Abstract
The genomic distribution of the abundant eukaryotic d(GA·TC)n DNA microsatellite suggests that it could contribute to DNA recombination. Here, it is shown that this type of microsatellite DNA sequence enhances DNA recombination in SV40 minichromosomes, the rate of homologous DNA recombination increasing by as much as two orders of magnitude in the presence of a d(GA·TC)22 sequence. This effect depends on the region of the SV40 genome at which the d(GA·TC)22 sequence is cloned. It is high when the sequence is located proximal to the SV40 control region but no effect is observed when located 3.5 kb away from the SV40 ori. These results indicate that the recombination potential of d(GA·TC)n sequences is likely linked to DNA replication and/or transcription. The potential contribution of the structural properties of d(GA·TC)n sequences to this effect is discussed.
INTRODUCTION
A distinctive trait of the genomes of higher eukaryotes is its high content of repetitive DNA sequences. Microsatellite DNA sequences, formed by the tandem repetition of very simple monomer units, from 1 to 8 nt long, are fairly abundant in eukaryotic genomes. The simple repeating d(GA·TC)n microsatellite is especially abundant in eukaryotic genomes, accounting for ~0.4–0.5% of the total mammalian genome (1,2). Little is known, however, about its possible biological function(s). The genomic distribution of this microsatellite suggests that it could be involved in processes of genetic recombination. These microsatellite sequences are found at ‘hot-spots’ for genetic recombination (3–5). They are also frequently present in the vicinity of genes belonging to large multigene families and it was proposed that, by enhancing gene conversion events, they could contribute to the establishment, stabilization and/or maintenance of the sequence homogeneity of the multigene family (6–11). Little is known, however, about the direct contribution of d(GA·TC)n microsatellite sequences to DNA recombination. It was shown that when cloned into the DNA tumor virus SV40, they induce an increased genomic instability that could arise from an increased frequency of DNA recombination (12,13). In this paper, the determination of the direct contribution of d(GA·TC)n microsatellite sequences to somatic DNA recombination was addressed using SV40 minichromosomes as a model system that mimics the properties of eukaryotic chromatin (14).
MATERIALS AND METHODS
DNAs
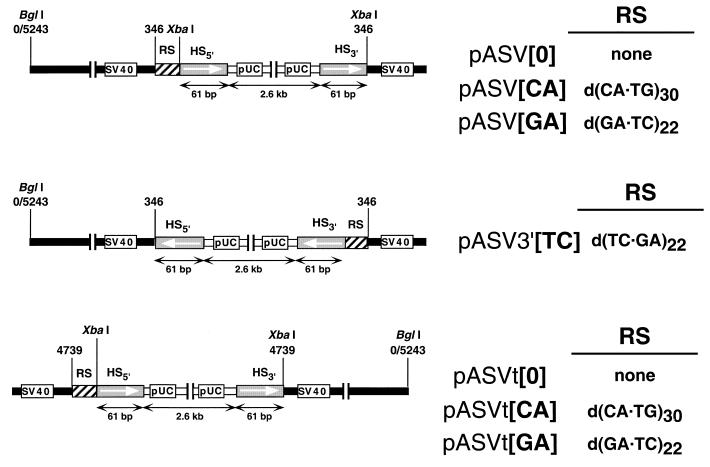
Figure 1 shows the different constructs used in these experiments. They were obtained by cloning, at the unique HpaII site (position 346) (pASV and pASV3′ vectors) or TaqI site (position 4739) (pASVt vectors) of SV40, a 2.6 kb long pUC18 derivative carrying a duplication of a modified polylinker missing the XbaI site. The genomic organization of these constructs is such that the non-viral DNA is flanked by two 61 bp long direct repeats (HS) arising from the duplicated polylinker. The sequence of the direct repeats is: 5′-AAG CTT GCA TGC CTG CAG GTC GAC TCT AGC TAG AGG ATC CCC GGG TAC CGA GCT CGA ATT C-3′. In pASV and pASVt vectors, microsatellite DNA sequences (RS) were located preceding the 5′ HS sequence. The sequences of the regions located between the SV40 sequence and HS5′ is: 5′-CGA CGG ATC CCA (RS) GGG ATC TCT AGA-3′ where RS corresponds to (CA)30 ([CA] constructs), (GA)22 ([GA] constructs) or no repeated sequence at all ([0] constructs). A short, 16 bp long, non-viral DNA fragment is located between HS3′ and the SV40 sequence. pASV3′[TC] was obtained by PCR amplification of the non-viral DNA insert of pASV[GA] that was then cloned at the HpaII site of SV40 in the reverse orientation with respect to pASV[GA]. As a consequence, in pASV3′[TC], the d(GA·TC)22 sequence precedes HS3′ and is inverted with respect to pASV[GA], the homopurine sequence being located on the lower strand. pASV3′[TC] also contains two short (40–50 bp) inverted repeats corresponding to the regions located between the primers used for the PCR amplification and the site of insertion of the non-viral DNA.
Figure 1.
Genomic organization of the constructs used in these experiments. All constructs contained a 2.6 kb long non-viral DNA insert flanked by 61 bp long direct repeats (HS), one of which is preceded by a repeated DNA sequence (RS). The BglI site at nucleotide position 0/5243 marks the position of the SV40 ori. See Materials and Methods for details.
Plaque assay
For the transfection experiments, CVI cells were grown on 60 mm diameter plates and transfected at ~90% confluence by the DEAE–dextran method (15), each plate receiving 50 ng of DNA in a solution containing 0.5 mg/ml of DEAE–dextran (Sigma). After transfection, cells were over-laid with 5 ml of media containing 0.9% agar and were fed every 3 days with the same media. After 10 days of transfection, plaques were visualized with 0.05% neutral red. The number of plaques was then counted every day until 17 days post-transfection. For the analysis of the genomic organization of the recombinants, individual plaques were isolated and used to prepare low-titer viral stocks to infect CV1 cells. Hirt extracts (16) were prepared when ~50% of the cells showed marked cytopathology. Viral DNAs were purified by CsCl gradient centrifugation and analyzed by restriction endonuclease cleavage and DNA sequencing.
RESULTS
The nucleosomal organization of the SV40 minichromosome imposes that DNA molecules exceeding in more than ~200 bp the actual size of the SV40 genome (5243 bp) would not be packed into the viral capside (14). As a consequence, oversized SV40 DNA molecules would not give rise to infective viral particles unless reduced by DNA recombination to a packagable size while preserving the SV40 regions that are essential for viral replication, transcription and encapsidation that is, most of the SV40 genome (14). This circumstance constitutes the basis of the experimental approach we used to determine the effects of d(GA·TC)n microsatellite DNA sequences on DNA recombination. The pASV constructs described in Figure 1 are 7850 bp long and, therefore, they cannot give rise by themselves to infective viral particles. In these constructs, the non-viral DNA insert is flanked by two short, 61 bp long, direct repeats (HS) so that recombination across them would reconstitute fully-infective wild-type SV40. Upon transfection into permissive CV1 cells, the formation of infective SV40 particles can be easily monitored by the plaque assay (17), providing a useful approach to estimate the frequency of recombination occurring across the HS sequences since each plaque arises from a single recombination event. In pASV plasmids, one of the HS sequences is preceded by either a d(CA·TG)30 ([CA] constructs) or d(GA·TC)22 ([GA] constructs) microsatellite DNA sequence, or no-repeated sequence at all ([0] constructs). Comparison of the relative plaquing efficiency of the different constructs would provide an estimate of the effects of the microsatellite sequences on the frequency of recombination across the HS sequences. Similar experiments have been performed by others to study DNA recombination in somatic cells (18–20).
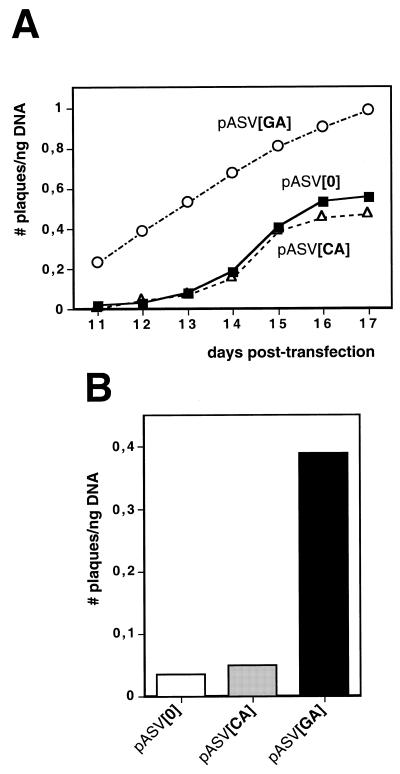
Figure 2 shows the plaquing efficiency, expressed as number of plaques obtained per ng of transfected DNA, of plasmids pASV[0], pASV[CA] and pASV[GA]. These constructs carry the non-viral DNA insert at the unique HpaII site of SV40 (Fig. 1, top), that occurs at position 346 at the 5′UTR region of the late mRNAs, just at the 3′ border of the SV40 control region (14). The plaquing efficiency was determined as a function of increasing post-transfection time, from 11 to 17 days (Fig. 2A). Plasmid pASV[GA] shows a higher plaquing efficiency than plasmids pASV[0] and pASV[CA]. This difference is higher at early stages of post-transfection, from 11 to 13 days, when only a few plaques are obtained with plasmids pASV[0] and pASV[CA]. At day 12 post-transfection, the plaquing efficiency of pASV[GA] is around 20 times higher than that of pASV[0] and pASV[CA] (Fig. 2B). The number of plaques obtained from plasmids pASV[0] and pASV[CA] increases from 14 to 16 days post-transfection. At these times of post-transfection, the number of plaques obtained with pASV[GA] also increases but to a lower rate likely because, in this case, a significantly lower number of cells are available to host new recombination events due to the early on-set of the formation of plaques. As a consequence, the difference in plaquing efficiency of pASV[GA] with respect to pASV[0] and pASV[CA] decreases with time of post-transfection. The results presented in Figure 2 correspond to a single experiment in which 400 ng of each of the three constructs were transfected to eight individual plates of subconfluent CV1 cells, each receiving 50 ng of DNA. As a consequence of the variable transfection efficiency, the actual plaquing efficiency of each construct varies from experiment to experiment but the differences in plaquing efficiency are always of the same magnitude. Table 1 shows the average plaquing efficiencies, at day 12 post-transfection, obtained from four individual series of transfections. As seen in Table 1, the plaquing efficiency of pASV[GA] is 18-fold higher than that of pASV[0]. On average, pASV[CA] also shows a modest higher plaquing efficiency, ~2-fold, than pASV[0]. Similar results were obtained with equivalent constructs missing the HS sequences but in which the non-viral DNA insert was flanked by direct repeats of the simple repeating microsatellite DNA sequences (RS). Also in this case, constructs carrying two copies of the d(GA·TC)22 sequence showed a higher plaquing efficiency than constructs carrying no RS sequences (8-fold) or two copies of the d(CA·GT)30 sequence (4-fold).
Figure 2.
(A) The plaquing efficiency, expressed as number of plaques per ng of transfected DNA, of pASV[GA] (circles), pASV[CA] (triangles) and pASV[0] (squares) is presented as a function of increasing days of post-transfection. (B) The plaquing efficiency at day 12 post-transfection is presented for pASV[GA], pASV[CA] and pASV[0].
Table 1. Relative recombination potential of pASV constructs.
| Plasmid | Totala | Homologousb | Non-homologousc |
|---|---|---|---|
| pASV[0] | 1 (29 ± 3) | 1 | 1 |
| pASV[CA] | 2 (61 ± 15) | 4 | 2 |
| pASV[GA] | 18 (512 ± 92) | 165 | 10 |
aThe relative recombination potential is expressed as the number of plaques obtained per ng of transfected DNA normalized to that of pASV[0]. The values correspond to the average of four independent series of experiments that, altogether, account for the transfection of 2400 ng of DNA of each of the constructs. Numbers in parentheses correspond to the actual recombination potential (number of plaques/ng DNA × 103) of each of the constructs.
bFor each construct, the potential of homologous recombination was determined by applying the proportion of recombinants originated by homologous recombination (see Table 2) to the total recombination potential. Values are normalized with respect to that of pASV[0].
cThe relative potential of non-homologous recombination was determined as described above, but taking into consideration the proportion of recombinants originated by non-homologous recombination (see Table 2).
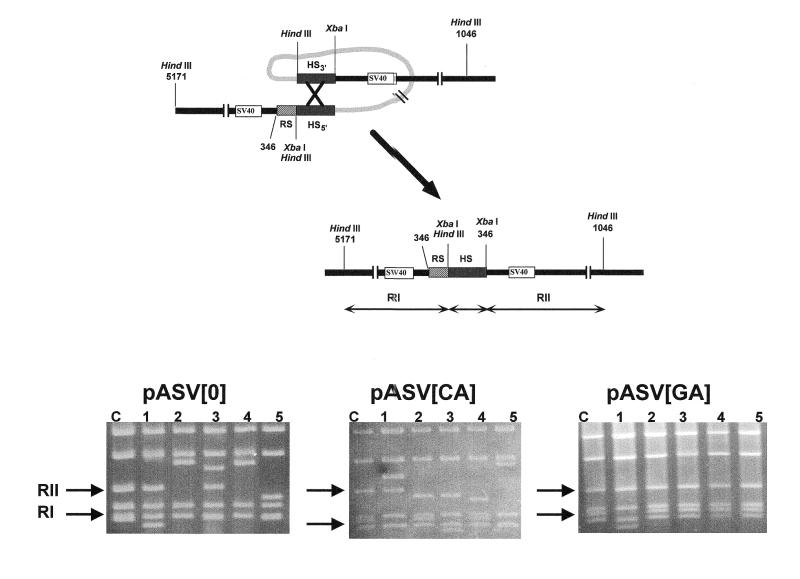
In pASV plasmids, infective SV40 particles could arise from homologous DNA recombination events occurring across the HS sequences or by non-homologous DNA recombination involving only one, or none, of the HS sequences and other sequences located nearby. To address the question as to what extent homologous DNA recombination across the HS sequences contributes to the generation of infective viral particles, the genomic organization of several SV40 recombinants obtained from each of the pASV constructs described above was determined by restriction endonuclease analysis and DNA sequencing. The two HS sequences contain a HindIII site, at their 5′ ends, and an XbaI site, which is located at the 5′ end in HS5′ but at the 3′ end in HS3′ (Fig. 1). Homologous DNA recombination across the HS sequences would result in molecules carrying a single HS sequence flanked by two XbaI sites (Fig. 3, top). As a consequence, the XbaI–HindIII restriction pattern of these recombinants will show a characteristic 700 bp long restriction fragment (RII), arising from cleavage at the 3′ XbaI site of the HS sequence and the HindIII site occurring at position 1046 on the SV40 genome, and a 500 bp long RI fragment, originating from digestion at the SV40 HindIII site at position 5171 and the 5′ XbaI, or HindIII, site of the HS sequence. The XbaI–HindIII restriction patterns of the original pASV constructs show RI and RII fragments of precisely these lengths, arising from cleavage at the XbaI sites located 5′ of HS5′ and 3′ of HS3′. Non-homologous DNA recombination events involving only one, or none, of the HS sequences would result in recombinant viruses showing restriction fragments RI and/or RII of variable lengths. Figure 3 (bottom) shows the XbaI–HindIII restriction patterns of several SV40 recombinants. In the case of the pASV[0] and pASV[CA] recombinants, fragments RI and RII are highly variable in length, as expected if generated by non-homologous DNA recombination events. On the contrary, many of the pASV[GA] recombinants show RI and RII fragments of the length expected if generated by homologous DNA recombination across the two HS sequences. Further analysis by DNA sequencing allows the determination of the precise exchange sites. Table 2 summarizes the results obtained from these analysis. The majority of the pASV[0] and pASV[CA] recombinants originate from non-homologous DNA recombination events that involved none, or only one, of the HS sequences with only a very minor proportion of the recombinants arising from homologous DNA recombination. On the other hand, in the case of the pASV[GA] recombinants, homologous DNA recombination accounts for the formation of a significant proportion of the recombinants, ~46%. For each construct, the potential of homologous DNA recombination can be estimated from the plaquing efficiency and the proportion of recombinants originated by homologous recombination. As summarized in Table 1, in the case of pASV[GA], the rate of homologous DNA recombination is increased by ~165-fold, with respect to pASV[0], while the rate of non-homologous exchanges increases only by ~10-fold. In the case of pASV[CA], homologous recombination is also slightly more favoured (Table 1). It must be noted, however, that the experimental approach used here allows only the identification of recombination events occurring at, or close to, the HS sequences. Non-homologous DNA recombination events giving rise to recombinants exceeding the maximal packagable size, or missing functionally important SV40 sequences, would not be detected. Therefore, while homologous DNA recombination across the HS sequences is precisely determined by this approach, the rate of non-homologous DNA recombination is likely to be underestimated.
Figure 3.
(Top) SV40 recombinants arising from homologous DNA recombination across the HS sequences contained a single HS sequence flanked by two XbaI sites. Cleavage with XbaI and HindIII gave rise to RI and RII fragments of 500 and 700 bp in length, respectively (see text for details). (Bottom) The XbaI–HindIII restriction patterns are presented for five independent recombinants (lanes 1–5) arising from pASV[0], pASV[CA] and pASV[GA]. Lanes C correspond to the restriction patterns obtained from the original vectors and the arrows indicate the positions corresponding to fragments RI and RII.
Table 2. Proportion of pASV recombinants arising from homologous and non-homologous DNA recombination events.
| Recombinants | pASV[0] | pASV[CA] | pASV[GA] |
|---|---|---|---|
| Totala | 21 | 19 | 24 |
| Homologousb | 1 (5%) | 2 (10%) | 11 (46%) |
| Non-homologousb | 20 (95%) | 17 (90%) | 13 (54%) |
aTotal number of recombinants analyzed.
bThe number of recombinants originated by homologous and non-homologous DNA recombination as determined by restriction endonuclease cleavage and DNA sequencing. The corresponding percentages are shown in parenthesis.
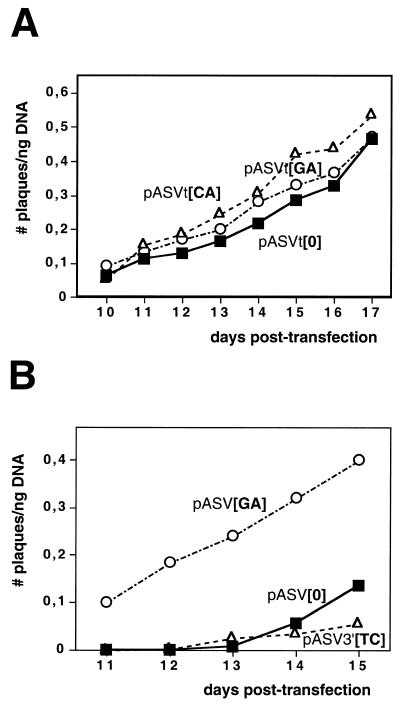
The results reported above indicate that d(GA·TC)n microsatellite DNA sequences can enhance homologous DNA recombination in SV40. This effect is, however, dependent on the region of the SV40 genome at which the d(GA·TC)n sequence was cloned. In pASVt constructs, the non-viral DNA insert was cloned at the unique TaqI site of SV40 (Fig. 1, bottom), which occurs at position 4739 within the intron of the SV40 T-antigen (14), instead of at the HpaII site as in pASV plasmids. Otherwise, the genomic organization of pASV and pASVt constructs is totally equivalent. As seen in Figure 4A, the plaquing efficiency of pASVt[GA], which carries a d(GA·TC)22 sequence, is not significantly different to those of pASVt[CA] or pASVt[0], which carry a d(CA·TG)30 sequence or no repeated RS sequence, respectively. A main difference between pASV and pASVt constructs refers to the relative position of the repeated RS sequence with respect to the SV40 control region. In pASV[GA], the d(GA·TC)22 sequence is proximal to the SV40 control region, ~350 bp away from the SV40 ori. On the other hand, in pASVt[GA], the d(GA·TC)22 sequence is located much more distal, at ~3500 bp from the SV40 ori. That proximity to the SV40 control region can influence the recombination potential of d(GA·TC)n sequences is also suggested by the fact that the plaquing efficiency of pASV3′[TC], in which the d(GA·TC)22 sequence is located at the HpaII site as in pASV plasmids but distal with respect to the SV40 control region as in pASVt constructs (Fig. 1, middle), is not significantly different to that of pASV[0] (Fig. 4B).
Figure 4.
(A) The plaquing efficiency of pASVt[GA] (circles), pASVt[CA] (triangles) and pASVt[0] (squares) is presented as a function of increasing days of post-transfection. (B) The plaquing efficiency of pASV3′[TC] (triangles), pASV[GA] (circles) and pASV[0] (squares) is presented as a function of increasing days of post-transfection.
DISCUSSION
The genomic distribution of the abundant d(GA·TC)n eukaryotic microsatellite suggests that it might be involved in processes of genetic recombination. In agreement with this hypothesis, we have shown here that d(GA·TC)n DNA sequences can enhance homologous DNA recombination in SV40 minichromosomes by as much as two orders of magnitude. Others had reported earlier that the most abundant d(CA·TG)n eukaryotic microsatellite is recombinogenic in SV40 (19,20), as well as in other eukaryotic systems (21–25). In the experiments described here, d(CA·TG)n sequences show, however, only a modest effect on DNA recombination. These differences are likely to arise from the different length of the regions of homology (HS) used to assess the recombination potential of d(CA·TG)n sequences in SV40, 61 bp in our vectors but 150–750 bp in those used by others (19,20). The high dependence of the frequency of recombination on the length of the homologous sequences strongly supports this interpretation.
Our results also indicate that, in SV40 minichromosomes, the recombination potential of d(GA·TC)n sequences depends on their localization on the viral genome. It is high when the sequences are located at the HpaII site, just at the 3′ border of the SV40 control region, but no effect is detected when they are located far away from the SV40 control region. In pASV3′[TC], the d(GA·TC)22 sequence is inverted with respect to pASV[GA] and, in addition to the two direct HS repeats, pASV3′[TC] carries two short inverted repeats (Materials and Methods). These two aspects could also influence the recombination potential of pASV3′[TC]. The SV40 control region is a highly active genomic region that contains the origin of viral replication, the early and late promoters, and the strong SV40 enhancer (14). Though further work is required before establishing the extent to which DNA replication and/or transcription determines the high recombination potential of pASV[GA], it is conceivable that proximity to such an active genomic region would have a strong effect on the rate of recombination. Increasing evidence indicates that, either due to a general increase in DNA accessibility or to specific pathways, both DNA replication and transcription induce a higher frequency of double-stranded-breaks and, therefore, facilitate DNA recombination (26,27). Interestingly, d(GA·TC)n microsatellite sequences are known to promote amplification, and integration, of large tandem arrays of U2 RNA genes only in the presence of active promoter elements (6). Furthermore, it was shown that alternating d(GA·TC)n sequences arrest DNA replication both in vitro and in vivo (28–30). Stalled replication forks are known to induce DNA repair and recombination (27).
Alternating d(GA·TC)n DNA sequences are structurally polymorphic, showing a high tendency to form triple-stranded DNA conformations (31–33). Though there is no direct evidence for the formation of triplex DNA at the d(GA·TC)22 sequence of pASV[GA], it is tempting to speculate that their recombination potential is actually linked to the structural properties of the microsatellite sequence. Similar experiments, performed both in bacterial and mammalian cells, with other homopurine·homopyrimidine sequences showed that their effects on DNA recombination correlate with their potential to form triple-stranded DNA (34,35). Interestingly, it was shown that, in bacterial cells, a d(G·C)n DNA sequence forms triple-stranded DNA, and enhances homologous DNA recombination, only when located at a transcriptionally active site (35). Moreover, formation of triplex-DNA appears to account for the potentially recombinogenic arrest of DNA replication that occurs at d(GA·TC)n sequences (36). Local increases on negative supercoiling associated to transcription, and/or replication, could facilitate the formation of triple-stranded DNA. Formation of triplex-DNA could promote strand-invasion events and, therefore, stimulate DNA recombination. In addition, it was shown that the formation of triplex-DNA arrests branch migration in vitro, suggesting also a contribution to DNA recombination (37).
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr J. Bernués for helpful discussions. We acknowledge the contribution of Drs R. Beltrán and M. J. Melia at the very initial phases of the project. This work was financed by grants from the Spanish DGES (PB96-812) and the CIRIT of the Generalitat de Catalunya (SGR97-55). A.B was a recipient of a doctoral fellowship from the CIRIT. This work was carried out within the framework of the Centre de Referència en Biotecnologia of the Generalitat de Catalunya.
REFERENCES
- 1.Manor H., Rao,B.S. and Martin,R.G. (1988) Abundance and degree of dispersion of genomic d(GA)n.d(TC)n sequences. J. Mol. Evol., 27, 96–101. [DOI] [PubMed] [Google Scholar]
- 2.Birnboim H.C., Sederoff,R.R. and Paterson,M.C. (1979) Distribution of polypyrimidine·polypurine segments in DNA from diverse organisms. Eur. J. Biochem., 98, 301–307. [DOI] [PubMed] [Google Scholar]
- 3.Collier D.A., Griffin,J.A. and Wells,R.D. (1988) Non-B right-handed DNA conformations of homopurine.homopyrimidine sequences in the murine immunoglobulin Cα switch region. J. Biol. Chem., 263, 7397–7405. [PubMed] [Google Scholar]
- 4.Richards J.E., Gilliam,A.C., Shen,A., Tucker,P.W. and Blattner,F.R. (1983) Unusual sequences in the murine immunoglobulin µ–δ heavy-chain region. Nature, 306, 483–487. [DOI] [PubMed] [Google Scholar]
- 5.Weinreb A., Collier,D.A., Birshtein,B.K. and Wells,R.D. (1990) Left-handed Z-DNA and intramolecular triplex formation at the site of an unequal sister chromatid exchange. J. Biol. Chem., 265, 1352–1359. [PubMed] [Google Scholar]
- 6.Bailey A.D., Pavelitz,T. and Weiner,A.M. (1998) The microsatellite sequence (CT)n.(GA)n promotes stable chromosomal integration of large tandem arrays of functional human u2 small nuclear RNA genes. Mol. Cell. Biol., 18, 2262–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey A.D., Li,Z., Pavelitz,T. and Weiner,A.M. (1995) Adenovirus type 12-induced fragility of the human RNU2 locus requires U2 small nuclear RNA transcriptional regulatory elements. Mol. Cell. Biol., 15, 6246–6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hentschel C.C. (1982) Homocopolymer sequences in the spacer of sea urchin histone gene repeat are sensitive to S1 nuclease. Nature, 295, 714–716. [DOI] [PubMed] [Google Scholar]
- 9.Htun H., Lund,E. and Dahlberg,J.E. (1984) Human U1 genes contain an unusually sensitive nuclease S1 cleavage site within the conserved 3′-flanking region. Proc. Natl Acad. Sci. USA, 81, 7288–7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mason A.J., Evans,B.A., Cox,D.R., Shine,J. and Richards,R.I. (1983) Structure of mouse kallikrein gene family suggest a role in specific processing of biologically active peptides. Nature, 303, 300–307. [DOI] [PubMed] [Google Scholar]
- 11.Sekiya Y., Kuchino,Y. and Nishimura,S. (1981) Mammalian tRNA genes: nucleotide sequence of rat genes for tRNAAsp, tRNAGly and tRNAGlu. Nucleic Acids Res., 9, 2239–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernués J., Beltrán,R. and Azorín,F. (1991) SV40 recombinants carrying a d(CT·GA)22 sequence show increased genomic instability. Gene, 108, 269–274. [DOI] [PubMed] [Google Scholar]
- 13.Stringer J.R. (1985) Construction of a viable simian virus 40 variant that carries a poly[d(GT).d(CA)] insertion. J. Virol., 53, 698–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tooze J. (1981) DNA Tumour Viruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 15.Lai L.J. (1980) Mapping SV40 mutants by marker rescue. Methods Enzymol., 65, 811–816. [DOI] [PubMed] [Google Scholar]
- 16.Hirt B. (1967) Selective extraction of polyoma virus DNA. J. Mol. Biol., 26, 365–369. [DOI] [PubMed] [Google Scholar]
- 17.Mertz J.E. and Berg,P. (1974) Defective simian virus 40 genomes: isolation and growth of individual clones. Virology, 62, 112–124. [DOI] [PubMed] [Google Scholar]
- 18.Subramani S. and Berg,P. (1983) Homologous and non-homologous recombination in monkey cells. Mol. Cell. Biol., 3, 1040–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stringer J.R. (1985) Recombination between poly[d(GT).d(CA)] sequences in simian virus 40-infected cells. Mol. Cell. Biol., 5, 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bullock P., Miller,J. and Botchan,M. (1986) Effects of poly[d(pGpT).d(pApC)] and poly[d(pCpG).d(pCpG)] repeats on homologous recombination in somatic cells. Mol. Cell. Biol., 6, 3948–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sargent R.G., Merrihew,R.V., Nairn,R., Adair,G., Meuth,M. and Wilson,J. (1996) The influence of (GT)29 microsatellite sequence in homologous recombination in the hamster adenine phosphoribosyltransferase gene. Nucleic Acids Res., 24, 746–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Treco D., Thomas,B. and Arnheim,N. (1985) Recombination hot spot in human β-globin gene cluster: meiotic recombination of human DNA fragments in Saccharomyces cerevisiae. Mol. Cell. Biol., 5, 2029–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Treco D. and Arnheim,N. (1986) The evolutionarily conserved repetitive sequence d(TG.AC)n promotes reciprocal exchange and generates unusual recombinant tetrads during yeast meiosis. Mol. Cell. Biol., 6, 3934–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wahls W.P., Wallace,L.J. and Moore,P.D. (1990) The Z-DNA motif d(TG)30 promotes reception of information during gene conversion events while stimulating homologous recombination in human cells in culture. Mol. Cell. Biol., 10, 785–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gendrel C.-G., Boulet,A. and Dutreix,M. (2000) (CA/GT)n microsatellites affect homologous recombination during yeast meiosis. Genes Dev., 14, 1261–1268. [PMC free article] [PubMed] [Google Scholar]
- 26.Nicolas A. (1998) Relationship between transcription and initiation of meiotic recombination: toward chromatin accessibility. Proc. Natl Acad. Sci. USA, 95, 87–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haber J.E. (1999) DNA recombination: the replication connection. Trends Biochem. Sci., 24, 271–275. [DOI] [PubMed] [Google Scholar]
- 28.Samadashwily G.M., Dayn,A. and Mirkin,S.M. (1993) Suicidal nucleotide sequences for DNA polymerization. EMBO J., 12, 4975–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao B.S., Manor,H. and Marin,R.G. (1988) Pausing in simian virus 40 DNA replication by a sequence containing (dG-dA)27.(dT-dC)27. Nucleic Acids Res., 16, 8077–8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lapidot A., Baran,N. and Manor,H. (1989) (dT-dC)n and (dG-dA)n tracts arrest single-stranded DNA replication in vitro. Nucleic Acids Res., 17, 883–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernués J. and Azorín,F. (1995) Triple-stranded DNA. Nucleic Acids Mol. Biol., 9, 1–21. [Google Scholar]
- 32.Frank-Kamenestkii M.D. and Mirkin,S.M. (1995) Triplex DNA structures. Annu. Rev. Biochem., 64, 65–95. [DOI] [PubMed] [Google Scholar]
- 33.Mirkin M.S. and Frank-Kamenestkii,M.D. (1994) H-DNA and related structures. Annu. Rev. Biophys. Biomol. Struct., 23, 541–576. [DOI] [PubMed] [Google Scholar]
- 34.Rooney S.M. and Moore,P.D. (1995) Antiparallel, intramolecular triplex DNA stimulates homologous recombination in human cells. Proc. Natl Acad. Sci. USA, 92, 2141–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohwi Y. and Panchenko,Y. (1993) Transcription-dependent recombination induced by triple-helix formation. Genes Dev., 7, 1766–1778. [DOI] [PubMed] [Google Scholar]
- 36.Baran N., Lapidot,A. and Manor,H. (1991) Formation of DNA triplexes accounts for arrests of DNA synthesis at d(TC)n and d(GA)n tracts. Proc. Natl Acad. Sci. USA, 88, 507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benet A. and Azorín,F. (1999) The formation of triple-stranded DNA prevents spontaneous branch-migration. J. Mol. Biol., 294, 851–857. [DOI] [PubMed] [Google Scholar]