Abstract
The first and, to date, only extrachromosomal circular replicon identified in the spirochete Leptospira is the LE1 prophage from Leptospira biflexa. The 74-kb LE1 genome has a GC content of 36%, which is similar to the GC content of Leptospira spp. Most of the 79 predicted open reading frames (ORFs) showed no similarities to known ORFs. However 21 ORFs appeared to be organized in clusters that could code for head and tail structural proteins and immunity repressor proteins. In addition, the pattern of gene expression showed that several LE1 genes are expressed specifically either in LE1 prophage or in L. biflexa late after infection. Since the LE1 prophage replicates autonomously as a circular replicon in L. biflexa, we were able to engineer an L. biflexa-Escherichia coli shuttle vector from a 5.3-kb DNA fragment of LE1 (Saint Girons et al., J. Bacteriol. 182:5700-5705, 2000), opening this genus to genetic manipulation. In this study, base compositional asymmetry confirms the location of the LE1 replication region and suggests that LE1 replicates via a bidirectional Θ-like replication mechanism from this unique origin. By subcloning experiments, the replication region can be narrowed down to a 1-kb region. This minimal replication region consists of a rep encoding a protein of 180 amino acids. Upstream from rep, putative partitioning genes, called parA and parB, were found to be similar to the par loci in Borrelia plasmids. A significant increase of plasmid stability in L. biflexa can be seen only when both parA and parB are present. These results enable the construction of new shuttle vectors for studying the genetics of Leptospira spp. This study will also contribute to a better knowledge of phages unrelated to lambdoid phages.
The Spirochaetales order of the domain Bacteria has a deep branching lineage (29) and is composed of both pathogenic and saprophytic species (14). Bacteriophage-like particles have been observed in association with a number of spirochete genera, including Borrelia, Brachyspira (Serpulina), and Leptospira (9). The LE1 leptophage has been previously isolated from sewage water, together with phages LE3 and LE4, and found to be specific for the saprophyte Leptospira biflexa (37). The study of the LE1 leptophage by electron microscopy revealed a polyhedral head and a contractile tail, which are characteristics of group A1 morphology in the family Myoviridae (37). Moreover, the LE1 leptophage was shown to be temperate and to behave as a circular plasmid within L. biflexa (36). Autonomous replication rather than integration into the host genome, although rare, is shared by other temperate phages such as P1 and N15 from Escherichia coli and ΦBB-1 from the spirochete Borrelia burgdorferi (10, 19, 33). A 5.3-kb region was previously identified as the LE1 replication region, allowing the construction of the first Leptospira-E. coli shuttle vector (36). This breakthrough in Leptospira genetics was followed by the first demonstration of gene knockout in Leptospira (30).
The last few years have witnessed remarkable progress in the attempt to understand the molecular biology of spirochetes. At present, seven spirochetal genomes have been sequenced. However, the genetics of this intriguing group of bacteria is still at a very early stage compared to that of other bacterial species. Since little information is available on spirochetal phages, we were interested to undertake a functional study of LE1. In this paper, we focus on the analysis of the 74-kb nucleotide sequence of the LE1 phage. Transcription studies in the LE1 lysogen and in a bacterium infected by LE1 allowed us to characterize functions necessary for the structure of the phage or for its replication as a plasmid. The elucidation of the LE1 genome provides insights into genome organization, evolution, and classification of phages in a phage with considerable potential for the genetic manipulation of Leptospira spp.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
L. biflexa serovar Patoc strain Patoc 1 (Leptospira National Reference Center, Institut Pasteur, Paris, France) was grown at 30°C in EMJH (11, 21) liquid medium or on 1% agar plates. L. biflexa serovar Patoc strain Patoc 1 containing the LE1 prophage was further designated L. biflexa strain 3c. E. coli was grown in Luria-Bertani (LB) medium. When necessary, kanamycin and spectinomycin were added at 40 μg ml−1.
Phage manipulations and sequencing of the LE1 genome.
Titer was determined by the soft agar overlay method as previously described (37). Large quantities of high-titer lysates of leptophage LE1 were produced by infection of exponentially growing L. biflexa cells with a multiplicity of 0.01 as previously described (37). Phage DNA was extracted three times with phenol followed by ethanol precipitation. Sequencing was performed by shotgun as previously described (7). Briefly, purified DNA was sheared by nebulization, end repaired by using T4 DNA polymerase (Boehringer Mannheim), and then ligated to BstXI adaptors (Invitrogen). The ligation mixture was fractionated by agarose gel electrophoresis and fragments from 1 kb to 3 kb were ligated into BstXI-digested pcDNA2.1 (Invitrogen); 768 recombinant plasmids were used as templates for cycle sequencing reactions consisting of 35 cycles (96°C for 30 seconds; 50°C for 15 seconds; and 60°C for 4 min). Samples were precipitated and loaded onto a 96-lane, 4% polyacrylamide gel, and electrophoresis was performed on a model ABI PRISM 377 automatic DNA sequencer (ABI) for 10 h. In the closure phase, sequencing reactions were performed using PCR products as templates and custom-made oligonucleotides as primers. The sequence was assembled by using PHRED (12, 13) and PHRAP (P. Green, unpublished); for the editing, CONSED (17) was used.
mRNA detection with RT-PCR.
L. biflexa was grown to 2 × 108 bacteria/ml in 200 ml EMJH medium, adsorbed to LE1 phage at a multiplicity of infection of 0.1 for 20 min at 30°C without shaking and then with shaking for 20 min at 30°C. Aliquots (100 ml) were removed at 45 min (early RNA) and 120 min (late RNA) and mixed immediately with 120 μl cold 0.2 M NaCl/0.002 M sodium azide. Total RNA was then extracted as previously described (3). RNA was also prepared from similar amounts of L. biflexa and L. biflexa 3c growing exponentially in EMJH medium. Reverse transcription (RT) of RNA was carried out as described by the manufacturer's instructions (SuperScript One Step RT-PCR with Platinum Taq, Gibco-BRL, Rockville, MD).
PCR primer pairs (primer nucleotide sequences available are on request) were used to amplify the transcripts corresponding to selected ORFs from LE1 (Table 1). Each 50-μl RT-PCR contained 0.005 μg RNA, reaction buffer, sense and antisense primers (at a final concentration of 0.2 μM), RT-Platinum Taq Mix (Gibco-BRL). cDNA synthesis was performed in a MJ Research model PTC 100 thermal cycler using one cycle at 50°C for 30 min, then 94°C for 2 min, followed by 35 cycles of 94°C for 15 seconds, 55°C for 30 seconds, and 72°C for 1 min. Ten microliters were subjected to electrophoresis through a 1.5% agarose gel and stained with ethidium bromide. Absence of genomic DNA in samples RNA preparation was verified by substituting the RT/Taq mix by 2 units of Taq DNA polymerase (Amersham Biosciences, Little Chalfont, England) in the reaction.
TABLE 1.
Putative bacteriophage LE1 genes
| ORFa | Positions | Size (aa) | Orientation | Putative function, reference organism (accession no.) (P value) and/or Pfam domain (E-value)b |
|---|---|---|---|---|
| 1 | 247-984 | 245 | + | ParA, Borrelia burgdorferi (NP_045595) (8e-10) |
| ParA family ATPase* (5.1e-07) | ||||
| 2 | 981-1541 | 186 | + | Chromosome segregation ATPases, Methanosarcina barkeri (ZP_00297007) (0.008) |
| 3 | 1976-2458 | 180 | + | gp26 Burkholderia phage (NP_944256) (3e-06) |
| 4 | 2791-3639 | 282 | + | Chromosome partitioning protein Spo0J, Pseudomonas aeruginosa (AAG08947) (3e-07) |
| ParB-like nuclease domain* (3.7e-09) | ||||
| 5 | 3636-4196 | 186 | + | M protein trans-acting positive regulator, Streptococcus pyogenes SSI-1 (NP_802988) (0.039) |
| 6 | 5028-4222 | 268 | − | Putative lysogenic conversion protein, bacteriophage P2-EC45 (CAD54899) (0.004) |
| 7 | 5311-5012 | 99 | − | Helix-turn-helix protein* (0.049) |
| 8 | 5156-5897 | 213 | + | Unknown |
| 9 | 5894-7585 | 563 | + | Uncharacterized phage protein, Desulfovibrio vulgaris (YP_011917) (6e-32) |
| Putative terminase large subunit, bacteriophage Aaphi23 (NP_852753) (9e-09) | ||||
| 10 | 8204-9874 | 556 | + | Putative portal protein, Staphylococcus phage K (YP_024471) (1e-15) |
| 11 | 9901-10506 | 201 | + | Unknown |
| 12 | 10520-11470 | 316 | + | NTX, Clostridium botulinum D phage (BAA75084) (0.082) |
| 13 | 11470-11931 | 153 | + | Unknown |
| 14 | 11931-13085 | 384 | + | Unknown |
| 15 | 13115-19051 | 1971 | + | DNA methylase, Cytophaga hutchinsonii (ZP_00309105) (6e-78) |
| Helicase conserved C-terminal domain* (0.00033) | ||||
| 16 | 19035-19985 | 316 | + | ORF3, bacteriophage A511 (CAA62541) (0.18) |
| 17 | 20392-20846 | 184 | + | Unknown |
| 18 | 21028-21219 | 63 | + | Viral matrix protein* (1) |
| 19 | 21213-22001 | 262 | + | Unknown |
| 20 | 21919-22308 | 129 | + | Unknown |
| 21 | 22309-22722 | 137 | + | Unknown |
| 22 | 22709-23542 | 277 | + | Unknown |
| 23 | 23544-24275 | 243 | + | bZIP transcription factor* (0.0029) |
| 24 | 24275-25828 | 517 | + | Major capsid protein, Staphylococcus phage Twort (AAQ62708) (1e-19) |
| 25 | 25878-28535 | 885 | + | Unknown, Sinorhizobium meliloti phage PBC5 (AAL49565) (6e-39) |
| Tail protein, bacteriophage bIL286 (AAK08339) (3e-04) | ||||
| 26 | 28575-31187 | 859 | + | Hypothetical protein, Lactococcus phage BK5-T (CAC80156) (2e-05) |
| ParB-like nuclease domain* (1.8e-09) | ||||
| 27 | 31184-31942 | 252 | + | Unknown |
| 28 | 31939-32715 | 258 | + | Hantavirus glycoprotein* (0.82) |
| 29 | 32753-33004 | 83 | + | Unknown |
| 30 | 33005-34789 | 594 | + | Putative major tail sheath protein, Staphylococcus phage K (YP_024479) (6e-08) |
| 31 | 34793-35242 | 149 | + | Fork head domain* (0.27) |
| 32 | 35245-35679 | 144 | + | Unknown |
| 33 | 35582-35923 | 113 | + | Unknown |
| 34 | 35926-37632 | 568 | + | Putative tape measure protein, bacteriophage A118 (CAB53802) (0.004) |
| 35 | 38204-37638 | 188 | − | Unknown |
| 36 | 38558-38280 | 92 | − | Unknown |
| 37 | 39880-38735 | 381 | − | Hypothetical protein, S. meliloti (NP_384604.1) (0.001) |
| ATP dependent DNA ligase domain* (0.24) | ||||
| 38 | 39883-40413 | 176 | − | Hypothetical protein, Fremyella diplosiphon (AAT41947) (2e-11) |
| 39 | 43418-40635 | 927 | − | Putative DNA methyltransferase, Corynebacterium diphtheriae (NP_940094) (e-142) |
| Methylase* (0.83) | ||||
| 40 | 43539-44228 | 229 | + | Unknown |
| 41 | 44228-44605 | 125 | + | Virion host shutoff protein* (0.7) |
| 42 | 46110-44602 | 502 | − | Glycosyl hydrolase family* (0.49) |
| 43 | 47951-46107 | 614 | − | Orf130, Lactobacillus plantarum bacteriophage LP65 (YP_164765) (0.002) |
| LysM domain* (0.024) | ||||
| 44 | 47958-49205 | 415 | + | Hypothetical protein KgORF62, Staphylococcus phage K (YP_024492) (3e-09) |
| 45 | 49202-49840 | 212 | + | Unknown |
| 46 | 49848-51473 | 543 | + | Unknown |
| 47 | 51481-52023 | 180 | + | Unknown |
| 48 | 52080-52745 | 221 | + | Unknown |
| 49 | 52742-53392 | 216 | + | Putative transglycosylase, bacteriophage SPBc2 (NP_046584) (8e-09) |
| Peptidase family M23/M37* (2.9e-06) | ||||
| 50 | 53389-53547 | 52 | + | Unknown |
| 51 | 53519-53941 | 133 | + | Poly(A) polymerase regulatory subunit* (0.65) |
| 52 | 53938-54267 | 109 | + | Unknown |
| 53 | 54662-54264 | 132 | + | Unknown |
| 54 | 55126-54860 | 88 | − | Unknown |
| 55 | 55472-55104 | 102 | − | Unknown |
| 56 | 55626-55417 | 69 | − | Unknown |
| 57 | 56499-56215 | 94 | − | Unknown |
| 58 | 56808-56506 | 99 | − | Unknown |
| 59 | 57057-56815 | 80 | − | Unknown |
| 60 | 57968-57054 | 304 | − | Triosephosphate isomerase* (0.27) |
| 61 | 58147-57965 | 60 | − | Unknown |
| 62 | 58369-58160 | 69 | − | Single-stranded DNA-binding protein, Proteus marinus (NP_876175) (0.038) |
| Single-strand binding protein family* (1.2e-06) | ||||
| 63 | 58983-58366 | 205 | − | Hypothetical protein, bacteriophage CP-1639 (CAH2338) (2e-17) |
| 64 | 59573-59163 | 136 | − | Unknown |
| 65 | 60564-59932 | 210 | − | Unknown |
| 66 | 61049-60597 | 150 | − | Unknown |
| 67 | 61906-61046 | 286 | − | DNA adenine methyltransferase, Pseudomonas aeruginosa phage F116 (YP_164275) (1e-31) |
| D12 class N6 adenine-specific DNA methylase* (0.00012) | ||||
| 68 | 62160-61903 | 85 | − | Unknown |
| 69 | 62650-62126 | 174 | − | Unknown |
| 70 | 64119-62614 | 501 | − | DEAD box family helicase, Streptococcus pyogenes phage M1 (NP_268909) (7e-63) |
| Helicase conserved C-terminal domain* (1.1e-11) | ||||
| 71 | 65048-64116 | 310 | − | DNA methyltransferase, Magnetococcus sp. (ZP_00288199) (1e-12) |
| ParB-like nuclease domain* (0.00019) | ||||
| 72 | 65451-65155 | 96 | − | Unknown |
| 73 | 67762-65423 | 779 | − | Hypothetical nuclease SbcCD/C subunit, Ralstonia metallidurans (YP_161700) (2e-38) |
| Putative exonuclease, Staphylococcus phage K (YP_024504) (8e-20) | ||||
| 74 | 68922-67759 | 387 | − | Nuclease SbcCD/D subunit, Dehalococcoides ethenogenes (YP_181520) (2e-09) |
| DNA repair exonuclease* (1.4e-09) | ||||
| 75 | 69872-68919 | 317 | − | Peptidase family M20/M25/M40* (0.64) |
| 76 | 70817-70515 | 100 | − | Unknown |
| 77 | 70920-71300 | 126 | + | Predicted transcriptional regulator, L. interrogans (NP_713411) (3e-06) |
| Repressor cI, bacteriophage phi-80 (P14819) (0.01) | ||||
| Helix-turn-helix* (2e-08) | ||||
| 78 | 73009-71825 | 394 | − | Unknown |
| 79 | 73540-73016 | 174 | − | Transcription regulator, Listeria monocytogenes (EAL08490) (1e-09) |
| Immunity region protein, bacteriophage phi105 (BAA19319) (3e-04) |
ORFs used for RT-PCR analysis during the three stages of the LE1 life cycle are underlined (see Fig. 4).
Similarities (even low) with phage-related proteins are indicated in boldface.
DNA manipulations and plasmid construction.
Genomic DNA of L. biflexa was extracted as previously described (30). For Southern blot analysis, digested DNA was subjected to electrophoresis overnight, transferred onto a nylon membrane, and hybridized with [α-32P]dATP-labeled probe under stringent conditions as previously described (30). All PCR amplifications were achieved using one cycle of denaturation (94°C, 5 min), followed by 35 cycles of amplification consisting of denaturation (94°C, 30 s), annealing (55°C, 30 s), and primer extension (72°C, 1 min), and a final cycle of extension of 10 min at 72°C.
The nucleotide sequence of LE1 replication region was amplified with primer pairs Le9-Le5, Le9-Le10, Le2-Le5, Le2-Le4, Le2-Le3, and Le1-Le5 (positions indicated in Fig. 2 and Table 2) and inserted into pCR2.1-TOPO by using the TOPO TA cloning kit (Invitrogen Life Technologies, Carlsbad, CA). After gel purification of PvuII digestion, DNA fragments containing the LE1 replication region were inserted into the dephosphorylated SmaI site of pGKm, which is derived from pGEM-7Zf(+) (Promega, Madison, WI). The promoterless rep gene was amplified with primers NDrep and Le4 (Table 2) and cloned into pCR2.1-TOPO, released with NdeI and NsiI restriction enzymes, and then inserted into the NdeI and PstI sites of plasmid pFGB containing the promoter of Borrelia burgdorferi flgB (3), resulting in plasmid pGKPfrep (Fig. 2). Site-directed mutagenesis (Quick-change mutagenesis kit, Stratagene, La Jolla, CA) was done with the primer pairs MUTDR1-MUTDR1 and MUTIR1-MUTIR2 (Table 2) to generate plasmids pGKBLDrm and pGKBLIrm, respectively. Amplified fragments le2-le3 and flg5-le4 (Table 2) were cloned into the SmaI site of plasmid pGKBLIrm, generating plasmids pLILe24 and pLIPfr, respectively (Fig. 2).
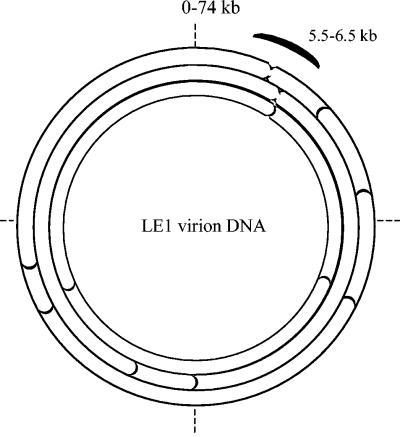
FIG. 2.
Restriction map of the LE1 virion DNA for the enzymes EagI, BglII, and BssHII (from the outer to the inner arc). The positions of ends of linear DNA molecules are indicated.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| Le1 | GAGCAAAAAAGATAAGCGCC |
| Le2 | AAAAAGGAAACGCGTTGCCG |
| Le3 | GTATCCATAATCCTGTTCTC |
| Le4 | GGTCATTGTCTGATTGTGGG |
| Le5 | CGCATCGAATTCTTTTTGGG |
| Le9 | TTACAGATTCTATTTTTTGG |
| Le10 | TTTGTCTATTTCGACAATAC |
| Le11 | GAAATCGGCAACGCGTTTCC |
| NDrep | CATATGAGAACAGATTATGG |
| Flg5 | TAATACCCGAGCTTCAAGGAA |
| MUTDR1 | CTCCTCCATCTTGATCCCCTATGTCTCGAGATGTGCTTGACTTATTATGTC |
| MUTDR2 | GACATAATAAGCAAGCACATCTCGAGACATAGGGGATCAAGATGGAGGAG |
| MUTIR1 | GAAAATTTGGCAATTTAGTGAAGCTGGCCGCATCCAATGATCCAAACAAAAAAATC |
| MUTIR2 | GATTTTTTGTTTGGATCATTGGATGCGGCCAGCTTCACTAAATTGCCAAATTTTC |
Plasmids from E. coli were recovered using a Qiaprep Spin miniprep kit (QIAGEN GmbH, Hilden, Germany). L. biflexa cells were electrotransformed as described previously (36). DNA sequencing was performed on plasmid constructs in order to check the constructions.
Shuttle vector stability in L. biflexa.
L. biflexa cells containing plasmid constructs (Fig. 2) were first grown in selective liquid medium. Cells in late exponential growth phase were diluted 1,000-fold in liquid medium without the antibiotic and grown at 30°C. After 48 h and 96 h, corresponding to approximately 10 and 20 generations, respectively, cultures were plated on nonselective medium, and 100 colonies were replicated to selective and nonselective plates to determine the frequency of plasmid loss based on the percentage of kanamycin-sensitive colonies.
Sequence analysis.
The DNA sequence of LE1 was analyzed by the CAAT-box package (15) for the presence of ORFs, which consisted of at least 50 codons. A total of 79 ORFs (Table 1) were thus predicted. Cumulative diagrams of AT and CG skew were generated as previously described (31). Nucleic acid and deduced amino acid sequences were also analyzed by using the BLAST software at the National Center for Biotechnology Institute (1), the Pfam protein families database (2), and programs from the Genetics Computer Group software package (Genetics Computer Group, University of Wisconsin, Madison, WI).
Nucleotide sequence accession number.
The GenBank accession number of the nucleotide sequence of LE1 from L. biflexa is BX571876.
RESULTS
Nucleotide sequence of the LE1 genome.
The LE1 phage genome was assembled into a single large contig of 73,623 bp. The average GC content (36 mol% GC) is identical to that of the L. biflexa genome (39). A total of 79 ORFs (Table 1) were predicted, of which 50 (63%) showed no similarity to proteins in the nonredundant protein database from the NCBI. Figure 1A corresponds to the prophage genome linearized arbitrarily at the beginning of ORF1 (parA homolog), which is close to the putative origin of replication (36).
FIG. 1.
Schematic representation of the LE1 genome. A. Genome sequence of leptophage LE1. The position and direction of transcription of the predicted ORFs are indicated by arrows. The filled circle is a potential rho-independent terminator. B. AT, GC, and cumulative skew diagrams of the LE1 genome. The analysis began at the designated nucleotide sequence position 1, corresponding to the LE1 replication region.
An important question refers to the topology of the LE1 virion DNA, knowing that it was circular as a prophage (36). When genomic DNA of the LE1 circular prophage remained trapped in the wells under different pulsed-field gel electrophoresis conditions (36), virion DNA migrated in pulsed-field gels like a 74-kb linear DNA molecule. The restriction map of DNA originating from the phage particle was compared with that originating from the lysogen by Southern blot analysis with LE1 as a probe. The results, using enzymes EagI, BglII, and BssHI, showed systematically an extra band when the DNA originates from the lytic cycle, in agrement with a linear configuration versus a circular one (Fig. 2). In addition, it was found that virion DNA molecules contain submolar fragments (data not shown) that could be due to the mechanism of headful packaging (20).
Further restriction analysis suggests that the extremities of the linear phage DNA are localized in the region between 5.5 and 6.5 kb (Fig. 2), upstream genes encoding the putative terminase and portal proteins (Fig. 1A), which are involved in the formation of mature DNA ends (4). We did not find sequence patterns in the LE1 region between 5.5 and 6.5 kb except 6- to 9-bp direct repeats that could determine the cleavage site of LE1. Overnight ligation of the virion DNA or heating HindIII and SacI restriction fragments for 10 min at 75°C did not change the restriction profiles of the virion DNA (data not shown), suggesting that LE1 does not contain cohesive ends (cos-containing phage) and that it may use a pac-type packaging mechanism (4).
Identification of the Rep protein and prediction of a Θ-type replication of leptophage LE1.
We analyzed the GC and AT skews (24) to locate the replication origin of the LE1 genome. Our results showed a V-shaped graph with obvious minimum cumulative skew point at the region which we have previously shown to contain the origin of replication (36) and maximum cumulative skew point occurring at half of the sequence length (approximately at position 38 kb) that could correspond to the terminus of replication (Fig. 1B). Similar to diagrams of cumulative skew obtained with other circular replicon (31), these results suggest that LE1 prophage replicates as a circular replicon by a theta-type mechanism in a bidirectional way. However, local change of the skew pattern occurred between kb 43 and 54. This is correlated with changes in the direction of transcription of putative genes in this region (Fig. 1A), suggesting that DNA rearrangements may be implicated in producing this distorted skew pattern.
By subcloning experiments, the smallest cloned fragment capable of sustaining autonomous replication in L. biflexa was found to be 948 bp in size (Fig. 3A). This minimal replication origin contains ORF3 (543 bp), designated the rep gene, and its promoter. The deduced 180-amino-acid sequence of rep showed weak but significant similarities to proteins of phages infecting both gram-negative and gram-positive bacteria, such as proteins from Burkholderia cepacia phage Bcep22 (40%, 41/101 amino acids), Lactococcus lactis phage phi31.1 (29%, 29/98 amino acids), Lactococcus phage BK5-T (28%, 24/85 amino acids), Salmonella enterica serovar Typhimurium phage ST64B (26%, 18/67 amino acids), and Shigella flexneri phage V (26%, 18/67 amino acids) (Fig. 3B). Among these homologs, proteins from phages BK5-T, ST64B, and V were annotated as putative replication proteins, and the biological function of other related proteins has not been assigned. The LE1 Rep protein also contains a putative helix-turn-helix motif (Fig. 3B) which is a DNA-binding motif that is characteristic of Rep proteins (8).
FIG. 3.
Identification of replication and partition functions of bacteriophage LE1. A. The ability of each plasmid construct to yield L. biflexa transformants and the stability of the plasmid without selective pressure are indicated on the right. ≪-≫ indicates that the plasmid was not replicative in L. biflexa. Plasmid stability in L. biflexa was evaluated as described in Material and Methods. The primers used to generate each DNA fragment are indicated. Boxes indicate site-directed mutagenesis of direct repeats (DR) and inverted repeats (IR), and nucleotide substitutions are written in uppercase. ND, not determined. B. Sequence alignment of Burkholderia cepacia phage Bcep22 protein (Bcep2), Lactococcus lactis phage phi31.1 (phi31), Lactococcus phage BK5-T (BK5), Salmonella enterica serovar Typhimurium phage ST64B (ST64B), Shigella flexneri phage V (phage V), and L. biflexa phage LE1 Rep (LE1) proteins. Residues conserved in at least four homologous proteins are shaded.
Replication origins of bacterial plasmids usually have multiple repeat sequences, called iterons, for binding the plasmid-specific replication initiator protein (8). Another characteristic feature of replication origins is the presence of AT-rich regions, which are known to be the sites of strand separation prior to recruitment of the replication machinery (8). Analysis of the nucleotide sequence of the minimal replication region suggests that the rep promoter, containing one direct repeat (DR) and one inverted repeat (IR) as well as several AT-rich regions (36), could be the LE1 replication origin.
To analyze the importance of these repeats, site-specific mutations were introduced in the direct and inverted repeats without any additional change in the replication region (Fig. 3A). While mutations in the DR have no effect on plasmid replication, mutations in the IR abolish plasmid replication (Fig. 3A). It has to be noted that the IR is located 60 bp upstream from the rep start codon and mutations in this region could affect rep expression rather than the initiation of replication. By cloning a transcriptional fusion between the B. burgdorferi flgB promoter and the LE1 rep coding sequence in pGKbLIrm, we were able to restore plasmid replication in L. biflexa, suggesting that the point mutations incorporated into the IR may alter transcription signals and rep expression. The iterons DR and IR may therefore not act as Rep binding sites.
A previous study has shown that the rep coding sequence of lactococcal bacteriophage TP901-1 contains repeats which act as the origin of replication (28). Since the LE1 rep coding sequence may function as an origin of replication, plasmid pGKPfrep harboring the transcriptional fusion between the B. burgdorferi flgB promoter and the LE1 rep coding sequence was tested for its ability to replicate in L. biflexa (Fig. 3A). Surprisingly, kanamycin-resistant colonies appeared after electrotransformation with pGKPfrep but 1 week later in comparison to colonies obtained with the original shuttle vector pGKLep1. This result suggests that the LE1 rep coding sequence under the control of the flgB promoter is able to induce autonomous plasmid replication in L. biflexa. However, additional signals may be required to confer optimal replication.
LE1 partition locus is related to parAB of Borrelia burgdorferi plasmids.
Upstream of rep, two possible ORFs, ORF1 and ORF2, encoding proteins of 245 and 186 amino acids were present. The deduced amino acid sequences of ORF1 exhibited similarities with homologs of the ParA partition protein of B. burgdorferi linear plasmids Lp36 (34%, 80/231 amino acids), lp28-1 (33%, 51/152 amino acids), and lp28-3 (29%, 64/220 amino acids). Similarities with ParA chromosome partitioning protein of L. interrogans (31%, 81/256 amino acids), Pseudomonas putida (29%, 56/190 amino acids), and Chromobacterium violaceum (24%, 64/258 amino acids) were also found.
Partitioning systems can be divided into two types, the Walker-type ATPase and the actin-like ATPase (16). The LE1 ParA contains a deviant Walker A motif, KGGXXKT/S, located between amino acids 10 and 16 (22). Partition systems usually consists of parA, a downstream gene called parB, and a centromere-like site, called parS. All three elements are necessary for plasmid stability. The LE1 parA downstream gene encodes a 186-amino-acid protein without significant similarities in the databases. Since parB genes are known to be less conserved, ORF2 may therefore constitute the parB of a par locus. Although the centromere-like site, called parS, is usually a highly repeated sequence near parAB, no obvious parS site was identified in the LE1 replication region. A phylogenetic analysis of Walker-type ParA ATPase-containing loci has shown that partition loci can be clustered in distinct subgroups (16). Based on the ParA and ParB sizes of LE1 as well as on protein similarities, it appears that the LE1 partition locus belongs to the subgroup containing the B. burdorgferi plasmids. This is in agreement with the phylogenetic relationship of the Leptospira and Borrelia genera.
To determine whether the putative par locus of LE1 can act as a partition locus, we compared the segregational stability of the L. biflexa-E. coli shuttle vectors with the par locus deleted and the replicative vector carrying the putative parA gene alone or both the parA and parB genes. The presence of both genes dramatically increased plasmid stability in L. biflexa (Fig. 3A), indicating that the parA and parB genes act together as a partition locus.
To establish a versatile vector system to facilitate genetic analysis, we have constructed L. biflexa-E. coli shuttle vectors where insertional inactivation of the lacZ gene can be used to identify cloned inserts by blue/white colony screening in E. coli (Fig. 4). A vector containing a gene encoding resistance to spectinomycin that allows complementation of kanamycin-resistant clones has also been developed (Fig. 4). In the absence of selective pressure, vectors pGKBLe24 and pGSBLe24 containing only the rep gene were not stable, and vector pGKBLe94 harboring the parA and parB genes was stable (Fig. 4).
FIG. 4.
Schematic of the L. biflexa-E. coli shuttle vectors pGKBLe24, pGSBLe24, and pGKBLe94. KmR, kanamycin resistance cassette; SpcR, spectinomycin-resistance cassette. Unique restriction sites are indicated.
Structural components of LE1.
ORF24 shared 41 and 40% similarity with the major capsid protein from Staphylococcus phage Twort and Listeria monocytogenes bacteriophage A511 (25), respectively, at the amino acid level. In addition, ORF24 from LE1 was expressed late after infection (see below). The C-terminal sequence of ORF25 (from residues 564 to 841) was similar to the tail gene product (1,640 amino acids) of Lactococcus lactis phage bIL286 (38% similarity) (5). Similarly, ORF30 and ORF34 shared significant protein similarities with phage tail proteins (Table 1). The rightward-oriented ORFs located upstream of ORF24 to the leftward-oriented ORF35 could represent a module related to phage head and tail morphogenesis (Fig. 1A). There is a rho-independent terminator of transcription predicted immediately downstream ORF24 (Fig. 1A), which could separate the tail and head genes.
Repressor cI and other phage-related genes.
The products of two open reading frames (ORF76 and ORF77), which are transcribed in opposite directions, were assumed to be the functional homologs of the lambdoid proteins Cro and cI, respectively. This was based on their sizes (100 and 126 amino acids, respectively), their transcription in opposite directions with an intergenic region of 97 nucleotides (compared with 100 bp between cro and cI), and the similarities shown by ORF77 to repressor cI of the lambda-like phage phi80 (58% similarity) (27). The N terminus of ORF77 presents a helix-turn-helix motif from amino acids 18 to 62, typical of DNA binding proteins. In addition, ORF77 was specifically expressed in the lysogen (see below) and may therefore be involved in the genetic switch of LE1. Although ORF76 does not show any significant similarity to Cro, it may be the functional homolog of Cro, a competitor of cI for binding at the operators that regulate the lytic-lysogenic pathways.
We thus identified three sets of inverted repeats of 24, 21, and 15 nucleotides in the intergenic region between ORF77 and ORF76 which may play the role of an operator. ORF79 (174 amino acids) was similar to an immunity repressor protein from bacteriophage phi105 (51% similarity) (38). ORF9 and ORF10 shared protein similarity with terminase and portal proteins, respectively, which are essential components of the DNA-packaging process (4). Since phages usually contain one large and one small subunit of terminase proteins (4), we suspect that ORF8 encodes the small subunit of the multimeric terminase. Finally, ORFs 15, 39, and 67 had similarity with methylases (Table 1). Analysis of ORFs 15 and 67 revealed an NPPYSR motif (from amino acids 647 to 652 and from 184 to 189, respectively) that is highly conserved among Dam DNA (N-6-adenine) methylase and is involved in binding of the S-adenosylmethionine substrate (23).
Pattern of gene expression during the LE1 life cycle.
Of the 79 ORFs, 21 were chosen for further study based on their similarities and tentative functions. Figure 5 shows one set of data for the 21 ORFs tested at the three different stages of the life cycle of LE1. ORF24 and ORF25, corresponding to the capsid and tail, respectively, are poorly expressed early after infection but strongly expressed late after infection and not expressed in the lysogen, which is characteristic of phage structural proteins. Some ORFs were expressed specifically in the lysogen, such as ORFs 4, 5, and 37 (orphans) and ORF77, encoding the putative repressor of the phage, supporting its role in the genetic switch of LE1. ORFs 1 to 3, involved in the partition and replication of the prophage, were expressed at the three stages of the LE1 life cycle. Other ORFs which are genes of unknown function evenly distributed throughout the genome were also expressed at the three stages of the LE1 life cycle.
FIG. 5.
LE1 transcription analysis by RT-PCR. The numbers above each lane represent a selection of potential ORFs (see Table 1) used for RT-PCR. The three different stages of the LE1 life cycle (lysogen, early, and late steps of the lytic stage) for which mRNA was extracted are indicated above the panels.
Since DNA packaging is the last stage of phage development, it is surprising to find that the expression of ORF10, which encodes the putative portal protein, was not down-regulated during lysogenic growth. This is also true for the downstream genes of unknown functions, ORF16 and ORF20, that may be part of the late operon of LE1, together with ORF10. This result could be due to spontaneous induction (lytic growth) in a minority of cells of the lysogen.
DISCUSSION
Since its description (37), phage LE1 has been the subject of studies which focused on the sensitivity of some L. biflexa strains to LE1 (6) and characterization of the LE1 replication region (36). This study shows that the complete nucleotide sequence of the LE1 leptophage is organized in modules containing a set of genes which carry out a biological function such as replication, morphogenesis, and immunity. Transcriptional analysis of LE1 genes also shows differential expression during infection or as a prophage. The genetic organization of the locus containing the putative cI repressor suggests that LE1 uses a lysogeny control system similar to that of phage lambda.
Unexpectedly, our results demonstrate the circularity of LE1 DNA during the lysogenic cycle and its linearity when encapsidated in the leptophage. An initial step in the packaging of LE1 DNA may be the cleavage of concatemeric LE1 DNA at the pac site, generating a linear LE1 virion molecule. In agreement with previous findings in pac-containing phages (4), the pac site of LE1 is adjacent to the genes encoding the putative terminase and portal proteins, which are involved in recognition and cleavage at the pac site (4). However, we do not have direct experimental evidence of the nucleotide sequence of the terminal DNA fragments of the LE1 linear genomic form. Previous hybridization experiments (37) and sequence comparison with the L. interrogans genomes completed to date (26, 34) suggest that the LE1 leptophage or related phages are not present as prophages or as remnants of phage in the Leptospira genome. In fact, few phage-related sequences were identified in the whole Leptospira genome (34). The weak similarities found for most of the LE1 genes in the databases show that Myoviridae, and phages in general, are more diverse than their morphologies suggest (35). Since we do not yet have a precise idea about the genetic complexity of the global phage population, the availability of more phage nucleotide sequences should provide new insights into the biology and evolution of phages.
Plasmids have been widely used in bacteria as genetic tools. However, until the discovery of leptophage LE1 (37), no replicons other than the chromosomes have been characterized in Leptospira spp. In this study, genetic determinants of LE1 replication and maintenance were clearly identified. The major stability systems identified for low-copy-number plasmids are active partition systems or toxin-antitoxin systems. Interestingly, three distinct toxin-antitoxin loci were identified in the L. interrogans genome (32, 40). However, there is no indication of such a system in LE1. On the other hand, a functional partition locus containing parA and parB genes was found immediately downstream of rep.
The L. biflexa-E. coli shuttle vector derived from LE1 has already proved useful in studies of gene function in the saprophytes L. biflexa and L. meyeri (3, 18, 30). In this study, our results enable the construction of new shuttle vectors for the genetics of Leptospira spp. (Fig. 3). L. biflexa may represent a good cloning host for members of the spirochete phylum. Indeed, data obtained with E. coli revealed that most of the spirochetal promoters functioned poorly in this genetic background. Among spirochetes, L. biflexa appears as a fast-growing bacterium and grows in aerobic conditions; it could therefore allow the functional analysis of genes from slow-growing or noncultivatable spirochetes. L. biflexa can be transformed at a high rate (100,000 transformants per μg of DNA) using kanamycin or spectinomycin resistance as a selectable marker.
Future studies may include the construction of new tools such as expression vectors for the use of L. biflexa as a surrogate spirochetal cloning host. A better understanding of the molecular mechanisms which determine LE1 packaging may also allow the development of an efficient in vitro packaging system for gene manipulation in Leptospira.
Acknowledgments
We thank C. Ottone for technical help, C. Buchrieser for her contribution to the initial recording of the sequencing data, J. R. Lobry for determination of the GC skew, D. Yelton for experiments on virion DNA, and S. Casjens, N. Charon, G. Baranton, and T. Stanton for their encouragement. We also thank the anonymous reviewers for their contribution in improving our manuscript.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, K. L. Howe, and E. L. L. Sonnhammer. 2000. The Pfam protein families database. Nucleic Acids Res. 28:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauby, H., I. Saint Girons, and M. Picardeau. 2003. Construction and complementation of the first auxotrophic mutant in the spirochaete Leptospira meyeri. Microbiology 149:689-693. [DOI] [PubMed] [Google Scholar]
- 4.Black, L. W. 1989. DNA packaging in dsDNA bacteriophages. Annu. Rev. Microbiol. 43:267-292. [DOI] [PubMed] [Google Scholar]
- 5.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL-1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenot, A., C. Werts, C. Ottone, N. Sertour, N. W. Charon, D. Postic, G. Barabton, and I. Saint Girons. 2001. First evidence for a restriction-modification system in Leptospira sp. FEMS Microbiol. Lett. 201:139-143. [DOI] [PubMed] [Google Scholar]
- 7.Buchrieser, C., C. Rusniok, L. Frangeul, E. Couve, A. Billault, F. Kunst, E. Carniel, and P. Glaser. 1999. The 102-kilobase pgm locus of Yersinia pestis: sequence analysis and comparison of selected regions among different Yersinia pestis and Yersinia pseudotuberculosis strains. Infect. Immun. 67:4851-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.del Solar, G., R. Giraldo, M. J. Ruiz-Echevarria, M. Espinosa, and R. Diaz-Orejas. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eggers, C. H., S. Casjens, S. F. Hayes, C. F. Garon, C. J. Damman, D. B. Oliver, and D. S. Samuels. 2000. Bacteriophages of spirochetes. J. Mol. Microbiol. Biotechnol. 2:365-373. [PubMed] [Google Scholar]
- 10.Eggers, C. H., and D. S. Samuels. 1999. Molecular evidence for a new bacteriophage of Borrelia burgdorferi. J. Bacteriol. 181:7308-7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellinghausen, H. C., and W. G. McCullough. 1965. Nutrition of Leptospira pomona and growth of 13 other serotypes: fractionation of oleic albumin complex and a medium of bovine albumin and polysorbate 80. Am. J. Vet. Res. 26:45-51. [PubMed] [Google Scholar]
- 12.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 13.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 14.Faine, S., B. Adler, C. Bolin, and P. Perolat. 1999. Leptospira and leptospirosis, 2nd ed. Medisci, Melbourne, Australia.
- 15.Frangeul, L., P. Glaser, C. Rusniok, C. Buchrieser, E. Duchaud, P. Dehoux, and F. Kunst. 2004. CAAT-box, contigs-assembly and annotation tool-box for genome sequencing projects. Bioinformatics 20:790-797. [DOI] [PubMed] [Google Scholar]
- 16.Gerdes, K., J. Moller-Jensen, and R. B. Jensen. 2000. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 37:455-466. [DOI] [PubMed] [Google Scholar]
- 17.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 18.Guégan, R., J. M. Camadro, I. Saint Girons, and M. Picardeau. 2003. Leptospira spp. possess a complete heme biosynthetic pathway and are able to use exogenous heme sources. Mol. Microbiol. 49:745-754. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda, H., and J.-I. Tomizawa. 1968. Prophage P1, an extrachromosomal replication unit. Cold Spring Harbor Symp. Quant. Biol. 33:791-798. [DOI] [PubMed] [Google Scholar]
- 20.Jackson, E. N., D. A. Jackson, and R. J. Deans. 1978. EcoRI analysis of bacteriophage P22 DNA packaging. J. Mol. Biol. 118:365-368. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, R. C., and V. G. Harris. 1967. Differentiation of pathogenic and saprophytic leptospires. J. Bacteriol. 94:27-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koonin, E. V. 1993. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J. Mol. Biol. 229:1165-1174. [DOI] [PubMed] [Google Scholar]
- 23.Kossykh, V. G., S. L. Schlagman, and S. Hattman. 1993. Conserved sequence motif DPPY in region IV of the phage T4 Dam DNA-(N-6 adenine) methyltransferase is important for S-adenosylmethionine binding. Nucleic Acids Res. 21:4659-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lobry, J. R. 1996. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 13:660-665. [DOI] [PubMed] [Google Scholar]
- 25.Loessner, M. J., and S. Scherer. 1995. Organization and transcriptional analysis of the Listeria phage A511 late gene region comprising the major capsid and tail sheath protein genes cps and tsh. J. Bacteriol. 177:6601-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nascimento, A. L., A. I. Ko, E. A. Martins, C. B. Monteiro-Vitorello, P. L. Ho, D. A. Haake, S. Verjovski-Almeida, R. A. Hartskeerl, M. V. Marques, M. C. Oliveira, C. F. Menck, L. C. Leite, H. Carrer, L. L. Coutinho, W. M. Degrave, O. A. Dellagostin, H. El-Dorry, E. S. Ferro, M. I. Ferro, L. R. Furlan, M. Gamberini, E. A. Giglioti, A. Goes-Neto, G. H. Goldman, M. H. Goldman, R. Harakava, S. M. Jeronimo, I. L. Junqueira-de-Azevedo, E. T. Kimura, E. E. Kuramae, E. G. Lemos, M. V. Lemos, C. L. Marino, L. R. Nunes, R. C. de Oliveira, G. G. Pereira, M. S. Reis, A. Schriefer, W. J. Siqueira, P. Sommer, S. M. Tsai, A. J. Simpson, J. A. Ferro, L. E. Camargo, J. P. Kitajima, J. C. Setubal, and M. A. Van Sluys. 2004. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J. Bacteriol. 186:2164-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogawa, T., H. Ogawa, and J. Tomizawa. 1988. Organization of the early region of bacteriophage phi 80. Genes and proteins. J. Mol. Biol. 202:537-550. [DOI] [PubMed] [Google Scholar]
- 28.Ostergaard, S., L. Brondsted, and F. K. Vogensen. 2001. Identification of a replication protein and repeats essential for DNA replication of the temperate lactococcal bacteriophage TP901-1. Appl. Environ. Microbiol. 67:774-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paster, E. J., and F. E. Dewhirst. 2001. Phylogenetic foundation of spirochetes, p. 5-9. In J. Garcia-Lara (ed.), The spirochetes. Molecular and cellular biology. Horizon Press, Wymondham, United Kingdom.
- 30.Picardeau, M., A. Brenot, and I. Saint Girons. 2001. First evidence for gene replacement in Leptospira spp. Inactivation of L. biflexa flaB results in non-motile mutants deficient in endoflagella. Mol. Microbiol. 40:189-199. [DOI] [PubMed] [Google Scholar]
- 31.Picardeau, M., J. R. Lobry, and B. J. Hinnebusch. 2000. Analyzing DNA strand compositional asymmetry to identify candidates replication origins of Borrelia burgdorferi linear and circular chromosomes. Genome Res. 10:1594-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Picardeau, M., S. Ren, and I. Saint Girons. 2001. Killing effect and antitoxic activity of the Leptospira interrogans toxin-antitoxin system in Escherichia coli. J. Bacteriol. 183:6494-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravin, V., N. Ravin, S. Casjens, M. E. Ford, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequence and analysis of the atypical temperate bacteriophage N15. J. Mol. Biol. 299:53-73. [DOI] [PubMed] [Google Scholar]
- 34.Ren, S., G. Fu, X. Jiang, R. Zeng, H. Xiong, G. Lu, H. Q. Jiang, Y. Miao, H. Xu, Y. Zhang, X. Guo, Y. Shen, B. Q. Qiang, X. Q., A. Danchin, I. Saint Girons, R. L. Somerville, Y. M. Weng, M. Shi, Z. Chen, J. G. Xu, and G. P. Zhao. 2003. Unique and physiological and pathogenic features of Leptospira interrogans revealed by whole genome sequencing. Nature 422:888-893. [DOI] [PubMed] [Google Scholar]
- 35.Rohwer, F. 2003. Global phage diversity. Cell 113:141. [DOI] [PubMed] [Google Scholar]
- 36.Saint Girons, I., P. Bourhy, C. Ottone, M. Picardeau, D. Yelton, R. W. Hendrix, G. P., and N. W. Charon. 2000. The LE1 bacteriophage replicates as a plasmid within Leptospira biflexa. Construction of an L. biflexa-Escherichia coli shuttle vector. J. Bacteriol. 182:5700-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saint Girons, I., D. Margarita, P. Amouriaux, and G. Baranton. 1990. First isolation of bacteriophages for a spirochaete: potential genetic tools for Leptospira. Res. Microbiol. 141:1131-1138. [DOI] [PubMed] [Google Scholar]
- 38.Shearvin, K. E., I. B. Dodd, and J. B. Egan. 2002. The helix-turn-Helix motif of the coliphage 186 immunity repressor binds to two distinct recognition sequences. J. Biol. Chem. 277:3186-3194. [DOI] [PubMed] [Google Scholar]
- 39.Yasuda, P. H., A. G. Steigerwalt, K. R. Sulzer, A. F. Kaufmann, F. Rogers, and D. J. Brenner. 1987. Deoxyribonucleic acid relatedness between serogroups and serovars in the family Leptospiraceae with proposals for seven new Leptospira species. Int. J. Syst. Bacteriol. 37:407-415. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, Y. X., J. Li, X. K. Guo, C. Wu, B. Bi, S. X. Ren, C. F. Wu, and G. P. Zhao. 2004. Characterization of a novel toxin-antitoxin module, VapBC, encoded by Leptospira interrogans chromosome. Cell Res. 14:208-216. [DOI] [PubMed] [Google Scholar]