Abstract
IncHI plasmids encode multiple-antibiotic resistance in Salmonella enterica serovar Typhi. These plasmids have been considered to play a relevant role in the persistence and reemergence of this microorganism. The IncHI1 plasmid R27, which can be considered the prototype of IncHI plasmids, is thermosensitive for transfer. Conjugation frequency is highest at low temperature (25 to 30°C), decreasing when temperature increases. R27 codifies an H-NS-like protein (open reading frame 164 [ORF164]) and an Hha-like protein (ORF182). The H-NS and Hha proteins participate in the thermoregulation of gene expression in Escherichia coli. Here we investigated the hypothetical role of such proteins in thermoregulation of R27 conjugation. At a nonpermissive temperature (33°C), transcription of several ORFs in both transfer region 1 (Tra1) and Tra2 from R27 is upregulated in cells depleted of Hha-like and H-NS-like proteins. Both chromosome- and plasmid-encoded Hha and H-NS proteins appear to potentially modulate R27 transfer. The function of R27-encoded Hha-like and H-NS proteins is not restricted to modulation of R27 transfer. Different mutant phenotypes associated with both chromosomal hha and hns mutations are compensated in cells harboring R27.
The role of architectural proteins in the organization and function of the bacterial chromosome is now well established. Among these proteins, the H-NS family of proteins in gram-negative bacteria (45) has deserved extensive research efforts in the last years. Best characterized in different enteric bacteria such as Escherichia coli or Salmonella enterica serovar Typhi, the H-NS protein is a relevant example of a global modulator exerting its effects, overwhelmingly negative, in response to different environmental signals, such as osmolarity or temperature (for a recent review, see reference 10). When interacting with DNA, H-NS binds preferentially to curved DNA (38, 48). To repress transcription, H-NS binds in many instances to two distal curved targets surrounding the promoter region. Binding to DNA is followed by oligomerization and subsequent alteration of DNA structure (11, 13, 27, 40).
H-NS oligomerization depends upon the N-terminal domain of the protein, extending up to residue 65 (3, 12). H-NS is able not only to generate homodimers and -oligomers but to interact with other proteins. Generation of heterodimers and -oligomers with its paralogue StpA is a well-documented process (21, 22, 47). H-NS also interacts with members of a family of small proteins (molecular mass, about 8 kDa), the Hha/YmoA family (35, 36). Hha and YmoA were initially described in E. coli and Yersinia enterocolitica as thermomodulators of the expression of virulence factors (4, 32, 34). Interaction of Hha and H-NS was evidenced when assessing the biological role of Hha as a modulator of the operon encoding the expression of the E. coli toxin α-hemolysin (36). In fact, an Hha-H-NS complex modulates the expression of the hly operon (27). Further studies have shown interaction between various members of both families. YmoA interacts with Y. enterocolitica H-NS (35), and Hha and its E. coli paralogue YdgT interact with StpA. Interaction of Hha/YdgT with StpA prevents proteolytic degradation of this latter protein (39).
Genomic analysis of conjugative plasmids has shown that many of them harbor genes encoding H-NS-like and Hha-like proteins (10, 26). In some instances, copies of both hns and hha are present in the plasmid. In IncM plasmid R446, both hns-like (open reading frame 4 [ORF4]) and hha-like (ORF5) ORFs overlap. The IncHI1 plasmid R27 has been extensively studied in the last years (15, 16, 24), and its complete nucleotide sequence has been obtained and annotated, revealing that it contains two ORFs encoding putative H-NS-like and Hha-like proteins (42) (Fig. 1). IncHI plasmids are low-copy-number conjugative plasmids that encode multiple-antibiotic resistance in S. enterica serovar Typhi (14). Plasmids belonging to the IncHI group have been considered to play a relevant role in the persistence and reemergence of this pathogen (20). Prevalence of these plasmids can be at least partially due to their ability to be horizontally transferred in natural environments. It is remarkable that IncHI plasmids are thermosensitive for transfer (29). R27 transfer follows the same regulatory pattern of other IncHI plasmids, and conjugation frequency is highest at low temperatures (25 to 30°C), decreasing when the temperature increases. Considering the well-described role of both Hha and H-NS proteins in thermoregulation of gene expression in E. coli (27, 36), we decided to investigate the hypothetical participation of R27-encoded Hha-like and H-NS-like proteins in thermoregulation of plasmid conjugation. The present study shows that, in fact, both chromosomal and plasmid hha-like and hns-like alleles can participate in thermomodulation of R27 conjugation. Furthermore, the interplay of functions also affects the host cell physiology: R27-encoded Hha-like and H-NS-like proteins restore the loss of resident Hha and H-NS functions.
FIG. 1.
Comparison of the amino acid sequences of R27 ORF182 and E. coli protein Hha and of R27 ORF164 and E. coli protein H-NS by using the ClustalW 1.81 program of the European Bioinformatics Institute.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids are listed in Table 1. The different strains were grown in Luria-Bertani (LB) medium (10 g/liter NaCl, 10 g/liter tryptone, 5 g/liter yeast extract) or in Penassay broth (1.5 g/liter meat extract, 1.5 g/liter yeast extract, 5 g/liter peptone, 1 g/liter glucose, 3.5 g/liter NaCl, 1.32 g/liter KH2PO4, 4.82 g/liter K2HP4 · 3H2O). The antibiotics used were kanamycin (25 μg/ml), ampicillin (50 μg/ml), chloramphenicol (50 μg/ml), tetracycline (15 μg/ml), and rifampin (50 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| 5K | F−hsdR hsdM thr thi leu lacZ | 23 |
| 5K Rf | 5K Rifr | 37 |
| 5K hns | 5K hns::Ampr | This work |
| Hha3 | 5K hha::Tn5phoA | 17 |
| Hha3 hns | Hha3 hns::Ampr | This work |
| S17-1 λpir | Tpr SmrrecA thi pro hsdR H+ | 31 |
| S614 | CSH50 Δbgl-AC11 ΔlacZ(pro+) hns::Ampr | 8 |
| E. coli R27 | Rifr Tcr | 42 |
| Plasmids | ||
| R27 | IncHI1 Tcr | 18 |
| R27 hha | R27 hha::mini-Tn5Km1 | This work |
| R27 hns | R27 Δhns | This work |
| R27 hha hns | R27 hha::mini-Tn5Km1 Δhns | This work |
| pUTmini-Tn5Km1 | Apr Kmr ori R6K mini-Tn5Km1 | 7 |
| pKD3 | Template plasmid | 5 |
| pKD46 | Red helper plasmid Apr | 5 |
| pCP20 | FLP helper plasmid Apr Cmr | 5 |
| pANN202-312 | hlyC hlyA hlyC hlyD Cmr | 17 |
Genetic manipulations.
5K hns and Hha3 hns strains were obtained by P1 vir-mediated transduction (30) of strains 5K and Hha3, respectively, with the hns::Ampr allele from strain S614.
To construct plasmid R27-hha, we performed mini-Tn5Km1 mutagenesis in order to obtain the insertion of mini-Tn5Km1 (7) into ORF182 of plasmid R27, corresponding to the hha-like ORF. We tested the insertion by PCR amplification with primer pair Tn5-3/R27-5 or Tn5-3/R27-3 (Table 2). In addition, Southern hybridization of genomic DNA isolated from strain 5K R27 hha with a probe corresponding to mini-Tn5Km1 labeled with digoxigenin showed that plasmid R27 hha contains only a mini-Tn5Km1 copy. A PCR analysis using primer pair R27-5/R27-3 and an elongation time that does not permit complete amplification of mini-Tn5Km1 confirmed that cells contained only R27 harboring the mutant hha allele.
TABLE 2.
Oligonucleotides used in this work
| Oligo-nucleotide | Sequence |
|---|---|
| R27-3 | 5′ GGGGAGCTCATCTTCATATTACTGACGG 3′ |
| R27-5 | 5′ GGGGAGCTCATTTAATGAAATTTCGTAAGT 3′ |
| Tn5-3 | 5′ GCTACTTGTGTATAAGAGTCAG 3′ |
| 164P1 | 5′ CAAAGTATTCTATTGATTTTATTTATTACTCTTAGTGAGGGTGTAGGCTGGAGCTGCTTC 3′ |
| 164P2 | 5′ GTGGCTTTTGAACTGAGCTGTTCGCGATTTTAATTTCAACCATATGAATATCCTCCTAGT 3′ |
| 164H1 | 5′ GATTTTATTTATTACTCTTAGTGA 3′ |
| 164H2 | 5′ GAACTGAGCTGTTCGTCGCGATT 3′ |
| PtraH | 5′ TTTTAAAAGTGGCACTATGCGT 3′ |
| traH-5 | 5′ GTCTAAATCGAAGCTATTAAAG 3′ |
| traH-3 | 5′ TCATTGTGCCACCTCTGATT 3′ |
| PtraJ | 5′ CCGACAATCTGGCGCAGT 3′ |
| traJ-5 | 5′ GCGGATAATTCTGCTCGTG 3′ |
| traJ-3 | 5′ GTTTTTGAAAGTTAGACCACA 3′ |
| PtrhR | 5′ TTTCACGAACGGGACTTTTT 3′ |
| trhR-5 | 5′ CAGAATCAGATAACATCACG 3′ |
| trhR-3 | 5′ TCAATAATCCTTCAAGAGCAC 3′ |
| PtrhA | 5′ CGTTTGGCCAGAAAGCATTT 3′ |
| trhA-5 | 5′ GGAACTGACATTGAATACTAA 3′ |
| trhA-3 | 5′ TCACAGAGGAATACCAGCAT 3′ |
| PtrhE | 5′ CGTGAAAATCGGTGTTTAGTA 3′ |
| trhE-5 | 5′ GAAACTTCTCAGCAGGTTAAA 3′ |
| trhE-3 | 5′ AGTTACCTATTGACGGGCGG 3′ |
| 16S-1 | 5′ GCTGCATGGCTGTCGTCAG 3′ |
| 16S-3 | 5′ CGCAGGTTCCCCTACGGTT 3′ |
| HlyS | 5′ CAGACCACACCTGGAAAAAC 3′ |
| HlyE | 5′ GACTTCGCTTGGAATTCGCA 3′ |
| HlyP | 5′ GTCATGCGTGGCGACATTGA 3′ |
| HlyH | 5′ GGGCTTCACTGCGAAATTCA 3′ |
| 5oriT | 5′ CGCGCATTTTTCCCTAAGGT 3′ |
| 123-3 | 5′ AGCATTGACATAAAGAAAAACTTG 3′ |
Plasmid R27 hns was constructed by one-step inactivation using PCR products as previously described (5). We used the sequence of ORF164 (corresponding to the hns-like ORF) to define the corresponding primers. The antibiotic resistance of plasmid pKD3 (Cm) was amplified using primers 164P1 and 164P2 (Table 2), corresponding to sequences P1 and P2 of plasmid pKD3, with homology extensions corresponding to nucleotides 148181 to 148220 and 148669 to148630 of the plasmid R27 sequence (accession no. NC_002305, 41). The PCR product was DpnI digested, purified, and used to electroporate strain 5K(R27) carrying plasmid pKD46 grown at 30°C in the presence of arabinose at 10 mM, conditions that allowed expression of Red recombinase. Recombinants were selected at 37°C in LB medium containing chloramphenicol and then tested for the presence of pKD46. A recombinant clone obtained was named 5K(R27 hns-Cm). Then, the Cmr was eliminated by transforming the mutant strain with plasmid pCP20, a thermosensitive plasmid that encodes the FLP recombinase (temperature inducible). By selection at 43°C in the absence of antibiotics, we obtained strain 5K(R27 hns), carrying a deletion of ORF164 of plasmid R27. As described for ORF182 mutation, we confirmed the deletion by PCR amplification with primer pair 164H1 and 164H2 (Table 2). Plasmid R27 hha hns was obtained by P1 vir-mediated transduction of strain 5K(R27 hns) with the hha::Mini-Tn5Km1 allele from strain 5K(R27 hha).
Mating experiments.
Mating assays were performed as previously described (44), using as a recipient strain E. coli 5K Rf. Briefly, cultures of donor and recipient strains were grown overnight without shaking at different temperatures (25 or 33°C) in Penassay broth. To 1 ml of preincubated Penassay broth were added 0.4 ml of the recipient strain culture and 0.1 ml of the donor strain culture. The mating mixture was incubated at the corresponding temperature, without shaking, for 2 h and then plated in MacConkey agar (Sharlab) supplemented with the corresponding markers. The plates were incubated at 37°C, and the mating frequency was calculated as the number of transconjugants per donor cell.
Total RNA isolation.
To be used in reverse transcription (RT)-PCR assays and microarray experiments, total RNA from different strains was isolated by using an RNeasy mini kit with RNA Protect Bacteria reagent (QIAGEN) according to the manufacturer's instructions. Total RNA was quantified using the GenQuant II RNA/DNA calculator (Amersham Biosciences). When necessary, DNA was eliminated with RQ1 RNase-free DNase (Promega).
RT-PCR.
To quantify the mRNA levels of different genes corresponding to transfer region 1 (Tra1) and Tra2 of R27, we used Ready-to-Go RT-PCR beads (Amersham Biosciences). The primer pairs used are described in Table 2 (traH-5/traH-3, traJ-5/traJ-3, trhE-5/trhE-3, trhR-5/trhR-3, and trhA-5/trhA-3) The RT-PCR was carried out in a Perkin-Elmer Gene Amp 2400 thermal cycle. The RNA was reverse transcribed for 1 h at 42°C. To inactivate the reverse transcriptase, samples were incubated at 95°C for 5 min. The amplification was accomplished by 40 cycles of denaturation for 30 s at 95°C, annealing for 30 s at 54°C, and extension for 30 s at 72°C. The RT-PCR was terminated by a final extension for 10 min at 72°C. The PCR products were analyzed by 2% agarose gel electrophoresis. 16S rRNA was used as the internal control (using primers 16S-1 and 16S-3 [Table 2]). First, saturation curves with increasing amounts of total RNA were performed to determine the interval of lineal increase in the relative amount of RT-PCR product and total RNA (data not shown).
Gel retardation assays.
Gel retardation assays were performed as described previously (27). The DNA fragments used, corresponding to genes traJ, traH, trhR, trhE, and trhA, including the 5′ upstream regions, and the oriT region, were obtained by PCR amplification using primer pairs PtraJ/traJ-3, PtraH/traH-3, PtrhR/trhR-3, PtrhE/trhE-3, PtrhA/trhA-3, and 5oriT/123-3, respectively (Table 2). As a competitor DNA, we used fragments corresponding to the regulatory region of plasmid pHly152, S/E (2,302 bp), S/P (1,167 bp), and S/H (427 bp), that have been shown not to be preferential targets for H-NS (27). These fragments were PCR amplified using primer pairs HlyS/HlyE, HlyS/HlyP, and HlyS/HlyH (Table 2).
Synthesis of specific probes and preparation of glass slides.
The specific probes used in this work for the microarray assay were synthesized by MWG on the basis of the nucleotide sequence of each ORF that we expected to detect (Table 3). The 5′-amino-modified 50-mer oligonucleotides were dissolved in Universal Spotting Solution (Corning) to a final concentration of 40 μM. Then probes were spotted onto UltraGAPII-coated slides with enhanced aminosilane (Corning) by using a QArray (GENETIX) arrayer. We also printed resuspension solutions and human and Arabidopsis thaliana probes as negative controls. Finally, the Microarray ScoreCard reagents (Amersham) were also spotted. These reagents consist of control spotting samples and control mRNA solutions (spike mixes) and are used as follows for validation of microarray experiments: RNA labeling, hybridization uniformity, detection limits, dynamic range, and expression ratios from each hybridized slide. Each slide contains 16 replicates of the probes.
TABLE 3.
Oligonucleotide probes used in this work
| Probe | Sequence (5′-3′) | Nucleotidesa | Gene |
|---|---|---|---|
| R27-01 | TCCGGGCGGCGGCAGGACCTGACTAGGTTATCTGAAAACCTGACCAATCA | 111870-113285 | trhH |
| R27-02 | AAGAACAACTGGCAACACTCGATAATCCGCCGCCGTCAATAGGTAACTAA | Complement (36519-37304) | trhE |
| R27-03 | CAAACGATTAGACTTATTCAAGGAGTGGAGGGACGCAAATGGCTGCGGAT | Complement (100712-101815) | R0118 |
| R27-04 | AATTCCCAGGTTCATTTGATTCCAAACAAATTGTAGTCACTCAATCATTT | 21987-22508 | R0017 |
| R27-05 | GCCATCATACCTCTATTCCATGAGGAGATCGGTAAATGACTCGATTAACG | 110782-110910 | trhX |
| R27-06 | GAATAAAAACGGTATTTACACACAAAACACTGCACCACTTAACCTGGATT | 20208-21155 | trhO |
| R27-07 | TTTGATTACTGGTTAAAACAAAGTGTCGAACCCTTACTCAAAAACCCATT | Complement (32115-32867) | htdT |
| R27-08 | AAGAGCGTCAGACTCGTGTTACTACGCGCCCAAGCCTGATTAAAAGATAA | Complement (35284-36516) | trhK |
| R27-09 | GAATCGATAATTACCGCCCTTCGCCTATCTGAGTCTAAGAGCGGTCAGTG | Complement (16077-16640) | htdK |
| R27-10 | GGTCTAAACCGTCTTGTGGACTACTATACGTCAATAGCTGAGCAGCAGTG | Complement (33494-34852) | trhB |
| R27-11 | CTTCGATATCAGTACCATACAGAAAACGACTATGAGGACTGAAAATGATT | 21155-21994 | R0016 |
| R27-12 | CGTCCACGTCATCGGCAATAGCACGGTATTCCCAGGAAGAGTTAGATTAG | 109710-110222 | trhY |
| R27-13 | GCGGACCCAGTGAACTATCTGAATGTGGGTGGAAGTTTAGGAGGTGCATA | 110834-111880 | trhF |
| R27-14 | AAGCTGAAATACGTGGTATTGGAACAAAAATCGCGGCAGAGGAGAATTGA | Complement (9378-10886) | trhW |
| R27-15 | ATTGGGAATTGCACTCAACCGTTTGTGGTCTAACTTTCAAAAACTTATGT | Complement (100063-100725) | traJ |
| R27-16 | GTGAGTATGGTCAGAAAGGATTCCCGGAAAAAACTCTGAAATCAAAATAA | 4082-4954 | R0004 |
| R27-17 | TTAGTAGTCGTTACTTTACCGATCCATTTATCAGAAACCTCTATTCTTGA | Complement (37316-37633) | trhL |
| R27-18 | AACGCTTAAAGGTGGACGGAAAAAAATCGGCATAGATAACTATGTTTACA | Complement (8181-9128) | trhU |
| R27-19 | ATATGCAACAGCAACAGCGAGCAACGAAAGCAGTTTCGTCAAACGGTATT | Complement (31155-32105) | trhV |
| R27-20 | ACGCTCAGGAAAACTTTTACAGGCCGATACTGGAGTACAGAAGTGCCTCA | Complement (101815-103899) | traG |
| R27-21 | TTTTCGGCAGCACTGCGACACTGTCCAAAAACCTTAATCGTGGGAAGCAA | Complement (11659-15879) | R0009 |
| R27-22 | TAATGGAATTACTGCGTTACGACACTATGTTTACATGCCACCGTCAGGAT | 108907-109707 | trhR |
| R27-23 | AAGGATAATGAAGTTATGTCCGGTACATGGTTCTTTAAGAAGAGAGTATG | Complement (16637-17071) | htdF |
| R27-24 | CCCCGTTAAACGTGACCAATGACAGTAATGCAGCAGCGGTTCATATAGAT | 22505-22897 | trhG |
| R27-25 | TGAAAGTTAAGCGATTCCAGCAACCAAAGAATGGAGTTAATAATGGACAT | Complement (34842-35282) | R0030 |
| R27-26 | CGGCGAAGCAATTGAGATTCTGTACGAACAGGAAGTTGCGAGCAAATCAG | Complement (28465-31146) | trhC |
| R27-27 | TGAATAATACAAATGAACCATATCAGTCGAGAATTGAGAAAGCTAAAAGC | 113294-117283 | trhG |
| R27-28 | GTGAGGTACAATCTCTTCCAGGCGAACCTAAAGAAAGTGAGATTCGATAA | Complement (32973-33485) | R0028 |
| R27-29 | CTGACGCAGTCATGTATTGAAAAGTACGGAACGCAGCTAAATGCGCCTGA | Complement (103899-106934) | traI |
| R27-30 | AACTGGGACGAAAAGTCCGTCAATACTCAAAATCAGAGGTGGCACAATGA | Complement (107664-108149) | traH |
| R27-31 | TTGGGGGGATCACCGCCGACAATTTGCTGTATCAACTCAGGCTTATGTGC | Complement (17061-17513) | htdA |
| R27-32 | GGTCAACTTATGTAGATCCAATTACAGGTAAGGAAATACCAAAATATTAA | Complement (4982-8158) | trhN |
| R27-33 | GTGAAATACGGCATACCTGCCGAGACCATTGAAAGAGAATTAACTATTCC | Complement (10873-11385) | trhP |
| R27-34 | AATTGAAAGAGGCTGCGAAAAGTAGAACAGAATCCCCTTTGACAGAATAA | Complement (2116-3900) | trhI |
| R27-35 | ATGCTGGTTATGGCAAACGGTGAAAAAATTATCAGCTCGTTCCTGGATGC | Complement (37685-38038) | trhA |
| zwf | AATCTCGATGTCAATGACACTGCTGCATTCAGCCGTCTCGGCGCGATGCT | Complement (1932863-1934338)b | zwf |
| rRNA16S | TGTGATTCATGACTGGGGTGAAGTCGTAACAAGGTAACCGTAGGGGAACC | 223771-225312b | rrsH |
Printed microarrays were placed in a dissector for up 48 h. Then spotted oligonucleotides were immobilized by applying 600 mJ of UV energy to the printed surface and stored in a dry environment at room temperature until use.
mRNA labeling.
Relative mRNA levels were determined by parallel two-color hybridization to oligonucleotide microarrays prepared as described above. Labeling of mRNAs was done by using the CyScribe Post-Labeling kit (Amersham Biosciences) according to the manufacturer's manual, with some changes. To validate and normalize experimental data in microarrays, in each labeling reaction mixture we used 10 μg of total RNA plus 1 μl of Spike Mix (Lucidea Universal ScoreCard; Amersham Biosciences). We used specific primers to label the ORFs to be detected (25 pM each) (Table 4) and oligo(dT) to label the controls used (as indicated by the supplier).
TABLE 4.
Specific oligonucleotides used in this work for labeling of mRNAs corresponding to ORFs of plasmid R27 and controls
| Region and oligonucleotide | Sequence (5′-3′) |
|---|---|
| Tra 1 | |
| TraJ | ACATAAGTTTTTGAAAGTTA |
| R0118 | TTATCCGCAGCCATTTG |
| TraG | TGAGGCACTTCTGTACT |
| TraI | AGGCGCATTTAGCTGC |
| TraH | TCATTGTGCCACCTCTG |
| TrhR | ATCCTGACGGTGGCAT |
| TrhY | CTAATCTAACTCTTCCTG |
| TrhX | CGTTAATCGAGTCATTTAC |
| TrhF | TATGCACCTCCTAAACTT |
| TrhH | TGATTGGTCAGGTTTTCA |
| TrhG | TCATTCAGCCAGCTTTTA |
| Tra 2 | |
| TrhI | TTATTCTGTCAAAGGGGAT |
| R0004 | TTATTTTGATTTCAGAGTTTT |
| TrhN | ATATTTTGGTATTTCCTTAC |
| TrhU | TGTAAACATAGTTATCTATG |
| TrhW | CAATTCTCCTCTGCCG |
| TrhF | AATAGTTAATTCTCTTTCAAT |
| R0009 | TTTTGCTTCCCACGATT |
| HtdK | CACTGACCGCTCTTAG |
| HtdF | CATACTCTCTTCTTAAAGA |
| HtdA | GCACATAAGCCTGAGTT |
| TrhO | AATCCAGGTTAAGTGGT |
| R0016 | AATCATTTTCAGTCCTCAT |
| R0017 | AAATGATTGAGTGACTACA |
| TrhZ | ATCTATATGAACCGCTG |
| TrhC | CTGATTTGCTCGCAACT |
| TrhV | AATACCGTTTGACGAAAC |
| HtdT | AATGGGTTTTTGAGTAAG |
| R0028 | TCAATTATCGAATCTCACT |
| TrhB | CACTGCTGCTCAGCTA |
| R0030 | ATGTCCATTATTAACTCCA |
| TrhK | TTATCTTTTAATCAGGCTT |
| TrhE | TTAGTTACCTATTGACGG |
| TrhL | CCATCAAGAATAGAGGTT |
| TrhA | GCATCCAGGAACGAG |
| Positive controls | |
| rrsH | GGTTCCCCTACGGTA |
| zwf | ACTCAGCATCGCGCC |
Hybridization.
Prior to hybridization, microarrays were immersed in a solution containing 150 ml of 1-methyl-2-pyrrolidinone, 3 g of succinic anhydride, and 17 ml of 0.2 M sodium borate. Then slides were prehybridized at 50°C for 20 min in a buffer containing 1% bovine serum albumin, 3.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and 0.1% sodium dodecyl sulfate (SDS).
Labeled cDNAs were combined with hybridization buffer (50% formamide, 2.5× Denhardt's solution, 6× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA at pH 7.7], 0.5% SDS).
Hybridization and washes were performed using the automatic system Lucidea SlidePro (Amersham). The hybridization was allowed to proceed for 15 h at 42°C. The arrays were then sequentially washed with medium-stringency buffer (2× SCC, 0.1% SDS), high-stringency buffer (0.1× SCC, 0.1%SDS), and postwash buffer (0.1× SSC) and air dried.
Each microarray was scanned using the Axon4000B scanner, and image quantification was done with GenepixPro software.
Data analysis.
Data obtained from the scanner were preprocessed by doing first a background correction and then an intensity-dependent normalization based on control spots.
The analysis was performed using a two-factor gene-to-gene linear model approach (43). Genes were selected as being differentially expressed if several criteria such as t statistic, B statistic, and n-fold change ratio indicated so. The analysis was performed using the open-source software limma (http://bioinf.wehi.edu.au/limma/).
Measurement of hemolysin production.
Hemolysin in the culture supernatants was assayed by measuring the hemolytic activity as previously described (33).
Protein purification.
Hha and H-NS proteins from E. coli were purified as described previously (35, 36).
RESULTS
Both the chromosome- and plasmid-encoded hns and hha alleles affect thermoregulation of R27 transfer.
To assess the role of the R27 hns and hha genes in plasmid transfer, we obtained R27 derivatives harboring hns, hha, or hns hha mutations. E. coli strain 5K harboring either wild-type (WT) R27 or its mutant derivatives was used as the donor in mating assays performed at either 25°C or 33°C (Table 5). Transfer results showed that independent R27 hha and R27 hns mutations slightly derepressed conjugation at 33°C. When both mutant alleles were combined in the same plasmid DNA molecule, the conjugation frequency at 33oC was further derepressed. Attending the high homology shown between the R27 plasmid-encoded hha and hns genes and the chromosome-encoded alleles, we hypothesized that the effect of a double knockout of the plasmid hns and hha alleles would be at least partially masked by the presence of the resident chromosomal hha and hns genes. We therefore decided to transfer both WT R27 and its hha, hns, and hns hha derivatives to strains Hha3, 5K hns, and Hha3 hns. Transconjugants were then used as donors in mating experiments carried out at both 25 and 33°C (Table 5). When WT R27 was used as the donor, independent chromosomal hha and hns alleles slightly increased the conjugation frequency at the nonpermissive temperature. Again, when both chromosomal mutations were combined, a moderate increase in the conjugation frequency was apparent. We then tested R27 conjugation in a double plasmid and chromosomal hha hns background. When strain Hha3 hns harboring R27 hha hns was used as the donor, a very significant increase in the conjugation frequency at 33°C was obtained. In fact, when grown at 33°C, strain Hha3 hns(R27 hha hns) transferred its plasmid at a frequency higher than strain 5K(R27) grown at 25°C. It is thus apparent that both the Hha and H-NS proteins, irrespective of their chromosomal or plasmid origin, play a very relevant role in thermoregulation of R27 conjugation. A further increase in temperature to 37°C resulted in a decrease in the conjugation frequency. In fact, conjugation could only be obtained when strain Hha3 hns(R27 hha hns) was used as the donor. The conjugation frequency was 5.5 × 10−6.
TABLE 5.
Effects of hns and hha mutations on temperature-dependent conjugation of R27
| Donor strain | Conjugation frequency at:
|
|
|---|---|---|
| 25°C | 33°C | |
| 5K(R27) | 2.6 × 10−3 | <10−9 |
| 5K(R27 hha) | 3.6 × 10−3 | 1.1 × 10−7 |
| 5K(R27 hns) | 2.8 × 10−3 | 0.8 × 10−7 |
| 5K(R27 hha hns) | 3.0 × 10−3 | 2.7 × 10−6 |
| Hha3(R27) | 1.5 × 10−4 | 5.2 × 10−6 |
| 5K hns(R27) | 2.0 × 10−2 | 2.5 × 10−5 |
| Hha3 hns(R27) | 4.0 × 10−2 | 5.0 × 10−5 |
| Hha3 hns(R27 hha hns) | 1 | 1.0 × 10−2 |
Transcription of several genes encoded in R27 Tra1 and Tra2 is enhanced in an hns hha genetic background.
The transfer genes of plasmid R27 are contained within two separate regions, termed Tra1 and Tra2 (41, 42). Tra1 contains the origin of transfer, a coupling protein, and nine essential genes encoding Mpf and relaxosome proteins (24). Among other ORFs, Tra2 contains 11 Mpf proteins that are essential for R27 transfer (25). We decided to test if the Hha and H-NS proteins influence the transcription of genes in both Tra1 and Tra2. The DNA microarray procedure was used for that purpose. A microarray including all ORFs from Tra1 and Tra2 was designed (see Materials and Methods for details), and RNA isolated from cultures of strains 5K(R27) and Hha3 hns(R27 hha hns) grown both at 25 and 33°C was hybridized. The results of the transcriptional studies are shown in Table 6. Significant differences in expression could be determined for several transfer determinants at 33°C (Table 6). With respect to Tra1, upregulation in hns hha cells was observed for six ORFs, encoding Mpf proteins ThrR, ThrY, and TrhF; the putative coupling protein TraG; the putative transcriptional regulator TraJ; and the putative relaxosome protein TraH. With respect to Tra2, upregulation was apparent for 11 ORFs. Four of them (trhV, trhK, trhE, and trhB) code for Mpf proteins. Other ORFs are the pilin determinant trhA and the putative transfer regulator trhZ. Expression of the rest of the ORFs from both Tra1 and Tra2 could be detected, but expression in WT and hha hns cells was not statistically significantly different. At 25°C, upregulation of different transfer determinants was also apparent. For Tra1, in addition to ORFs upregulated at 33°C, two other ORFs, encoding the coupling protein TrhH and the putative relaxase TraI, were also upregulated. For Tra2, in addition to the ORFs upregulated at 33°C, statistically significant upregulation was also evidenced for the ORFs encoding Mpf proteins TrhU, TrhP, and TrhC and the putative transfer repressor HtdA. The other two ORFs upregulated encode proteins not directly related to the conjugation process.
TABLE 6.
Effect of an hha hns genetic background on transcription of ORFs belonging to R27 Tra1 and Tra2a
| Tra 1 | Ratio at 33°C | Tra 2 | Ratio at 33°C | Tra 1 | Ratio at 25°C | Tra 2 | Ratio at 25°C |
|---|---|---|---|---|---|---|---|
| trhR | 13.8 | trhV | 6.9 | traH | 7.3 | trhA | 21.6 |
| trhY | 11.5 | trhK | 4.5 | trhF | 5.1 | trhE | 9.3 |
| trhF | 1.9 | trhE | 4.3 | traG | 3.7 | trhV | 8.9 |
| traG | 1.9 | trhA | 3.7 | trhY | 3.3 | trhK | 7.6 |
| traJ | 1.7 | htdT* | 3.1 | trhR | 3.0 | R0028* | 6.2 |
| traH | 1.5 | R0009* | 3.0 | traJ | 2.5 | htdT* | 5.0 |
| trhX* | 1.5 | trhI* | 2.6 | trhH | 2.1 | R0009* | 4.8 |
| trhH | 1.3 | R0028* | 2.6 | traI | 2.1 | trhI* | 4.5 |
| trhG | 1.3 | trhB | 2.4 | trhG | 1.4 | R0004* | 4.5 |
| traI | 1.2 | trhZ | 2.2 | R0118* | 1.4 | htdA | 4.2 |
| R0118* | 1.1 | htdK* | 2.0 | trhX* | 1.2 | trhP | 4.0 |
| trhU | 1.9 | trhC | 3.8 | ||||
| trhN | 1.9 | trhB | 3.8 | ||||
| R0004* | 1.8 | trhZ | 3.0 | ||||
| htdA | 1.7 | trhU | 2.8 | ||||
| trhP | 1.7 | R0030* | 2.7 | ||||
| trhC | 1.6 | htdK* | 2.4 | ||||
| trhO | 1.5 | trhN | 1.3 | ||||
| R0030* | 1.3 | trhO | 1.2 | ||||
| htdF* | 1.2 | R0016* | 1.1 | ||||
| R0017* | 1.2 | trhW | 1.1 | ||||
| R0016* | 1.1 | trhL | 1.0 | ||||
| trhL | 0.9 | htdF* | 1.0 | ||||
| trhW | 0.8 | R0017* | 0.8 |
We decided to confirm these microarray data by performing RT-PCR transcriptional analysis of some of the genes exhibiting significant transcriptional differences in the array analysis (traH, traJ, and trhR from Tra1 and trhA and trhE from Tra1). Again, depletion of chromosome- and plasmid-encoded Hha and H-NS proteins increased transcription of the genes assayed (Fig. 2A). When cells were grown at 25°C, significant differences in transcription could be evidenced. At the nonpermissive temperature (33°C), for most of the genes tested, transcription was not apparent in WT cells but were significant in hha hns cells.
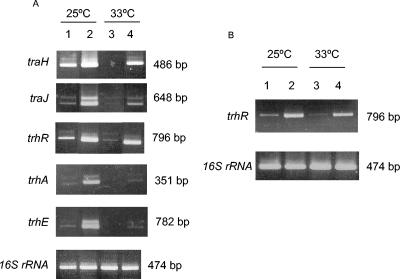
FIG. 2.
(A) RT-PCR analysis of transcription of the traH, traJ, and trhR (Tra1) genes and the trhA and trhE (Tra2) genes from plasmid R27 in WT and hha hns backgrounds. Cells were grown at 25°C (lanes 1 and 2) or 33°C (lanes 3 and 4). Lanes 1 and 3, strain 5K(R27); lanes 2 and 4, strain Hha3-hns(R27 hha hns). (B) RT-PCR analysis of transcription of trhR in cells lacking Hha-like proteins. Cells were grown at 25°C (lanes 1 and 2) and at 33°C (lanes 3 and 4). Lanes 1 and 3, strain 5K(R27); lanes 2 and 4, strain Hha3 ydgT(R27 hha). RT-PCR of 16S rRNA was used as a control to confirm equivalent quantities of template loading.
We also used the RT-PCR approach to evaluate transcription of tra genes in cells lacking Hha-like proteins. It has been recently shown that the Hha paralogue YdgT can, at least partially, compensate for the loss of Hha in E. coli (39). Transcription of tra genes was therefore assessed in strain Hha3 ydgT(R27 hha) at 25 and 33°C. When considering trhR, it is apparent that depletion of Hha-like proteins results in a significant increase in transcription (Fig. 2B). Similar results were obtained for trhE, traH, traJ, and trhA (data not shown).
H-NS binds preferentially to the oriT region and to DNA fragments including putative regulatory regions of different tra genes.
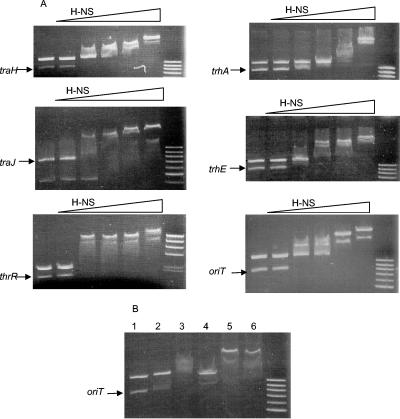
We also tested the ability of H-NS to specifically bind to different sequences of both Tra1 and Tra2. Considering that the fine details of the transcriptional organization of both Tra1 and Tra2 are not known, we used as targets DNA fragments corresponding to the traJ, traH, trhR, trhE, and trhA genes, including 5′ upstream sequences. Moreover, we also tested binding of H-NS to the oriT region. Competitive electrophoretic band shift assays were performed with protein-DNA mixtures containing as target DNA the corresponding R27 sequence and, as a nonspecific target, a DNA fragment corresponding to the regulatory region of the hemolysin operon of plasmid pHly152. These fragments have been shown not to be preferential targets for H-NS (27). Results obtained (Fig. 3A) showed (i) preferential binding of H-NS to the oriT region; (ii) preferential binding to fragments corresponding to traH, trhA, traJ, and trhE; and (iii) nonspecific binding to the trhR fragment.
FIG. 3.
(A) Effect of increasing amounts of H-NS protein (0.1625, 0.325, 0.4875, 0.65, and 1.3 μg) on the electrophoretic mobility of DNA fragments corresponding to the promoter regions of the traH, traJ, and trhR (Tra1) genes, the trhA and trhE (Tra2) genes, or the oriT region. Arrows point to the R27 DNA fragment used in each test. (B) Effect of H-NS protein plus Hha protein on the electrophoretic mobility of a DNA fragment corresponding to the oriT region. Lane 1, no protein added; lane 2, Hha protein added (48.44 μg); lane 3, Hha protein (48.44 μg) and H-NS protein (0.325 μg) added; lane 4, H-NS protein (0.325 μg) added; lane 5, Hha (48.44 μg) and H-NS (0.65 μg) proteins added; lane 6, H-NS protein (0.65 μg) added. As nonspecific competitor DNA, an S/E (in thrR assay), S/P (in traH, trhA, trhE, and oriT assays), or S/H (in traJ assay) fragment corresponding to the upstream regulatory region of the hly operon of plasmid pHly152 was used (see text for details). The DNA standard used was a 100-bp ladder (Biotools) in all cases, except for the thrR assay, where λ/HindIII (Biotools) was used.
We also tested how the presence of Hha would affect the formation of low-migration protein-DNA complexes with H-NS and the oriT region. At low H-NS concentrations, the presence of Hha allowed the formation of low-migration protein-DNA complexes different from those generated by H-NS or Hha alone (Fig. 3B). As could be shown for the hemolysin operon (36), the presence of Hha in H-NS/DNA mixtures appears to contribute to the generation of higher-order oligomers.
R27-encoded H-NS-like and Hha-like proteins can play a role as molecular backups for host-encoded H-NS and Hha proteins.
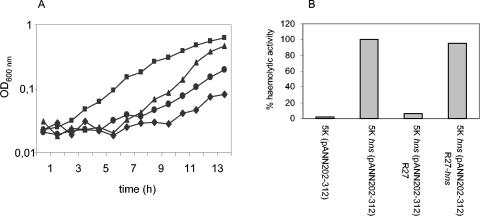
Being aware of the interplay between chromosome- and plasmid-encoded Hha and H-NS proteins to modulate transcriptional control of Tra1 and Tra2 from R27, we decided to test if plasmid-encoded Hha and H-NS proteins could as well participate in modulating chromosome-encoded host functions. For that purpose we decided to test complementation of independent chromosomal hns and hha mutations by WT R27. A global physiological perturbation in E. coli caused by the hns mutation is the impairment of growth at low temperature (25°C) (9). Therefore, we investigated the effect of the R27 plasmid on the thermosensitivity of an hns strain. We compared the growth of strains 5K, 5K hns, 5K hns(R27), and 5K hns(R27 hns) in LB medium at 25°C (Fig. 4A). The severe influence of the hns mutation on the growth rate of strain 5K growing at low temperature was compensated when plasmid R27 was present. Inactivation of the R27 hns gene resulted in cold-sensitive growth. We also tested the ability of R27-encoded H-NS protein to modify another hns-associated phenotype, overexpression of the E. coli hemolysin operon. Hemolysin production was evaluated in E. coli strains 5K(pANN202-312), 5K hns(pANN202-312), 5K hns(pANN202-312 R27), and 5K hns(pANN202-312 R27hns). As expected, the plasmidic hns+ allele restored regulation of the hly genes in strain 5K hns (Fig. 4B).
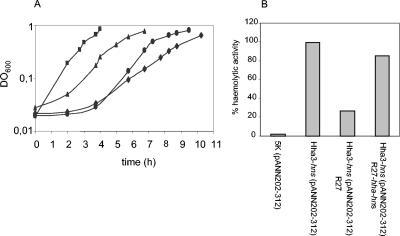
FIG. 4.
Plasmid R27 compensates for some hns-induced phenotypes. (A) Growth at low temperature of strains 5K (squares), 5K hns (circles), 5K hns(R27) (triangles), and 5K hns(R27 hns) (diamonds). (B) Hemolytic activity in culture supernatants of the mutant and WT strains transformed with hemolytic plasmid pANN202-312. The hemolytic activity of strain 5K hns(pANN202-312), referred to as 100%, was 1,327 U. OD600 nm, optical density at 600 nm.
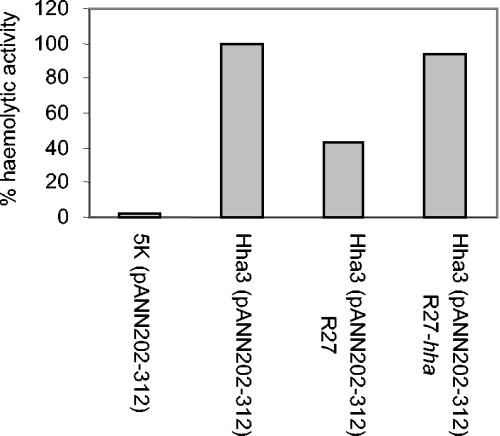
We also assessed if the R27 hha+ allele compensated for the hha mutation. Again, we used as a model the expression of the hly genes. Hemolysin production was evaluated in strains 5K(pANN202-312), Hha3(pANN202-312), Hha3(pANN202-312 R27), and Hha3(pANN202-312 R27hha). As expected, acquisition of WT plasmid R27 by strain Hha3 significantly reduced hemolysin expression (Fig. 5). This effect was not obtained when plasmid R27 hha was used.
FIG. 5.
Plasmid R27 compensates for the deregulation of hemolysin production that hha mutants show. Hemolytic activity in culture supernatants of strains 5K, Hha3, Hha3(R27), and Hha3(R27 hha) transformed with hemolytic plasmid pANN202-312. The hemolytic activity of strain Hha3(pANN202-312), referred to as 100%, was 436 U.
Finally, R27 was tested for complementation of both hha and hns mutations. Compared to WT E. coli cells, hha hns cells harboring plasmid pANN202-312 suffer a dramatic increase in hemolysin production (36). We therefore tested acquisition of plasmid R27 by hha hns cells to restore hemolysin production and growth restriction. With respect to growth in LB medium, R27 significantly enhanced the growth rate of strain Hha3 hns(pANN202-312), although not to the same levels of strain 5K(pANN202-312) (Fig. 6A). With respect to hemolysin production, plasmid R27 accounted for a fourfold reduction in hemolysin production (Fig. 6B). Again, repression of hemolysin production was not obtained when plasmid R27 hha hns was used.
FIG. 6.
Plasmid R27 compensates for hha-hns-induced deregulation of hemolysin expression (B) and for alterations of the growth rate at 37°C (A). The strains tested were 5K(pANN202-312) (squares), Hha3 hns(pANN202-312) (circles), Hha3 hns(pANN202-312 R27) (triangles), and Hha3 hns(pANN202-312 R27 hha hns) (diamonds). Hemolytic activity of strain Hha3 hns(pANN202-312), referred to as 100%, was 7,630 U. OD600, optical density at 600 nm.
DISCUSSION
Thermoregulation of plasmid transfer is a feature common to IncHI plasmids (29). In this study, we used plasmid R27 as a model to analyze the mechanisms underlying temperature-dependent IncH plasmid transfer. We took advantage of the fact that the availability of its complete DNA sequence had led to the identification of two ORFs that encode, respectively, Hha-like and H-NS-like proteins. The genetic analysis initially performed in this work by obtaining plasmid derivatives of R27 harboring deletions in either hha-like, hns-like, or both genes and testing how these mutant alleles influence thermoregulation of plasmid transfer suggested that both proteins participate in the thermoregulation process. Similar results were obtained by assessing the effect of temperature on WT R27 transfer with donor cells harboring chromosomal hha and hns mutations. Nevertheless, the actual network modulating R27 temperature-dependent transfer could be best evidenced when a genetic background with no functional copies of genes encoding Hha-like or H-NS-like proteins was used. The possibility that either chromosome- and R27-encoded H-NS proteins (or Hha proteins) may play a role in thermoregulation of R27 transfer is consistent with the previously reported results for the Sfh protein encoded by the IncHI plasmid in S. flexneri 2a strain 2457T; this H-NS-like plasmid-encoded protein interacts with host-encoded H-NS and StpA, generating heterodimers. Cross-regulation is also apparent (2).
The role of Hha/H-NS in the transcription of R27 Tra1 and Tra2 ORFs was assessed initially by designing a gene array including all ORFs from both regions. For those genes showing differential expression either at 25°C or at 33°C, upregulation in an hha hns genetic background was the rule. The influence of the hha hns genetic background on transcription of tra genes at 33°C appears more dramatic when the RT-PCR approach is used. Compared to that of the hha hns double mutant, transcription of tra genes at 25°C is significantly reduced in WT cells. At high temperature (33°C), the increase in the transcription of trhA, trhE, and traH is more dramatic (Fig. 2). Altogether, it is apparent that the Hha and H-NS proteins downregulate tra genes at both 25 and 33°C. Deregulation of R27 transfer genes in hha hns mutants at 25°C is not surprising. In fact, in both hns and hha mutants, deregulation is in some instances apparent under permissive conditions as well (1, 36). This probably reflects the fact that in some instances H-NS and Hha molecules partially repress gene expression under permissive conditions (i.e., hemolysin at high temperature) because permissive conditions in the laboratory probably are not equivalent to fully permissive conditions in a natural environment. Altogether, it is apparent that the array analysis showed a global transcriptional effect of the hha hns alleles and that effects on specific genes require additional approaches to better evaluate transcription. The detailed regulatory cascade by which H-NS and Hha influence transcription of the different genes must await the characterization of the precise transcriptional organization of both Tra1 and Tra2, which is not known yet in detail.
Band shift assays suggest interaction of the Hha-H-NS complex at different sites of both Tra1 and Tra2. The facts that many of the genes exhibiting Hha-H-NS-dependent expression belong to different transcriptional units and that H-NS shows preferential binding to different sequences suggests multiple DNA-protein interactions along both Tra1 and Tra2. It is remarkable that one of the sequences to which H-NS preferentially binds is oriT. Altogether, our results suggest that, in addition to interacting with different promoter regions corresponding to transcriptional units from both Tra1 and Tra2, Hha and H-NS also might interact with oriT sequences. We have previously shown that members of the Hha family of proteins bind with low affinity to DNA but with high affinity to members of the H-NS family of proteins (35, 36, 37). We also showed that, for the hemolysin operon, addition of Hha to H-NS-DNA mixtures did not modify the affinity of H-NS for its target sequences. Instead, higher-order heterooligomeric complexes were formed (35). By assaying in this work binding of H-NS/Hha to the R27oriT region, similar results were obtained: the presence of Hha facilitates the generation of higher-order heterooligomers that interact with DNA.
It is remarkable that strain 5K hha hns(R27 hha hns) transfers its plasmid at high efficiency at 33°C but shows a reduced conjugation frequency at 37°C. It is thus apparent that, in addition to influencing Hha- and H-NS-mediated transcriptional repression of transfer genes, temperature must also affect other aspects of the mating process. Whereas it has been shown that H pili synthesized at 27°C remain morphologically stable at 37°C (28), it has been also reported that a short shift (1 min) to 37°C of a mating culture significantly reduces the R27 transfer frequency (42). The mechanisms underlying tight temperature-dependent regulation of R27 transfer include N-NS-Hha-mediated repression of the expression of several tra genes and, most likely, structural alterations of one or more proteins of the conjugation system.
Participation of H-NS (with or without Hha) in thermoregulation processes has usually been correlated with downregulation of gene expression at low temperature. A relationship among temperature, DNA flexibility, and the ability of H-NS to bind to promoter sequences at low temperature (about 25°C) and hence repress transcription was proposed by Falconi et al. (13). Such a model was supported by our observations concerning thermoregulation of the hemolysin operon by H-NS and Hha (27). The results we present here show that these proteins are also able to repress transcription at high temperature. Recently, it has been shown that H-NS-mediated repression of the rovA and inv genes of Yersinia pseudotuberculosis occurs at 37°C and induction occurs at 28°C (19). It is thus relevant to gain insight into the structure and properties of the different N-NS-DNA (or N-NS-Hha-DNA) complexes that account for transcriptional repression in different promoters under apparently opposite physical stimuli (high and low temperatures).
Modulation of plasmid-borne genes by H-NS is a well-documented fact (for a review, see reference 10). On the other hand, participation of nucleoid-associated proteins in the regulation of plasmid conjugation is currently being investigated. The role of host-encoded H-NS in silencing F transfer gene expression when cells enter the stationary phase has been recently reported (46). It is thus reasonable to expect that, in many instances, either the host-encoded nucleoid-associated proteins, those encoded by plasmids, or both may play relevant roles modulating, among other functions, those related to plasmid transfer. In addition, as happens in the case of plasmid R27, plasmid-encoded nucleoid-associated proteins may participate in the modulation of chromosomal genes. Here we report that plasmid-encoded Hha- and H-NS-like proteins had the ability to cross talk with chromosomal genes. Recently, it was shown that Shigella flexneri strain 2a harbors an IncHI plasmid that encodes an H-NS like protein (Sfh), which was found to interact and generate heterodimers with both the chromosome-encoded H-NS and StpA proteins. The plasmid-encoded sfh gene can compensate for hns null mutations (2). It has been also shown that, when S. flexneri cells enter the stationary phase, H-NS intracellular levels increase and Sfh levels decrease (6). This suggests a different regulatory pattern for H-NS and Sfh. Regulation of R27-encoded hns and hha genes remains to be elucidated. If, as happens in S. flexneri 2a strain 24575, regulation of the expression of both determinants could be different from that of its corresponding chromosomal alleles, acquisition of plasmid R27 might result in alterations of some of the host cell central regulatory networks. The presence of additional copies of the H-NS/Hha system may eventually modify the regulatory responses of the bacterial cell. It is tempting to speculate that, in addition to different well-characterized virulence functions, many conjugative plasmids provide its hosts with hitherto cryptic functions (i.e., genes encoding global modulators such as Hha/H-NS) that may account for subtle changes in some of the central regulatory networks, thus modifying the efficiency of the bacterial cell to adapt to or grow in complex environments, such as those that pathogens face throughout the invasion process.
Acknowledgments
This work was supported by grants from the Ministerio de Ciencia y Tecnología (BMC2001 3499), from the Generalitat de Catalunya (2001 SGR 00100), and from the Fondo de Investigación Sanitaria (FIS G03/25 COLIRED-O157). Núria Forns was the recipient of a FI grant from the Generalitat de Catalunya.
We acknowledge Diane Taylor for providing E. coli strain R27. We thank Lidia Sevilla (Transcriptomics Plataform, SCT-PCB, University of Barcelona) for technical assistance in microarray experiments and Alex Sánchez for help in data analysis.
REFERENCES
- 1.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7-17. [DOI] [PubMed] [Google Scholar]
- 2.Beloin, C., P. Deighan, M. Doyle, and C. J. Dorman. 2003. Shigella flexneri 2a strain 2457T expresses three members of the H-NS-like protein family: characterization of the Sfh protein. Mol. Gen. Genomics 270:66-77. [DOI] [PubMed] [Google Scholar]
- 3.Bloch, V., Y. Yang, E. Margeat, A. Chavanieu, M. T. Augé, B. Robert, S. Arold, S. Rimsky, and M. Kochoyan. 2003. The H-NS dimerization domain defines a new fold contributing to DNA recognition. Nat. Struct. Biol. 10:212-218. [DOI] [PubMed] [Google Scholar]
- 4.Cornelis, G. R., C. Sluiters, I. Delor, D. Gelb, K. Kaninga, C. Lambert de Rouvroit, M.-P. Sory, J.-C. Vanooteghem, and T. Michiels. 1991. ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol. Microbiol. 5:1023-1034. [DOI] [PubMed] [Google Scholar]
- 5.Datsenko, K. A., and L. B. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deighan, P., C. Beloin, and C. J. Dorman. 2003. Three-way interactions among the Sfh, StpA and H-NS nucleoid-associated proteins of Shigella flexneri 2a strain 2457T. Mol. Microbiol. 48:1401-1416. [DOI] [PubMed] [Google Scholar]
- 7.De Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dersch, P., K. Schmidt, and E. Bremer. 1993. Synthesis of the Escherichia coli K-12 nucleoid-associated DNA-binding protein H-NS is subjected to growth-phase control and autoregulation. Mol. Microbiol. 8:875-889. [DOI] [PubMed] [Google Scholar]
- 9.Dersch, P., S. Kneip, and E. Bremer. 1994. The nucleoid-associated DNA-binding protein H-NS is required for the efficient adaptation of Escherichia coli K12 to a cold environment. Mol. Gen. Genet. 245:255-259. [DOI] [PubMed] [Google Scholar]
- 10.Dorman, C. J. 2004. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2:391-400. [DOI] [PubMed] [Google Scholar]
- 11.Dorman, C. J., and P. Deighan. 2003. Regulation of gene expression by histone-like proteins in bacteria. Curr. Opin. Genet. Dev. 13:179-184. [DOI] [PubMed] [Google Scholar]
- 12.Esposito, D., A. Petrovic, R. Harris, S. Ono, J. F. Eccleston, A. Mbabaali, I. Haq, C. F. Higgins, J. C. D. Hinton, R. C. Driscoll, and J. E. Ladbury. 2002. H-NS oligomerization domain structure reveals the mechanism for high order self-association of the intact protein. J. Mol. Biol. 324:841-850. [DOI] [PubMed] [Google Scholar]
- 13.Falconi, M., B. Colonna, G. Prosseda, G. Micheli, and C. O. Gualerzi. 1998. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of VirF promoter to transcriptional repressor H-NS. EMBO J. 17:7033-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fica, A., M. E. Fernandez-Beros, L. Aron-Hott, A. Rivas, D′Ottone, K., J. Chumpitaz, J. M. Guevara, M. Rodríguez, and F. Cabello. 1997. Antibiotic-resistant Salmonella typhi from two outbreaks: few ribotypes and IS200 types harbour IncHI1 plasmids. Microb. Drug Resist. 3:339-343. [DOI] [PubMed] [Google Scholar]
- 15.Gilmour, M. W., J. E. Gunton, T. E. Lawley, and D. E. Taylor. 2003. Interaction between the IncHI1 plasmid R27 coupling protein and type IV secretion system: TraG associates with the coiled-coil mating pair formation protein TrhB. Mol. Microbiol. 49:105-116. [DOI] [PubMed] [Google Scholar]
- 16.Gilmour, M. W., T. D. Lawley, M. M. Rooker, P. J. Newnham, and D. E. Taylor. 2001. Cellular location and temperature-dependent assembly of IncHI1 plasmid R27-encoded TrhC-associated conjugative transfer protein complexes. Mol. Microbiol. 42:705-715. [DOI] [PubMed] [Google Scholar]
- 17.Godessart, N., F. J. Muñoa, M. Regué, and A. Juárez. 1988. Chromosomal mutations that increase the production of a plasmid-encoded hemolysin in Escherichia coli. J. Gen. Microbiol. 134:2779-2787. [DOI] [PubMed] [Google Scholar]
- 18.Grindley, N. D. F., J. N. Grindley, and E. S. Anderson. 1972. R factor compatibility groups. Mol. Gen. Genet. 119:287-297. [DOI] [PubMed] [Google Scholar]
- 19.Heroven, A. K., G. Nagel, H. J. Tran, S. Parr, and P. Dersch. 2004. RovA is autoregulated and antagonizes H-NS-mediated silencing of invasin and rovA expression in Yersinia pseudotuberculosis. Mol. Microbiol. 53:871-888. [DOI] [PubMed] [Google Scholar]
- 20.Ivanoff, B., and M. M. Levine. 1997. Typhoid fever: continuing challenges from resilient bacterial foe. Bull. Inst. Pasteur 95:129-142. [Google Scholar]
- 21.Johansson, J., and B. E. Uhlin. 1999. Differential protease-mediated turnover of H-NS and StpA revealed by a mutation altering protein stability and stationary survival of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:10776-10781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansson, J., S. Eriksson, B. Sondén, S. N. Wai, and B. E. Uhlin. 2001. Heteromeric interactions among nucleoid-associated bacterial proteins: localization of StpA stabilizing regions in H-NS of Escherichia coli. J. Bacteriol. 183:2343-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juárez, A., M. Hartlein, and W. Goebel. 1984. Study of regulation and transport of hemolysin by using fusion of the β-galactosidase gene (lacZ) to hemolysin genes. J. Bacteriol. 160:161-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawley, T. D., M. W. Gilmour, J. E. Gunton, L. J. Standeven, and D. E. Taylor. 2002. Functional and mutational analysis of conjugative transfer region 1 (Tra1) from the IncHI1 plasmid R27. J. Bacteriol. 184:2173-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawley, T. D., M. W. Gilmour, J. E. Gunton, D. M. Tracz, and D. E. Taylor. 2003. Functional and mutational analysis of conjugative transfer region 2 (Tra2) from the IncHI1 plasmid R27. J. Bacteriol. 185:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madrid, C., J. M. Nieto, and A. Juárez. 2002. Role of the Hha/YmoA family of proteins in the thermoregulation of the expression of virulence factors. Int. J. Med. Microbiol. 291:425-432. [DOI] [PubMed] [Google Scholar]
- 27.Madrid, C., J. M. Nieto, S. Paytubi, M. Falconi, C. O. Gualerzi, and A. Juárez. 2002. Temperature- and H-NS-dependent regulation of a plasmid-encoded virulence operon expressing Escherichia coli hemolysin. J. Bacteriol. 184:5058-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maher, D., R. Sherburne, and D. E. Taylor. 1993. H-pilus assembly kinetics determined by electron microscopy. J. Bacteriol. 175:2175-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maher, D., and D. E. Taylor. 1993. Host range and transfer efficiency of incompatibility group HI plasmid. Can. J. Microbiol. 39:581-587. [DOI] [PubMed] [Google Scholar]
- 30.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Miller, V., and J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mouriño, M., C. Madrid, C. Balsalobre, A. Prenafeta, F. J. Muñoa, J. Blanco, M. Blanco, J. E. Blanco, and A. Juárez. 1996. The Hha protein as a modulator of expression of virulence factors in Escherichia coli. Infect. Immun. 64:2881-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mouriño, M., F. J. Muñoa, C. Balsalobre, P. Diaz, C. Madrid, and A. Juárez. 1994. Environmental regulation of α-hemolysin expression in Escherichia coli. Microb. Pathog. 16:249-259. [DOI] [PubMed] [Google Scholar]
- 34.Nieto, J. M., M. Carmona, S. Bolland, Y. Jubete, F. de la Cruz, and A. Juárez. 1991. The hha gene modulates hemolysin expression in Escherichia coli. Mol. Microbiol. 5:1285-1293. [DOI] [PubMed] [Google Scholar]
- 35.Nieto, J. M., C. Madrid, E. Miquelay, J. L. Parra, S. Rodríguez, and A. Juárez. 2002. Evidence for direct protein-protein interaction between members of the enterobacterial Hha/YmoA and H-NS families of proteins. J. Bacteriol. 184:629-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nieto, J. M., C. Madrid, A. Prenafeta, E. Miquelay, C. Balsalobre, M. Carrascal, and A. Juárez. 2000. Expression of the hemolysin operon in Escherichia coli is modulated by a nucleoid-protein complex that includes the proteins Hha and H-NS. Mol. Gen. Genet. 263:349-358. [DOI] [PubMed] [Google Scholar]
- 37.Nieto, J. M., A. Prenafeta, E. Miquelay, S. Torrades, and A. Juárez. 1998. Sequence, identification and effect on conjugation of the rmoA gene of plasmid R100-1. FEMS Microbiol. Lett. 169:50-56. [DOI] [PubMed] [Google Scholar]
- 38.Owen-Hughes, T. A., G. D. Pavitt, D. S. Santos, J. M. Sidebotham, C. S. J. Hulton, J. C. D. Hinton, and C. F. Higgins. 1992. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell 71:255-265. [DOI] [PubMed] [Google Scholar]
- 39.Paytubi, S., C. Madrid, N. Forns, J. M. Nieto, C. Balsalobre, B. E. Uhlin, and A. Juárez. 2004. YdgT, the Hha paralogue in Escherichia coli forms heteromeric complexes with H-NS and StpA. Mol. Microbiol. 54:251-263. [DOI] [PubMed] [Google Scholar]
- 40.Rimsky, S. 2004. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr. Opin. Microbiol. 7:109-114. [DOI] [PubMed] [Google Scholar]
- 41.Rooker, M. M., C. Sherburne, T. D. Lawley, and D. E. Taylor. 1999. Characterization of the Tra2 region of the IncHI1 plasmid R27. Plasmid 41:226-239. [DOI] [PubMed] [Google Scholar]
- 42.Sherburne, C. K., T. D. Lawley, M. W. Gilmour, F. R. Blattner, V. Burland, E. Grotbeck, D. J. Rose, and D. E. Taylor. 2000. The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 28:2177-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smyth, G. K. 2004. Linear models and empirical Bayer methods for assessing differential expression in microarrays experiments. Stat. Appl. Genet. Mol. Biol. vol. 3, no. 1, article 3. [Online.] www.bepress.com/sagmb/vol3/iss1/art3. [DOI] [PubMed]
- 44.Taylor, D. E., and J. G. Levine. 1980. Studies of temperature-sensitive transfer and maintenance of H incompatibility group plasmids. J. Gen. Microbiol. 116:475-484. [DOI] [PubMed] [Google Scholar]
- 45.Tendeng, C., and P. N. Bertin. 2003. H-NS in gram-negative bacteria: a family of multifaceted proteins. Trends Microbiol. 11:511-518. [DOI] [PubMed] [Google Scholar]
- 46.Will, W. R., J. Lu, and L. S. Frost. 2004. The role of H-NS in silencing F transfer gene expression during entry into stationary phase. Mol. Microbiol. 54:769-781. [DOI] [PubMed] [Google Scholar]
- 47.Williams, R. M., S. Rimsky, and H. Buc. 1996. Probing the structure, function, and interactions of the Escherichia coli H-NS and StpA proteins by using dominant negative derivatives. J. Bacteriol. 178:4335-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamada, H., S. Muramatsu, and C. Sasakawa. 1990. An Escherichia coli protein that preferentially binds to sharply curved DNA. J. Biochem. 108:420-425. [DOI] [PubMed] [Google Scholar]