data.
- 56.Viswanathan, P., and L. Kroos. 2003. cis Elements necessary for developmental expression of a Myxococcus xanthus gene that depends on C signaling. J. Bacteriol. 185:1405-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Viswanathan, P., and L. Kroos. Unpublished data.
- 58.Wu, L. J., and J. Errington. 1994. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science 264:572-575. [DOI] [PubMed] [Google Scholar]
- 59.Xu, D., C. Yang, and H. B. Kaplan. 1998. Myxococcus xanthus sasN encodes a regulator that prevents developmental gene expression during growth. J. Bacteriol. 180:6215-6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang, C., and H. B. Kaplan. 1997. Myxococcus xanthus sasS encodes a sensor histidine kinase required for early developmental gene expression. J. Bacteriol. 179:7759-7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yanisch-Perron, C., J. Vieria, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences M13 mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 62.Yanofsky, C., T. Platt, I. P. Crawford, B. P. Nichols, G. E. Christie, H. Horowitz, M. VanCleemput, and A. M. Wu. 1981. The complete nucleotide sequence of the tryptophan operon of Escherichia coli. Nucleic Acids Res. 9:6647-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoder, D., and L. Kroos. 2004. Mutational analysis of the Myxococcus xanthus Ω4400 promoter region provides insight into developmental gene regulation by C signaling. J. Bacteriol. 186:661-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoder, D., and L. Kroos. 2004. Mutational analysis of the Myxococcus xanthus Ω4499 promoter region reveals shared and unique properties in comparison with other C-signal-dependent promoters. J. Bacteriol. 186:3766-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]