Abstract
The impossibility of obtaining a rho null mutant and sensitivity to bicyclomycin have indicated that rho is essential for the viability of Caulobacter crescentus. Transcription gene fusions of sequences with serial deletions of the rho 5′ untranslated region (5′-UTR) with a lacZ reporter gene indicated that rho is autoregulated at the level of attenuation of transcription in the 5′-UTR.
The importance of Rho-dependent termination in bacterial gene expression is well established for genes that are regulated by termination of transcription in the leader sequences, as described for the tna operon from Escherichia coli (15, 33, 35). Studies with E. coli, Rhodobacter sphaeroides, and Micrococcus luteus showed that rho is an essential gene in these bacteria (6, 8, 26) but not in Bacillus subtilis and Staphylococcus aureus (29, 34). The current model proposes that Rho loads onto nascent mRNAs at a cytosine-rich region that lacks a strong secondary structure, known as the Rho utilization (rut) site (1, 23, 30). It has been proposed that once bound, a hexameric Rho complex hydrolyzes ATP, moves directionally 5′-3′ along the RNA to the site of transcription, and with its helicase function, disengages the message from the paused RNA polymerase and the DNA template, separating the transcription complex (4, 17, 22, 27, 31, 32).
The Rho factor was described to be autogenously regulated in E. coli and B. subtilis via an attenuation mechanism that occurs in Rho-dependent terminators located within the mRNA leader of the rho gene (2, 12, 20). The ATPase activity of Rho in several bacteria has been described and characterized (10, 11, 12, 16, 25), but autoregulation by Rho-dependent terminators in bacteria other than E. coli and B. subtilis is yet to be characterized.
In a previous work from our group, a rho mutation generated by a Tn5 insertion (strain SP3710) caused deficiency in survival to high salt concentration and acidic pH and resulted in an extreme sensitivity to oxidative stress (13). Experiments showed that transcription of the C. crescentus rho gene increases in the rho mutant strain, which is indicative of a negative autoregulatory circuit (13).
The Rho factor is essential for C. crescentus.
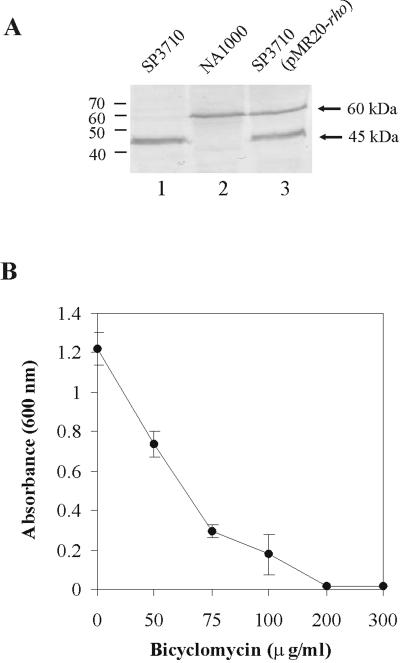
The Rho protein in SP3710 was interrupted after residue Gly-112, immediately before the first RNA binding motif (13). Immunoblotting assays of cell extracts probed with a polyclonal Rho antiserum showed that SP3710 still produces a 45-kDa truncated polypeptide but does not present the complete form of the Rho protein of the parental strain, NA1000, except when it is complemented in trans with the rho gene (Fig. 1A). The C. crescentus Rho protein has a predicted molecular mass of 52.8 kDa but migrates with an apparent mass of 60 kDa in sodium dodecyl sulfate-polyacrylamide gel. The 45-kDa protein should have all the residues essential for RNA binding at the high-affinity primary RNA binding site (5, 32), but the missing N-terminal 112 residues may have a role in protein function, probably in the interaction of the subunits to form the active hexamer, as proposed for E. coli (32). In order to generate an independent rho null mutant, a 3.4-kb fragment containing the rho gene was amplified by PCR with primers Rho4 and Rho5 (Table 1) as described in reference 13 and cloned into plasmid pNPTS138. This plasmid carries a sacB gene and a kanamycin resistance (Kmr) gene and does not replicate in Caulobacter. A 1.0-kb fragment containing the rho coding region was replaced by a spectinomycin resistance cassette (ΩSpr) (28). Two clones (1S and 4S) were selected after the first recombination event (Spr and Kmr clones), in plates containing 5 μg/ml kanamycin and 20 μg/ml spectinomycin. These clones were grown in plates containing 3% sucrose and spectinomycin to select for cells in which the plasmid was deleted from the chromosome, leaving the disrupted copy of the gene. The colonies obtained were then analyzed for their kanamycin resistance pattern, as shown in Table 2. The chromosomal copy of the rho gene was lost only in clones 1S and 4S, which carry a copy of the rho gene on a plasmid (pMR20-Rho), generating Spr/kanamycin-sensitive (Kms) clones. These results indicate that the rho gene is essential for the viability of C. crescentus as described for other gram-negative bacteria, such as E. coli and Rhodobacter sphaeroides and the gram-positive Micrococcus luteus (6, 8, 26). These results also indicate that the truncated form of Rho in SP3710 must have some remaining activity.
FIG. 1.
(A) Immunoblot of C. crescentus cell extracts with polyclonal anti-Rho antiserum. Lane 1, extract from strain SP3710; lane 2, extract from strain NA1000; lane 3, extract from SP3710 containing the pMR20-Rho plasmid. (B) Analysis of the sensitivity of C. crescentus NA1000 cells to bicyclomycin. Growth was determined by measuring the optical density at 600 nm after 48 h at 30°C of cultures in peptone-yeast extract medium containing increasing concentrations of bicyclomycin: 0, 50, 75, 100, 200, and 300 μg/ml.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′-3′)a |
|---|---|
| Rho1 | CGGTCATGGCAAGACTCAAG |
| Rho2 | TTGAATTCATGACCGAAGACACCGAAAACC |
| Rho3 | CATAAGCTTTCAGGTGTTCATCGACTGG |
| Rho4 | GCCAAGCTTCGAATGGACCGTCTGTTACC |
| Rho5 | TGGAATTCATCGACACCTATCACCGATC |
| Rho6 | GTCCGCGTGGTTTGCATCCACAG |
| Rho7 | CAGCTCCTGCAGGGACATGGAC |
| Rho8 | GGTTTGGATCCACAGTCGGTTC |
| Rho9 | GCGAATTCTTCCTGCTGCAGCCCTG |
| Rho10 | GGGGATCCCAGGACGGGCGCCAGTC |
| Rho11 | CAGGATCCTCTGTTGACGATGACGC |
| Rho12 | GGGGATCCCGGTCATGGCAAGACTC |
| Rho14 | CAGAATTCGAGCTTGCGGCGGCGGC |
Underlined nucleotides indicate restriction sites incorporated into oligonucleotides.
TABLE 2.
Antibiotic resistance profile of the colonies after the second recombination event
| Strain | No. of colonies
|
||
|---|---|---|---|
| Spr/Kms | Spr/Kmr | Total | |
| 1S | 0 | 73 | 73 |
| 1S(pMR20-Rho) | 26 | 51 | 77 |
| 4S | 0 | 79 | 79 |
| 4S(pMR20-Rho) | 15 | 68 | 83 |
The antibiotic bicyclomycin is a specific inhibitor of Rho activity in E. coli (36) and prevents the growth of several gram-negative bacteria at low dosages (24). In vitro assays have shown that bicyclomycin inhibits the ATPase activity of Rho from E. coli (18, 36), M. luteus (25), B. subtilis (12), and Streptomyces lividans (11), but its antimicrobial activity is found only against gram-negative bacteria, with the exception of M. luteus (25, 26). The growth of C. crescentus NA1000 was tested in the presence of increasing concentrations of bicyclomycin (Fig. 1B), and it was verified that 100 μg/ml was the minimal concentration tested where there was still growth and 200 μg/ml abolished it completely. These results further confirm that the rho gene is essential for the viability of C. crescentus and that the truncated form of Rho in strain SP3710 is probably functional enough to ensure viability, although the cells are still severely affected (13). The minimal bicyclomycin concentrations that prevented growth were 75 μg/ml for E. coli DH5α (11) and 90 μg/ml for M. luteus (26). However, higher bicyclomycin concentrations did not affect the growth of bacteria for which Rho is not essential, such as B. subtilis (300 μg/ml) (12) and Streptomyces lividans ZX7 (600 μg/ml) (11).
Sequences downstream of the rho promoter are required for autoregulation.
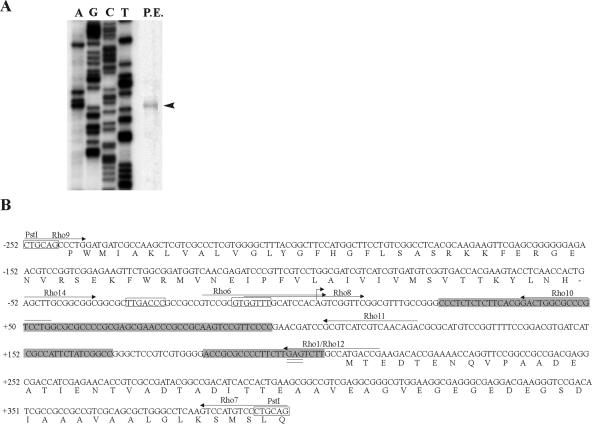
The transcription start site of the rho gene was determined by primer extension analysis using 50 μg of total RNA from the NA1000 strain and primer Rho1 (Table 1), labeling with [γ-32P]ATP, and extension with SuperScript III reverse transcriptase (Invitrogen) (Fig. 2A). The rho mRNA presented a 206-nt-long 5′ untranslated region (5′-UTR). Such long UTRs were also described for the rho genes from E. coli (255 nucleotides [nt]) and B. subtilis (at least 293 nt), suggesting that a long leader sequence is important for the regulation of this gene (12, 20, 29).
FIG. 2.
(A) Determination of the transcription start site of the rho gene. Primer extension analysis was carried out with 50 μg total RNA from exponential-phase cells (P.E.). Primer Rho1 was 5′-end labeled with 32P, extended with reverse transcriptase to determine the transcription start site (arrowhead), and also used in the sequencing reaction (shown on the left). (B) Nucleotide sequence of the rho regulatory region. The bent arrow indicates the transcription start site and was designated position +1, and the −35/−10 sequences are boxed. C-rich sequences are shaded, the ribosome-binding site is double underlined, and the PstI restriction sites are indicated. The arrows indicate the positions of each oligonucleotide used for primer extension, transcriptional fusions, and RNA gel mobility shift assays.
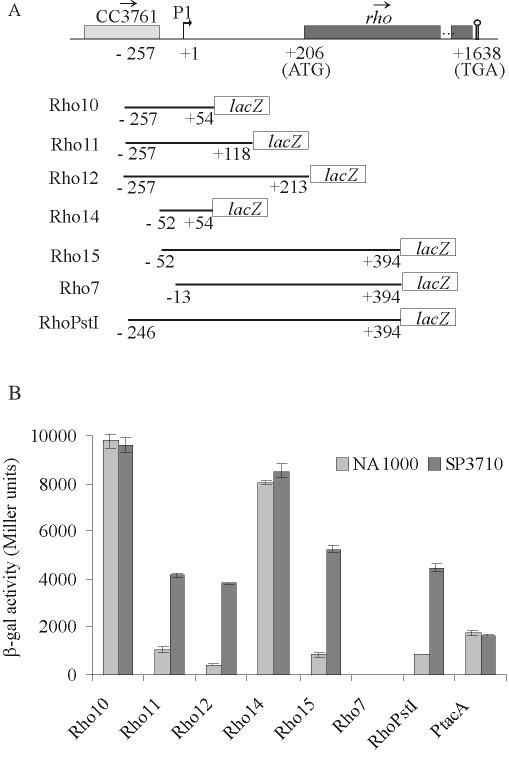
Different fragments containing the rho promoter and the 5′-UTR were amplified by PCR with the following primers (Table 1): Rho10 (Rho9/Rho10), Rho11 (Rho9/Rho11), Rho12 (Rho9/Rho12), Rho14 (Rho14/Rho10), Rho15 (Rho14/Rho7), and Rho7 (Rho8/Rho7) (Fig. 3A). The fragments were cloned in plasmid pRKlacZ290 (7), generating transcriptional fusions with the lacZ gene, and β-galactosidase activity was measured by the method of Miller (21).
FIG. 3.
Analysis of expression driven by rho regulatory regions. (A) Map (not to scale) of the region surrounding the rho gene in C. crescentus. Boxes indicate open reading frames, and the arrows above them show the direction of transcription. Coordinates in nucleotides are relative to the first nucleotide of the rho transcription start site (bent arrow), determined by primer extension analysis (+1). The promoter is indicated as P1. The stem and loop indicate a putative Rho-independent transcription terminator. Below the map, each transcriptional fusion construct is indicated, where coordinates indicate the extent of the regulatory rho region cloned in front of the reporter lacZ gene. (B) β-Galactosidase activity of each construct in the NA1000 and SP3710 strains. The results are in Miller units (21) and are the average of results of at least three independent assays.
The fragment containing the promoter and 5′-UTR regions of rho (RhoPstI) yielded about 850 β-galactosidase units in NA1000 and 4,500 units in SP3710, indicating that autoregulation occurs in this construct (Fig. 3B). As a control, the tacA gene (19), which is not regulated by Rho, showed similar levels of expression in both strains. On the other hand, constructs containing either the promoter only (Rho14) or the promoter and 54 bp of the 5′-UTR (Rho10) showed much higher β-galactosidase activities in both strains, indicating that the autogenous regulation is lost in these constructs. These results indicate that the autoregulation does not occur at the level of transcription initiation and that the rho gene has a strong promoter.
Both Rho11 and Rho12 constructs showed β-galactosidase activities similar to that of RhoPstI, restoring the autoregulation. These results suggested that attenuation sites could be present in the region between nt +54 and +213, but there was still a possibility that a second promoter could exist in this region. Rho7, which lacks the −35 region of the rho promoter but keeps the complete 5′-UTR and 187 bp of the Rho coding region, showed no β-galactosidase activity, confirming that there is no other promoter downstream of position −13. When the complete promoter was present at the 5′ end (Rho15), it showed normal levels of activity. Overall, the results indicated that transcription attenuation sites are present between nucleotides +54 and +213, which is similar to what was described for rho genes from E. coli and B. subtilis (12, 20).
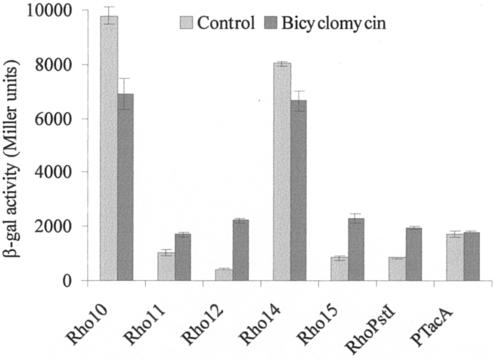
These results were confirmed by measuring rho expression in the presence of 100 μg/ml bicyclomycin for 3 h. As shown in Fig. 4, a pattern of rho expression was obtained for NA1000 with bicyclomycin that was similar to those presented for SP3710 with all the constructs. These results confirm that the presence of Rho causes a decrease in the transcription of its own gene and that this effect is dependent on the 5′-UTR downstream of position +54.
FIG. 4.
Analysis of expression driven by rho regulatory regions in the presence of bicyclomycin. Expression was determined from NA1000 cells harboring the respective constructs described in Fig. 3 after incubation at 30°C for 3 h in the presence or absence of 100 μg/ml bicyclomycin. The β-galactosidase (β-gal) activities are expressed in Miller units (21) and are the averages of results of at least three independent assays.
Binding of Rho to the 5′-UTR of rho mRNA.
The results above suggested that there would be attenuators between position +54 and position +213 (Fig. 2B). This region is notably cytidine/uridine rich and is a strong candidate to contain rut sites, similar to those proposed for enteric bacteria (1, 3, 9, 23). The binding of Rho to this region was tested by an RNA electrophoretic mobility shift assay. The rho gene was amplified by PCR, using primers Rho2 and Rho3 (Table 1), and the products were cloned into pProEX-HT (Invitrogen). Expression of His-Rho in E. coli DH5α was obtained after induction with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 h, and the protein was purified in Ni-nitrilotriacetic acid agarose columns (QIAGEN).
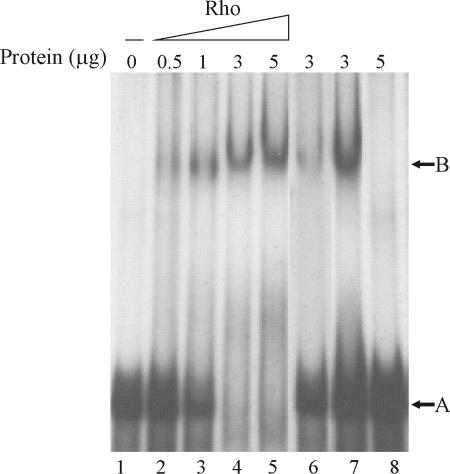
A probe extending from primer Rho6 to primer Rho12 was transcribed in vitro by T7 RNA polymerase in the presence of [α-32P]UTP from a pGEM-T Easy plasmid (Promega). An RNA binding assay was carried out essentially as described in reference 14, using an RNA template (5 × 105 cpm) and purified His-Rho protein (dialyzed against the binding buffer). Incubation of the probe with different amounts of purified His-Rho protein (Fig. 5) caused retardation in its migration (lanes 2 to 5). The entire amount of probe was shifted with 3 μg/ml of Rho protein (lane 4). The same unlabeled RNA fragment was able to compete efficiently for binding (compare lanes 4 and 6), but tRNA was not (compare lanes 4 and 7). As a control, the assay was carried out with 5 μg/ml of bovine serum albumin, and no shift in the band was observed (lane 7). The interaction of Rho protein with the rho 5′-UTR mRNA confirms the presence of rut sites in this region and indicates that expression of the C. crescentus rho gene is autoregulated by attenuation of transcription.
FIG. 5.
RNA electrophoretic mobility shift assay of rho mRNA. The indicated amount of purified Rho was incubated with 70 ng of 32P-labeled RNA fragment for 20 min, and the complexes were resolved in a 5% polyacrylamide gel. Lane 1, RNA probe; lanes 2 to 5, increasing amounts of Rho (0.5, 1, 3, and 5 μg, respectively) added to the RNA probe; lane 6, the probe incubated with 3 μg of Rho and 0.7 μg of the same nonradioactive RNA competitor; lane 7, the probe incubated with 3 μg of Rho and 1 μg of tRNA; lane 8, the probe incubated with bovine serum albumin (5 μg).
Acknowledgments
We thank Suely L. Gomes for critical reading of the manuscript, M. R. K. Alley for pNPTS138, and Fujisawa Pharmaceuticals Co. (Japan) for providing the bicyclomycin sample.
This work was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP grant 2002/05762-1). During the course of this work, V.C.S.I. was supported by a fellowship from FAPESP. M.V.M. is partly supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
REFERENCES
- 1.Alifano, P., F. Rivellini, D. Limauro, C. B. Bruni, and M. S. Carlomagno. 1991. A consensus motif common to all Rho-dependent prokaryotic transcription terminators. Cell 64:553-563. [DOI] [PubMed] [Google Scholar]
- 2.Barik, S., P. Bhattacharya, and A. Das. 1985. Autogenous regulation of transcription termination factor Rho. J. Mol. Biol. 182:495-508. [DOI] [PubMed] [Google Scholar]
- 3.Bektesh, S. L., and J. P. Richardson. 1980. A rho-recognition site on phage lambda cro-gene mRNA. Nature 283:102-104. [DOI] [PubMed] [Google Scholar]
- 4.Brennan, C. A., A. J. Dombroski, and T. Platt. 1987. Transcription termination factor rho is an RNA-DNA helicase. Cell 48:945-952. [DOI] [PubMed] [Google Scholar]
- 5.Chen, X., and B. L. Stitt. 2004. The binding of C10 oligomers to Escherichia coli transcription termination factor Rho. J. Biol. Chem. 279:16301-16310. [DOI] [PubMed] [Google Scholar]
- 6.Das, A., D. Court, and S. Adhya. 1976. Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor Rho. Proc. Natl. Acad. Sci. USA 73:1959-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gober, J. W., and L. Shapiro. 1992. A developmentally regulated Caulobacter flagellar promoter is activated by 3′ enhancer and IHF binding elements. Mol. Biol. Cell 3:913-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomelsky, M., and S. Kaplan. 1996. The Rhodobacter sphaeroides 2.4.1 rho gene: expression and genetic analysis of structure and function. J. Bacteriol. 178:1946-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hart, C. M., and J. W. Roberts. 1994. Deletion analysis of the lambda tR1 termination region. Effect of sequences near the transcript release sites, and the minimum length of rho-dependent transcripts. J. Mol. Biol. 237:255-265. [DOI] [PubMed] [Google Scholar]
- 10.Ingham, C. J. 1999. Characterisation of the enzymatic and RNA-binding properties of the Rhodobacter sphaeroides 2.4.1. Rho homologue. Biochim. Biophys. Acta 1446:115-125. [DOI] [PubMed] [Google Scholar]
- 11.Ingham, C. J., I. S. Hunter, and M. C. Smith. 1996. Isolation and sequencing of the rho gene from Streptomyces lividans ZX7 and characterization of the RNA-dependent NTPase activity of the overexpressed protein. J. Biol. Chem. 271:21803-21807. [DOI] [PubMed] [Google Scholar]
- 12.Ingham, C. J., J. Dennis, and P. A. Furneaux. 1999. Autogenous regulation of transcription termination factor Rho and the requirement for Nus factors in Bacillus subtilis. Mol. Microbiol. 31:651-663. [DOI] [PubMed] [Google Scholar]
- 13.Italiani, V. C. S., L. F. G. Zuleta, and M. V. Marques. 2002. The transcription termination factor Rho is essential for oxidative stress survival in Caulobacter crescentus. Mol. Microbiol. 44:181-194. [DOI] [PubMed] [Google Scholar]
- 14.Jiang, W., Y. Hou, and M. Inouye. 1997. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272:196-202. [DOI] [PubMed] [Google Scholar]
- 15.Konan, K. V., and C. Yanofsky. 2000. Rho-dependent transcription termination in the tna operon of Escherichia coli: roles of the boxA sequence and the rut site. J. Bacteriol. 182:3981-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowery, C., and J. P. Richardson. 1977. Characterization of the nucleoside triphosphate phosphohydrolase (ATPase) activity of RNA synthesis termination factor p. I. Enzymatic properties and effects of inhibitors. J. Biol. Chem. 252:1375-1380. [PubMed] [Google Scholar]
- 17.Lowery-Goldhammer, C., and J. P. Richardson. 1974. An RNA-dependent nucleoside triphosphate phosphohydrolase (ATPase) associated with rho termination factor. Proc. Natl. Acad. Sci. USA 71:2003-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magyar, A., X. Zhang, H. Kohn, and W. R. Widger. 1996. The antibiotic bicyclomycin affects the secondary RNA binding site of Escherichia coli transcription termination factor Rho. J. Biol. Chem. 271:25369-25374. [DOI] [PubMed] [Google Scholar]
- 19.Marques, M. V., S. L. Gomes, and J. W. Gober. 1997. A gene coding for a putative sigma 54 activator is developmentally regulated in Caulobacter crescentus. J. Bacteriol. 179:5502-5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsumoto, Y., K. Shigesada, M. Hirano, and M. Imai. 1986. Autogenous regulation of the gene for transcription termination factor Rho in Escherichia coli: localization and function of its attenuators. J. Bacteriol. 166:945-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, J. M. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Morgan, W. D., D. G. Bear, and P. H. von Hippel. 1983. Rho-dependent termination of transcription. I. Identification and characterization of termination sites for transcription from the bacteriophage lambda PR promoter. J. Biol. Chem. 258:9553-9564. [PubMed] [Google Scholar]
- 23.Morgan, W. D., D. G. Bear, B. L. Litchman, and P. H. von Hippel. 1985. RNA sequence and secondary structure requirements for rho-dependent transcription termination. Nucleic Acids Res. 13:3739-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishida, M., Y. Mine, T. Matsubara, S. Goto, and S. Kuwahara. 1972. Bicyclomycin, a new antibiotic. 3. In vitro and in vivo antimicrobial activity. J. Antibiot. 25:582-593. [PubMed] [Google Scholar]
- 25.Nowatzke, W. L., and J. P. Richardson. 1996. Characterization of an unusual Rho factor from the high G+C gram-positive bacterium Micrococcus luteus. J. Biol. Chem. 271:742-747. [DOI] [PubMed] [Google Scholar]
- 26.Nowatzke, W. L., E. Keller, G. Koch, and J. P. Richardson. 1997. Transcription termination factor Rho is essential for Micrococcus luteus. J. Bacteriol. 179:5238-5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oda, T., and M. Takanami. 1972. Observations on the structure of the termination factor rho and its attachment to DNA. J. Mol. Biol. 71:799-802. [DOI] [PubMed] [Google Scholar]
- 28.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 29.Quirk, P. G., E. A. Dunkley, P. Lee, and T. A. Krulwich. 1993. Identification of a putative Bacillus subtilis rho gene. J. Bacteriol. 175:647-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richardson, J. P. 1990. Rho-dependent transcription termination. Biochim. Biophys. Acta 1048:127-138. [DOI] [PubMed] [Google Scholar]
- 31.Richardson, J. P. 2002. Rho-dependent termination and ATPases in transcript termination. Biochim. Biophys. Acta 1577:251-260. [DOI] [PubMed] [Google Scholar]
- 32.Skordalakes, E., and J. M. Berger. 2003. Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell 114:135-146. [DOI] [PubMed] [Google Scholar]
- 33.Stewart, V., R. Landick, and C. Yanofsky. 1986. Rho-dependent transcription termination in the tryptophanase operon leader region of Escherichia coli K12. J. Bacteriol. 166:217-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Washburn, R. S., A. Marra, A. P. Bryant, M. Rosenberg, and D. R. Gentry. 2001. rho is not essential for viability or virulence in Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1099-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanofsky, C., and V. Horn. 1995. Bicyclomycin sensitivity and resistance affect Rho factor-mediated transcription termination in the tna operon of Escherichia coli. J. Bacteriol. 177:4451-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zwiefka, A., H. Kohn, and W. R. Widger. 1993. Transcription termination factor rho: the site of bicyclomycin inhibition in Escherichia coli. Biochemistry 32:3564-3570. [DOI] [PubMed] [Google Scholar]