Abstract
In 1995, the Institute for Genomic Research completed the genome sequence of a rough derivative of Haemophilus influenzae serotype d, strain KW20. Although extremely useful in understanding the basic biology of H. influenzae, these data have not provided significant insight into disease caused by nontypeable H. influenzae, as serotype d strains are not pathogens. In contrast, strains of nontypeable H. influenzae are the primary pathogens of chronic and recurrent otitis media in children. In addition, these organisms have an important role in acute otitis media in children as well as other respiratory diseases. Such strains must therefore contain a gene repertoire that differs from that of strain Rd. Elucidation of the differences between these genomes will thus provide insight into the pathogenic mechanisms of nontypeable H. influenzae. The genome of a representative nontypeable H. influenzae strain, 86-028NP, isolated from a patient with chronic otitis media was therefore sequenced and annotated. Despite large regions of synteny with the strain Rd genome, there are large rearrangements in strain 86-028NP's genome architecture relative to the strain Rd genome. A genomic island similar to an island originally identified in H. influenzae type b is present in the strain 86-028NP genome, while the mu-like phage present in the strain Rd genome is absent from the strain 86-028NP genome. Two hundred eighty open reading frames were identified in the strain 86-028NP genome that were absent from the strain Rd genome. These data provide new insight that complements and extends the ongoing analysis of nontypeable H. influenzae virulence determinants.
In 1995 Haemophilus influenzae strain Rd, a rough derivative of H. influenzae serotype d strain KW20 (strain Rd hereafter), became the first free-living organism to have its genome sequenced to completion (34). Importantly, this also helped establish the large-scale shotgun approach, mated with the utilization of a scaffolding library and computer-assisted assembly, as a rational and expeditious approach for the sequencing of small bacterial genomes. Strain Rd was chosen as the prototypic bacterium for complete genome sequencing as it has a genome size representative of other bacteria and a G+C content close to that of the human genome. Additionally, at the time of sequencing, a physical map of the strain Rd genome did not exist, so this genome was a good test for the approach of shotgun sequencing, scaffolding, and assembly (34).
Although strain Rd is the exemplar organism for the current small-genome sequencing rationale and an important model organism for studying H. influenzae biology, strain Rd is a poor model for the study of pathogenicity caused by members of the genus Haemophilus. Serotype b strains of H. influenzae cause invasive diseases, for example, meningitis, and nontypeable H. influenzae (NTHi) strains principally have a role in localized respiratory disease, particularly in otitis media, acute sinusitis, and community-acquired pneumonia and have important consequences in patients with chronic obstructive pulmonary disease or cystic fibrosis (59, 74, 95, 100, 108). Strain Rd, however, is a derivative of a serotype d strain. Serotype d strains are rarely associated with disease (23, 44, 94, 102).
Because one of the most useful sets of data in the study of an organism's biology is its genomic sequence, a number of investigations have identified and characterized genes found in H. influenzae type b strains, H. influenzae biogroup Aegyptius strains or in nontypeable strains that are not present in strain Rd (13, 17, 30, 60, 64, 82, 106). Previously we carried out partial analyses and comparisons of the genomes of two NTHi strains, 1885MEE and 86-028NP, that were isolated from the middle ear and nasopharynx, respectively, of children with chronic otitis media (72). A genomic DNA-based microarray approach was employed to compare the genomes of strains 1885MEE and Rd as well as the genomes of strains 1885MEE and 86-028NP. In concert, a bioinformatics approach was used to compare the published genome of strain Rd with that of strain 86-028NP, which had been sequenced to threefold coverage. These analyses suggested that the genomes of strains 1885MEE and 86-028NP are more similar to each other than to the genome of strain Rd. Moreover, both analytical approaches identified a number of genes present in the NTHi strains that were absent from strain Rd. These included genes encoding proteins involved in protection against reactive oxygen species (tsaA), with possible roles in adhesion, biofilm formation, and pH regulation (tnaA), and a member of a family of virulence-associated autotransporters (lav).
Although this analysis was of great utility, due to of the fragmentary nature of the data available, it was desirable to expand this analysis. We thus completed sequencing the genome of strain 86-028NP and compared the completed genome to the genome of strain Rd. This allowed us to identify genes common to both strains and, importantly, strain 86-028NP-specific genes that may be involved in virulence. As strain 86-028NP has been extensively characterized, both in chinchilla models of otitis media and at the genetic level, having a full understanding of the genetic make-up of strain 86-028NP will complement existing data as well as allow a better understanding of the processes involved in the pathogenesis of otitis media and, by extension, other diseases in which H. influenzae is involved.
MATERIALS AND METHODS
Choice of strain sequenced.
Previous studies have indicated that NTHi strains are heterogeneous with respect to outer membrane protein profiles (8, 75), enzyme allotypes (76), and lipooligosaccharide genes (18). Furthermore, attempts to subtype NTHi did not identify a suitable candidate for genomic sequencing. We therefore chose the low-passage isolate strain 86-028NP, which was recovered from the nasopharynx of a child with chronic otitis media. This strain has subsequently been well characterized both in vitro (6, 46), in chinchilla models of otitis media (5, 58, 63, 111), and partially at the genomic level (72).
Library construction.
Chromosomal DNA was prepared from strain 86-028NP using Puregene reagents (Gentra Systems, Minneapolis, MN). For the initial shotgun sequencing of the genome, libraries of 1 to 2 kb and 2 to 4 kb of genomic DNA were constructed in pUC18 as previously described (72). For the scaffolding library, genomic DNA was manually sheared into a mean fragment size of 40 kb using a Hamilton syringe. After end repair, fragments were fractionated using a 0.7% low-melting-temperature agarose gel. Fragments larger than 30 kb were excised, and an in-gel ligation to pEpiFOS-5 was performed. The ligation mixture recovered from the gel was packaged into lambda phage in vitro and used to transfect EPI100 cells (Epicentre, Madison, WI).
Sequencing.
For the shotgun portion of the sequencing, cycle-sequencing reactions were run using PE Big-Dye terminators and universal primers (M13 forward and reverse) as previously described (72). To end-sequence the scaffolding library, the plasmid was first purified using an R.E.A.L. Prep 96 plasmid kit (QIAGEN Inc., Valencia, CA), then amplified using a TempliPhi DNA amplification kit (Amersham Biosciences Corp., Piscataway, NJ) before running reactions using PE Big-Dye terminators and pEpiFOS-5 forward and reverse sequencing primers (Epicentre, Madison, WI). The reactions for the clean-up portions of the project were run using PE Big-Dye terminators and custom primers (Integrated DNA Technologies, Coralville, IA). Excess dye terminators were removed with Sephadex G50 columns in a 96-well format, and sequence was determined on either an ABI 3700 or an ABI 3100 capillary electrophoresis DNA sequencer (Applied Biosystems, Foster City, CA).
Genome closure.
Paired end sequences from the scaffolding library and PCR were used to order the contigs and to add sequence in areas of low sequence coverage. Paired custom primers (Integrated DNA Technologies, Coralville, IA) were designed to bind at the ends of each contig as well as regions flanking areas of low sequence coverage. The intervening regions were amplified with a standard PCR protocol (98) using Taq polymerase (Roche Diagnostics, Indianapolis, IN) and sequenced on both strands. rRNA operons and the high-molecular-weight gene clusters were completely sequenced using clones from the scaffolding library as templates.
Assembly.
Phred/Phrap was used for data assembly, employing the default assembly parameters (32, 33, 36) as described (72). Assemblies were checked using the paired-end sequence data from 507 clones using the Seqman II program from the DNASTAR suite.
Data analyses.
Coding regions were identified using Glimmer2 (v2.13) trained on the 1,178 longest open reading frames (ORFs) identified by the Glimmer2 long-orfs program (25). Automated annotation by similarity was done by searching the Glimmer ORF set against the strain Rd proteome, the SwissProt database, the NCBI COGs database, and the KEGG database. The strain Rd database was compared bidirectionally with the strain 86-028NP ORF set using Tricross to determine high-confidence regions of similarity and to produce the dotplot comparison of genome organization (87).
The automatically predicted annotation information was further manually curated using Artemis (97) for visualization and demarcation of genomic regions of interest, and a custom FileMaker Pro database was generated which was then used to apply manual revisions and archive data related to the functional assignment. FASTA analyses were used for the primary automated comparisons. The strong synteny between the strain 80-028NP and strain Rd genomes allowed assignment of a function to the majority of the genes automatically, with similarity held to 90% or better at the amino acid level for matching. The nearly one-to-one mapping from the strain 86-028NP genome to the strain Rd genome was confirmed by assembly of the strain Rd ORFs onto the strain 86-028NP genome sequence and the reverse assembly of the strain 86-028NP ORFs onto the Rd genome, using the SeqMan program with the assembly criterion of 80% identity at the nucleotide level.
Manual BLAST analyses were used to explore the potential function of ORFs that did not show strong similarity to known genes. Manual curation of the automatic assignments was carried out to conform annotations to the current literature and repair the few places where the automated algorithm was easily led astray (notably the high-molecular-weight gene clusters, the hemoglobin-binding proteins, and the hsd gene clusters, whose high family similarity confounds automated assignment).
The tRNA genes were identified by tRNAscan-SE v1.11 (61). The rRNA operons were identified based on 16, 23, and 5S rRNA similarity with strain Rd and the ClustalW alignment of the neighborhoods containing these genes to determine the boundaries of the semiconserved regions.
Nucleotide sequence accession number.
The genome sequence is available from GenBank, where it has been assigned accession number CP000057. A list of the ORFs that are unique to each strain is given in Tables S1 and S2 at our website, www.microbial-pathogenesis.org/H.influenzae.86028/.
RESULTS AND DISCUSSION
Comparison between the genomes of NTHi strain 86-028NP and H. influenzae strain Rd.
The genomic sequence of strain 86-028NP contains 1,913,428 bp. This is approximately 4% larger than the strain Rd genome (1,830,137 bp) (34). There are also a larger number of genes in strain 86-028NP, 1,821 compared to 1,743 in strain Rd. A graphical representation of the strain 86-028NP genome is shown in Fig. 1. The gene complement was compared to that of strain Rd using the Seqman program in the DNASTAR suite. With 80% identity at the nucleotide level as a cutoff value, 280 ORFs were identified in the 86-028NP genome that were absent from the strain Rd genome, and 169 ORFs were identified in the strain Rd genome that are absent from the strain 86-028NP genome.
FIG. 1.
Schematic diagram of the organization of the H. influenzae strain 86-028NP genome. NTHI0001 is at the top of the figure, and increasing coordinates as annotated proceed clockwise around the circle. From the periphery in, the diagram indicates the following information. Labeled arrows indicate the identity, position, and direction of key gene loci. The maroon and orange double band indicates ORF location and coding direction. Clockwise-facing (positive strand as annotated) ORFs are indicated in the outer maroon band, while counterclockwise-facing ORFs are indicated in the inner, orange band. tRNA and rRNA operon positions and directions are indicated by the ring of purple and cyan arrows, purple for tRNAs and cyan for rRNA operons. The triple band of dark red, green, and dark blue demarcations indicates the major predicted COG classification for the corresponding ORF. Dark red indicates information storage and processing, green indicates cellular processes and signaling, and blue indicates metabolism. The prevailing gene order, in comparison to the strain Rd genome in a sliding window 10 ORFs wide, is indicated by the colored and offset background. Yellow inset regions are highly similar to strain Rd and follow the same gene order. Pink outset regions are highly similar to strain Rd and are in the reverse gene order from the strain 86-028NP genome. Regions with neither yellow nor pink background have no significant similarity to strain Rd.
Strain 86-028NP, like strain Rd, has six ribosomal operons. Using tRNAscan-SE v1.11, 58 tRNA genes were identified in the strain 86-028NP genome, representing the 20 common amino acids. The tRNA-Glu, tRNA-Ala, and tRNA-Ile genes were located in spacer regions between the 16S and 23S rRNA genes. A tRNA gene containing the UCA anticodon was also identified. This anticodon corresponds to an opal stop codon and is typically associated with an opal-suppressing tRNA that incorporates selenocysteine. The tRNA is adjacent to two genes encoding selB (NTHI0836), a Sec tRNA-specific elongation factor, and selA (NTHI0835), the enzyme that converts serine to dehydroalanine preparatory to forming selenocysteine by incorporation of selenium (35). The selD gene (NTHI0297), encoding selenophosphate synthetase, was also identified. The importance of this selenocysteine system is evidenced by the coding sequence for the alpha subunit of formate dehydrogenase (NTHI0007) containing an in-frame TGA stop codon that is presumably read as a selenocysteine codon. The in-frame TGA stop codon was previously noted in the current annotation of the strain Rd formate dehydrogenase gene (GenPept accession P46448).
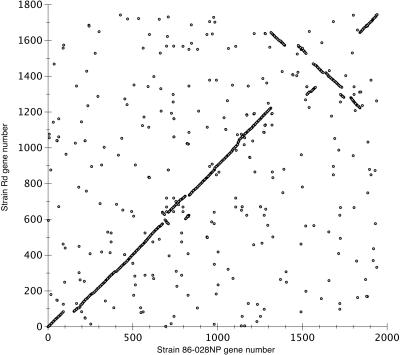
A gross comparison between the genomes involving analysis of the gene order of strain 86-028NP and that of strain Rd reveals a single major rearrangement in the form of a large inversion (Fig. 1 and 2). This 471-kb inversion represents almost 25% of the strain 86-028NP genome and is bounded by NTHI1391 and NTHI1394 (homologues of HI1218 and HI1645, respectively) and by NTHI1949 and NTHI1950 (homologues of HI1219 and HI1647, respectively). HI1219 and HI1646 are partially duplicated genes in strain Rd annotated as cmkA and cmkB (cytidylate kinases). One cmk gene (NTHI1949) is present in strain 86-028NP with a small cmk-like fragment between NTHI1391 and NTHI1394. Several clones from the scaffolding library overlap each end of the inversion in the 86-028NP genome, validating our assembly. Within this large inversion are several insertions, the largest of which are approximately 13 kb, 27 kb, and 51 kb in size. These regions contain predominantly hypothetical and conserved hypothetical genes as well as a number of homologues of phage genes. For example, the 27-kb insertion contains remnants of HP1- and HP2-like phage genes. The largest insert is bounded by homologues of integrase genes. In strain Rd, a mu-like phage is localized to this region (67). This phage is not present in the strain 86-028NP genome. Also within the large inverted region is a 21-kb inversion that restores synteny with the Rd genome.
FIG. 2.
Tricross dotplot diagram of the gene organization of strain 86-028NP compared to strain Rd. Each Tricross two-way hit between strain 86-028NP and strain Rd ORFs attaining a translation-to-translation FASTA E-score of 10−16 or better, is plotted in the diagram.
In addition to the large inversion, strain 86-028NP has other regions of divergence from colinearity with the strain Rd genome. These include nine regions greater than 5 kb, which contain sequences with no apparent homology to DNA that is present in strain Rd. Two of these regions contain the high-molecular-weight adhesins that are discussed below. Hypothetical genes predominate in six of the unique regions. The ninth region is approximately 56 kb in size. It lies between NTHI0100 and NTHI0165. BLASTn analysis indicates that genes in this region, designated ICEHin86-028NP, have high homology to genes in the H. influenzae type b plasmid ICEHin1056 (66). ICEHin1056 is a member of an extended family of genomic islands that are defined by a series of common core genes (66). ICEHin86-028NP possesses homologues of 45 ICEHin1056 ORFs. These include ORFs near the 5′ end of ICEHin86-028NP, including the defined core genes, that primarily encode proteins with putative roles in plasmid replication and conjugation and ORFs near the 3′ end that primarily encode conserved hypothetical proteins with motifs that suggest that they may be either membrane associated or exported. Notably, ICEHin86-028NP lacks the genes encoding proteins involved in tetracycline, chloramphenicol, and β-lactam resistance found in ICEHin1056. Scattered within ICEHin86-028NP are a transposase, resolvases, and a putative integrase regulator, suggesting that ICEHin86-028NP is a composite element derived from several mobile genetic elements.
ICEHin1506 has a sequence designated as an attP site 5′ of the first gene. In strain 86-028NP, a perfect copy of this attP site is present 5′ to NTHI0101 and a copy of this attP site, with a single nucleotide change, is present 3′ of NTHI0164. attP sites are implicated in the incorporation of mobile genetic elements into bacterial chromosomes to form genomic islands, possibly suggesting a mechanism by which this large section of genetic material became integrated into the strain 86-028NP genome (27). ICEHin86-028NP has a G+C content of 39%, lower than any of the other related genomic islands and close to strain 86-028NP's overall genome G+C content of 38%. This implies a long-term genomic association for this element. The presence of this element with its complement of genes homologous to those in ICEHin1506 (27), which are thought to encode membrane-associated and secreted proteins, may have important implications for the virulence of strain 86-028NP.
Several members of the family Pasteurellaceae, including Haemophilus ducreyi, Pasteurella multocida, and Actinobacillus actinomycetemcomitans, produce well-characterized protein toxins. In contrast, H. influenzae does not appear to produce protein toxins, and genes encoding putative protein toxins were not identified in the strain 86-028NP genome. In H. influenzae, the genes encoding glycosyltransferases, responsible for endotoxin biosynthesis, and genes encoding proteins that give the bacteria enhanced “fitness” during the process of infection have generally been considered virulence determinants. These genes include those that encode adhesins, the heme and haemoglobin binding proteins, as well as the genes that encode proteins that protect against oxidative stress. In the following sections, we discuss these genes, among others, in strain 86-028NP and note whether there are homologues in strain Rd.
Contingency genes.
H. influenzae has a limited number of two-component regulatory systems and other global regulators. Moxon and coworkers have argued that loci termed “simple contingency loci” provide an alternative mechanism for regulating gene expression, thus increasing the fitness of an organism by contributing to that organism's ability to rapidly respond to changing environmental conditions. These loci contain short tandem sequence repeats either within or 5′ to a coding region. During DNA replication, addition or loss of a repeat within a reading frame results in an alteration in the reading frame. When localized 5′ to a coding region, addition or loss of a repeat results in a change in promoter activity (11). Loci containing simple sequence repeats have been studied extensively in H. influenzae, for example (50). Several of the loci described in the following sections as phase variable contain simple sequence repeats.
Adhesins.
Strain 86-028NP possesses a number of genes whose products' primary function is in adherence to host cells (Table 1). One of these, the outer membrane protein P5, has previously been identified and its function was carefully dissected (54, 58, 77-79, 101). Recently, we demonstrated that strain 86-028NP possesses a gene cluster containing four genes that are homologues of pilABCD from strain Rd, Actinobacillus pleuropneumoniae, and Pasteurella multocida (3, 28, 96, 107). These genes, together with the comE gene and genes yet to be identified, encode a type IV pilus that has a role in adherence of strain 86-028NP to nasopharyngeal tissues (57).
TABLE 1.
Strain 86-028NP genes encoding proteins with a role in adherence
| NTHi no.a | HI no.b | Gene name | Function | Contingency repeats |
|---|---|---|---|---|
| 354 | hap | Adhesion and penetration protein Hap | ||
| 406 | 296 | pilD | Putative type 4 prepilin-like protein specific leader peptidase (EC 3.4.23.43) | |
| 407 | 297 | pilC | Putative type IV pilin secretion protein | |
| 408 | 298 | pilB | Putative type IV pilin secretion protein | |
| 409 | 299 | pilA | Type IV pilin subunit protein | |
| 1332 | 164 | ompP5 | Outer membrane protein P5 (OMP P5-homologous adhesin) | |
| 1448 | hmw2C | HMW2C, putative glycosyltransferase involved in glycosylation of HMW1A and HMW2A | ||
| 1449 | hmw2B | HMW2B, OMP-85-like protein required for HMW1A and HMW2A secretion | ||
| 1450 | hmw2A | HMW2A, high-molecular-weight adhesin 2 | ATCTTTC repeated 23 times; 5′ of gene | |
| 1983 | hmw1A | HMW1A, high-molecular-weight adhesin 1 | ATCTTTC repeated 17 times, 5′ of gene | |
| 1984 | hmw1B | HMW1B, OMP-85-like protein required for secretion of HMW1A and HMW2A | ||
| 1985 | hmw1C | HMW1C, putative glycosyltransferase involved in glycosylation of HMW1A and HMW2A |
Assigned locus number for each strain 86-028NP gene listed.
Locus number assigned by TIGR for the corresponding Rd homolog.
Strain 86-028NP possesses two high-molecular-weight (HMW) adhesin gene clusters that are absent in strain Rd. The high-molecular-weight adhesins were first characterized in NTHi strain 12, which has two HMW gene clusters, each encoding three proteins (HMWA, HMWB, and HMWC). HMWA is the structural component of the adhesin, HMWB has a role in transmembrane translocation, and HMWC is required for glycosylation of HMWA (7, 9, 37, 110). Similarly, strain 86-028NP's two HMW gene clusters contain homologues of the hmwA, hmwB, and hmwC genes in the same gene context as in strain 12 (16). The HMW1A and HMW2A proteins from strain 86-028NP are 72% identical, with the major area of divergence, including a 41-amino-acid insertion in HMW2A, toward the C termini. The paired HMWB and HMWC proteins from strain 86-028NP are 99% identical. The sequence ATCTTTC is repeated 17 times upstream of hmw1A and 23 times upstream of hmw2A. In strain 12, 16 repeats of this sequence are found 5′ of each hmw gene cluster (7).
Hap is an autotransported protein with a domain homologous to the catalytic domain of immunoglobulin A1 proteases. The NTHI0354 gene encodes a protein with 83% identity to Hap from NTHi strain N187 (109). Strain 86-028NP, along with other NTHi strains that possess HMW1 and HMW2, lacks the gene encoding Hia, another Haemophilus adhesin (10). Strain 86-028NP also lacks the hif gene cluster, encoding the hemagglutinating pilus, as we previously reported (72).
Lipooligosaccharide synthesis.
The structure, biosynthesis, and role in virulence of H. influenzae lipooligosaccharide has been studied extensively. Table 2 contains a list of genes involved in lipooligosaccharide biosynthesis. Strain 86-028NP has the full complement of genes required to synthesize the heptose-2-keto-3-deoxyoxtulosonic acid-lipid A portion of lipooligosaccharide. The lgtF and lpsA genes encode glycosyltransferases that add glucose and glucose or galactose to heptose residues 1 and 3, respectively. Both of these genes are present in the strain 86-028NP genome, and therefore it is likely that carbohydrate chains can be extended from the heptose 1 and heptose 3 residues of the strain 86-028NP lipooligosaccharide (49).
TABLE 2.
Strain 86-028NP genes encoding proteins with a role in lipooligosaccharide biosynthesis
| NTHi no.a | HI no.b | Gene name | Function | Contingency repeats |
|---|---|---|---|---|
| 68 | 58 | kdsB | 3-Deoxy-d-manno-octulosonic acid cytidylyltransferase | |
| 69 | 59 | lpxK | Tetraacyldisaccharide 4′-kinase | |
| 72 | 60 | msbA | Lipid A export ATP-binding protein msbA | |
| 296 | 199 | msbB | Lipid A biosynthesis (KDO)2-(lauroyl)-lipid IVA acyltransferase | |
| 365 | 258 | lgtC | UDP-galactose-lipooligosaccharide galactosyltransferase | GACA repeated 10 times, in frame |
| 366 | 260 | orfM | Xanthosine triphosphate pyrophosphatase | |
| 367 | 260.1 | kdkA | 3-Deoxy-d-manno-octulosonic acid kinase | |
| 368 | 261 | opsX | ADP-heptose-lipooligosaccharide heptosyltransferase I | |
| 383 | 275 | lpt6 | PE-tn-6-lipooligosaccharide phosphorylethanolamine transferase | |
| 471 | 351 | galE | UDP-glucose 4-epimerase | |
| 472 | 352 | lic3A | CMP-Neu5Ac-lipooligosaccharide alpha-2-3-sialyltransferase | CAAT repeated 18 times, in frame |
| 512 | 391 | Predicted acyltransferase | AGCA repeated 8 times, in frame | |
| 649 | 523 | waaQ | ADP-heptose-lipooligosaccharide heptosyltransferase III | |
| 677 | 550 | lic2A | UDP-galactose-lipooligosaccharide galactosyltransferase | CAAT repeated 14 times, in frame |
| 772 | 652 | kdtA | 3-Deoxy-d-manno-octulosonic acid transferase | |
| 773 | 653 | lgtF | UDP-glucose-lipooliqosaccharide glucosyltransferase | |
| 892 | 735 | lpxH | UDP-2,3-diacylglucosamine hydrolase | |
| 899 | 740 | pgmB | Phosphoglucomutase | |
| 913 | lex2B | UDP-glucose-lipooliqosaccharide glucosyltransferase | ||
| 926 | 765 | lpsA | Lipooligosaccharide glycosyltransferase | |
| 976 | 812 | qalU | UTP-glucose-1-phosphate uridylyltransferase | |
| 1034 | lic3A2 | CMP-Neu5Ac-lipooligosaccharide alpha-2-3-sialyltransferase | CAAT repeated 18 times, in frame | |
| 1037 | 873 | rmlB | dTDP-glucose 4,6-dehydratase | |
| 1082 | 915 | lpxD | UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase | |
| 1180 | 1005 | Predicted PE-lipooligosaccharide phosphorylethanolamine transferase | ||
| 1220 | 1060 | lpxB | Lipid-A-disaccharide synthase | |
| 1222 | 1061 | lpxA | Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase | |
| 1224 | 1064 | Predicted PE-lipooligosaccharide phosphorylethanolamine transferase | ||
| 1272 | 1105 | rfaF | ADP-heptose-lipooligosaccharide heptosyltransferase II | |
| 1278 | 1114 | rfaD | ADP-l-glycero-d-manno-heptose-6-epimerase | |
| 1312 | 1144 | lpxC | UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase | |
| 1350 | 1181 | lpcA | Phosphoheptose isomerase | |
| 1474 | 1578 | lgtD | Putative UDP-GlcNAc-lipooligosaccharide N-acetylglucosamine glycosyltransferase | |
| 1576 | 1557 | kdsA | Phospho-2-dehydro-3-deoxyoctonate aldolase and 3-deoxy-d-manno-octulosonic add 8-phosphate synthetase | |
| 1594 | 1540 | licD | Phosphorylcholine transferase | |
| 1595 | 1539 | licC | Protein LicC, CTP-phosphocholine cytidylyltransferase | |
| 1596 | 1538 | licB | Protein LicB, putative choline uptake protein | |
| 1597 | 1537 | licA | Protein LicA, choline kinase | CAAT repeated 15 times, in frame |
| 1606 | 1527 | htrB | Lipid A biosynthesis lauroyl acyltransferase | |
| 1607 | 1526 | rfaE | ADP-heptose synthase | |
| 1664 | 1337 | mrsA | Predicted phosphomannomutase | |
| 1750 | Putative glycosyltransferase, glycosyltransferase family 8 protein | GACA repeated 14 times, in frame | ||
| 1769 | Putative glycosyltransferase | CCAA repeated 17 times, out of frame | ||
| 1891 | 1279 | slaB | CMP-Neu5Ac synthetase | |
| 1921 | 1244 | Possible polysaccharide biosynthesis protein | ||
| 2002 | 1695 | lsgF | Putative UDP-galactose-lipooligosaccharide galactosyltransferase | |
| 2003 | 1696 | lsgE | Putative UDP-galactose-lipooligosaccharide galactosyltransferase | |
| 2004 | 1697 | lsgD | Putative UDP-GlcNAc-lipooligosaccharide N-acetylglucosaminyl glycosyltransferase | |
| 2005 | 1698 | lsgC | Putative UDP-galactose-lipooligosaccharide galactosyltransferase | |
| 2006 | 1699 | lsgB | CMP-N-acetylneuramlnate-beta-galactosamide-alpha-2,3-sialyltransferase | |
| 2007 | 1700 | lsgA | Putative lipooligosaccharide flippase | |
| 2025 | 1716 | wecA | Undecaprenyl-phosphate alpha-N-acetylglucosaminyl 1-phosphate transferase |
Assigned locus number for each strain 86-028NP gene listed.
Locus number assigned by TIGR for the corresponding Rd homolog.
In the serotype b strain RM153, the lic2C gene encodes a glucosyltransferase that adds glucose to heptose 2 (49). In the strain 86-028NP genome, this gene contains a frame shift. The phase-variable lic2A and licA genes, encoding a galactosyltranferase and choline kinase, respectively, are present in the strain 86-028NP genome (45, 48, 117). The lex2B gene, which encodes a glucosyltransferase in the serotype b strain DL42 as well as a number of other serotypeable strains, is present in the strain 86-028NP genome (41, 53). Five-prime to the lex2B gene in strain DL42 is the short phase-variable lex2A gene. In strain 86-028NP, this gene is out of frame compared to the DL42 sequence (GenBank accession U05670), due to the loss of one tetranucleotide repeat and a 5-bp deletion.
Recently, Hood and coworkers described a locus in strain Rd, designated hmg, that contains HI0866 through HI0874 (51). With the exception of a homologue of rmlB, these genes are absent from the strain 86-028NP genome. This includes the siaA gene, which encodes a sialyltransferase recently shown to be important in biofilm formation in NTHi strain 2019 (40, 56). Two copies of a homologue of the lic3A gene, encoding an alternative sialyltransferase, were identified in the strain 86-028NP genome (47, 56), as well as a copy of the lsgB gene, which encodes another sialyltransferase (56).
Iron acquisition.
H. influenzae strains have an absolute requirement for either heme or iron, together with protophorphyrin IX, the immediate precursor of heme (31, 118). Table 3 contains a list of genes involved in iron acquisition. Three hemoglobin and hemoglobin-haptoglobin binding proteins HgpA, HgpB, and HgpC were identified in H. influenzae type b strain HI689 (55, 69, 91). In strain HI689, these genes have CCAA tetranucleotide repeats and are known to be regulated by slip-strand mispairing. Two of these genes are present in strain 86-028NP. They both contain CCAA repeats; the hgpB gene is in frame, while the hgpC gene is out of frame. The derived amino acid sequence of a third gene that contains CCAA repeats is 45% identical to hgpA. We have designated this gene hgpD. This gene is out of frame.
TABLE 3.
Strain 86-028NP genes encoding proteins with a role in iron acquisition
| NTHi no.a | HI no.b | Gene name | Function | Contingency repeats |
|---|---|---|---|---|
| 177 | 97 | hitA | hFbpA, iron utilization periplasmic protein | |
| 179 | 98 | hitB | hFbpB, iron(III)-transport system permease protein | |
| 180 | 99 | hitC | hFbpC, iron utilization ATP-binding protein | |
| 202 | 113 | hemR | Hemin receptor | |
| 284 | 190 | fur | Ferric uptake regulation protein | |
| 369 | 262 | hxuC | Heme/hemopexin-binding protein C (heme:hemopexin utilization protein C) | |
| 370 | 263 | hxuB | Heme/hemopexin-binding protein B (heme:hemopexin utilization protein B) | |
| 371 | 264 | hxuA | Heme/hemopexin-binding protein A (heme:hemopexin utilization protein A) | |
| 477 | 359 | hfeD | Putative ABC-type chelated iron transport system, permease component | |
| 478 | 360 | hfeC | Putative ABC-type chelated iron transport system, permease component | |
| 479 | 361 | hfeB | Putative ABC-type chelated iron transport system, ATPase component | |
| 481 | 362 | hfeA | Putative periplasmic chelated iron binding protein | |
| 736 | hgpD | Hemoglobin-haptoglobin binding protein D (hemoglobin-haptoglobin utilization protein D) | CCAA repeated 17 times, out of frame | |
| 782 | 661 | hgpB | Hemoglobin-haptoglobin binding protein B (hemoglobin-haptoglobin utilization protein B) | CCAA repeated 12 times, in frame |
| 840 | 712 | hgpC | Hemoglobin-haptoglobin binding protein C (hemoglobin-haptoglobin utilization protein C) | CCAA repeated 20 times, out of frame |
| 1021 | 853 | hbpA | Heme-binding protein A (hemin-binding lipoprotein) | |
| 1168 | 994 | tbpA | Transferrin-binding protein 1 | |
| 1169 | 995 | tbpB | Transferrin-binding protein 2 | |
| 1329 | 1160 | hemH | Ferrochelatase | |
| 1390 | 1217 | hup | Heme utilization protein | |
| 2035 | 1728 | Mn2+ and Fe2+ transporter of the NRAMP family |
Assigned locus number for each strain 86-028NP gene listed.
Locus number assigned by TIGR for the corresponding Rd homolog.
Homologues of the hxuABC genes of H. influenzae type b that encode heme and heme-hemopexin complexes (19-21) as well as a homologue of the hemR receptor were identified. Strain 86-028NP also has the gene encoding the heme-binding lipoprotein HbpA (43). Downstream of hbpA is NTHI1022, a hypothetical gene whose product is a member of COG0748, a cluster that includes putative heme utilization proteins. A homologue of the hup gene, recently identified in H. influenzae type b, that encodes a general heme utilization protein was also identified (68).
In addition to the heme transport systems, iron transport systems were also identified. The hitABC genes encode the FbpABC proteins, respectively, members of a highly specific ferric iron ABC transport system that was elegantly characterized by complementing a siderophore-deficient Escherichia coli strain with the hitABC genes cloned from an H. influenzae type b strain (1). Transferrin-binding proteins 1 and 2 encoded by tbpAB (38, 39) as well as genes designated hfeABCD that are homologues of an ABC transport system involved in iron uptake, originally characterized in Yersinia pestis (12), were identified. This gene cluster is also present in strain Rd. NTHI2035 encodes a putative homologue of the NRAMP family of Mn2+ and Fe2+ transporters (92).
As noted above, H. influenzae can use iron, together with protoporphyrin IX, as a source of heme for growth in vitro. The hemH gene encoding ferrochelatase, which catalyzes the incorporation of iron into protoporphyrin IX (99), was identified. The gene encoding the global regulator, Fur, was also identified (2, 105).
Oxidative stress.
Although necessary for growth, the active acquisition of iron can have deleterious effects on bacterial cells. Through the Fenton reaction, iron can react with hydrogen peroxide and generate highly reactive hydroxyl radicals. These products have profound effects, including lipid peroxidation and damage to both iron-containing enzymes and DNA (52). The best-known defense system against hydroxyl radicals consists of superoxide dismutases A and B, which convert highly reactive superoxide to hydrogen peroxide, which is then converted, by catalase, into water and oxygen (26).
Strains 86-028NP and Rd contain the sodA gene (NTHI1251) but lack the sodB gene. Both strains also possess a catalase gene, hktE (NTHI1099) (14), the oxyR gene (NTHI0704), encoding a primary regulator of genes involved in protection against oxidative stress (62, 83), and the gene encoding a chimeric peroxidase termed Prx/Grx that has a glutathione-dependent role in protection against small alkyl hydroperoxides (81, 115, 116). We previously identified NTHI0212, a gene encoding a homologue of the P. multocida peroxiredoxin, TsaA, that is absent in strain Rd (72). Strain 86-028NP, however, lacks AhpF, a dedicated alkyl hydroperoxide reductase known to be involved in the reduction of TsaA in Salmonella spp. (84).
Further protection against oxidative stress may be afforded by the ferritin-like proteins encoded by the ftnA and ftnB (NTHI1773 and NTHI1772, respectively) genes. Overexpression of these proteins was shown to protect an iron-overloaded E. coli fur mutant against oxidative damage (113). A conserved hypothetical gene, NTHI1817, encodes a protein with homology to a DNA-binding ferritin-like protein. This is a member of the Dps family of nonspecific DNA binding proteins, which in Salmonella enterica have roles in protection against oxidative stress, both in the presence of iron and during phagocytosis, and are important for virulence in a murine model of Salmonella infection (42). In E. coli, Dps was shown to preferentially bind iron that had been oxidized by hydrogen peroxide, thus having an important role in abrogating the production of hydroxyl radicals generated via the Fenton reaction (120).
Secretion.
In addition to the Sec system, strain 86-028NP has genes that encode the TatA, TatB, and TatC proteins, cytoplasmic membrane-associated proteins that are involved in Sec-independent transport of proteins with twin arginines in their signal peptides (NTHI0279, NTHI0280, and NTHI0282) (15, 119). As previously reported, strain 86-028NP possesses NTHI0585, the gene encoding the autotransported protein Lav (72). This protein is absent in strain Rd, present in Neisseria spp., and appears, within Haemophilus, to be restricted to pathogenic strains (24). Strain 86-028NP also has the gene encoding an immunoglobulin A protease (NTHI1164) (86), and as noted above, the gene encoding the Hap adhesin. Both are proteins of the autotransporter class. As described above, the HMW adhesins are members of the two-partner secretion pathway group of proteins.
Outer membrane proteins.
A number of outer membrane protein (OMP)-encoding genes have been identified by homology to those in other Haemophilus isolates. These include the major OMPs that were all originally identified in H. influenzae type b; the surface-expressed P1 (NTHI0522), the porin P2 (NTHI0225), the phosphomonoesterase and heme transporter P4 (NTHI0816), the adhesin P5 (NTHI1332) and the lipoprotein P6 (NTHI0501) (70, 71, 73, 88-90). Strain 86-028NP also shares a number of minor OMPs with other Haemophilus strains. These include D15 and the transferrin binding proteins from H. influenzae type b as well as a homologue of OMP26, which was identified in NTHi strain 289 (29, 38, 39, 112). All have subsequently been characterized in NTHi strains and analyzed as potential vaccine candidates (4, 22, 65, 74, 85).
Restriction enzymes systems.
Strain 86-028NP lacks the HindII and HindIII type II restriction systems (34, 80, 104). In contrast, genes encoding the HaeII system that was originally identified in H. aegyptius (103) are present in the strain 86-028NP genome but absent in strain Rd. Both strain 86-028NP and strain Rd have Hsd type I restriction systems encoding a methyltransferase (HsdM), a sequence recognition protein (HsdS), and a restriction enzyme (HsdR) (93). These genes are adjacent in the strain Rd genome (HI1285-HI1287). The 86-028NP genome contains three hsd-like loci that each contain four genes. One hsd system is encoded by NTHI1838 to NTHI1843. In this gene cluster, NTHI1841 encodes a hypothetical protein. A second hsd-like locus is encoded by NTHI0314 to NTHI0318. In this gene cluster, NTHI0316 encodes a putative anticodon nuclease. This hsd-like system may be similar to the prr system in E. coli (114). A third hsd locus is encoded by NTHI0188 to NTHI0193. In this gene cluster, NTHI0190 encodes a predicted transcriptional regulator with a helix-turn-helix domain.
Summary.
We have sequenced and annotated the genome of strain 86-028NP, a clinical isolate of a nontypeable H. influenzae strain isolated from a child with chronic otitis media. The genome is approximately 1.91 kb in size, slightly larger than the strain Rd genome. We identified a number of regions of gross genome rearrangement relative to the strain Rd genome as well as a number of genes specific to strain 86-028NP. These data will be instrumental to the long-term goal of increasing our understanding of the pathogenesis of diseases caused by NTHi.
Acknowledgments
This work was supported by National Institutes of Health grants R01 DC03915 (to L.O.B.) and R01 DC005980 (to R.S.M.). The DNA Sequencing Core at Columbus Children's Research Institute was supported, in part, by National Institutes of Health grant K12 HD43372.
REFERENCES
- 1.Anderson, D. S., P. Adhikari, A. J. Nowalk, C. Y. Chen, and T. A. Mietzner. 2004. The hFbpABC transporter from Haemophilus influenzae functions as a binding-protein-dependent ABC transporter with high specificity and affinity for ferric iron. J. Bacteriol. 186:6220-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 3.Bakaletz, L. O., B. D. Baker, J. A. Jurcisek, A. Harrison, L. A. Novotny, J. E. Bookwalter, R. Mungur, and R. S. Munson, Jr. 2005. Demonstration of Type IV pilus expression and a twitching phenotype by Haemophilus influenzae. Infect. Immun. 73:1635-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakaletz, L. O., S. J. Barenkamp, J. Eskola, B. Green, X. X. Gu, T. Harada, T. Heikkinen, P. Karma, J. O. Klein, Y. Kurono, G. Mogi, T. F. Murphy, P. L. Ogra, J. A. Patel, M. Suzuki, and N. Yamanaka. 2002. Recent advances in otitis media. 7. Vaccine Ann. Otol. Rhinol. Laryngol. Suppl. 188:82-94. [PubMed] [Google Scholar]
- 5.Bakaletz, L. O., E. R. Leake, J. M. Billy, and P. T. Kaumaya. 1997. Relative immunogenicity and efficacy of two synthetic chimeric peptides of fimbrin as vaccinogens against nasopharyngeal colonization by nontypeable Haemophilus influenzae in the chinchilla. Vaccine 15:955-961. [DOI] [PubMed] [Google Scholar]
- 6.Bakaletz, L. O., B. M. Tallan, T. Hoepf, T. F. DeMaria, H. G. Birck, and D. J. Lim. 1988. Frequency of fimbriation of nontypable Haemophilus influenzae and its ability to adhere to chinchilla and human respiratory epithelium. Infect. Immun. 56:331-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barenkamp, S. J., and E. Leininger. 1992. Cloning, expression, and DNA sequence analysis of genes encoding nontypeable Haemophilus influenzae high-molecular-weight surface-exposed proteins related to filamentous hemagglutinin of Bordetella pertussis. Infect. Immun. 60:1302-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barenkamp, S. J., R. S. Munson, Jr., and D. M. Granoff. 1982. Outer membrane protein and biotype analysis of pathogenic nontypable Haemophilus influenzae. Infect. Immun. 36:535-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barenkamp, S. J., and J. W. St Geme 3rd. 1994. Genes encoding high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae are part of gene clusters. Infect. Immun. 62:3320-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barenkamp, S. J., and J. W. St Geme 3rd. 1996. Identification of a second family of high-molecular-weight adhesion proteins expressed by non-typable Haemophilus influenzae. Mol. Microbiol. 19:1215-1223. [DOI] [PubMed] [Google Scholar]
- 11.Bayliss, C. D., D. Field, and E. R. Moxon. 2001. The simple sequence contingency loci of Haemophilus influenzae and Neisseria meningitidis. J. Clin. Investig. 107:657-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bearden, S. W., T. M. Staggs, and R. D. Perry. 1998. An ABC transporter system of Yersinia pestis allows utilization of chelated iron by Escherichia coli SAB11. J. Bacteriol. 180:1135-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergman, N. H., and B. J. Akerley. 2003. Position-based scanning for comparative genomics and identification of genetic islands in Haemophilus influenzae type b. Infect. Immun. 71:1098-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bishai, W. R., H. O. Smith, and G. J. Barcak. 1994. A peroxide/ascorbate-inducible catalase from Haemophilus influenzae is homologous to the Escherichia coli katE gene product. J. Bacteriol. 176:2914-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolhuis, A., J. E. Mathers, J. D. Thomas, C. M. Barrett, and C. Robinson. 2001. TatB and TatC form a functional and structural unit of the twin-arginine translocase from Escherichia coli. J. Biol. Chem. 276:20213-20219. [DOI] [PubMed] [Google Scholar]
- 16.Buscher, A. Z., K. Burmeister, S. J. Barenkamp, and J. W. St Geme 3rd. 2004. Evolutionary and functional relationships among the nontypeable Haemophilus influenzae HMW family of adhesins. J. Bacteriol 186:4209-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang, C. C., J. R. Gilsdorf, V. J. DiRita, and C. F. Marrs. 2000. Identification and genetic characterization of Haemophilus influenzae genetic island 1. Infect. Immun. 68:2630-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cody, A. J., D. Field, E. J. Feil, S. Stringer, M. E. Deadman, A. G. Tsolaki, B. Gratz, V. Bouchet, R. Goldstein, D. W. Hood, and E. R. Moxon. 2003. High rates of recombination in otitis media isolates of non-typeable Haemophilus influenzae. Infect. Genet. Evol. 3:57-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cope, L. D., R. P. Love, S. E. Guinn, A. Gilep, S. Usanov, R. W. Estabrook, Z. Hrkal, and E. J. Hansen. 2001. Involvement of HxuC outer membrane protein in utilization of hemoglobin by Haemophilus influenzae. Infect. Immun. 69:2353-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cope, L. D., S. E. Thomas, Z. Hrkal, and E. J. Hansen. 1998. Binding of heme-hemopexin complexes by soluble HxuA protein allows utilization of this complexed heme by Haemophilus influenzae. Infect. Immun. 66:4511-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cope, L. D., R. Yogev, U. Muller-Eberhard, and E. J. Hansen. 1995. A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzae type b. J. Bacteriol. 177:2644-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cripps, A. W., and J. Kyd. 2003. Bacterial otitis media: current vaccine development strategies. Immunol. Cell Biol. 81:46-51. [DOI] [PubMed] [Google Scholar]
- 23.Daines, D. A., L. A. Cohn, H. N. Coleman, K. S. Kim, and A. L. Smith. 2003. Haemophilus influenzae Rd KW20 has virulence properties. J. Med. Microbiol. 52:277-282. [DOI] [PubMed] [Google Scholar]
- 24.Davis, J., A. L. Smith, W. R. Hughes, and M. Golomb. 2001. Evolution of an autotransporter: domain shuffling and lateral transfer from pathogenic Haemophilus to Neisseria. J. Bacteriol. 183:4626-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Demple, B. 1991. Regulation of bacterial oxidative stress genes. Annu. Rev. Genet. 25:315-337. [DOI] [PubMed] [Google Scholar]
- 27.Dimopoulou, I. D., J. E. Russell, Z. Mohd-Zain, R. Herbert, and D. W. Crook. 2002. Site-specific recombination with the chromosomal tRNA(Leu) gene by the large conjugative Haemophilus resistance plasmid. Antimicrob. Agents Chemother. 46:1602-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doughty, S. W., C. G. Ruffolo, and B. Adler. 2000. The type 4 fimbrial subunit gene of Pasteurella multocida. Vet. Microbiol. 72:79-90. [DOI] [PubMed] [Google Scholar]
- 29.El-Adhami, W., J. M. Kyd, D. A. Bastin, and A. W. Cripps. 1999. Characterization of the gene encoding a 26-kilodalton protein (OMP26) from nontypeable Haemophilus influenzae and immune responses to the recombinant protein. Infect. Immun. 67:1935-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erdos, G., S. Sayeed, P. Antalis, F. Z. Hu, J. Hayes, J. Goodwin, R. Dopico, J. C. Post, and G. D. Ehrlich. 2003. Development and characterization of a pooled Haemophilus influenzae genomic library for the evaluation of gene expression changes associated with mucosal biofilm formation in otitis media. Int. J. Pediatr. Otorhinolaryngol. 67:749-755. [DOI] [PubMed] [Google Scholar]
- 31.Evans, N. M., D. D. Smith, and A. J. Wicken. 1974. Haemin and nicotinamide adenine dinucleotide requirements of Haemophilus influenzae and Haemophilus parainfluenzae. J. Med. Microbiol. 7:359-365. [DOI] [PubMed] [Google Scholar]
- 32.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 33.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 34.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 35.Forchhammer, K., W. Leinfelder, and A. Bock. 1989. Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein. Nature 342:453-456. [DOI] [PubMed] [Google Scholar]
- 36.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 37.Grass, S., A. Z. Buscher, W. E. Swords, M. A. Apicella, S. J. Barenkamp, N. Ozchlewski, and J. W. St Geme 3rd. 2003. The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involved in lipooligosaccharide biosynthesis. Mol. Microbiol. 48:737-751. [DOI] [PubMed] [Google Scholar]
- 38.Gray-Owen, S. D., S. Loosmore, and A. B. Schryvers. 1995. Identification and characterization of genes encoding the human transferrin-binding proteins from Haemophilus influenzae. Infect. Immun. 63:1201-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gray-Owen, S. D., and A. B. Schryvers. 1995. Characterization of transferrin binding proteins 1 and 2 in invasive type b and nontypeable strains of Haemophilus influenzae. Infect. Immun. 63:3809-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greiner, L. L., H. Watanabe, N. J. Phillips, J. Shao, A. Morgan, A. Zaleski, B. W. Gibson, and M. A. Apicella. 2004. Nontypeable Haemophilus influenzae strain 2019 produces a biofilm containing N-acetylneuraminic acid that may mimic sialylated O-linked glycans. Infect. Immun. 72:4249-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griffin, R., A. D. Cox, K. Makepeace, J. C. Richards, E. R. Moxon, and D. W. Hood. 2003. The role of lex2 in lipopolysaccharide biosynthesis in Haemophilus influenzae strains RM7004 and RM153. Microbiology 149:3165-3175. [DOI] [PubMed] [Google Scholar]
- 42.Halsey, T. A., A. Vazquez-Torres, D. J. Gravdahl, F. C. Fang, and S. J. Libby. 2004. The ferritin-like Dps protein is required for Salmonella enterica serovar Typhimurium oxidative stress resistance and virulence. Infect. Immun. 72:1155-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanson, M. S., C. Slaughter, and E. J. Hansen. 1992. The hbpA gene of Haemophilus influenzae type b encodes a heme-binding lipoprotein conserved among heme-dependent Haemophilus species. Infect. Immun. 60:2257-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heath, P. T., R. Booy, H. J. Azzopardi, M. P. Slack, J. Fogarty, A. C. Moloney, M. E. Ramsay, and E. R. Moxon. 2001. Non-type b Haemophilus influenzae disease: clinical and epidemiologic characteristics in the Haemophilus influenzae type b vaccine era. Pediatr. Infect. Dis. J. 20:300-305. [DOI] [PubMed] [Google Scholar]
- 45.High, N. J., M. E. Deadman, and E. R. Moxon. 1993. The role of a repetitive DNA motif (5′-CAAT-3′) in the variable expression of the Haemophilus influenzae lipopolysaccharide epitope alpha Gal(1-4)beta Gal. Mol. Microbiol. 9:1275-1282. [DOI] [PubMed] [Google Scholar]
- 46.Holmes, K. A., and L. O. Bakaletz. 1997. Adherence of non-typeable Haemophilus influenzae promotes reorganization of the actin cytoskeleton in human or chinchilla epithelial cells in vitro. Microb. Pathog. 23:157-166. [DOI] [PubMed] [Google Scholar]
- 47.Hood, D. W., A. D. Cox, M. Gilbert, K. Makepeace, S. Walsh, M. E. Deadman, A. Cody, A. Martin, M. Mansson, E. K. Schweda, J. R. Brisson, J. C. Richards, E. R. Moxon, and W. W. Wakarchuk. 2001. Identification of a lipopolysaccharide alpha-2,3-sialyltransferase from Haemophilus influenzae. Mol. Microbiol. 39:341-350. [DOI] [PubMed] [Google Scholar]
- 48.Hood, D. W., A. D. Cox, W. W. Wakarchuk, M. Schur, E. K. Schweda, S. L. Walsh, M. E. Deadman, A. Martin, E. R. Moxon, and J. C. Richards. 2001. Genetic basis for expression of the major globotetraose-containing lipopolysaccharide from H. influenzae strain Rd (RM118). Glycobiology 11:957-967. [DOI] [PubMed] [Google Scholar]
- 49.Hood, D. W., M. E. Deadman, A. D. Cox, K. Makepeace, A. Martin, J. C. Richards, and E. R. Moxon. 2004. Three genes, lgtF, lic2C and lpsA, have a primary role in determining the pattern of oligosaccharide extension from the inner core of Haemophilus influenzae LPS. Microbiology 150:2089-2097. [DOI] [PubMed] [Google Scholar]
- 50.Hood, D. W., M. E. Deadman, M. P. Jennings, M. Bisercic, R. D. Fleischmann, J. C. Venter, and E. R. Moxon. 1996. DNA repeats identify novel virulence genes in Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 93:11121-11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hood, D. W., G. Randle, A. D. Cox, K. Makepeace, J. Li, E. K. Schweda, J. C. Richards, and E. R. Moxon. 2004. Biosynthesis of cryptic lipopolysaccharide glycoforms in Haemophilus influenzae involves a mechanism similar to that required for O-antigen synthesis. J. Bacteriol. 186:7429-7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Imlay, J. A. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57:395-418. [DOI] [PubMed] [Google Scholar]
- 53.Jarosik, G. P., and E. J. Hansen. 1994. Identification of a new locus involved in expression of Haemophilus influenzae type b lipooligosaccharide. Infect. Immun. 62:4861-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang, Z., N. Nagata, E. Molina, L. O. Bakaletz, H. Hawkins, and J. A. Patel. 1999. Fimbria-mediated enhanced attachment of nontypeable Haemophilus influenzae to respiratory syncytial virus-infected respiratory epithelial cells. Infect. Immun. 67:187-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin, H., Z. Ren, P. W. Whitby, D. J. Morton, and T. L. Stull. 1999. Characterization of hgpA, a gene encoding a haemoglobin/haemoglobin-haptoglobin-binding protein of Haemophilus influenzae. Microbiology 145:905-914. [DOI] [PubMed] [Google Scholar]
- 56.Jones, P. A., N. M. Samuels, N. J. Phillips, R. S. Munson, Jr., J. A. Bozue, J. A. Arseneau, W. A. Nichols, A. Zaleski, B. W. Gibson, and M. A. Apicella. 2002. Haemophilus influenzae type b strain A2 has multiple sialyltransferases involved in lipooligosaccharide sialylation. J. Biol. Chem. 277:14598-14611. [DOI] [PubMed] [Google Scholar]
- 57.Jurcisek, J. A., B. D. Baker, R. S. Munson, Jr., and L. O. Bakaletz. 2005. The type IV pilus of Haemophilus influenzae is important for both maintaining adherence to the nasopharynx as well as biofilm formation in the middle ear of the chinchilla host, abstr. D-097. Abstracts of the 105th General Meeting of the American Society for Microbiology. American Society for Microbiology, Washington, D.C.
- 58.Kennedy, B. J., L. A. Novotny, J. A. Jurcisek, Y. Lobet, and L. O. Bakaletz. 2000. Passive transfer of antiserum specific for immunogens derived from a nontypeable Haemophilus influenzae adhesin and lipoprotein D prevents otitis media after heterologous challenge. Infect. Immun. 68:2756-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kilpi, T., E. Herva, T. Kaijalainen, R. Syrjanen, and A. K. Takala. 2001. Bacteriology of acute otitis media in a cohort of Finnish children followed for the first two years of life. Pediatr. Infect. Dis. J. 20:654-662. [DOI] [PubMed] [Google Scholar]
- 60.Li, M. S., J. L. Farrant, P. R. Langford, and J. S. Kroll. 2003. Identification and characterization of genomic loci unique to the Brazilian purpuric fever clonal group of H. influenzae biogroup aegyptius: functionality explored using meningococcal homology. Mol. Microbiol. 47:1101-1111. [DOI] [PubMed] [Google Scholar]
- 61.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maciver, I., and E. J. Hansen. 1996. Lack of expression of the global regulator OxyR in Haemophilus influenzae has a profound effect on growth phenotype. Infect. Immun. 64:4618-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mason, K. M., R. S. Munson, Jr., and L. O. Bakaletz. 2003. Nontypeable Haemophilus influenzae gene expression induced in vivo in a chinchilla model of otitis media. Infect. Immun. 71:3454-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McGillivary, G., A. P. Tomaras, E. R. Rhodes, and L. A. Actis. 2005. Cloning and sequencing of a genomic island found in the Brazilian purpuric fever clone of Haemophilus influenzae biogroup aegyptius. Infect. Immun. 73:1927-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McMichael, J. C., and B. A. Green. 2003. Vaccines for Moraxella catarrhalis and non-typeable Haemophilus influenzae. Curr. Opin. Investig. Drugs 4:953-958. [PubMed] [Google Scholar]
- 66.Mohd-Zain, Z., S. L. Turner, A. M. Cerdeno-Tarraga, A. K. Lilley, T. J. Inzana, A. J. Duncan, R. M. Harding, D. W. Hood, T. E. Peto, and D. W. Crook. 2004. Transferable antibiotic resistance elements in Haemophilus influenzae share a common evolutionary origin with a diverse family of syntenic genomic islands. J. Bacteriol. 186:8114-8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morgan, G. J., G. F. Hatfull, S. Casjens, and R. W. Hendrix. 2002. Bacteriophage Mu genome sequence: analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J. Mol. Biol. 317:337-359. [DOI] [PubMed] [Google Scholar]
- 68.Morton, D. J., A. Smith, Z. Ren, L. L. Madore, T. M. Vanwagoner, T. W. Seale, P. W. Whitby, and T. L. Stull. 2004. Identification of a haem-utilization protein (Hup) in Haemophilus influenzae. Microbiology 150:3923-3933. [DOI] [PubMed] [Google Scholar]
- 69.Morton, D. J., P. W. Whitby, H. Jin, Z. Ren, and T. L. Stull. 1999. Effect of multiple mutations in the hemoglobin- and hemoglobin-haptoglobin-binding proteins, HgpA, HgpB, and HgpC, of Haemophilus influenzae type b. Infect. Immun. 67:2729-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Munson, R., Jr., and S. Grass. 1988. Purification, cloning, and sequence of outer membrane protein P1 of Haemophilus influenzae type b. Infect. Immun. 56:2235-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Munson, R. S., Jr., and D. M. Granoff. 1985. Purification and partial characterization of outer membrane proteins P5 and P6 from Haemophilus influenzae type b. Infect. Immun. 49:544-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Munson, R. S., Jr., A. Harrison, A. Gillaspy, W. C. Ray, M. Carson, D. Armbruster, J. Gipson, M. Gipson, L. Johnson, L. Lewis, D. W. Dyer, and L. O. Bakaletz. 2004. Partial analysis of the genomes of two nontypeable Haemophilus influenzae otitis media isolates. Infect. Immun. 72:3002-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Munson, R. S., Jr., J. L. Shenep, S. J. Barenkamp, and D. M. Granoff. 1983. Purification and comparison of outer membrane protein P2 from Haemophilus influenzae type b isolates. J. Clin. Investig. 72:677-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murphy, T. F. 2003. Respiratory infections caused by non-typeable Haemophilus influenzae. Curr. Opin. Infect. Dis. 16:129-134. [DOI] [PubMed] [Google Scholar]
- 75.Murphy, T. F., K. C. Dudas, J. M. Mylotte, and M. A. Apicella. 1983. A subtyping system for nontypable Haemophilus influenzae based on outer-membrane proteins. J. Infect. Dis. 147:838-846. [DOI] [PubMed] [Google Scholar]
- 76.Musser, J. M., S. J. Barenkamp, D. M. Granoff, and R. K. Selander. 1986. Genetic relationships of serologically nontypable and serotype b strains of Haemophilus influenzae. Infect. Immun. 52:183-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Novotny, L. A., and L. O. Bakaletz. 2003. The fourth surface-exposed region of the outer membrane protein P5-homologous adhesin of nontypable Haemophilus influenzae is an immunodominant but nonprotective decoying epitope. J. Immunol. 171:1978-1983. [DOI] [PubMed] [Google Scholar]
- 78.Novotny, L. A., J. A. Jurcisek, M. E. Pichichero, and L. O. Bakaletz. 2000. Epitope mapping of the outer membrane protein P5-homologous fimbrin adhesin of nontypeable Haemophilus influenzae. Infect. Immun. 68:2119-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Novotny, L. A., M. E. Pichichero, P. A. Denoel, C. Neyt, S. Vanderschrick, G. Dequesne, and L. O. Bakaletz. 2002. Detection and characterization of pediatric serum antibody to the OMP P5-homologous adhesin of nontypeable Haemophilus influenzae during acute otitis media. Vaccine 20:3590-3597. [DOI] [PubMed] [Google Scholar]
- 80.Nwankwo, D. O., L. S. Moran, B. E. Slatko, P. A. Waite-Rees, L. F. Dorner, J. S. Benner, and G. G. Wilson. 1994. Cloning, analysis and expression of the HindIII R-M-encoding genes. Gene 150:75-80. [DOI] [PubMed] [Google Scholar]
- 81.Pauwels, F., B. Vergauwen, F. Vanrobaeys, B. Devreese, and J. J. Van Beeumen. 2003. Purification and characterization of a chimeric enzyme from Haemophilus influenzae Rd that exhibits glutathione-dependent peroxidase activity. J. Biol. Chem. 278:16658-16666. [DOI] [PubMed] [Google Scholar]
- 82.Pettigrew, M. M., B. Foxman, C. F. Marrs, and J. R. Gilsdorf. 2002. Identification of the lipooligosaccharide biosynthesis gene lic2B as a putative virulence factor in strains of nontypeable Haemophilus influenzae that cause otitis media. Infect. Immun. 70:3551-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pomposiello, P. J., and B. Demple. 2001. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 19:109-114. [DOI] [PubMed] [Google Scholar]
- 84.Poole, L. B., A. Godzik, A. Nayeem, and J. D. Schmitt. 2000. AhpF can be dissected into two functional units: tandem repeats of two thioredoxin-like folds in the N-terminus mediate electron transfer from the thioredoxin reductase-like C-terminus to AhpC. Biochemistry 39:6602-6615. [DOI] [PubMed] [Google Scholar]
- 85.Poolman, J. T., L. Bakaletz, A. Cripps, P. A. Denoel, A. Forsgren, J. Kyd, and Y. Lobet. 2000. Developing a nontypeable Haemophilus influenzae (NTHi) vaccine. Vaccine 19(Suppl. 1):S108-S115. [DOI] [PubMed] [Google Scholar]
- 86.Poulsen, K., J. Reinholdt, and M. Kilian. 1992. A comparative genetic study of serologically distinct Haemophilus influenzae type 1 immunoglobulin A1 proteases. J. Bacteriol. 174:2913-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ray, W. C., R. S. Munson, Jr., and C. J. Daniels. 2001. Tricross: using dot-plots in sequence-id space to detect uncataloged intergenic features. Bioinformatics 17:1105-1112. [DOI] [PubMed] [Google Scholar]
- 88.Reidl, J., and J. J. Mekalanos. 1996. Lipoprotein e(P4) is essential for hemin uptake by Haemophilus influenzae. J. Exp. Med. 183:621-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reilly, T. J., D. L. Chance, and A. L. Smith. 1999. Outer membrane lipoprotein e (P4) of Haemophilus influenzae is a novel phosphomonoesterase. J. Bacteriol. 181:6797-6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reilly, T. J., B. A. Green, G. W. Zlotnick, and A. L. Smith. 2001. Contribution of the DDDD motif of H. influenzae e (P4) to phosphomonoesterase activity and heme transport. FEBS Lett. 494:19-23. [DOI] [PubMed] [Google Scholar]
- 91.Ren, Z., H. Jin, D. J. Morton, and T. L. Stull. 1998. hgpB, a gene encoding a second Haemophilus influenzae hemoglobin- and hemoglobin-haptoglobin-binding protein. Infect. Immun. 66:4733-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Richer, E., P. Courville, I. Bergevin, and M. F. Cellier. 2003. Horizontal gene transfer of “prototype” Nramp in bacteria. J. Mol. Evol. 57:363-376. [DOI] [PubMed] [Google Scholar]
- 93.Roberts, R. J., M. Belfort, T. Bestor, A. S. Bhagwat, T. A. Bickle, J. Bitinaite, R. M. Blumenthal, S. Degtyarev, D. T. Dryden, K. Dybvig, K. Firman, E. S. Gromova, R. I. Gumport, S. E. Halford, S. Hattman, J. Heitman, D. P. Hornby, A. Janulaitis, A. Jeltsch, J. Josephsen, A. Kiss, T. R. Klaenhammer, I. Kobayashi, H. Kong, D. H. Kruger, S. Lacks, M. G. Marinus, M. Miyahara, R. D. Morgan, N. E. Murray, V. Nagaraja, A. Piekarowicz, A. Pingoud, E. Raleigh, D. N. Rao, N. Reich, V. E. Repin, E. U. Selker, P. C. Shaw, D. C. Stein, B. L. Stoddard, W. Szybalski, T. A. Trautner, J. L. Van Etten, J. M. Vitor, G. G. Wilson, and S. Y. Xu. 2003. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 31:1805-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rodriguez, C. A., V. Avadhanula, A. Buscher, A. L. Smith, J. W. St. Geme III, and E. E. Adderson. 2003. Prevalence and distribution of adhesins in invasive non-type b encapsulated Haemophilus influenzae. Infect. Immun. 71:1635-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roman, F., R. Canton, M. Perez-Vazquez, F. Baquero, and J. Campos. 2004. Dynamics of long-term colonization of respiratory tract by Haemophilus influenzae in cystic fibrosis patients shows a marked increase in hypermutable strains. J. Clin. Microbiol. 42:1450-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ruffolo, C. G., J. M. Tennent, W. P. Michalski, and B. Adler. 1997. Identification, purification, and characterization of the type 4 fimbriae of Pasteurella multocida. Infect. Immun. 65:339-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 98.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 99.Schlor, S., M. Herbert, M. Rodenburg, J. Blass, and J. Reidl. 2000. Characterization of ferrochelatase (hemH) mutations in Haemophilus influenzae. Infect. Immun. 68:3007-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sethi, S., and T. F. Murphy. 2001. Bacterial infection in chronic obstructive pulmonary disease in 2000: a state-of-the-art review. Clin. Microbiol. Rev. 14:336-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sirakova, T., P. E. Kolattukudy, D. Murwin, J. Billy, E. Leake, D. Lim, T. DeMaria, and L. Bakaletz. 1994. Role of fimbriae expressed by nontypeable Haemophilus influenzae in pathogenesis of and protection against otitis media and relatedness of the fimbrin subunit to outer membrane protein A. Infect. Immun. 62:2002-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Skoczynska, A., M. Lewandowska, A. Klarowicz, and W. Hryniewicz. 2005. Prevalence and serotype distribution of encapsulated Haemophilus influenzae isolates from patients with lower respiratory tract infections in Poland. J. Clin. Microbiol. 43:938-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Slatko, B. E., R. Croft, L. S. Moran, and G. G. Wilson. 1988. Cloning and analysis of the HaeIII and HaeII methyltransferase genes. Gene 74:45-50. [DOI] [PubMed] [Google Scholar]
- 104.Smith, H. O., and G. M. Marley. 1980. Purification and properties of HindII and HindIII endonucleases from Haemophilus influenzae Rd. Methods Enzymol. 65:104-108. [DOI] [PubMed] [Google Scholar]
- 105.Smoot, L. M., E. C. Bell, J. H. Crosa, and L. A. Actis. 1999. Fur and iron transport proteins in the Brazilian purpuric fever clone of Haemophilus influenzae biogroup aegyptius. J. Med. Microbiol. 48:629-636. [DOI] [PubMed] [Google Scholar]
- 106.Smoot, L. M., D. D. Franke, G. McGillivary, and L. A. Actis. 2002. Genomic analysis of the F3031 Brazilian purpuric fever clone of Haemophilus influenzae biogroup aegyptius by PCR-based subtractive hybridization. Infect. Immun. 70:2694-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stevenson, A., J. Macdonald, and M. Roberts. 2003. Cloning and characterisation of type 4 fimbrial genes from Actinobacillus pleuropneumoniae. Vet. Microbiol. 92:121-134. [DOI] [PubMed] [Google Scholar]
- 108.St. Geme, J. W., III 2000. The pathogenesis of nontypable Haemophilus influenzae otitis media. Vaccine 19(Suppl. 1):S41-50. [DOI] [PubMed] [Google Scholar]
- 109.St. Geme, J. W., III, M. L. de la Morena, and S. Falkow. 1994. A Haemophilus influenzae IgA protease-like protein promotes intimate interaction with human epithelial cells. Mol. Microbiol. 14:217-233. [DOI] [PubMed] [Google Scholar]
- 110.St. Geme, J. W., III, and S. Grass. 1998. Secretion of the Haemophilus influenzae HMW1 and HMW2 adhesins involves a periplasmic intermediate and requires the HMWB and HMWC proteins. Mol. Microbiol. 27:617-630. [DOI] [PubMed] [Google Scholar]
- 111.Suzuki, K., and L. O. Bakaletz. 1994. Synergistic effect of adenovirus type 1 and nontypeable Haemophilus influenzae in a chinchilla model of experimental otitis media. Infect. Immun. 62:1710-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Thomas, W. R., M. G. Callow, R. J. Dilworth, and A. A. Audesho. 1990. Expression in Escherichia coli of a high-molecular-weight protective surface antigen found in nontypeable and type b Haemophilus influenzae. Infect. Immun. 58:1909-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Touati, D., M. Jacques, B. Tardat, L. Bouchard, and S. Despied. 1995. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: protective role of superoxide dismutase. J. Bacteriol. 177:2305-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tyndall, C., J. Meister, and T. A. Bickle. 1994. The Escherichia coli prr region encodes a functional type IC DNA restriction system closely integrated with an anticodon nuclease gene. J. Mol. Biol. 237:266-274. [DOI] [PubMed] [Google Scholar]
- 115.Vergauwen, B., F. Pauwels, and J. J. Van Beeumen. 2003. Glutathione and catalase provide overlapping defenses for protection against respiration-generated hydrogen peroxide in Haemophilus influenzae. J. Bacteriol. 185:5555-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vergauwen, B., F. Pauwels, M. Vaneechoutte, and J. J. Van Beeumen. 2003. Exogenous glutathione completes the defense against oxidative stress in Haemophilus influenzae. J. Bacteriol. 185:1572-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Weiser, J. N., M. Shchepetov, and S. T. Chong. 1997. Decoration of lipopolysaccharide with phosphorylcholine: a phase-variable characteristic of Haemophilus influenzae. Infect. Immun. 65:943-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.White, D. C., and S. Granick. 1963. Hemin biosynthesis in Haemophilus. J. Bacteriol. 85:842-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yen, M. R., Y. H. Tseng, E. H. Nguyen, L. F. Wu, and M. H. Saier, Jr. 2002. Sequence and phylogenetic analyses of the twin-arginine targeting (Tat) protein export system. Arch. Microbiol. 177:441-450. [DOI] [PubMed] [Google Scholar]
- 120.Zhao, G., P. Ceci, A. Ilari, L. Giangiacomo, T. M. Laue, E. Chiancone, and N. D. Chasteen. 2002. Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli. J. Biol. Chem. 277:27689-27696. [DOI] [PubMed] [Google Scholar]