Abstract
DNA has recently been described as a major structural component of the extracellular matrix in biofilms. In streptococci, the competence-stimulating peptide (CSP) cell-to-cell signal is involved in competence for genetic transformation, biofilm formation, and autolysis. Among the genes regulated in response to the CSP are those involved in binding and uptake of extracellular DNA. We show in this study that a functional DNA binding-uptake system is involved in biofilm formation. A comGB mutant of Streptococcus mutans deficient in DNA binding and uptake, but unaffected in signaling, showed reduced biofilm formation. During growth in the presence of DNase I, biofilm was reduced in the wild type to levels similar to those found with the comGB mutant, suggesting that DNA plays an important role in the wild-type biofilm formation. We also showed that growth in the presence of synthetic CSP promoted significant release of DNA, with similar levels in the wild type and in the comGB mutant. The importance of the DNA binding-uptake system in biofilm formation points to possible novel targets to fight infections.
Bacteria in natural environments are most often found attached to surfaces, embedded in an extracellular matrix (9). In the biofilm mode, bacteria exhibit increased resistance to antimicrobials and to host defense systems (25). The composition of the extracellular matrix includes polysaccharides and proteins. More recently, extracellular DNA has also been implicated as a major structural component of biofilms (26, 31, 38).
Both gram-positive and gram-negative bacteria communicate through quorum-sensing signals to coordinate population behavior. For Streptococcus pneumoniae, DNA release was recently shown to be activated by the quorum-sensing circuit involved in competence development for natural transformation (27, 35). This genetically programmed physiological state is triggered in streptococci by the competence-stimulating signal peptide (CSP). CSP is recognized by the sensor kinase receptor ComD, which autophosphorylates and transfers a phosphoryl group to the ComE response regulator. The gene for the CSP is comC, which in most transformable streptococci is organized in an operon together with comD and comE (15). Phosphorylated ComE activates expression of the comCDE operon and other early competence genes encoding, for instance, the CSP exporter ComAB.
Late competence genes, such as those involved in the binding and uptake of extracellular DNA, are regulated by the alternative sigma factor ComX. Expression of ComX is activated by phosphorylated ComE. In S. pneumoniae, more than a hundred genes are regulated by ComX (10, 33).
Gram-positive and gram-negative bacteria share several homologous proteins of the DNA binding-uptake machinery, related to type II secretion systems and type IV pili (7). In gram-positive bacteria, comGA and comGB homologues are predicted to encode a traffic NTPase and a polytopic membrane protein, respectively. ComGC shows homology to PilE of Neisseria gonorrhoeae, a major pseudopilin. Three minor pseudopilins are encoded by comGD, comGE, and comGG. The function of the comG apparatus is probably to span the thick cell wall and bring exogenous DNA to a membrane DNA receptor. A single strand of the bound DNA is then transported through a membrane channel, whereas the other strain is degraded and released extracellularly. Unlike in gram-negative bacteria, efficient binding and uptake of DNA is not sequence specific in gram-positive microorganisms. The comG operon in streptococci has also received other designations, such as cgl (30), cilD (5), and comY (22).
In S. pneumoniae, DNA release triggered by competence induction involves cell lysis by the autolytic amidase LytA (27, 35) and the autolytic lysozyme LytC (27). DNA release continues after competence shutoff and is maximum in stationary phase (27). The autolysis induced by CSP may provide a source of DNA during competence development.
There is increasing evidence that cell-to-cell communication through quorum-sensing systems may influence biofilm formation by both gram-positive and gram-negative bacteria (12). It has been demonstrated, for several streptococci, that the ability to form biofilms is affected by the quorum-sensing system involved in development of competence for natural transformation (19-21, 31, 39). Mutants of Streptococcus gordonii (21) and Streptococcus mutans (19, 20, 39) deficient in competence development exhibit altered biofilm phenotypes. Direct evidence for a role of CSP in favoring the biofilm mode of growth in Streptococcus intermedius has also been presented (31).
S. mutans is a cariogenic bacteria found in the oral cavity, and it depends on the biofilm mode of growth for colonization. In the present study we found that a synthetic CSP (SCSP) specific for S. mutans induced biofilm formation and DNA release in S. mutans wild type. We investigated whether the effects on biofilm formation and DNA release involved the same regulatory mechanisms as the system controlling competence. We also investigated whether the effect on biofilm formation was associated with the DNA binding-uptake system, which expression is activated during competence development. We found that the DNA binding-uptake system may play an important role in biofilm formation.
MATERIALS AND METHODS
Bacterial strains and media.
The S. mutans strains used (Table 1) were grown in Trypticase soy broth (TSB; Becton Dickinson, Sparks, Md.) or Todd-Hewitt broth (THB; Difco Laboratories, Detroit, Mich.) supplemented with yeast extract (THY) or with horse-serum (TH-HS). Incubations were done at 37°C aerobically or in a 5% CO2 aerobic atmosphere. Antibiotics were added at the following concentrations: erythromycin at 20 μg/ml and kanamycin at 500 μg/ml. The strains were stored at −70°C in 15% glycerol. Before each experiment, the cells were grown on THB agar plates and incubated for 48 h.
TABLE 1.
Strains used in this study
| Strain | Genotype | Descriptiona |
|---|---|---|
| LT11 | UA159 derivative | |
| SM015 | LT11 comGB′::pSF151::′comGB | Insertion-duplication mutagenesis with pSF151 containing a Tsp509I-digested comYB fragment; Kanr |
| SM017 | LT11 comX′::pSF151::′comX | Insertion-duplication mutagenesis with pSF151 containing a Tsp509I digested comX fragment; Kanr |
| SM016 | LT11 comE′::pSF151::′comE | Insertion-duplication mutagenesis with pSF151 containing a Tsp509I digested comE fragment; Kanr |
| SM001 | LT11 ΔcomC::PcKan | Insertion-deletion replacement of comC with synthetic Kan amplicon; Kanr |
| SM003 | LT11 ΔcomD::PcKan | Insertion-deletion replacement of comD with synthetic Kan amplicon; Kanr |
Kanr, kanamycin resistance.
Synthetic CSP.
CSP was synthesized by MedProbe (SCSP; amino acid sequence: NH2-SGSLSTFFRLFNRSFTQALGK-COOH), with a purity grade of >95%. The lyophilized SCSP was reconstituted in distilled water at 12 mM and stored in small aliquots at −20°C. SCSP was used at a final concentration of 1 μM, except for the titration experiment, in which concentrations from 32 nM to 1 μM were tested.
Construction of mutants.
Sequence information was obtained from the S. mutans UA159 genome (1). The targeted sequences for insertion-inactivation or deletion are illustrated (Fig. 1). The sizes and locations of the PCR amplicons in the genome were verified by in silico analyses (4). The sequences of the oligonucleotide primers used are listed in Table 2.
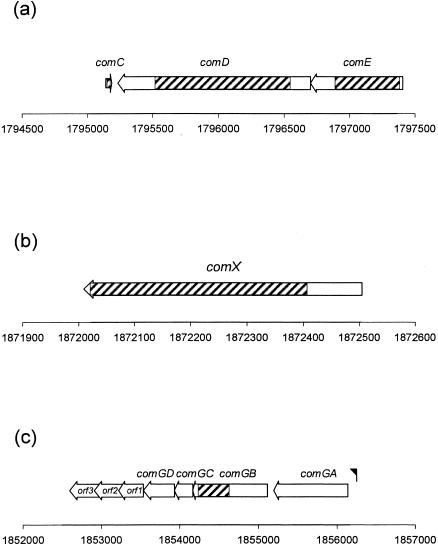
FIG. 1.
Organization of the comCDE (a), comX (b), and comG (c) gene clusters in S. mutans UA159. The numbers correspond to the localization of the genes (bp) in the S. mutans UA159 genome sequence. ▨, Targeted sequences for insertion-inactivation or deletion. The black flag indicates the putative consensus promoter sequence recognized by ComX. orf, Open reading frame.
TABLE 2.
Synthetic oligonucleotides used for targeted deletion or disruption and confirmation of insertion
| Target | Gene IDa | Type of inactivation | Oligonucleotide and sequence (5′ to 3′)b |
|---|---|---|---|
| comGB | 1029176 | Disruption | FP042, AACAGCTTATTATGCGCGTGAATG |
| FP043, AAAGTGTTCCCATGTTTCCTCAGC | |||
| comX | 1029188 | Disruption | FP044, GTGAAGATTGGCAACAAGAGG |
| FP045, TCACATGTTCCATTCTTTCCC | |||
| comE | 1029114 | Disruption | FP053, GACGTCTTGAAACCACCATTGCA |
| FP054, TGCCGTAGAATTCAATCCGTTCA | |||
| comC | 1029111 | Deletion | FP009, TATGGACCAAGAAATGCTGT |
| FP010, AGG/CGCGCCGATAATCTCTAATTCATCAGTCTT | |||
| FP011, AGGCCGG/CCAGAAGTTTTACACAAGCTTTG | |||
| FP012, TGGAGTTGCTTGATTTCATT | |||
| comD | 1029115 | Deletion | FP053, GACGTCTTGAAACCACCATTGCA |
| FP013, AGG/CGCGCCCGTAACAGCAATCATTATCAGAAA | |||
| FP014, AGGCCGG/CCAATCGCGGTATTGGTTTAGCA | |||
| FP012, TGGAGTTGCTTGATTTCATT | |||
| Kan cassette | FP001, AGG/CGCGCCGTTTGATTTTTAATG | ||
| FP002, AGGCCGG/CCATCGATACAAATTCCTC | |||
| FP037, TCATTTTCTCCCACCAGCTT; confirm insertion | |||
| FP038, GCGCCTACGAGGAATTTGTA; confirm insertion | |||
| pSF151 | FP039, AGCGGATAACAATTTCACACAGGA; confirm insertion |
S. mutans UA159 gene identification number (ID).
Restriction sites are in boldface: AscI, GG/CGCGCC; and FseI, GGCCGG/CC.
Mutants with deletion of the comC and comD genes were constructed by the PCR ligation mutagenesis method, in which two PCR amplified flanking sequences of the gene are ligated to a resistance cassette, and used for transformation (18). AscI or FseI restriction sites were incorporated into the oligonucleotide primers used to generate the flanking DNA fragments and the resistance cassette. The kanamycin resistance cassette KANT (AY334019) was PCR amplified from plasmid pR410 (33) (a kind gift from D. A. Morrison) with the primer pair FP001 and FP002 (FP001-002). The primer pairs FP009-010 and FP011-012 were used to amplify the upstream and downstream flanking regions of comC, respectively. The comD flanking regions were amplified by the primer pairs FP053-013 and FP014-009. After restriction enzyme digestion with AscI and FseI, two separate reactions with the upstream or downstream amplicons were ligated to KANT. The two ligation products were then mixed and PCR amplified with the distal primers FP009-012 for comC and FP053-009 for comD. The resultant amplicons were used to transform S. mutans LT11, as described below. The gene deletion in each mutant was confirmed by PCR with distal primers and internal primers for KANT. The primer pairs FP009-037 and FP038-012 were used for confirmation of the comC deletion, and the primer pairs FP053-037 and FP038-012 were used for confirmation of the comD deletion.
The comGB, comX, and comE genes were inactivated by insertion-duplication, using the suicide vector pSF151 (36). Internal sequences of comGB, comX, and comE were amplified with the primer pairs FP042-043, FP044-045, and FP053-54, respectively. The three amplicons contained Tsp509I restriction sites. Tsp509I-digested amplicons and EcoRI-digested pSF151were ligated and used for S. mutans transformation as described below. Southern hybridization was used to confirm insertion inactivation. We also conducted PCR with the FP039 internal primer for pSF151 and with the forward or reverse primers designed to amplify the target genes (Table 1). Both Southern hybridization and sequence of the amplicons confirmed the sites of inactivation.
Transformation assay.
For targeted deletion or disruption of defined genes, the cells were grown in TH-HS, and transformation was conducted as previously described (32), except that aerobic growth conditions were used. We have found that competence development under aerobic conditions is at least twofold more efficient than in 5% CO2 aerobic atmosphere (data not shown). Increased transformation efficiency during aerobic growth has also been reported for S. pneumoniae (11). Transformants were selected by growth on THB agar plates containing kanamycin. In the assays described below in which the wild type was compared to the mutants, kanamycin was not added to the liquid cultures to avoid a possible effect of the antibiotic on bacterial growth. We found that in the presence or absence of kanamycin the comG and comX mutants lost their transformability, and the comE and comD mutants had similar levels of transformation, indicating that the mutations by insertion-duplication were stable.
To assess competence development by the mutants and the wild type, overnight cells grown in THY were diluted 1:200 in TSB and exposed to the plasmid pVA838 expressing erythromycin resistance (final concentration, 1 μg/ml) (24) in the presence or absence of SCSP. The cells were incubated for 2 h under aerobic conditions, followed by 2 h in 5% CO2, and subsequently plated. The transformants were selected by growth on THB agar containing erythromycin.
Biofilm assay.
Cells grown overnight in THY were diluted 1:200, and 500 μl was inoculated into wells of polystyrene flat-bottom 24-well microtiter plates as previously described (31). The plates were incubated at 37°C aerobically for 2 h and then in a 5% CO2 aerobic atmosphere for 24 or 48 h. The biofilms were washed twice with distilled water and suspended in 500 μl of fresh TSB by scraping the bottom and lateral walls of the wells with a disposable cell scraper adapted to fit into the wells. In separate wells, total growth was measured by collecting the biofilm and planktonic cells together in the 500 μl of medium used during incubation. To disperse clumps and chains, the bacterial suspensions were gently sonicated for 10 s at 4°C. The optical densities at 600 nm (OD600) of the suspended cells were measured.
To investigate the role of extracellular DNA during biofilm formation, the wild type and the comGB mutant were grown in the absence or presence of 576 U of DNase I grade I (Roche)/ml. This concentration was chosen based on titration experiments in the presence of SCSP showing maximum inhibitory effect on wild-type biofilm formation at DNase I concentrations of >384 U/ml. Without SCSP, maximum effect was observed with a concentration as low as 96 U/ml.
The biofilms formed by the wild type and the comGB mutant were subjected to scanning electron microscopy. Biofilms were grown in the presence or absence of SCSP as described above, except that a polystyrene disk (Nunc) was immersed in each well before inoculation. After 48 h, the disks were removed, rinsed, fixed, and dehydrated as previously described (31).
DNA release assay.
The bacterial cultures grown in the microtiter plates or in glass tubes were centrifuged (10,000 × g, 5 min, 4°C). The supernatants were diluted 1:10 with Tris-EDTA buffer. The amount of double-stranded DNA released was determined by using the PicoGreen fluorescent dye system (Molecular Probes) with lambda DNA as the standard according to the producer's recommended protocol.
DNA binding and uptake assay.
One microliter of uniformly 3H-labeled Escherichia coli genomic DNA (25 μCi/ml; DuPont-NEN, Boston, Mass.), corresponding to 50 ng (ca. 6,000 disintegrations per minute [dpm]), was added to 1 ml of competent cells grown in TH-HS as previously described (22). After 15 min, the cells were chilled on ice and centrifuged (10,600 × g, 1 min, 4°C). The measurement of DNA binding and cell uptake was conducted as previously described (2), with minor modifications. Briefly, the cell pellets were washed twice to remove unbound DNA. The pellet was then resuspended in 100 μl of TSB and treated with 48 U of DNase I grade I (Roche) for 10 min at 37°C to release and break down bound, unincorporated DNA. After centrifugation (10,600 × g, 1 min, 4°C), the pellets were resuspended in 100 μl of TSB. The resuspended pellets and supernatants were transferred to separate flasks containing 10 ml of scintillation fluid. Radioactivity was measured as the number of dpm in a liquid scintillation analyzer (Packard 1900 TR; Packard Instrument Company, Meriden, Conn.).
RESULTS
Relationship between SCSP concentration and activity.
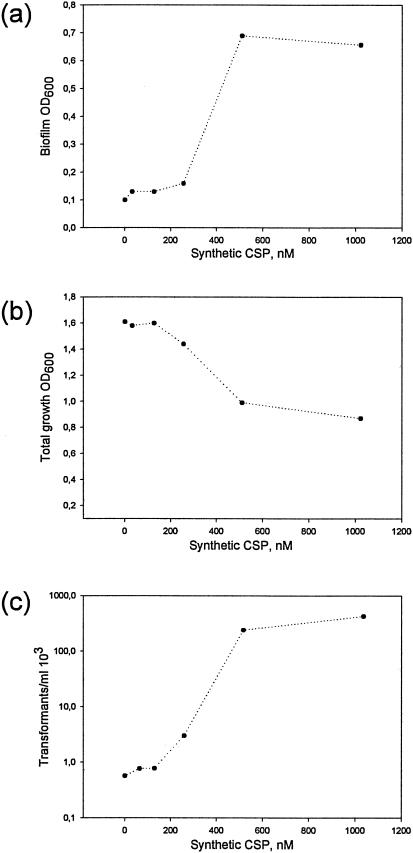
To determine the stimulatory effect of SCSP on competence development and biofilm formation, as well as a possible effect on growth (the sum of planktonic and biofilm cells in the wells), wild-type cells grown in TSB were exposed to SCSP concentrations from 32 nM to 1 μM (Fig. 2). Competence development in streptococci is growth phase dependent and usually develops at early to mid-log phases of growth, whereas biofilm may only be assessed after a certain time. In the present study we therefore measured competence at early log phase and biofilm after 24 h under similar growth conditions. Biofilm formation in the presence of 1 μM SCSP was 85% higher than without SCSP (Fig. 2a), concomitant with a reduction by 50% in the total amount of cells (Fig. 2b). The number of transformants after growth in the presence of 1 μM SCSP was increased 800 times relative to cells not exposed to SCSP (Fig. 2c).
FIG. 2.
Titration of SCSP stimulating activity. Biofilm formation (a) and growth (b) of the wild type (LT11) was monitored after 24 h by measuring the cell density (OD600). The competence stimulating activity (c) determined as erythromycin-resistant transformants on THB agar containing the selective antibiotic after exposure of 1 ml of culture cells to 1 μg of pVA838.
Response of signaling-defective mutants to SCSP.
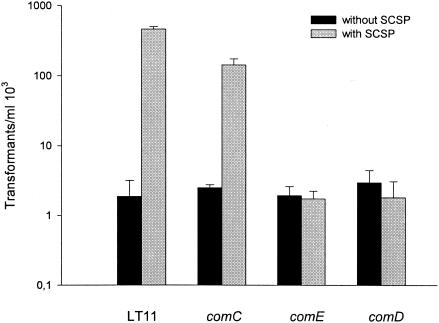
Conditions that allow exogenous activation of competence by SCSP but not endogenous activation provide the means for direct assessment of CSP functions. We have previously found that S. intermedius grown in TSB are not spontaneously competent but that competence is induced by addition of S. intermedius SCSP. The present results showed that SCSP stimulated competence in S. mutans as well, but transformation occurred also without addition of SCSP. We asked, therefore, whether competence in the absence of SCSP was dependent on the CSP regulatory circuit. We also investigated whether the effects of SCSP on biofilm formation, competence, and growth involved the same CSP regulatory system. We inactivated the comC, comD, comE, and comX genes in S. mutans. Inactivation of comC, comD, or comE did not affect the ability of the cells to undergo transformation (Fig. 3) in the absence of SCSP. These results confirm previous findings that in S. mutans transformation may occur in the absence of endogenous CSP production or a functional ComDE two-component system (19). As observed previously (20), inactivation of comX resulted in complete loss of transformability, both in the presence and in the absence of SCSP. In S. pneumoniae, competence was shown to occur in comE deletion mutants that overexpress ComX, supporting the notion that comX alone may be sufficient to support competence development without the participation of the CSP response circuit (ComCDE) (23).
FIG. 3.
Transformation of the wild type (LT11) and the comC (SM001), comE (SM016), and comD (SM003) mutants in the absence or presence of 1 μM SCSP. Transformants were selected on THB agar containing the selective antibiotic erythromycin, after exposure of 1 ml of culture cells to 1 μg of pVA838. The results for the comGB (SM015) and comX (SM017) mutants are not shown since transformation was completely abolished. The results are mean values and standard errors from two independent experiments.
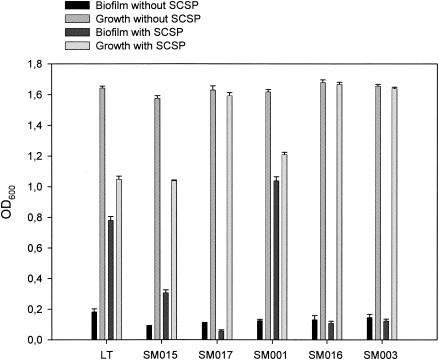
In the presence of SCSP, both biofilm formation and transformation were stimulated in the comC mutant. Induction of competence was, however, not completely restored by growth of the comC mutant in the presence of SCSP (Fig. 3). Thus, in the presence of SCSP, the number of transformants in the comC mutant was 30% of that of the wild type. Moreover, in the presence of SCSP the comC mutant formed 25% more biofilm than the wild type, whereas growth was inhibited by only 25% compared to 40% in the wild type (Fig. 4). Thus, the difference in biofilm formation between the comC mutant and the wild type in the presence of SCSP may be related to a higher SCSP inhibitory effect on growth of the wild type than the mutant.
FIG. 4.
Biofilm formation and growth by the wild type (LT11) and the comGB (SM015), comX (SM017), comC (SM001), comE (SM016), and comD (SM003) mutants in the absence or presence of 1 μM SCSP monitored after 48 h by measuring cell density (OD600). The results are mean values and standard errors from three independent experiments with two parallels.
The SCSP stimulatory effect on biofilm formation (Fig. 4) and transformation (Fig. 3) observed in the wild type were abolished in the comD and comE mutants. This is in accordance with the role of the two-component regulatory system ComDE in monitoring the CSP concentration in the environment during competence.
Like the comD and comE mutants, growth of the comX mutant was not affected by SCSP (Fig. 4). Notably, the comX mutant formed 34% less biofilm than the wild type in the absence of SCSP (Fig. 4), supporting a possible involvement of ComX in biofilm formation (20). Intriguingly, the comX mutant formed less biofilm in the presence than in the absence of SCSP. The com regulon in S. pneumoniae consists of more than hundred genes (10, 33). Proteases and chaperones are among the upregulated genes, reflecting the gross shift of protein synthesis during competence development. Thus, inactivation of regulators involved in competence, including comDE and comX, is likely to interfere with the expression of several genes, and the possibility of large pleiotropic effects affecting biofilm formation might not be excluded. Our results showing that SCSP stimulated biofilm formation of the wild type supported, however, the involvement of CSP signaling in biofilm formation. We considered, therefore, that the reduced biofilm formation by the the comX mutant could be related, at least in part, to a specific effect on the expression of ComX regulated genes. Genes of the DNA binding-uptake system are among those induced by ComX during competence. We hypothesized that the DNA binding-uptake system could be involved in the reduced biofilm formation by the comX mutant.
Importance of the DNA binding-uptake system in biofilm formation.
The DNA binding-uptake system is a multiprotein complex; thus, the loss of any component may result in disruption of the complex and loss of function (8). In the present study, we inactivated comGB, one of the first sequences of the DNA binding-uptake system to become available from the S. mutans UA149 genome sequence. The comGB gene is the second of at least seven ORFs in the putative comG operon (Fig. 1c). The first ORF shows 58% similarity with the Bacillus subtilis ComGA traffic NTPase and 80% similarity with the S. pneumoniae ComGA. The consensus promoter sequence, CGAATA, which is recognized by ComX in S. pneumoniae, is found upstream of S. mutans comGA. The S. mutans ComGB has 47% similarity with the ComGB polytopic membrane protein of B. subtilis and 76% similarity with the S. pneumoniae ComGB. Downstream of ComGB are homologues to the ComGC and ComGD pseudopilins, followed by orf1, orf2, and orf3 with homology to minor pseudopilins. In B. subtilis, the major and minor pseudopilins are processed by a prepilin peptidase and assembled into the pseudopilus. The traffic NTPase ComGA and the membrane protein ComGB participate in this process. The pseudopilus crosses the cell wall and allows the extracellular DNA to access its membrane-bound receptor, ComEA (7).
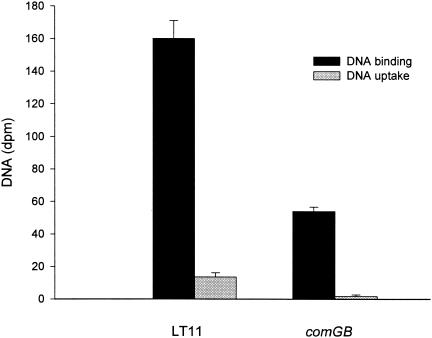
We investigated whether the comGB mutant was affected in DNA binding and uptake. Inactivation of comGB resulted in complete loss of transformability, both in the presence and in the absence of SCSP. We also measured the uptake and binding of 3H-labeled DNA and found that the uptake of DNA in the comGB mutant was reduced by 88% (DNase I resistant) and the DNA binding was reduced by 66% compared to the wild type (Fig. 5). Also, in other streptococci, DNA binding (2, 22) and uptake (22) are not completely abolished after inactivation of comG genes. It is possible that alternative mechanisms for DNA binding and uptake independent of competence may exist in streptococci. In S. mutans, for instance, a gene encoding deoxyribose aldolase involved in the catabolism of deoxynucleosides was recently identified (13).
FIG. 5.
DNA binding and uptake in the wild type (LT11) and the comGB mutant (SM015) exposed to 3H-labeled E. coli DNA. The results were measured as dpm. The results are mean values and standard errors from two independent experiments with two parallels.
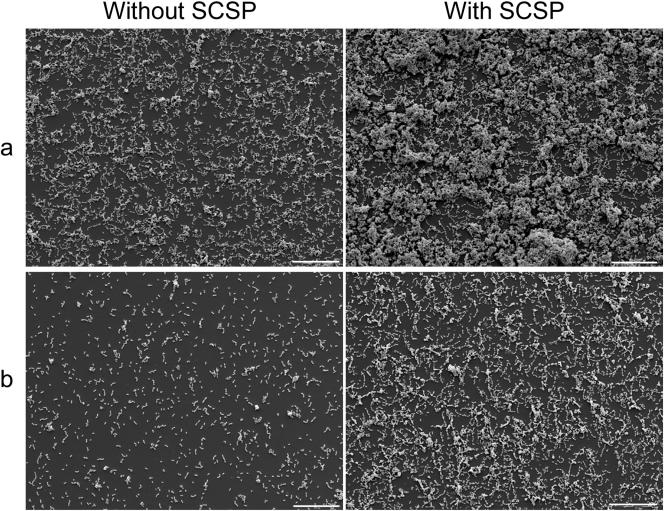
Scanning electron microscopy of the biofilms indicated that the wild type formed a denser biofilm than the comGB mutant both in the absence and in the presence of SCSP (Fig. 6). In the quantitative assay using cell density values of suspended biofilms, we found that comGB formed 70 or 51% less biofilm than the wild type in the presence or absence of SCSP, respectively (Fig. 4). Measurements using safranin staining of the cells confirmed the differences in biofilm formation between the wild type and the comGB mutant (data not shown). The fact that the comGB mutant formed less biofilm than the comD and comE mutants is not surprising, since the comDE mutants were transformed in the absence of SCSP. This indicates that the comG genes were being expressed in the comD and comE mutants.
FIG. 6.
Scanning electron microscopy of biofilms of S. mutans wild type (LT11) (a) and the comGB mutant (SM015) (b) in the absence or presence of 1 μM SCSP. Scale bar, 20 μm.
The inhibitory effect of SCSP on growth was similar in the comGB mutant and in the wild type (Fig. 4). This was in contrast to the comD, comE, and comX mutants in which SCSP had no inhibitory effect on growth. Thus, the results indicate that the mechanism(s) involved in growth inhibition by SCSP is not related to the DNA binding-uptake system.
Extracellular DNA is important during biofilm formation in the presence of a functional DNA binding-uptake system.
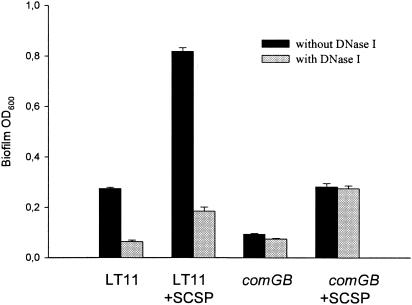
The results of the experiments described above indicated that the DNA binding-uptake system was involved in biofilm formation. Thus, extracellular DNA is expected to play an important role during biofilm formation in the wild type. On the other hand, the deficiency to bind and take up DNA suggests that, in the comGB mutant, extracellular DNA may not be involved or may play a less important role during biofilm formation. We tested these hypotheses by growing the wild type and the comGB mutant in the presence or absence of DNase I, with or without SCSP (Fig. 7). During growth in the presence of DNase I, biofilm was reduced in the wild type to levels similar to those found with the comGB mutant. Interestingly, DNase I had no effect on biofilm formation by the comGB mutant, either in the presence or absence of SCSP.
FIG. 7.
Effect of DNase I on biofilm formation by the wild type (LT11) and the comGB mutant (SM015) in the absence or presence of 1 μM SCSP. Biofilm formation was monitored after 48 h by measuring the cell density of resuspended cells (OD600). DNase I was added at 960 U/ml. The results are mean values and standard errors from two independent experiments with two parallels.
Effect of SCSP on DNA release.
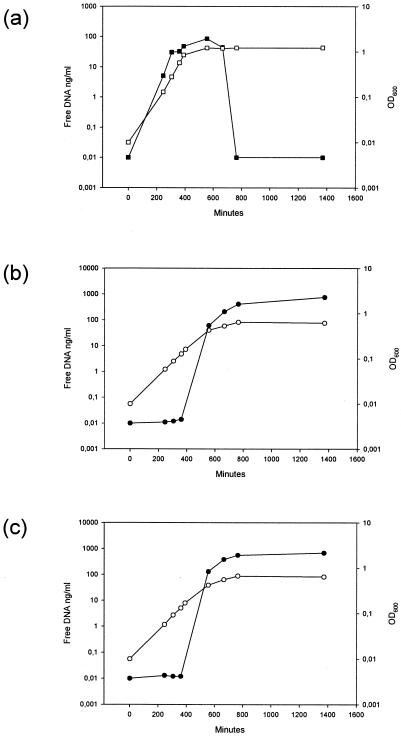
The results presented above indicated that extracellular DNA may play an important role during S. mutans biofilm formation. In S. pneumoniae, cell lysis was recently shown to be stimulated by the CSP regulatory circuit. We first sought to determine whether the CSP could also stimulate DNA release in S. mutans and whether the stimulation was similar in the wild type and the comGB mutant. In the absence of SCSP, the extracellular DNA concentration in the wild type increased up to 85 ng/ml at early stationary phase and then dropped to undetectable levels within the next 3.5 h (Fig. 8a). In the presence of SCSP, a distinct kinetic pattern was observed (Fig. 8b). A sudden increase in the concentration of free DNA occurred in the late exponential growth phase. In the early stationary phase, the amount of free DNA corresponded to 400 ng of DNA/ml and reached 760 ng/ml by the late stationary phase (Fig. 8b). According to our calculation this may represent 20 to 30% of the total DNA in the cultures. During growth in the presence of 2% choline and SCSP, the concentration of free DNA measured at early stationary phase was 130 ng/ml. This was only 30% of that obtained in the absence of choline. Release of DNA by the comGB mutant grown in the presence of SCSP (Fig. 8c) was similar to the wild type, except during the exponential growth phase in which values were 1.3 to 2.1 higher than for the wild type.
FIG. 8.
Concentration of free DNA during culture growth of the wild type (LT11) in the absence (a) or presence (b) of 1 μM SCSP and of the comGB mutant (SM015) in the presence of 1 μM SCSP (c). Solid symbols indicate the concentration of free DNA (ng/ml), and open symbols indicate the cell density (OD600) as a function of time.
We also sought to determine whether the CSP regulatory circuit was involved in the increased release of DNA. We measured the concentration of free DNA in the wells after 48 h biofilm growth of the wild type, as well as of the comC, comD, comE, comX, and comGB mutants. In the absence of SCSP, no free DNA was found in the wild type or the mutants. In the presence of SCSP, the amount of free DNA was 428 ng/ml (SE 24, n = 4), 555 ng/ml (SE 25, n = 4), and 516 ng/ml (SE 12, n = 4) in the wild type, comGB mutant, and comC mutant, respectively. No free DNA was found in the comD, comE, and comX mutants grown in the presence of SCSP. Thus, the results indicate that the CSP regulatory circuit is involved in the DNA release during growth of the wild type, comGB mutant, and comC mutant in the presence of SCSP.
DISCUSSION
In this study we found that SCSP stimulated S. mutans biofilm formation, competence, and DNA release, using a similar regulatory system. In the absence of SCSP, competence developed independent of the CSP response circuit, although at lower levels than in the presence of SCSP. We found that inactivation of comX resulted in a mutant deficient in biofilm formation and with complete loss of transformability. We hypothesized that the DNA binding-uptake system, whose expression is regulated by ComX, might be involved in the mechanism of S. mutans biofilm formation. We disrupted the DNA binding-uptake system by inactivation of comGB. The comGB mutant exhibited total loss of transformability and was significantly affected in DNA binding and uptake. Biofilm formation by the comGB mutant was significantly reduced both in the presence and in the absence of SCSP, whereas growth was similarly inhibited by SCSP in both the wild type and the comGB mutant. We found that extracellular DNA played an important role during biofilm formation in the wild type but not in the comGB mutant.
A link between competence development and DNA release was recently described in S. pneumoniae (27, 34, 35). In the first study, release of DNA was measured by the ability of cell-free filtrates of competent antibiotic-resistant cells to induce competence in cells sensitive to the antibiotic. In the last two studies, β-galactosidase activity was used as an indicator of cell lysis. Competence assays provide a measurement of the biologically active fraction of DNA, but not all of the released DNA by the resistant cells may be taken up by the sensitive cells. In our study we used a direct quantitative approach, the Pico-green method, for detection of double-stranded DNA. We found that SCSP induced significant DNA release in S. mutans. We investigated the ability of the wild type and comGB mutant to provide a source of exogenous DNA that could be used during biofilm formation. In the presence of SCSP, the amount of free DNA increased exponentially during growth of the wild type and the comGB mutant. Except during late stationary phase, a higher amount of free DNA was found in the comGB mutant than in the wild type. Notably, the addition of DNase I inhibited biofilm formation by the wild type, both in the presence and in the absence of SCSP, whereas biofilm formation by the comGB mutant was not affected. The residual biofilm formation by the wild type in the presence of DNase I was similar to that achieved by the comGB mutant. In Bacillus subtilis, all of the genes in the comG operon are required for DNA binding (8). It is likely, therefore, that all of the homologues to comG genes may be involved in DNA binding by S. mutans. In Pseudomonas aeruginosa, the type IV pilus homologue to DNA binding-uptake systems is involved in biofilm formation (28) and was recently shown to bind DNA (37). No evidence of natural transformation has been observed in P. aeruginosa. It is, therefore, possible that DNA binding by type IV pilus, but not uptake, may play a role in P. aeruginosa biofilm formation. We have initiated studies to separate the processes of DNA binding and uptake to determine the relative importance of each process in S. mutans biofilm formation.
DNA release during competence of S. pneumoniae is related to the ability of the cells to express autolysins with choline-binding activity, including lytA (27, 35) and lytC (27). Search of the S. mutans genome for autolysin homologues revealed the presence of a putative autolysin sharing 25% homology with the S. pneumoniae LytA in the autolytic amidase domain. In our study, choline had an inhibitory effect on DNA release by S. mutans, supporting a possible involvement of autolysins in the mechanism of DNA release.
We found that the response to SCSP resulting in increased biofilm formation and transformation levels involved the CSP regulatory circuit. Inactivation of comD and comE encoding the two-component system that monitors the CSP concentration abolished the effects of SCSP on biofilm formation, growth, competence, and DNA release. In addition, the SCSP response seems to be species specific, since growth in the presence of S. intermedius SCSP has no effect on S. mutans biofilm formation (31).
It is interesting that under conditions similar to those used in the present investigation, S. intermedius competence could be induced by concentrations of its specific SCSP of <20 nM, without an effect on growth (31). Also, in S. pneumoniae, maximum competence is observed with concentrations close to 20 nM (14). In the present study, concentrations of >160 nM were required before a significant increase in S. mutans transformation and biofilm formation was observed. The requirement for relatively high SCSP concentrations in S. mutans has been previously observed (39). The authors found that biofilm formation by a comC mutant of strain GS5 could be restored to levels equivalent to those of the wild type by ca. 1 μM SCSP but not by 100 nM. In the present study, the levels of response to SCSP varied between the wild type and the comC mutant. SCSP stimulation of competence, as well as inhibition of growth, was more pronounced in the wild type. Moreover, in the presence of SCSP, the comC mutant formed more biofilm than did the wild type. This may be related to the SCSP inhibitory effect on growth being higher in the wild type than in the mutant. The inability to restore completely the wild-type level of biofilm formation (20) or competence (19) in a comC mutant of S. mutans NG8 has been reported previously. In our study we found that the lack of full complementation of the comC mutant was not related to the SCSP concentration, since the stimulation of biofilm formation and the inhibitory effect on growth of the comC mutant leveled off at concentrations of >2 μM (data not shown). Development of competence in streptococci depends on several conditions, including CSP concentration, growth phase, and cell density. Unlike the wild type, the comC mutant does not produce endogenous CSP. Thus, conditions similar to those found in the wild type may be difficult to reproduce in the comC mutant by addition of SCSP.
In S. intermedius and S. pneumoniae, the genes encoding the CSP, the kinase receptor, and the cognate response regulator are organized in an operon. In S. mutans, however, the promoter for the CSP-encoding gene is distinct from the promoter for the kinase receptor and the response regulator. Inactivation of the genes encoding the histidine kinase or the response regulator results in the complete abolishment of competence in S. pneumoniae (29). In our study, inactivation of comC, comD, or comE did not affect the ability of S. mutans to undergo transformation in the absence of SCSP. This confirmed previous findings that in S. mutans transformation may occur in the absence of endogenous CSP production or a functional ComDE two-component system (3, 19). In agreement with previous reports on S. mutans and S. pneumoniae, the comX mutant was completely deficient in transformation. It seems therefore that in S. mutans ComX is sufficient to support competence development without the involvement of the CSP response circuit (ComCDE). However, activation of the CSP response circuit by SCSP increased the transformation of the wild type by 800 times.
Several roles have been ascribed to extracellular DNA. In P. aeruginosa, DNA is an important structural component of the extracellular matrix. Extracellular DNA also plays an important role during S. intermedius biofilm formation. Our results indicate that this is also the case in S. mutans. Other roles for extracellular DNA include stimulation of inflammatory cytokine production and lymphocyte proliferation (6, 16, 17). Our study suggests that the DNA binding-uptake system may be an important component in the mechanism of biofilm formation in S. mutans. Homologues with the DNA binding-uptake system are widespread in both gram-positive and gram-negative bacteria. It will be interesting to see whether such homologues are also implicated in biofilm formation in other bacteria.
Acknowledgments
We thank Steinar Stølen for scanning electron microscopic analysis.
This study was supported by a postdoctoral fellowship from The Norwegian Research Council.
REFERENCES
- 1.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berge, M., M. Moscoso, M. Prudhomme, B. Martin, and J. P. Claverys. 2002. Uptake of transforming DNA in gram-positive bacteria: a view from Streptococcus pneumoniae. Mol. Microbiol. 45:411-421. [DOI] [PubMed] [Google Scholar]
- 3.Bhagwat, S. P., J. Nary, and R. A. Burne. 2001. Effects of mutating putative two-component systems on biofilm formation by Streptococcus mutans UA159. FEMS Microbiol. Lett. 205:225-230. [DOI] [PubMed] [Google Scholar]
- 4.Bikandi, J., R. San Millan, A. Rementeria, and J. Garaizar. 2004. In silico analysis of complete bacterial genomes: PCR, AFLP-PCR, and endonuclease restriction. Bioinformatics 20:798-799. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, E. A., S. Y. Choi, and H. R. Masure. 1998. A competence regulon in Streptococcus pneumoniae revealed by genomic analysis. Mol. Microbiol. 27:929-939. [DOI] [PubMed] [Google Scholar]
- 6.Chatellier, S., and M. Kotb. 2000. Preferential stimulation of human lymphocytes by oligodeoxynucleotides that copy DNA CpG motifs present in virulent genes of group A streptococci. Eur. J. Immunol. 30:993-1001. [DOI] [PubMed] [Google Scholar]
- 7.Chen, I., and D. Dubnau. 2004. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2:241-249. [DOI] [PubMed] [Google Scholar]
- 8.Chung, Y. S., and D. Dubnau. 1998. All seven comG open reading frames are required for DNA binding during transformation of competent Bacillus subtilis. J. Bacteriol. 180:41-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 10.Dagkessamanskaia, A., M. Moscoso, V. Henard, S. Guiral, K. Overweg, M. Reuter, B. Martin, J. Wells, and J. P. Claverys. 2004. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol. Microbiol. 51:1071-1086. [DOI] [PubMed] [Google Scholar]
- 11.Echenique, J. R., S. Chapuy-Regaud, and M. C. Trombe. 2000. Competence regulation by oxygen in Streptococcus pneumoniae: involvement of ciaRH and comCDE. Mol. Microbiol. 36:688-696. [DOI] [PubMed] [Google Scholar]
- 12.Greenberg, E. P. 2003. Bacterial communication: tiny teamwork. Nature 424:134.. [DOI] [PubMed] [Google Scholar]
- 13.Han, T. K., Z. Zhu, and M. L. Dao. 2004. Identification, molecular cloning, and sequence analysis of a deoxyribose aldolase in Streptococcus mutans GS-5. Curr. Microbiol. 48:230-236. [DOI] [PubMed] [Google Scholar]
- 14.Håvarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Håvarstein, L. S., R. Hakenbeck, and P. Gaustad. 1997. Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J. Bacteriol. 179:6589-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klinman, D. M., A. K. Yi, S. L. Beaucage, J. Conover, and A. M. Krieg. 1996. CpG motifs present in bacteria DNA rapidly induce lymphocytes to secrete interleukin 6, interleukin 12, and interferon gamma. Proc. Natl. Acad. Sci. USA 93:2879-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krieg, A. M., A. K. Yi, S. Matson, T. J. Waldschmidt, G. A. Bishop, R. Teasdale, G. A. Koretzky, and D. M. Klinman. 1995. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 374:546-549. [DOI] [PubMed] [Google Scholar]
- 18.Lau, P. C., C. K. Sung, J. H. Lee, D. A. Morrison, and D. G. Cvitkovitch. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193-205. [DOI] [PubMed] [Google Scholar]
- 19.Li, Y. H., P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183:897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, Y. H., N. Tang, M. B. Aspiras, P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lunsford, R. D., and A. G. Roble. 1997. comYA, a gene similar to comGA of Bacillus subtilis, is essential for competence-factor-dependent DNA transformation in Streptococcus gordonii. J. Bacteriol. 179:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo, P., H. Li, and D. A. Morrison. 2003. ComX is a unique link between multiple quorum sensing outputs and competence in Streptococcus pneumoniae. Mol. Microbiol. 50:623-633. [DOI] [PubMed] [Google Scholar]
- 24.Macrina, F. L., J. A. Tobian, K. R. Jones, R. P. Evans, and D. B. Clewell. 1982. A cloning vector able to replicate in Escherichia coli and Streptococcus sanguis. Gene 19:345-353. [DOI] [PubMed] [Google Scholar]
- 25.Mah, T. F., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 26.Matsukawa, M., and E. P. Greenberg. 2004. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186:4449-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moscoso, M., and J. P. Claverys. 2004. Release of DNA into the medium by competent Streptococcus pneumoniae: kinetics, mechanism and stability of the liberated DNA. Mol. Microbiol. 54:783-794. [DOI] [PubMed] [Google Scholar]
- 28.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 29.Pestova, E. V., L. S. Håvarstein, and D. A. Morrison. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21:853-862. [DOI] [PubMed] [Google Scholar]
- 30.Pestova, E. V., and D. A. Morrison. 1998. Isolation and characterization of three Streptococcus pneumoniae transformation-specific loci by use of a lacZ reporter insertion vector. J. Bacteriol. 180:2701-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petersen, F. C., D. Pecharki, and A. A. Scheie. 2004. Biofilm mode of growth of Streptococcus intermedius favored by a competence-stimulating signaling peptide. J. Bacteriol. 186:6327-6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petersen, F. C., and A. A. Scheie. 2000. Genetic transformation in Streptococcus mutans requires a peptide secretion-like apparatus. Oral Microbiol. Immunol. 15:329-334. [DOI] [PubMed] [Google Scholar]
- 33.Peterson, S. N., C. K. Sung, R. Cline, B. V. Desai, E. C. Snesrud, P. Luo, J. Walling, H. Li, M. Mintz, G. Tsegaye, P. C. Burr, Y. Do, S. Ahn, J. Gilbert, R. D. Fleischmann, and D. A. Morrison. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51:1051-1070. [DOI] [PubMed] [Google Scholar]
- 34.Steinmoen, H., E. Knutsen, and L. S. Håvarstein. 2002. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc. Natl. Acad. Sci. USA 99:7681-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steinmoen, H., A. Teigen, and L. S. Håvarstein. 2003. Competence-induced cells of Streptococcus pneumoniae lyse competence-deficient cells of the same strain during cocultivation. J. Bacteriol. 185:7176-7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tao, L., D. J. LeBlanc, and J. J. Ferretti. 1992. Novel streptococcal-integration shuttle vectors for gene cloning and inactivation. Gene 120:105-110. [DOI] [PubMed] [Google Scholar]
- 37.van Schaik, E. J., C. L. Giltner, G. F. Audette, D. W. Keizer, D. L. Bautista, C. M. Slupsky, B. D. Sykes, and R. T. Irvin. 2005. DNA binding: a novel function of Pseudomonas aeruginosa type IV pili. J. Bacteriol. 187:1455-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida, A., and H. K. Kuramitsu. 2002. Multiple Streptococcus mutans genes are involved in biofilm formation. Appl. Environ. Microbiol. 68:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]