Abstract
The Staphylococcus aureus plasmid pI258 cadCA operon encodes a P-type ATPase, CadA, that confers resistance to the heavy metals Cd(II), Zn(II), and Pb(II). Expression of this heavy-metal efflux pump is regulated by CadC, a homodimeric repressor that dissociates from the cad operator/promoter upon binding of Cd(II), Pb(II), or Zn(II). CadC is a member of the ArsR/SmtB family of metalloregulatory proteins. Here we report the X-ray crystal structure of CadC at 1.9 Å resolution. The dimensions of the protein dimer are approximately 30 Å by 40 Å by 70 Å. Each monomer contains six α-helices and a three-stranded β-sheet. Helices 4 and 5 form a classic helix-turn-helix motif that is the putative DNA binding region. The α1 helix of one monomer crosses the dimer to approach the α4 helix of the other monomer, consistent with the previous proposal that these two regulatory metal binding sites for the inducer cadmium or lead are each formed by Cys-7 and Cys-11 from the N terminus of one monomer and Cys-58 and Cys-60 of the other monomer. Two nonregulatory metal binding sites containing zinc are formed between the two antiparallel α6 helices at the dimerization interface. This is the first reported three-dimensional structure of a member of the ArsR/SmtB family with regulatory metal binding sites at the DNA binding domain and the first structure of a transcription repressor that responds to the heavy metals Cd(II) and Pb(II).
The toxic heavy metals cadmium and lead are common environmental contaminants, both from natural and from anthropogenic sources. In response to heavy-metal toxicity, most organisms have evolved resistance mechanisms. One such mechanism is the cadCA operon of plasmid pI258 from Staphylococcus aureus, which confers resistance to Cd(II) and Pb(II), as well as excessive amounts of the required metal ion Zn(II) (21). The cadA gene product is a P-type Cd(II)/(Pb(II)/Zn(II)-translocating ATPase that extrudes Cd(II), Pb(II), and Zn(II) from cells and confers resistance by lowering the intracellular concentrations of these toxic soft-metal ions (21, 33).
The cadC gene product is a 27-kDa trans-acting, homodimeric repressor that negatively regulates expression of CadA (11). CadC is a member of the ArsR/SmtB family of metalloregulatory proteins (3, 37). Derepression involves dissociation from the cad operator/promoter following binding of a heavy-metal ion to a site on each monomer (11, 30), and both sites must be filled for derepression (29). This regulatory site, which we term a type 1 metal binding site, is composed of four cysteine residues: Cys-7, Cys-11, Cys-58, and Cys-60 (30). Only Cys-7, Cys-58, and Cys-60 are required for biological activity. Even though Cys-11 is not required, all four cysteine thiolates participate in Cd(II) binding as a tetrathiolate complex, with a Cd(II)-S distance of 2.5 Å (2). This type 1 site is related to the three-coordinate As(III) binding site in ArsR (28), in which two of the ligands, Cys-32 and Cys-34, correspond to Cys-58 and Cys-60 in CadC. From cross-linking experiments with heterodimers composed of mutants with different combinations of cysteine positions, we showed that each type 1 metal binding site is formed by Cys-7 and Cys-11 from one subunit and Cys-58 and Cys-60 from the other subunit (34). In contrast, the type 1 site in ArsR is composed of three cysteine residues contained within a single subunit. Thus, the type 1 metal sites in CadC are more complex than the ArsR type 1 metal binding sites.
Recently a second type of binding site for harder metals was identified at the interface of the two subunits of CadC (4) in the region of CadC that corresponds to the inducer binding site of SmtB, a Zn(II)-responsive homologue of CadC (31). To differentiate between the two types of sites, we designate the second type a type 2 metal binding site. From spectroscopic analysis, this is a Zn(II) or Co(II) site composed of oxygen and nitrogen ligands (4). Alignment of the protein sequences of SmtB and CadC suggests that the site in CadC might involve residues Asp-101, His-103, His-114, and Glu-117. We showed that a mutant CadC with an H103A substitution exhibited wild-type properties both in vivo and in vitro (34), and more recently we have shown that a mutant lacking all four residues responds to Cd(II), Pb(II), and Zn(II) in vivo (A. Kandegedara, J. Ye, and B. P. Rosen, unpublished data). Thus, the type 2 sites are not essential for CadC function. In contrast, the structure of SmtB without (8) and with (10) Zn(II) shows that it has only type 2 sites.
Here we report the X-ray crystal structure of CadC at 1.9 Å resolution, the first three-dimensional structure reported for a regulatory protein that uses a type 1 site to respond to the heavy metals cadmium and lead. The asymmetric unit contains two homodimers (termed dimera and dimerb). Each dimer has dimensions of approximately 30 Å by 40 Å by 70 Å. In each monomer there are six α-helices and a three-stranded β-sheet. CadC is a winged-helix repressor in which helices 4 and 5 form a classic helix-turn-helix DNA binding site (12, 16). The structure shows a unique feature of CadC in which the α1 helix of one monomer crosses the dimer to approach the α4′ helix of the other monomer, which would allow formation of a type 1 metal binding site in that location. In addition, apo-CadC contained two zinc ions bound at two type 2 sites between the α6 and α6′ helices of the dimerization interface, even though no metals were added during purification or crystallization. Each zinc ion is coordinated by Asp-101 and His-103 from one monomer and His-114′ and Glu-117′ from the other monomer. Thus, CadC has both type 1 and type 2 metal binding sites, but only the type 1 sites are required for metalloregulation. We propose that CadC is an evolutionary intermediate between ArsR, which uses only type 1 sites for metalloregulation, and SmtB, which uses only type 2 sites.
MATERIALS AND METHODS
Purification of CadC.
C11G CadC, which has wild-type metalloregulatory properties, was used for this study (30). This variant has improved solubility properties and is less prone to form intermolecular disulfide bonds than is CadC with a cysteine residue at position 11. Purified CadC was prepared from Escherichia coli strain BL21(DE3) pYSCM2, as described previously (30), with one additional purification step. Following S-Sepharose chromatography, CadC was concentrated anaerobically in a vacuum chamber and chromatographed on a Superdex-75 column using a buffer consisting of 50 mM morpholinepropanesulfonic acid (MOPS), 5 mM dithiothreitol, and 0.5 M NaCl, pH 7.0. All purification buffers were purged with argon for 1 h before use. Selenomethionine-containing (SeMet) CadC was prepared as follows. A culture of E. coli strain BL21(DE3) pYSCM2 grown overnight in LB medium (25) was centrifuged at 3,000 × g for 10 min at 4°C and suspended in an equal volume of M9 minimal medium (25) supplemented with 2 mM MgSO4, 0.2% (wt/vol) glucose, and 40 μg/ml kanamycin. One milliliter of the cell suspension was diluted into 1 liter of M9 medium and allowed to grow to mid-exponential phase at 37°C. At that point 100 mg each of lysine, phenylalanine, and threonine, 50 mg each of isoleucine, leucine, and valine, and 60 mg of l-(+)-selenomethionine (Sigma) were added. The culture was grown for 15 min at 37°C, following which the temperature was lowered to 20°C. CadC was induced by addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (final concentration). The culture was grown overnight at 20°C, and CadC was purified as described above.
The metal content of purified CadC was determined by inductively coupled plasma mass spectrometry (ICP-MS) (Perkin-Elmer ELAN 9000). All samples were dissolved in 2% analytical-grade HNO3 acid for analysis. Standard solutions were purchased from Perkin-Elmer for calibration. The concentration of purified CadC was determined using a calculated molar extinction coefficient at 280 nm of 6,585 M−1 cm−1 (14).
Crystallization conditions.
Crystals were grown at 23 ± 2°C in hanging drops. Purified CadC (4 μl, at 13.5 mg/ml) was added to 2 μl of a well solution containing 2 M (NH4)2SO4 and 0.1 M sodium citrate, pH 5.5, and 1 μl of 30% (vol/vol) methanol. The droplets were equilibrated with 0.6 ml of well solution. The crystals grew within 5 days, attaining dimensions of 0.6 by 0.15 by 0.15 mm3. Crystals of SeMet CadC were grown similarly, except that methanol was replaced with 5% glycerol. For mass analysis, crystals were dissolved and analyzed by matrix-assisted laser desorption ionization (MALDI) mass spectrometry at the Michigan Proteome Consortium at the University of Michigan.
Structure determination and refinement.
SeMet CadC crystallized in space group P43, with three dimers in the asymmetric unit. Data were collected at the Advanced Photon Source, BioCars beamline 14ID-B, at three different wavelengths (peak, 0.9793 Å; inflection point, 0.9795 Å; high-energy remote, 0.9563 Å) on a frozen crystal at 100 K. Data were processed to structure intensities with HKL2000 (22) and converted to structure amplitudes with CCP4, version 4.2.2 (24). The selenium substructure was solved with SHELXD (26). Twelve of the 18 theoretical sites were located. The selenium sites occur in pairs with about a 6.5-Å separation, corresponding to the signals from selenomethionine residues 108 and 109. Data were used to 2.5 Å resolution, and MLPHARE in the CCP4 suite was used to refine the sites and calculate phases. Using the selenomethionine sites as a starting point, noncrystallographic sixfold symmetry averaging and density modification by means of DM within CCP4 were used to generate an interpretable map. Refinement with Refmac5 (32) gave initial R and Rfree values of 23.8% and 29.6%, respectively.
Better diffracting crystals of native (non-SeMet) CadC were grown in space group P41, with two dimers in the asymmetric unit (Table 1). One of the SeMet dimers was used as a model for fitting native CadC by molecular replacement with MOLREP in the CCP4 suite. Refinement with REFMAC5 resulted in slightly high bond length errors (root mean square deviation [RMSD] on bond lengths, 0.03 Å). At that point the entire structure was refitted automatically using ARP/wARP, version 6.1.1, automated model building starting from the existing model phases and “free atom” refinement (23). Of the 488 possible residues (four chains of 122 residues apiece), ARP/wARP fit 371 residues in nine chains.
TABLE 1.
Data collection
| Measurement | Value for crystala
|
|
|---|---|---|
| Native CadC | SeMet CadC | |
| Space group | P41 | P43 |
| Cell dimensions (Å) | a = b = 116.5; c = 41.8 | a = b = 90.2; c = 147.8 |
| Beamline | 14-BM-C | 14-ID-B |
| Wavelength (Å)b | 0.9000 | 0.9795, 0.9793, 0.9563 |
| Resolution (Å) | 1.9 | 2.5, 2.5, 2.5 |
| No. of unique reflections | 40,691 | 40,682 40,488 40,784 |
| Completeness (%) | 95.6 (99.6)c | 98.8, 98.7, 98.8 |
| I/σ | 28.0 (8.0) | 26.3, 28.0, 26.0 |
| Redundancy | 3.5 (3.4) | 4.0, 4.0, 4.0 |
| Rsymd | 0.045 (0.225) | 0.047, 0.044, 0.050 |
Mean figure of merit (before/after density modification), 0.35/0.76.
Data in the SeMet CadC column were collected at three different wavelengths: peak, inflection point, and high-energy remote.
Last shell value is in parentheses.
Rsym = Σhkl(Σi|Ihkl,i − <Ihkl>|)/Σhkl,i<Ihkl>, where Ihkl,i is the intensity of an individual measurement of the reflection with Miller indices h, k, and l, and <Ihkl> is the mean intensity of that reflection.
The structure was completed using manual rebuilding with the help of simulated annealing omit electron density maps and refinement to the present R and Rfree of 0.205 and 0.261, respectively. The RMSDs of the bond lengths and angles were acceptable values of 0.019 Å and 1.8°, respectively. Ninety-seven percent of all residues are in the most favored regions, and none is in the disallowed region of the Ramachandran plot (18). N-terminal residues 1 to 10 were disordered in the P41 crystal form. In the P43 structure of SeMet CadC, N-terminal residues could be fit to residue 7 in three of the six monomers. Using noncrystallographic symmetry restraints, the structure was refined to R and Rfree values of 0.190 and 0.251, respectively. The statistics are shown in Table 2.
TABLE 2.
Refinement statistics
| Measurement | Value |
|---|---|
| Resolution (Å) | 50-1.9 (1.95-1.9) |
| I/σ cutoff | 0 |
| No. of residues | 404 |
| No. of ions | 3 |
| No. of protein atoms | 3,152 |
| No. of water molecules | 470 |
| R/Rfree (%)a | 20.5/26.1 (27.8/32.9) |
| RMSDs | |
| Bond (Å) | 0.019 |
| Angle (°) | 1.77 |
| Bonded B-factors (Å2), main chain/side | |
| chain | 1.26/3.39 |
Last shell value is in parentheses.
Protein structure accession number.
The native structure has been deposited in the Protein Data Bank with accession number PDB ID 1U2W.
RESULTS
Crystallization and X-ray structure analysis.
Wild-type CadC was difficult to prepare without formation of intermolecular disulfide bonds during purification. CadC C11G, which is fully functional (30), was found to be less prone to formation of disulfide bonds and so was used for crystallization. The mass of the crystallized CadC was determined by mass spectroscopy to be 13,733 Da, corresponding to a full-length monomer of 122 residues. Because the structure of SmtB (8) was not similar enough for molecular replacement to be used, the CadC structure was solved by multiwavelength anomalous dispersion (15) with the SeMet protein. The native CadC structure was refined to 1.9 Å resolution, and the SeMet structure was refined to 2.2 Å resolution.
Overall structure of CadC.
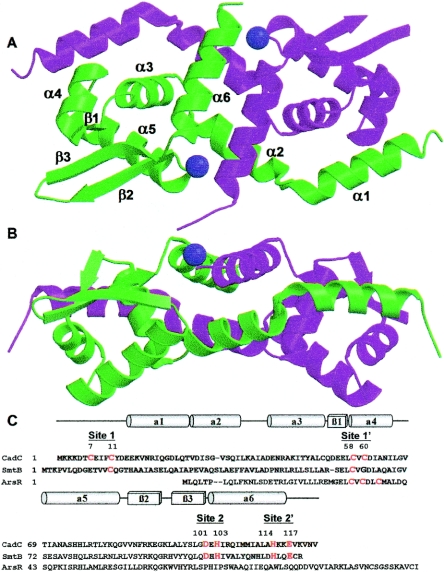
CadC is a homodimer (30), and in the native structure, the asymmetric unit contains two dimers, designated dimera and dimerb, with weak interactions between the dimers. The dimensions of each dimer are approximately 30 Å by 40 Å by 70 Å. Each monomer contains six α-helices (residues 15 to 26, 27 to 39, 42 to 52, 58 to 66, 69 to 83, and 102 to 116) and a three-stranded β-sheet (residues 55 to 57, 85 to 90, and 94 to 100), although the first β-strand is much shorter than the other two (Fig. 1). In the four molecules of the asymmetric unit, residues 11 to 89 and 94 to 119 of molecule 1, residues 11 to 117 of molecule 2 (molecules 1 and 2 form dimera), residues 17 to 114 of molecule 3, and residues 11 to 81 and 96 to 118 of molecule 4 (molecules 3 and 4 form dimerb) are visible in the electron density map. Residues 90 to 94 between β2 and β3 are less ordered, as shown by missing electron density or higher temperature factors. The RMSD between Cα atoms of the two dimers is 0.74 Å. The RMSDs between Cα atoms of molecule 4 and those of molecules 1, 2, and 3 are 0.66 Å, 0.66 Å, and 0.52 Å, respectively. The RMSDs between Cα atoms of molecule 3 and those of molecules 1 and 2 are 0.56 Å and 0.45 Å, respectively. The RMSD between Cα atoms of molecules 1 and 2 is 0.49 Å.
FIG. 1.
Structure of S. aureus pI 258 CadC. (A) Ribbon diagram of the CadC dimer with secondary structural units (N-α1-α2-α3-β1-α4-α5-β2-β3-α6-C). All figures were created with MolScript (17), Raster3D (19), and PyMOL (9). The two zinc ions are shown as purple spheres. (B) To illustrate the location of the type 2 metal binding sites, the dimer was rotated 90° relative to the orientation above. (C) Sequence alignment of CadC with SmtB and ArsR. The residues that comprise the metal binding sites are shown in red. The numbers are for the CadC type 1 and type 2 metal binding sites. Each site is composed of two residues from one monomer (no “prime”) and two residues from the other monomer (designated “prime”). The CadC secondary-structure elements are indicated above the sequence (cylinders, α-helices; rectangle, β-strands).
In the middle part of the dimer, helices α2, α3, and α6 from both monomers form the dimerization interface. The residues involved in dimerization are mainly hydrophobic. Residues Val-27, Ile-29, Val-32, Ile-35, Leu-36, and Ile-39 from α2, residue Ala-50 from α3, and residues Ile-104, Ile-107, Ile-110, Ala-111, Leu-112, and Ala-113 from α6 form the hydrophobic core. The metal ions in the dimerization interface probably provide extra stabilization (see below). The first helix, α1, which has not been observed in structures of CadC homologues, also contributed to dimerization, with interactions with α3′ and α4′ of the other subunit. The total buried area between the two monomers is about 4,500 Å2. If α1 and its N-terminal extension are excluded from the calculation, the buried area decreases to about 2,800 Å2.
Metal binding sites.
Two distinct types of metal binding sites in a CadC dimer have been proposed (4). The type 1 metal binding site, also designated α3N, is a thiolate-rich site composed of Cys-7 and Cys-11 in the N terminus and Cys-58 and Cys-60 in the first helix of the DNA binding site (2, 30). The type 2 metal binding site, also designated α5, is proposed to be an oxygen/nitrogen site at the dimer interface. Since CadC is shown here to have an extra N-terminal helix, the type 1 sites might have an alternate designation of α4N, and the type 2 sites could alternately be called the α6 sites, but for simplicity, they are designated simply type 1 and type 2 metal binding sites. A unique feature of CadC is that each of the two type 1 sites in the homodimer is formed from the N terminus of one subunit and the α4′ helix of the other subunit (1, 34). We have demonstrated by chemical cross-linking that the Cys-7 and Cys-11 of one subunit are spatially located near the Cys-58′ and Cys-60′ of the other subunit, consistent with formation of intersubunit metal sites (34). The structure shows that α1 of one monomer is adjacent to α4′ of the other subunit in both homodimers of the asymmetric unit, consistent with CadC having two intersubunit type 1 metal sites (Fig. 1).
Helix α1 and its N-terminal extension interact extensively with α3′ and α4′ of the other monomer (Fig. 2A). The residues involved in hydrophobic interactions are Val-17, Ile-20, and Leu-24 of one monomer and Tyr-49′, Ala-50′, Leu-57′, and Ile-65′ of the other monomer. There are four intersubunit hydrogen bonds between side chain atoms of Tyr-12, Asp-13, Glu-14, and Lys-16 of one subunit and Cys-58′, Asp-61′, Asn-64′, and Asp-54′ of the other subunit. Tyr-12 is close enough to Cys-58′ and Cys-60′ of the other monomer to form a hydrogen bond with the thiol of Cys-58′ (Fig. 2B). In the SeMet-substituted CadC structure, three of the six molecules in the asymmetric unit can be traced to Cys-7 (Fig. 2C). When these three structures are superimposed on each other, they show three different orientations (Fig. 2D). This suggests that the N terminus is flexible and can adopt a variety of orientations in the absence of metal. The type 1 metal binding site would form only when metal is available to bridge Cys-7, Cys-58′, and Cys-60′. Attempts to introduce metals by soaking the crystals with Cd(II), Pb(II), or Zn(II) resulted in cracking of the crystals or loss of resolution. A reasonable possibility is that Tyr-12, which is hydrogen bonded to Cys-58′, sterically hinders binding of metal within the crystal, since rotation about chi 1 of residue Tyr-12 is restricted sterically. Alternative approaches such as cocrystallization with metals or using mutants with substitutions of Tyr-12 will be attempted in the future.
FIG. 2.
Location of the type 1 metal binding sites. (A) The type 1 metal binding site is located at the interface of α3 and α4 from one monomer (magenta) and α1′ from the other monomer (green). Residues at the dimer interface are labeled. (B) The vicinity of the type 1 metal binding site. The 2Fo-Fc simulated annealing omit electron density map at the 1σ level is shown for residues 58 to 61 in helix α4 and residues 11 to 13 from the other monomer. A hydrogen bond is formed between the hydroxyl of Tyr-12 and the thiol of Cys-58. (C) Molecules 1, 2, and 4 of SeMet CadC structure can be traced up to Cys-7. The 2Fo-Fc simulated annealing omit electron density map at the 1σ level is shown for residues 7 to 10 of molecule 2. (D) The N termini of molecules 2 and 4 of the SeMet CadC structure are superimposed on molecule 1, showing that, without metal ions bound to the type 1 site, Cys-7 can assume multiple orientations. The side chains of Cys-7, Cys-58′, and Cys-60′ are shown.
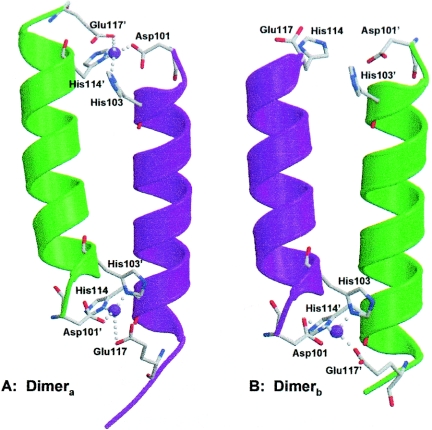
Even though no metals were added during purification of native CadC, two metal ions were observed in the electron density map between the α6 and α6′ helices of dimera (Fig. 3A). Dimerb contained only one electron density peak at the α6-α6′ dimerization interface (Fig. 3B). The α6-α6′ interface was less ordered in this dimer. To identify the metal bound at the interface, crystals were washed with crystallization buffer, dissolved in high-pressure chromatography-quality water, and analyzed by ICP-MS. The only metal found in stoichiometric amounts was zinc, which was present in a Zn(II)-to-CadC monomer molar ratio of 0.3 to 0.6 in several different crystals. If there were three zinc ions at 100% occupancy at the interfaces of two dimers in the crystal, a stoichiometry of 0.75 would be expected. However, some zinc is probably lost upon washing the crystals, and perhaps there was incomplete occupancy in the crystal. These results are consistent with endogenous zinc filling the type 2 sites in CadC.
FIG. 3.
Location of the type 2 metal binding sites. The two type 2 metal binding sites in each monomer are formed at the interface of the α6 and α6′ helices in each dimer. The asymmetry in the distribution of zinc is shown for the two dimers in the asymmetric unit. (A) Dimera has zinc (purple spheres) bound at both ends of the dimer interface via side chain ligands arranged tetrahedrally (donated by the side chains of Asp-101 and His-103 of one monomer [green] and His-114 and Glu-117 of the other monomer [magenta]). (B) Dimerb has only a single bound zinc, because the second type 2 site is more disordered, and Glu117 is not visible in the structure.
The four residues involved in these type 2 Zn(II) binding sites are Asp-101 and His-103 from CadC and His-114′ and Glu-117′ from CadC′ (Fig. 3). The coordination geometry is irregular. The four ligands are donated by Asp-101 OD2 and His-103 ND1 from one subunit and by His-114′ ND1 and Glu-117′ OE2 from the other. The distances from Zn(II) to the side chain ligands range from 1.9 Å to 2.3 Å.
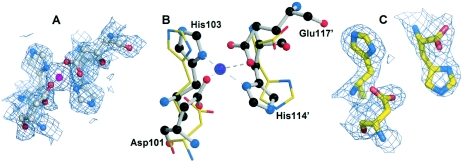
When the filled site of dimera was superimposed on the empty site of dimerb (Fig. 4B), local conformational differences were observed. This is clearly shown in the electron density maps (Fig. 4A and C). Without Zn(II) bound, the side chains on the empty site of dimerb were moved away, and Glu-117 was not visible in the electron density map, which suggests that Zn(II) may play a structural role in stabilizing the dimer. Even so, Zn(II) is not required, since, as mentioned above, a mutant lacking all four protein ligands is functional in vivo.
FIG. 4.
Comparison of a filled and an empty type 2 metal binding site. (A and C) Close-ups of the type 2 metal binding sites (shown is the 2Fo-Fc simulated annealing omit electron density map contoured at the 1σ level) with and without zinc (purple sphere), respectively. (B) Superposition of the empty (in dimerb) and filled (in dimera) zinc sites shows how the backbone and side chain atoms of the histidine and aspartate pairs differ. His-114 is the last residue that can be seen in dimerb. The filled site is shown as a ball-and-stick diagram, and the empty site as bonds.
Comparison of CadC with SmtB.
To gain insight into the evolution of the type 1 and type 2 metal binding sites, it is important to compare the structure of a repressor with only type 1 sites, such as ArsR, or a repressor with only type 2 sites, such as SmtB, with that of a transitional repressor that has both types of sites, such as CadC. While the structure of ArsR has not been determined, the structure of SmtB crystallized in the absence of DNA and inducer has been reported (8). As with CadC, each SmtB monomer has an α/β topology with five α-helices and a two-stranded β-hairpin. The DNA binding domains are helix-loop-helix structures located at each end of the elongated dimer. The dimer interface is composed of two N-terminal and two C-terminal helices that align antiparallel with their cognate helix from the other subunit. Recently the structures of the Zn-filled SmtB and a homologue, CzrA, were reported (10). Most significantly, binding of zinc at the two type 2 sites at the dimer interface resulted in a 2.4-Å movement of the recognition helix of the DNA binding domain such that the metalated form became more compact. In contrast, CzrA showed little change upon Zn(II) binding at the type 2 sites, and both forms were more similar to the Zn(II)-bound form of SmtB than to apo-SmtB.
Superposition of the CadC dimer with apo-SmtB reveals that the apo-CadC dimer is more compact, resembling more closely the metalated form of SmtB, consistent with CadC containing Zn-filled type 2 sites. The point-to-point distance between the Ser-74 Cα atoms on each subunit is 51.7 Å in apo-SmtB. The corresponding distance in apo-CadC from the Ala-71 Cα atoms on each monomer is 42.1 Å. The buried solvent-accessible area between subunits is 4,500 Å2 in apo-CadC, compared with 3,800 Å2 in apo-SmtB. This difference is due in part to contributions from the interactions between α1 and the other monomer in the CadC dimer. Interestingly, superposition of a CadC monomer with SmtB using α2, α3, and α6 of CadC and α1, α2, and α5 of SmtB shows that α2 and α6 of CadC at the intersubunit interface are superimposable with α1 and α5 of SmtB, while the rest of the CadC monomer appears to be rotated 20° relative to that of SmtB (Fig. 5). This suggests that the dimerization interface might be the most structurally conserved region in this family of proteins. In the superposition, the Ala-71 Cα atom in the α5 helix of CadC is 8.2 Å away from the analogous Ser-74 Cα atom of apo-SmtB, a distance greater than that between apo-SmtB and fully zinc-liganded α5-SmtB, which is an average of 2.4 Å (10). Thus, the apo-CadC dimer is more compact than either apo-SmtB or Zn2 α5-SmtB.
FIG. 5.
Comparison of the structures of CadC and SmtB. Stereo view of superposition of the CadC (magenta) and SmtB (gray) monomers. CadC and SmtB were overlapped using only helices 2, 3, and 6 (RMSD = 1.25 Å). The positions of Ala-71 of CadC and Ser-74 of SmtB are shown.
The DNA binding domain.
CadC is a winged-helix DNA binding protein. The two wings (W1 and W2), three α-helices (α3, α4, and α5) and three β-strands (β1, β2, and β3), arranged in the order α3-β1-α4-α5-β2-W1-β3-W2, form a typical winged-helix motif (12, 16). The putative DNA binding domain consists of a helix-turn-helix motif consisting of α4-turn-α5. The center-to-center distance between the recognition helices of the CadC dimmer is approximately 30 Å. When involved in DNA binding, the recognition helix, α5, might be present in the major groove of a duplex DNA and interact with the major groove, as suggested by other winged-helix protein-DNA cocrystal structures (7). The positively charged Arg-78 and Lys-82 on α5 are predicted to interact with phosphate groups on the DNA backbone. The wing W1, which is flexible with poor electron density in the structure, is predicted to interact with the adjacent minor groove. However, we cannot rule out the possibility that CadC might adopt another DNA binding mode. For example, in the human regulatory factor X1 (hRFX1) structure, in which W1 interacts with the major groove, the recognition helix makes only one minor groove contact (13). Residues 100 to 102 after the third β-strand form wing W2, which is immediately followed by the final helix, α6. Binding of Cd(II) in both type 1 sites results in dissociation of CadC from the cad operator/promoter DNA (30). Comparison of DNA binding domains of this CadC structure with one in which the two type 1 sites are filled will be necessary in order to elucidate how metal binding results in dissociation from the DNA.
DISCUSSION
The 117-residue ArsR from E. coli plasmid R773 was the first identified member of the ArsR/SmtB family of small metalloregulatory proteins (28, 35, 36) (Fig. 1C). Since this initial report, the number of members of this family listed in the National Center for Biotechnology Information database has grown to nearly 200, with at least 172 homologues in gram-positive and -negative bacteria and at least 25 homologues in Archaea. These include proteins that respond to As(III)/Sb(III) (ArsR) (35), Pb(II)/Cd(II)/Zn(II) (CadC) (11), Cd(II)/Pb(II) (CmtR) (5), Zn(II) (SmtB and ZiaR) (20), and Co(II)/Ni(II) (NmtR) (6). The focus of our studies has been on the nature of the binding sites for the inducing metals or metalloids in ArsR and CadC, and how the information of inducer binding is transformed into transcriptional activity. ArsR, CadC, and SmtB (and, by extrapolation, all members of the family) are homodimers that repress transcription by binding to DNA in the absence of an inducing metal ion. They dissociate from the DNA when metal is bound, resulting in expression of metal ion resistances.
A more detailed understanding of the relationship between metal binding and derepression requires knowledge of the structure of the repressors, in particular the metal and DNA binding sites. We have shown by a combination of biochemical and molecular genetic approaches that the As(III) binding site in ArsR is composed of two required cysteine residues, Cys-32 and Cys-34, and a third nonessential residue, Cys-37, that form an AsS3 site in which the three sulfur atoms are each 2.25 Å from the trivalent metalloid (27, 28). While the structure of ArsR has not been determined, the structure of SmtB crystallized in the absence of DNA with and without an inducer (8) has been reported. Each monomer of the homodimer has an α/β topology with five α-helices and a two-stranded β-hairpin. The DNA binding domains are helix-loop-helix structures located at each end of the elongated dimer. The dimer interface is composed of two N-terminal and two C-terminal helices that align antiparallel with their cognate helix from the other subunit. From a model of ArsR based on the SmtB structure, we can predict that the thiol ligands of Cys-32 and Cys-34 form a very simple type 1 metal binding site that is located at the start of the first helix of the DNA binding site on each monomer (37).
The two type 1 sites in CadC are similar to the ArsR inducer binding sites but are more complicated. Two of the ligands in the CadC type 1 sites, Cys-58 and Cys-60, correspond to Cys-32 and Cys-34 of the ArsR sites. Both the number and the spatial location of available protein ligands must be critical for selectivity (3, 34). ArsR is either S2 or S3 (depending on the availability of Cys-37) (27), while the site in CadC is S3 or S4 (depending on the availability of Cys-11) (2). CadC has an N-terminal extension that is absent in ArsR. This sequence contains Cys-7 and Cys-11. We predicted and subsequently confirmed by cross-linking experiments that each of the two binding sites is composed of Cys-7 and Cys-11 from one subunit and Cys-58′ and Cys-60′ of the other (34). From these data there can be no doubt that each of the two type 1 metal binding sites of CadC is composed of 1 or 2 cysteine residues (depending on whether Cys-11 is present) from the N terminus of one subunit and 2 cysteine residues of the DNA binding domain of the other subunit, a prediction that can be verified by crystallization of metalated CadC. The structure of apo-CadC extends to residue 7 and clearly shows that Cys-7 in one subunit can assume a number of orientations in the absence of metal. However, Cys-7 of one subunit is held in the proximity of the α3′ and α4′ helices by a series of hydrogen bonds and hydrophobic interactions. Thus, when metal is available, Cys-7 is programmed to form a metal binding site with Cys-58′ and Cys-60′.
How does metal binding result in transcriptional derepression? In our model of apo-ArsR, the sulfur atoms of the three cysteines are linearly arrayed along the first helix of the helix-loop-helix DNA binding domain, with more than 10 Å separating Cys-32 from Cys-37 (37). To form an S3 site, it is necessary to bring the three sulfur atoms to 2.25 Å from bound As(III), which would require a substantial conformational change in that helix. We hypothesized that this distorts the helix, resulting in dissociation from the operator/promoter site and transcription of the resistance genes. However, with free rotation around the carbon-sulfur bond, the distance between the sulfur atoms of Cys-58 and Cys-60 observed in the CadC structure can easily accommodate the geometry of a CdS4 site without distorting the DNA binding site. As an alternative, we propose that metalating the type 1 site brings the N terminus of CadC into close contact with α4, the first part of the helix-loop-helix DNA binding motif, making it inaccessible to the DNA. Thus, binding of Cd(II), Pb(II), or Zn(II) to CadC could sterically block interaction with the cad operator/promoter.
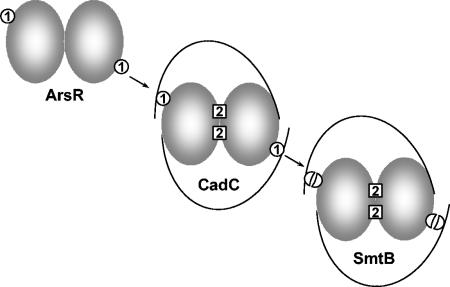
Giedroc and colleagues have proposed that the type 2 sites in CadC might be vestigial remnants of a functional site in the common ancestor of members of this family (4). While this proposal is reasonable, we consider it more likely that the founding members of the family had only an ArsR-like type 1 site and that the inducing metal interacted with the DNA binding domain to produce derepression (Fig. 6). ArsR repressors have only simple type 1 sites and none of the residues associated with the type 2 metal binding site. The As(III) binding site in ArsR is composed of residues from a monomer only and requires just two residues, Cys-32 and Cys-34. In contrast, the CadC type 1 site and the CadC and SmtB type 2 sites are all formed between subunits and require three or four residues. Application of Occam's razor would suggest that the simple site in ArsR would have evolved before the more complicated sites in homologues. CadC would represent a transitional form that evolved a variant and more complicated type 1 site by acquiring an N-terminal extension with additional cysteine residues. Its type 2 site at the dimer interface represents an evolutionary intermediate, with a nonregulatory structural role for Zn(II), compared with repressors such as SmtB and CzrA, which would have lost the function of the type 1 sites and acquired a regulatory role for the metal bound at the type 2 sites. Without a function, the type 1 site in SmtB would gradually be lost. Indeed, SmtB has remnants of type 1 sites, retaining residue Cys-14, which may correspond to one of the two cysteine residues in the N terminus of CadC, and the sequence L60CVGDL, which corresponds to the site 1 motif L57CVGDL in CadC and L31CVGDL in ArsR. Thus, while ArsR has no residues corresponding to the histidines and carboxylates that form the type 2 site, SmtB has a degenerate CdS4 type 1 site. A reasonable inference is that type 1 metal binding sites were present in the common ancestor of ArsR, CadC, and SmtB.
FIG. 6.
Model for the evolutionary history of type 1 and type 2 metal binding sites. The type 1 metal binding sites in ArsR are proposed to be the most ancient, because each site requires only two residues (Cys-32 and Cys-34) from a single subunit of the homodimer. In CadC, the equivalents of these two cysteine residues, Cys-58 and Cys-60, form more-complicated type 1 sites with residues Cys-7′ and Cys-11′ from the other subunit. CadC also has type 2 sites composed of residues Asp-101 and His-103 from one subunit and residues His-114′ and Glu-117′ from the other subunit. The equivalent type 2 sites in SmtB are composed of residues Asp-104 and His-106 from one subunit and His-117′ and Glu-120′ from the other subunit. SmtB also has residue Cys-14, which corresponds to either CadC residue Cys-7 or CadC residue Cys-11, and residue Cys-61′, which corresponds to CadC residue Cys-58′ and may represent a degenerate type 1 site.
Acknowledgments
This work was supported by U.S. Public Health Service grant AI45428.
REFERENCES
- 1.Busenlehner, L. S., J. L. Apuy, and D. P. Giedroc. 2002. Characterization of a metalloregulatory bismuth(III) site in Staphylococcus aureus pI258 CadC repressor. J. Biol. Inorg. Chem. 7:551-559. [DOI] [PubMed] [Google Scholar]
- 2.Busenlehner, L. S., N. J. Cosper, R. A. Scott, B. P. Rosen, M. D. Wong, and D. P. Giedroc. 2001. Spectroscopic properties of the metalloregulatory Cd(II) and Pb(II) sites of Staphylococcus aureus pI258 CadC. Biochemistry 40:4426-4436. [DOI] [PubMed] [Google Scholar]
- 3.Busenlehner, L. S., M. A. Pennella, and D. P. Giedroc. 2003. The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27:131-143. [DOI] [PubMed] [Google Scholar]
- 4.Busenlehner, L. S., T. C. Weng, J. E. Penner-Hahn, and D. P. Giedroc. 2002. Elucidation of primary (α3N) and vestigial (α5) heavy metal-binding sites in Staphylococcus aureus pI258 CadC: evolutionary implications for metal ion selectivity of ArsR/SmtB metal sensor proteins. J. Mol. Biol. 319:685-701. [DOI] [PubMed] [Google Scholar]
- 5.Cavet, J. S., A. I. Graham, W. Meng, and N. J. Robinson. 2003. A cadmium-lead sensing ArsR-SmtB repressor with novel sensory sites: complementary metal-discrimination by NMTR and CMTR in a common cytosol. J. Biol. Chem. 278:44560-44566. [DOI] [PubMed] [Google Scholar]
- 6.Cavet, J. S., W. Meng, M. A. Pennella, R. J. Appelhoff, D. P. Giedroc, and N. J. Robinson. 2002. A nickel-cobalt sensing ArsR-SmtB family repressor: contributions of cytosol and effector binding sites to metal selectivity. J. Biol. Chem. 277:38441-38448. [DOI] [PubMed] [Google Scholar]
- 7.Clark, K. L., E. D. Halay, E. Lai, and S. K. Burley. 1993. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364:412-420. [DOI] [PubMed] [Google Scholar]
- 8.Cook, W. J., S. R. Kar, K. B. Taylor, and L. M. Hall. 1998. Crystal structure of the cyanobacterial metallothionein repressor SmtB: a model for metalloregulatory proteins. J. Mol. Biol. 275:337-346. [DOI] [PubMed] [Google Scholar]
- 9.DeLano, W. L. 2001. The PyMOL user's manual. DeLano Scientific, San Carlos, Calif.
- 10.Eicken, C., M. A. Pennella, X. Chen, K. M. Koshlap, M. L. VanZile, J. C. Sacchettini, and D. P. Giedroc. 2003. A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins. J. Mol. Biol. 333:683-695. [DOI] [PubMed] [Google Scholar]
- 11.Endo, G., and S. Silver. 1995. CadC, the transcriptional regulatory protein of the cadmium resistance system of Staphylococcus aureus plasmid pI258. J. Bacteriol. 177:4437-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gajiwala, K. S., and S. K. Burley. 2000. Winged helix proteins. Curr. Opin. Struct. Biol. 10:110-116. [DOI] [PubMed] [Google Scholar]
- 13.Gajiwala, K. S., H. Chen, F. Cornille, B. P. Roques, W. Reith, B. Mach, and S. K. Burley. 2000. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature 403:916-921. [DOI] [PubMed] [Google Scholar]
- 14.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-326. [DOI] [PubMed] [Google Scholar]
- 15.Hendrickson, W. A., J. R. Horton, and D. M. LeMaster. 1990. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 9:1665-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huffman, J. L., and R. G. Brennan. 2002. Prokaryotic transcription regulators: more than just the helix-turn-helix motif. Curr. Opin. Struct. Biol. 12:98-106. [DOI] [PubMed] [Google Scholar]
- 17.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 18.Laskowski, R. A., J. A. Rullmann, M. W. MacArthur, R. Kaptein, and J. M. Thornton. 1996. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8:477-486. [DOI] [PubMed] [Google Scholar]
- 19.Merrit, E. A., and M. E. P. Murphy. 1994. Raster3D, version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D 50:869-873. [DOI] [PubMed] [Google Scholar]
- 20.Morby, A. P., J. S. Turner, J. W. Huckle, and N. J. Robinson. 1993. SmtB is a metal-dependent repressor of the cyanobacterial metallothionein gene smtA: identification of a Zn inhibited DNA-protein complex. Nucleic Acids Res. 21:921-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nucifora, G., L. Chu, T. K. Misra, and S. Silver. 1989. Cadmium resistance from Staphylococcus aureus plasmid pI258 cadA gene results from a cadmium-efflux ATPase. Proc. Natl. Acad. Sci. USA 86:3544-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 23.Perrakis, A., M. Harkiolaki, K. S. Wilson, and V. S. Lamzin. 2001. ARP/wARP and molecular replacement. Acta Crystallogr. D 57:1445-1450. [DOI] [PubMed] [Google Scholar]
- 24.Potterton, E., S. McNicholas, E. Krissinel, K. Cowtan, and M. Noble. 2002. The CCP4 molecular-graphics project. Acta Crystallogr. D 58:1955-1957. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y..
- 26.Schneider, T. R., and G. M. Sheldrick. 2002. Substructure solution with SHELXD. Acta Crystallogr. D 58:1772-1779. [DOI] [PubMed] [Google Scholar]
- 27.Shi, W., J. Dong, R. A. Scott, M. Y. Ksenzenko, and B. P. Rosen. 1996. The role of arsenic-thiol interactions in metalloregulation of the ars operon. J. Biol. Chem. 271:9291-9297. [DOI] [PubMed] [Google Scholar]
- 28.Shi, W., J. Wu, and B. P. Rosen. 1994. Identification of a putative metal binding site in a new family of metalloregulatory proteins. J. Biol. Chem. 269:19826-19829. [PubMed] [Google Scholar]
- 29.Sun, Y., M. D. Wong, and B. P. Rosen. 2002. Both metal binding sites in the homodimer are required for metalloregulation by the CadC repressor. Mol. Microbiol. 44:1323-1329. [DOI] [PubMed] [Google Scholar]
- 30.Sun, Y., M. D. Wong, and B. P. Rosen. 2001. Role of cysteinyl residues in sensing Pb(II), Cd(II), and Zn(II) by the plasmid pI258 CadC repressor. J. Biol. Chem. 276:14955-14960. [DOI] [PubMed] [Google Scholar]
- 31.Turner, J. S., P. D. Glands, A. C. Samson, and N. J. Robinson. 1996. Zn2+-sensing by the cyanobacterial metallothionein repressor SmtB: different motifs mediate metal-induced protein-DNA dissociation. Nucleic Acids Res. 24:3714-3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winn, M. D., G. N. Murshudov, and M. Z. Papiz. 2003. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 374:300-321. [DOI] [PubMed] [Google Scholar]
- 33.Wong, M. D., B. Fan, and B. P. Rosen. 2004. Bacterial transport ATPases for monovalent, divalent and trivalent soft metal ions, p. 159-178. In M. Futai, Y. Wada, and J. H. Kaplan (ed.), Handbook of ATPases. Wiley-VCH, Weinheim, Germany.
- 34.Wong, M. D., Y. F. Lin, and B. P. Rosen. 2002. The soft metal ion binding sites in the Staphylococcus aureus pI258 CadC Cd(II)/Pb(II)/Zn(II)-responsive repressor are formed between subunits of the homodimer. J. Biol. Chem. 277:40930-40936. [DOI] [PubMed] [Google Scholar]
- 35.Wu, J., and B. P. Rosen. 1991. The ArsR protein is a trans-acting regulatory protein. Mol. Microbiol. 5:1331-1336. [DOI] [PubMed] [Google Scholar]
- 36.Wu, J., and B. P. Rosen. 1993. Metalloregulated expression of the ars operon. J. Biol. Chem. 268:52-58. [PubMed] [Google Scholar]
- 37.Xu, C., and B. P. Rosen. 1999. Metalloregulation of soft metal resistance pumps, p. 5-19. In B. Sarkar (ed.), Metals and genetics. Plenum Press, New York, N.Y.