Abstract
Several flagellar genes in Helicobacter pylori are dependent on σ54 (RpoN) for their expression. These genes encode components of the basal body, the hook protein, and a minor flagellin, FlaB. A protein-protein interaction map for H. pylori constructed from a high-throughput screen of a yeast two-hybrid assay (http://pim.hybrigenics.com/pimriderext/common/) revealed interactions between σ54 and the conserved hypothetical protein HP0958. To see if HP0958 influences σ54 function, the corresponding gene was disrupted with a kanamycin resistance gene (aphA3) in H. pylori ATCC 43504 and the resulting mutant was analyzed. The hp0958:aphA3 mutant was nonmotile and failed to produce flagella. Introduction of a functional copy of hp0958 into the genome of the hp0958:aphA3 mutant restored flagellar biogenesis and motility. The hp0958:aphA3 mutant was deficient in expressing two σ54-dependent reporter genes, flaB′-′xylE and hp1120′-′xylE. Levels of σ54 in the hp0958 mutant were substantially lower than those in the parental strain, suggesting that the failure of the mutant to express the genes in the RpoN regulon and produce flagella was due to reduced σ54 levels. Expressing σ54 at high levels by putting rpoN under the control of the ureA promoter restored flagellar biogenesis and motility in the hp0958:aphA3 mutant. Turnover of σ54 was more rapid in the hp0958:aphA3 mutant than it was in the wild-type strain, suggesting that HP0958 supports wild-type σ54 levels in H. pylori by protecting it from proteolysis.
Helicobacter pylori, a member of the ɛ subdivision of the proteobacteria, colonizes the human gastric epithelium, which leads to a gastric inflammation that can progress to chronic gastritis, peptic ulcer, gastric cancer, or mucosa-associated lymphoma (6, 10, 12). H. pylori must be motile to colonize the gastric epithelium (13, 14), and motility by the bacterium occurs through the action of two to six polar flagella.
Flagellar biogenesis in H. pylori involves the coordinated expression of over 40 flagellar genes scattered throughout the genome and organized into 25 or more transcriptional units (1, 33). Transcriptional regulation of these flagellar operons in H. pylori is complex, involving all three σ factors found in the bacterium, σ80 (the primary σ factor in H. pylori), σ54 (RpoN), and σ28 (FliA) (7, 15, 19, 29, 30, 32). In this regard regulation of flagellar biogenesis in H. pylori is similar to that of Vibrio cholerae and Pseudomonas aeruginosa, which also require both σ54 and σ28 for expression of different classes of flagellar genes (11, 23, 34).
In H. pylori, σ80 is required for transcription of flagellar genes whose products are needed early in flagellar biogenesis and include basal body proteins and components of the flagellar protein export apparatus (30). H. pylori flagellar genes whose products are required midway through flagellar assembly are dependent on σ54 for their expression and include genes that encode the proximal rod proteins of the basal body (flgBC), the hook protein (flgE), hook-associated proteins (flgK and flgL), and a minor flagellin (flaB) (19, 29, 32). Finally, genes whose products are needed at the end of flagellar biogenesis are transcribed by σ28-RNA polymerase holoenzyme and include genes encoding the major flagellin (flaA), the filament cap protein (fliD), and flagellar protein chaperones (fliS and fliT) (15, 16, 19).
Transcription of σ54-dependent genes in H. pylori requires the activator FlgR, which belongs to the NtrC family of transcriptional activators (7, 29). FlgR is a response regulator of a two-component signal transduction system and must be phosphorylated by its cognate sensor kinase, FlgS, to activate transcription (2, 7, 19). Sensor kinases are often responsive to environmental or cellular signals, but it is not known if FlgS responds to such cues. Activators of σ54-RNA polymerase holoenzyme (σ54-holoenzyme) typically bind enhancers located upstream of the promoters of the genes which they activate (8, 25). After binding to the enhancer, the activator contacts σ54-holoenzyme bound at the promoter in a closed complex via looping of the DNA between the enhancer and promoter (26, 31). The activator hydrolyzes ATP and couples energy released from hydrolysis to stimulate the isomerization of the closed promoter complex to an open complex that is competent to initiate transcription (22, 35). Unlike most σ54-dependent activators, H. pylori FlgR lacks a DNA-binding domain and apparently contacts σ54-holoenzyme directly rather than through DNA looping to activate transcription (7).
In addition to the involvement of the FlgS/FlgR system in the regulation of σ54-dependent genes, other factors may regulate expression of genes of the RpoN regulon. A protein-protein interaction map for H. pylori was constructed using a high-throughput screen of a yeast two-hybrid system (24). The protein-protein interaction map indicated interactions between σ54 and the conserved hypothetical protein HP0958 (Hybrigenics PimRider database; http://pim.hybrigenics.com/pimriderext/common/). To determine if HP0958 influences σ54 function in H. pylori, we disrupted its corresponding gene with a cassette bearing a kanamycin resistance gene (aphA3) in H. pylori ATCC 43504 and analyzed the phenotype of the resulting mutant. The hp0958:aphA3 mutant was nonmotile and aflagellated, which appeared to result from reduced levels of σ54. The turnover rate of σ54 in the hp0958:aphA3 mutant was significantly higher than that in the parental strain, suggesting that HP0958 protects σ54 from proteolysis.
MATERIALS AND METHODS
Bacterial strains and media.
Escherichia coli DH5α [φ80 lacZΔ M15 recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 deoRΔ (lacZYA-argF)U169] was grown in Luria-Bertani broth at 37°C. H. pylori ATCC 43504 was grown on blood agar supplemented with 10% sheep blood or tryptic soy agar supplemented with 5% horse serum (TSA serum) and grown at 37°C under an atmosphere of 4% oxygen, 5% carbon dioxide, and 91% nitrogen. Serum-free medium was used to grow liquid cultures of H. pylori in brain heart infusion broth supplemented with 0.1% β-cyclodextrin (Sigma) as described previously (9). Motility agar plates consisted of Mueller-Hinton broth supplemented with 5% horse serum and contained 0.35% agar. Sterile toothpicks were used to inoculate the motility agar with H. pylori strains, and motility was scored after incubating the plates at 37°C under an atmosphere of 4% oxygen, 5% carbon dioxide, and 91% nitrogen for 4 to 5 days. When required, medium was supplemented with 30 μg/ml chloramphenicol, kanamycin, or tetracycline.
Protein purification.
Details for construction of plasmids used for overproduction of H. pylori proteins are available upon request. PCR products used to construct the expression vectors were sequenced at the Integrated Biotechnology Laboratories at the University of Georgia to verify that no errors had been introduced during amplification.
A DNA fragment bearing hp0958 was amplified by PCR from H. pylori 26695 genomic DNA and cloned into plasmid pJES489, which is a derivative of pMAL-c (New England Biolabs), to create a fusion of hp0958 and E. coli malE (encoding the maltose-binding protein [MBP]). MBP-HP0958 was expressed from this plasmid in E. coli DH5α by growing a 1-liter culture to an optical density of 0.3 and then adding isopropyl-β-d-thiogalactoside (1 mM final concentration). After 3.5 h, cells were harvested; resuspended in 50 mM Tris-acetate, pH 8.2, 200 mM KCl, 1 mM EDTA, and 1 mM dithiothreitol (DTT); and lysed with a French pressure cell at 7,000 lb/in2. The crude cell extract was centrifuged at 17,000 × g for 45 min, and the resulting supernatant liquid was applied to an amylose-agarose (New England Biolabs) affinity column that had been equilibrated previously with 20 mM Tris-HCl, pH 7.4, 5% (wt/vol) glycerol, 1 mM EDTA, 1 mM DTT, and 200 mM KCl (buffer A). MBP-HP0958 was eluted from the column with buffer A plus 10 mM maltose; dialyzed against 20 mM Tris-HCl, pH 8.8, 0.5 mM DTT, and 5% (wt/vol) glycerol (buffer C); and then applied to a High Trap Q anion-exchange column (5 ml; Amersham Biosciences) that had been equilibrated previously with buffer C. MBP-HP0958 was eluted from the column with a gradient to buffer C plus 1 M KCl at a salt concentration of ∼0.2 M.
H. pylori rpoN was amplified by PCR from H. pylori strain 26695 and cloned into pJES489 to create a chimeric malE-rpoN gene. MBP-σ54 protein was overproduced in E. coli DH5α and purified through the amylose-agarose affinity chromatography step as described for MBP-HP0958. Fractions containing the protein were pooled and dialyzed against 10 mM Tris-HCl, pH 8.0, 50 mM KCl, 0.1 mM EDTA, 1 mM DTT, 10 mM MgCl2, 10 mM maltose, and 5% (wt/vol) glycerol (buffer B). The dialyzed fractions were applied to a heparin-agarose column which was equilibrated with buffer B. After being washed with buffer B, MBP-σ54 was eluted from the column at ∼0.2 M KCl with a linear gradient to buffer plus 1 M KCl.
H. pylori flaB was amplified by PCR from H. pylori strain 26695 and cloned into pJES489 to create a chimeric malE-flaB gene. MBP-FlaB protein was overproduced in E. coli DH5α and purified as described for MBP-HP0958. Antisera directed against MBP-HP0958, MBP-σ54, or MBP-FlaB were raised in New Zealand White rabbits by Cocalico Labs, Reamstown, PA.
Construction of H. pylori mutant strains.
Mutant strains were constructed using suicide vectors derived from pGEM-T (Promega) and contained fragments of the genes targeted for mutagenesis. Targeted genes were amplified from H. pylori 26695 genomic DNA by PCR, cloned into pGEM-T, and disrupted with a 1.4-kb EcoRI fragment bearing a Campylobacter coli aphA3 (conferring resistance to kanamycin) cassette from plasmid pHP1 (17). Suicide vectors were introduced into H. pylori ATCC 43504 by natural transformation, and recombinants in which the chromosomal copy of the targeted gene had been replaced by the disrupted copy of the gene were selected on TSA serum supplemented with kanamycin. Insertion of the aphA3 cassette in the targeted gene was verified by PCR using a set of primers that flanked the site of insertion. An H. pylori rpoN mutant was constructed by introducing an EcoRI site ∼370 bp from the start codon of H. pylori rpoN using the QuickChange II site-directed mutagenesis kit (Stratagene) and cloning the aphA3 cassette into this site. An H. pylori hp0958 mutant was constructed following introduction of the aphA3 cassette within an NheI site, which resulted in disruption of the gene at codon 76. An H. pylori hp0959 mutant was generated by inserting the aphA3 cassette into a BamHI site located ∼450 bp downstream of the start codon of hp0959.
Complementation of hp0958 mutant.
A ∼1.5-kb PCR product that carried most of hp0959 and all of hp0958 was cloned into pGEM-T and sequenced. The cloned DNA fragment was introduced into the EcoRV site of plasmid pEU39Cm (21), which carries a copy of H. pylori hp0405 disrupted with a cassette containing C. coli chloramphenicol transacetylase (cat). The EcoRV site is present in hp0405 and is adjacent to the cat cassette. The resulting suicide vector was transformed into the hp0958:aphA3 mutant, and recombinants in which the chromosomal copy of hp0405 had been replaced with the disrupted gene carrying hp0959-hp0958 along with the cat cassette were selected on TSA serum supplemented with chloramphenicol. Introduction of hp0959-hp0958 into the hp0405 locus was verified by PCR using a set of primers in which one was internal to cat and the other was within hp0405. PCR was also used to confirm that this strain retained the aphA3 cassette in the hp0958 locus.
Overproduction of σ54 and HP0958 in H. pylori.
H. pylori rpoN was introduced into plasmid pPA (5) to place it under the control of the H. pylori ureA promoter. A ∼1.4-kb DNA fragment bearing the ureA promoter plus rpoN from the resulting plasmid was cloned into the EcoRV site of plasmid pEU39Cm. This plasmid was transformed into the hp0958:aphA3 mutant and into wild-type H. pylori to introduce the rpoN allele under the control of the ureA promoter into the hp0405 locus as described above.
Similarly, H. pylori hp0958 was placed under the control of the ureA promoter by introducing it into plasmid pPA. A ∼975-bp DNA fragment bearing the PureA-hp0958 allele was cloned into pEU39Cm, and the resulting plasmid was transformed into the wild-type or hp0958:aphA3 mutant strains to introduce the PureA-hp0958 allele into the hp0405 locus.
Construction of H. pylori reporter strains.
Reporter genes in which the H. pylori flaB promoter region (positions −67 to +26 relative to the transcriptional start site) or the H. pylori flaA promoter region (positions −126 to +47 relative to the transcriptional start site) was joined to promoterless Pseudomonas putida xylE (encoding catechol 2,3-dioxygenase) have been described previously (7). An orf1120′-′xylE reporter gene was constructed by cloning a DNA fragment corresponding to positions −70 to +44 relative to the transcriptional start site of the hp1120-flgK operon upstream of xylE. The flaB′-′xylE, hp1120′-′xylE, and flaA′-′xylE reporter genes were cloned into the EcoRV site of plasmid pEU39Cm to create suicide vectors that were used to introduce the reporter genes into the hp0405 locus of various H. pylori strains as described above.
To construct a strain with an rpoN′-′xylE reporter gene, the start codon of the P. putida xylE gene was joined in-frame to codon 61 of H. pylori rpoN that had been cloned previously in pGEM-T. A 1.3-kb fragment bearing C. coli cat was cloned immediately downstream of xylE in the EcoRI site that had been introduced previously in rpoN. The resulting suicide vector was transformed into wild-type H. pylori and the hp0958:aphA3 mutant. Recombinants in which the chromosomal copy of rpoN was replaced with the rpoN′-′xylE reporter and the cat cassette were selected on TSA serum supplemented with chloramphenicol and verified by PCR.
XylE assays.
Whole-cell XylE assays were carried out essentially as described previously (7). The rate of product (2-hydroxymuconic semialdehyde) formation was determined, and activities were expressed as μmoles product formed/min/108 H. pylori cells from at least 10 independent assays for each sample.
Electron microscopy.
H. pylori cells were grown on blood agar for 48 h at 37°C and then gently resuspended in phosphate-buffered saline, pH 7.4. Samples were negatively stained with 2% phosphotungstic acid (pH 7.0) on Formvar-coated copper grids and observed with a JEOL 100CK electron microscope (JEOL USA, Peabody, MA) at the Center for Advanced Ultrastructural Research at the University of Georgia. Micrographs were taken at an accelerating voltage of 80 kV.
Western blot analysis.
Immunoblotting with primary antibodies directed against MBP-HP0958, MBP-σ54, MBP-FlaB, or VacA (Austral Biologicals, San Ramon, CA) was done as described previously (7). The bound antibody was detected by enhanced chemiluminescence using peroxidase-coupled goat anti-rabbit antibody as the secondary antibody (MP Biomedicals Inc., Aurora, ID). Antibody directed against MBP-σ54 was affinity purified prior to use as follows. Purified histidine-tagged σ54 was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and visualized by staining with Ponceau red. The band was excised, and the membrane strip was rinsed in distilled water, blocked with a 2% nonfat dry milk solution, washed with Tris-buffered saline containing 0.05% Tween 20 (TBST), and incubated overnight with 20 ml of a 1:5 dilution of antiserum directed against MBP-σ54. The membrane strip was washed five times with TBST and incubated with an acidic buffer (0.2 M glycine, 0.2 M sodium chloride, 1% gelatin, pH 2.8) for 15 min to elute the antibody. The eluate was dialyzed overnight against a buffer containing 50 mM citric acid and 50 mM Na2HPO4, pH 5.5, followed by overnight dialysis against Tris-buffered saline.
σ54 protein stability assay.
H. pylori cultures were grown to mid-log phase in serum-free medium as described previously (9), at which point tetracycline was added to stop protein synthesis. Aliquots were removed at various times following tetracycline addition. Cells were recovered by centrifugation and resuspended in phosphate-buffered saline. Approximately 108 cells from each sample were lysed and analyzed by Western blotting.
To assay for secreted proteins, H. pylori cultures were grown to mid-log phase in serum-free medium as described previously (9). Cells were removed by centrifugation for 15 min at 20,000 × g at 4°C. The resulting supernatant liquids were filtered through a 0.22-μm-pore-size membrane filter to remove any residual bacteria. Extracellular proteins were precipitated using a modified trichloroacetic acid method as described previously (9), and approximately 20 μg of precipitated protein was analyzed by Western blotting.
RESULTS
Inactivation of hp0958 interferes with flagellar biogenesis.
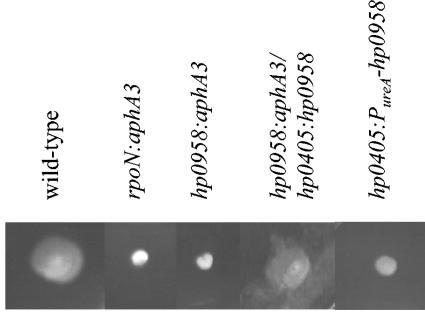
H. pylori hp0958 encodes a protein of unknown function that is predicted to be 254 amino acid residues in length. To determine if the interactions between HP0958 and σ54 observed in the yeast two-hybrid assay were of physiological relevance, hp0958 was disrupted by introducing a cassette bearing aphA3 into codon 76 of the gene in H. pylori ATCC 43504. The hp0958:aphA3 mutant was nonmotile when tested on motility agar (Fig. 1) as well as when examined microscopically. Since loss of motility could have resulted from defects of either flagellar assembly or function, the hp0958 mutant was examined by transmission electron microscopy for the presence of flagella. Sheathed polar flagella were readily apparent in the wild-type H. pylori strain but were absent in the hp0958 mutant (data not shown).
FIG. 1.
Motilities of H. pylori 43504 and various mutant derivatives. Cells were inoculated on semisolid motility agar plates with a sterile toothpick and incubated for 4 to 5 days at 37°C under microaerophilic conditions. Strains that were tested for motility were H. pylori 43504 (wild type), an rpoN:aphA3 mutant, an hp0958:aphA3 mutant, an hp0958:aphA3 mutant in which a functional copy of hp0958 was introduced into the hp0405 locus (hp0958:aphA3/hp0405:hp0958), and H. pylori 43504 bearing the PureA-hp0958 allele in the hp0405 locus (hp0405:PureA-hp0958).
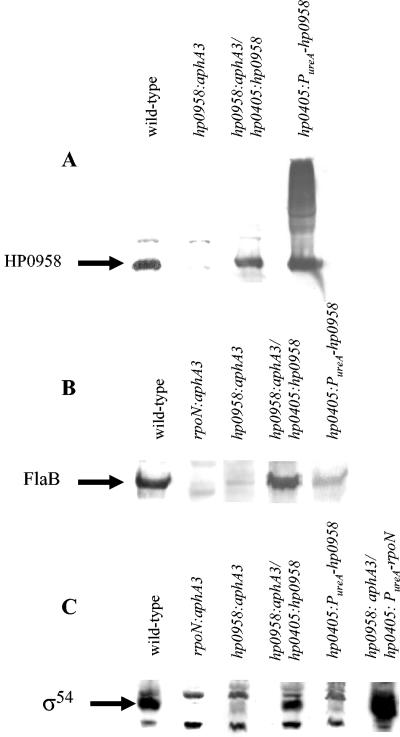
The gene immediately downstream of hp0958 is kdtA, which encodes 3-deoxy-d-manno-octulosonic acid transferase, an enzyme that is essential for lipopolysaccharide (LPS) biosynthesis (3, 4). Mutations in H. pylori that interfere with both LPS biosynthesis and flagellar biogenesis have been described previously (18), and so we wished to verify that the effect on flagellar biogenesis that we observed in the hp0958:aphA3 mutant was not due to polar effects on kdtA. Mass spectrometry analysis of purified LPS from the hp0958 mutant and wild-type strains revealed no differences between the two strains (data not shown), which argued strongly against the aphA3 cassette in hp0958 interfering with expression of kdtA or other genes downstream of hp0958. To confirm that disruption of hp0958 was responsible for the phenotype of the mutant strain, we complemented the hp0958:aphA3 mutant by introducing a functional copy of hp0958 in the mutant strain. The hp0958 gene along with most of the gene immediately upstream of hp0958 (hp0959) was introduced into the hp0958:aphA3 mutant at the hp0405 locus. H. pylori hp0405 encodes a predicted NifS-like protein of unknown function, and this locus was used previously to construct H. pylori merodiploid strains (20, 21). Disruption of hp0405 does not affect motility. Introduction of a functional copy of hp0958 into the hp0958:aphA3 mutant allowed expression of HP0958 at wild-type level (Fig. 2A) and restored flagellar synthesis and motility (Fig. 1). These results demonstrated that disruption of hp0958 was responsible for the lesion in flagellar biogenesis in the original mutant. Since hp0958 and hp0959 are separated by only 10 bp of DNA, the restoration of HP0958 expression to wild-type level in the complemented strain suggests that the promoter for hp0958 is in hp0959.
FIG. 2.
Western blot analysis of HP0958, FlaB, and σ54 in various H. pylori strains. For panels A and B, approximately 108 cells were lysed and loaded in each lane, while ∼4 × 108 cells were lysed and loaded in each lane in panel C. Membranes were probed with antiserum directed against MBP-HP0958 (A), MBP-FlaB (B), or MBP-σ54 (C). Relevant genotypes of the strains that were analyzed are indicated above the lanes and include H. pylori 43504 (wild type), an rpoN:aphA3 mutant, an hp0958:aphA3 mutant, an hp0958:aphA3 mutant complemented with a copy of hp0958 in the hp0405 locus (hp0958:aphA3/hp0405:hp0958), an hp0958:aphA3 mutant in which σ54 was overproduced (hp0958:aphA3/hp0405:PureA-rpoN), and H. pylori 43504 bearing a PureA-hp0958 allele in the hp0405 locus (hp0405:PureA-hp0958). Arrows indicate the positions of the proteins analyzed in each Western blot.
Homologs of HP0958 are found in several bacteria, including all of the ɛ-proteobacteria whose genomes have been sequenced to date (Helicobacter hepaticus, Campylobacter jejuni, and Wolinella succinogenes). In all of these representatives of ɛ-proteobacteria, the gene located immediately upstream of the hp0958 homolog shares homology with hp0959, and in W. succinogenes the two genes appear to be fused. This prompted us to investigate whether HP0959 also had a role in flagellar biogenesis. The aphA3 cassette was used to disrupt hp0959 in H. pylori ATCC 43504, and the resulting mutant expressed wild-type levels of HP0958 and was flagellated and motile (data not shown). These data indicate that, despite the apparent fusion of HP0958 and HP0959 homologs in W. succinogenes, HP0959 is not required for HP0958 function in H. pylori flagellar biogenesis.
Inactivation of HP0958 interferes with expression of flagellar genes in the RpoN regulon.
Given the interaction between σ54 and HP0958 in the yeast two-hybrid assay and our observation that HP0958 is required for flagellar biogenesis, we reasoned that HP0958 may influence σ54 function in H. pylori. To test this hypothesis, FlaB levels were assessed by Western blotting in H. pylori ATCC 43504 and the mutant strains. The flaB gene is part of the RpoN flagellar regulon (19, 29), and we observed that an rpoN mutant failed to accumulate FlaB (Fig. 2B). A faint cross-reacting protein band was seen just below where FlaB was expected to migrate in the lane with cell lysate from the rpoN mutant. FlaA shares 58.4% amino acid identity with FlaB and is slightly smaller than FlaB (53.3 kDa versus 53.9 kDa), and we infer that this cross-reacting band is FlaA. Consistent with this hypothesis, the FlaB antiserum cross-reacted weakly with purified histidine-tagged FlaA (data not shown). The level of FlaB in the hp0958:aphA3 mutant was dramatically reduced but was restored to wild-type level in the hp0958:aphA3 mutant carrying a functional copy of hp0958 in the hp0405 locus, as expected from the restoration of flagellar synthesis and motility (Fig. 2B).
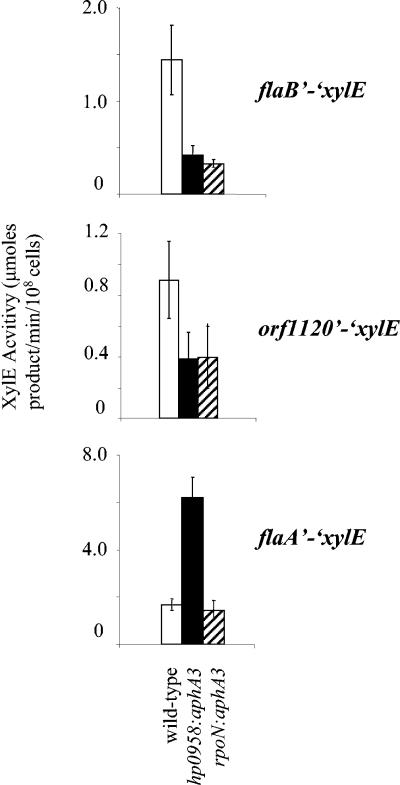
To determine if the reduced levels of FlaB in the hp0958:aphA3 mutant were due to decreased transcription of flaB, we introduced a flaB′-′xylE reporter gene into the hp0405 locus of the mutant. The flaB′-′xylE reporter gene was also introduced into the hp0405 locus of the rpoN:aphA3 and wild-type strains. Expression of the flaB′-′xylE reporter gene was reduced in the hp0958:aphA3 mutant to a level that was comparable to that in the rpoN:aphA3 mutant (Fig. 3), indicating that the reduced level of FlaB in the hp0958:aphA3 mutant was due to decreased transcription of flaB. Expression of another RpoN-dependent reporter gene, hp1120′-′xylE, was reduced in the hp0958 mutant (Fig. 3), suggesting that HP0958 is required for transcription of genes of the RpoN flagellar regulon. A σ28-dependent flaA′-′xylE reporter gene was introduced into the H. pylori strain to determine if HP0958 was required for expression of flagellar genes outside the RpoN regulon. Expression of the flaA′-′xylE reporter gene in the hp0958:aphA3 mutant was about threefold higher than that in the wild-type and rpoN:aphA3 mutant strains (Fig. 3), indicating that HP0958 is not required for expression of flagellar genes in the FliA regulon.
FIG. 3.
Expression of flaB′-′xylE, hp1120′-′xylE, and flaA′-′xylE reporter genes in various H. pylori strains. XylE activities were measured for the reporter genes indicated to the right of the graphs in H. pylori 43504 (wild type; open bars), an hp0958:aphA3 mutant (filled bars), and an rpoN:aphA3 mutant (striped bars). Values represent the averages of at least 10 assays, and standard deviations for these values are indicated by the error bars.
Inactivation of HP0958 results in a decreased level of σ54.
To determine if the decreased expression of flaB in the hp0958:aphA3 mutant was due to decreased activity or levels of σ54, σ54 levels in the hp0958:aphA3 mutant and its parental strain were compared. As shown in Fig. 3, the level of σ54 in the hp0958:aphA3 mutant was significantly reduced compared to the wild-type strain. Introduction of a functional copy of hp0958 in the hp0405 locus of the hp0958:aphA3 mutant restored the level of σ54 to close to wild-type level, demonstrating that HP0958 is required for the accumulation of a wild-type level of σ54.
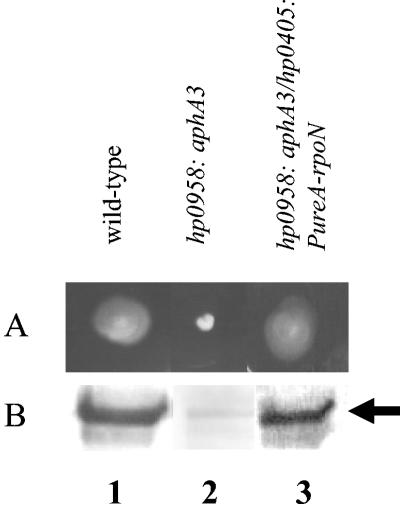
To test if overproduction of σ54 in the hp0958:aphA3 mutant would restore flagellar synthesis and motility, rpoN was placed under the control of the ureA promoter and introduced into the hp0405 locus of the hp0958:aphA3 mutant. The level of σ54 in this strain exceeded that of the wild-type strain (Fig. 2C) and restored motility and expression of flaB (Fig. 4). Taken together, these data suggest that HP0958 is required for accumulation, but not function, of σ54.
FIG. 4.
Overproduction of σ54 in the hp0958:aphA3 mutant restores motility and FlaB synthesis. (A) The motilities of H. pylori 43504 (wild type), an hp0958:aphA3 mutant, and an hp0958:aphA3 mutant in which σ54 was overproduced from the ureA promoter were assessed in semisolid agar. (B) H. pylori strains (∼108 cells) were analyzed by Western blotting for FlaB (indicated by arrow). Lane 1, H. pylori 43504; lane 2, hp0958:aphA3 mutant; lane 3, hp0958:aphA3 mutant bearing a PureA-rpoN allele in the hp0405 locus (hp0958:aphA3/hp0405:PureA-rpoN).
Overproduction of HP0958 interferes with its function.
In the initial attempt to complement the hp0958:aphA3 mutation, an hp0958 allele under the control of the ureA promoter (PureA-hp0958) was introduced into the hp0405 locus of the mutant, which failed to restore motility (data not shown). Introduction of the PureA-hp0958 allele into wild-type H. pylori interfered with motility (Fig. 1), and as with inactivation of hp0958, it resulted in decreased levels of FlaB and σ54 (Fig. 2B and C). The level of HP0958 in the strain with the PureA-hp0958 allele was ∼10-fold higher than the wild-type level, which was estimated by determining the lowest number of cells needed to visualize the protein by Western blotting (data not shown). We do not know why overproduction of HP0958 interfered with its function. Behavior of HP0958 overproduced in H. pylori in the sodium dodecyl sulfate-polyacrylamide gel was unusual in that much of the protein migrated with a reduced mobility (Fig. 2A). This may have resulted from cross-linking or other stable interactions between HP0958 monomers which could have interfered with activity of the protein.
HP0958 prevents the rapid turnover of σ54.
To address whether HP0958 exerts its control over σ54 accumulation posttranslationally, we introduced an rpoN′-′xylE reporter gene into the hp0958:aphA3 mutant and its parental strain. This rpoN′-′xylE reporter gene encoded a translational fusion in which xylE was joined in-frame with codon 61 of H. pylori rpoN. Expression of the rpoN′-′xylE reporter gene was indistinguishable in the two strains (0.47 ± 0.10 units XylE activity/108 cells for the parental strain versus 0.51 ± 0.07 units XylE activity/108 cells for the hp0958:aphA3 mutant), which argued that HP0958 affects σ54 levels in H. pylori at a posttranslational step.
To determine if HP0958 influences σ54 stability, σ54 levels were monitored in wild-type and hp0958:aphA3 mutant strains where rpoN was under the control of the ureA promoter. Strains were grown in serum-free medium to mid-log phase, at which time tetracycline was added to block translation. Samples were analyzed for σ54 at various times following the addition of tetracycline. Levels of σ54 in the hp0958:aphA3 mutant decreased with an apparent half-life of ∼33 min, whereas σ54 levels remained constant for at least 4 h in the wild-type strain (Fig. 5). In the absence of tetracycline, σ54 levels remained constant in the wild-type and the hp0958:aphA3 mutant strains over the entire course of the assay (data not shown). We were unable to detect σ54 in the extracellular proteins of the hp0958:aphA3 mutant or its parental strain, but in a positive control, the vacuolating cytotoxin VacA was detected in the extracellular proteins of both strains (data not shown). Taken together, these data suggest that the rapid decrease in σ54 levels in the hp0958:aphA3 mutant does not result from secretion but rather from protein degradation.
FIG. 5.
Comparison of σ54 stability in an hp0958:aphA3 mutant and its parental strain. Levels of σ54 in an hp0958:aphA3 mutant that overproduced σ54 from a PureA-rpoN allele (hp0958:aphA3/hp0405:PureA-rpoN) and its parental strain (hp0405:PureA-rpoN) were analyzed by Western blotting at various times following the addition of tetracycline. Approximately 108 cells were lysed and loaded in each lane. σ54 is indicated by the arrows.
DISCUSSION
We demonstrate here that HP0958 is required for the normal accumulation of σ54 in H. pylori, indicating that it may play a regulatory role in modulating σ54 levels under different growth or environmental conditions. HP0958 appears to influence σ54 levels by protecting it from proteolysis. Regulated turnover is a mechanism by which the levels of other σ factors are sometimes controlled. E. coli σS, for example, is degraded by ClpXP protease but is protected from proteolysis under certain conditions by the chaperone DnaK (27, 28). Like DnaK, HP0958 may function as a chaperone to protect σ54 from proteolysis, which would explain the interactions between HP0958 and σ54 in the yeast two-hybrid assay. Suppression of the hp0958:aphA3 mutation by overexpressing rpoN may be explained by high levels of σ54 overwhelming the proteolysis of the protein. Alternatively, HP0958 may modify σ54 to protect it from proteolysis. The suppression of the hp0958 mutation by overproducing σ54, however, suggests that any such potential modification of σ54 is not needed to convert it from an inactive to an active form. Thus, the only essential role that HP0958 appears to play in flagellar biogenesis is in maintaining σ54 levels capable of supporting efficient expression of the RpoN regulon. HP0958 may have an additional, nonessential role in regulating the FliA flagellar regulon, however, since expression of the flaA′-′xylE reporter gene was elevated in the hp0958 mutant but not in the rpoN mutant. HP0958 may affect flaA expression by influencing the level or activity of the anti-σ28 factor FlgM or another factor that inhibits expression of flaA.
We have not observed any other phenotypes for the hp0958:aphA3 mutant. HP0958 interacts with other H. pylori proteins in the yeast two-hybrid assay, including FliH (a regulator of the flagellar protein export apparatus), HP1462 (a putative secreted protein involved in motility), and TonB1 (a putative siderophore-mediated iron transport protein). Thus, HP0958 may have other roles in addition to maintaining a wild-type level of σ54. Since σ54 is required for flagellar biogenesis, the interactions between HP0958 and FliH are intriguing. Inactivation of fliH, however, does not affect σ54 levels in H. pylori (L. Pereira, unpublished data). Despite several attempts, we were unable to construct an hp0958/fliH double mutant, suggesting that this combination of mutations is lethal.
Comparison of the genome sequences of H. pylori, H. hepaticus, C. jejuni, and W. succinogenes reveals similar organization of flagellar genes and potential regulatory mechanisms controlling their expression. All of these ɛ-proteobacteria possess HP0958 homologs, and we anticipate that these proteins have roles in maintaining wild-type σ54 levels in these bacteria. FlgR is the only σ54-dependent activator present in H. pylori, H. hepaticus, and C. jejuni, suggesting that σ54 is dedicated for flagellar biogenesis in these bacteria. W. succinogenes, however, possesses two σ54-dependent activators, FlgR and NifA, which appear to be required for the expression of flagellar and nitrogen fixation genes, respectively. Thus, the HP0958 homolog in W. succinogenes may be required for flagellar biogenesis and nitrogen fixation.
In addition to the ɛ-proteobacteria, homologs of HP0958 are present in a number of bacteria from diverse phylogenic groups, including Aquifex aeolicus, Bacteroides thetaiotaomicron, Chlamydia trachomatis, Porphyromonas gingivalis, Prevotella intermedia, Thermoanaerobacter tengcongensis, Chlorobium tepidum, Borrelia burgdorferi, and Treponema pallidum. Many of these bacteria possess σ54, and the HP0958 homologs in these bacteria may have roles in modulating σ54 levels. Elucidating the mechanism by which HP0958 influences σ54 levels in H. pylori is certain to lead to a better understanding of the function of its homologs in other bacteria.
Acknowledgments
This work was funded by awards MCB-9974558 from the National Science Foundation and AI056036 from the National Institute of Allergy and Infectious Diseases to T.R.H.
We thank John Shields and Mark Farmer for technical assistance with electron microscopy. We also thank Anup Datta for LPS analysis.
REFERENCES
- 1.Alm, R. A., L.-S. L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smithe, B. Noonon, B. D. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, J. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. R. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Beier, D., and R. Frank. 2000. Molecular characterization of two-component systems of Helicobacter pylori. J. Bacteriol. 182:2068-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belunis, C. J., S. M. Clementz, S. M. Carty, and C. R. Raetz. 1995. Inhibition of lipopolysaccharide biosynthesis and cell growth following inactivation of the kdtA gene in Escherichia coli. J. Biol. Chem. 270:27646-27652. [DOI] [PubMed] [Google Scholar]
- 4.Belunis, C. J., and C. R. Raetz. 1992. Biosynthesis of endotoxins. Purification and catalytic properties of 3-deoxy-d-octolusonic acid transferase from Escherichia coli. J. Biol. Chem. 267:9988-9997. [PubMed] [Google Scholar]
- 5.Benoit, S., and R. J. Maier. 2003. Dependence of Helicobacter pylori urease activity on the nickel-sequestering ability of the UreE accessory protein. J. Bacteriol. 185:4787-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaser, M. J. 1993. Helicobacter pylori: microbiology of a ‘slow’ bacterial infection. Trends Microbiol. 1:255-260. [DOI] [PubMed] [Google Scholar]
- 7.Brahmachary, P., M. G. Dashti, J. W. Olson, and T. R. Hoover. 2004. Helicobacter pylori FlgR is an enhancer-independent activator of σ54-RNA polymerase holoenzyme. J. Bacteriol. 186:4535-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buck, M., S. Miller, M. Drummond, and R. Dixon. 1986. Upstream activator sequences are present in the promoters of nitrogen fixation genes. Nature (London) 320:374-378. [Google Scholar]
- 9.Bumann, D., S. Aksu, M. Wendland, K. Janek, U. Zimny-Arndt, N. Sabarth, T. F. Meyer, and P. R. Jungblut. 2002. Proteome analysis of secreted proteins of the gastric pathogen Helicobacter pylori. Infect. Immun. 70:3396-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cover, T. L., and M. J. Blaser. 1992. Helicobacter pylori and gastroduodenal disease. Annu. Rev. Med. 43:135-145. [DOI] [PubMed] [Google Scholar]
- 11.Dasgupta, N., M. C. Wolfgang, A. L. Goodman, S. K. Arora, J. Jyot, S. Lory, and R. Ramphal. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50:809-824. [DOI] [PubMed] [Google Scholar]
- 12.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eaton, K. A., D. R. Morgan, and S. Krakowka. 1989. Campylobacter pylori virulence factors in gnotobiotic piglets. Infect. Immun. 57:1119-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eaton, K. A., D. R. Morgan, and S. Krakowka. 1992. Motility as a factor in the colonisation of gnotobiotic piglets by Helicobacter pylori. J. Med. Microbiol. 37:123-127. [DOI] [PubMed] [Google Scholar]
- 15.Kim, J. S., J. H. Chang, S. I. Chung, and J. S. Yum. 1999. Molecular cloning and characterization of the Helicobacter pylori fliD gene, an essential factor in flagellar structure and motility. J. Bacteriol. 181:6969-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leying, H., S. Suerbaum, G. Geis, and R. Haas. 1992. Cloning and genetic characterization of a Helicobacter pylori flagellin gene. Mol. Microbiol. 6:2863-2874. [DOI] [PubMed] [Google Scholar]
- 17.McGee, D. J., F. J. Radcliff, R. L. Mendz, R. L. Ferrero, and H. L. Mobley. 1999. Helicobacter pylori rocF is required for arginase activity and acid protection in vitro but is not essential for colonization of mice or for urease activity. J. Bacteriol. 181:7314-7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merkx-Jacques, A., R. K. Obhi, G. Bethune, and C. Creuzenet. 2004. The Helicobacter pylori flaA1 and wbpB genes control lipopolysaccharide and flagellum synthesis and function. J. Bacteriol. 186:2253-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niehus, E., H. Gressmann, F. Ye, R. Schlapbach, M. Dehio, C. Dehio, A. Stack, T. F. Meyer, S. Suerbaum, and C. Josenhans. 2004. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol. Microbiol. 52:947-961. [DOI] [PubMed] [Google Scholar]
- 20.Olson, J. W., M. K. Johnson, and R. J. Maier. 2000. Characterization of the NifU and NifS Fe-S cluster formation proteins essential for viability in Helicobacter pylori. Biochemistry 39:16213-16219. [DOI] [PubMed] [Google Scholar]
- 21.Olson, J. W., N. S. Mehta, and R. J. Maier. 2001. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol. Microbiol. 39:176-182. [DOI] [PubMed] [Google Scholar]
- 22.Popham, D., D. Szeto, J. Keener, and S. Kustu. 1989. Function of a bacterial activator protein that binds to transcriptional enhancers. Science 243:629-635. [DOI] [PubMed] [Google Scholar]
- 23.Prouty, M. G., N. E. Correa, and K. E. Klose. 2001. The novel σ54- and σ28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39:1595-1609. [DOI] [PubMed] [Google Scholar]
- 24.Rain, J.-C., L. Selig, H. De Reuse, V. Battaglia, C. Reverdy, S. Simon, G. Lenzen, F. Petel, J. Wojcik, V. Schachter, Y. Chemama, A. Labigne, and P. Legrain. 2001. The protein-protein interaction map of Helicobacter pylori. Nature 409:211-215. [DOI] [PubMed] [Google Scholar]
- 25.Reitzer, L. J., and B. Magasanik. 1986. Transcription at glnA of E. coli is stimulated by activator bound to sites far from the promoter. Cell 45:785-792. [DOI] [PubMed] [Google Scholar]
- 26.Rippe, K., M. Guthold, P. H. von Hippel, and C. Bustamante. 1997. Transcriptional activation via DNA-looping: visualization of intermediates in the activation pathway of E. coli RNA polymerase σ54 holoenzyme by scanning force microscopy. J. Mol. Biol. 270:125-138. [DOI] [PubMed] [Google Scholar]
- 27.Rockabrand, D., K. Livers, T. Austin, R. Kaiser, D. Jensen, R. Burgess, and P. Blum. 1998. Roles of DnaK and RpoS in starvation-induced thermotolerance of Escherichia coli. J. Bacteriol. 180:846-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schweder, T., K. H. Lee, O. Lomovskaya, and A. Matin. 1996. Regulation of Escherichia coli starvation factor (σs) by ClpXP protease. J. Bacteriol. 178:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spohn, G., and V. Scarlato. 1999. Motility of Helicobacter pylori is coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J. Bacteriol. 181:593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spohn, G., and V. Scarlato. 2001. Motility, chemotaxis, and flagella, p. 239-257. In S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, D.C. [PubMed]
- 31.Su, W., S. Porter, S. Kustu, and H. Echols. 1990. DNA-looping and enhancer activity: association between DNA-bound NTRC activator and RNA polymerase at the bacterial glnA promoter. Proc. Natl. Acad. Sci. USA 87:5504-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suerbaum, S., C. Josenhans, and A. Labigne. 1993. Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flaB flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J. Bacteriol. 175:3278-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomb, J.-F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 34.Totten, P. A., J. C. Lara, and S. Lory. 1990. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J. Bacteriol. 172:389-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiss, D. S., J. Batut, K. E. Klose, J. Keener, and S. Kustu. 1991. The phosphorylated form of the enhancer-binding protein NTRC has an ATPase activity that is essential for activation of transcription. Cell 67:155-167. [DOI] [PubMed] [Google Scholar]