Abstract
Salmonella enterica can obtain pyridine from exogenous nicotinamide mononucleotide (NMN) by three routes. In route 1, nicotinamide is removed from NMN in the periplasm and enters the cell as the free base. In route 2, described here, phosphate is removed from NMN in the periplasm by acid phosphatase (AphA), and the produced nicotinamide ribonucleoside (NmR) enters the cell via the PnuC transporter. Internal NmR is then converted back to NMN by the NmR kinase activity of NadR. Route 3 is seen only in pnuC* transporter mutants, which import NMN intact and can therefore grow on lower levels of NMN. Internal NMN produced by either route 2 or route 3 is deamidated to nicotinic acid mononucleotide and converted to NAD by the biosynthetic enzymes NadD and NadE.
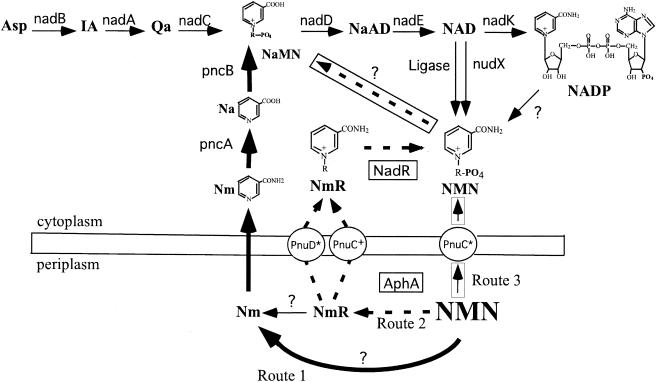
The pyridine cofactors NAD and NADP are electron carriers that are essential for both catabolic and biosynthetic redox reactions. In addition, NAD is used by bacterial DNA ligase to activate single-strand ends prior to joining (25, 49) and serves as a precursor of cofactor B12 (23). NAD can also serve as an acceptor of an acetyl group in protein deacetylation (38). Figure 1 shows pathways for synthesis, recycling, and assimilation of pyridines in Salmonella enterica and incorporates the conclusions drawn here.
FIG. 1.
NAD biosynthetic and recycling pathway of S. enterica serovar Typhimurium incorporating conclusions drawn from this study. The oxygen-stimulated pyridine nucleotide cycle is the cycle of reactions that take NADP through NMN to NaMN and back to NAD. The three routes for assimilation of exogenous NMN are indicated as follows: route one, large solid arrows; route 2, dashed arrows; and route 3, arrows in boxes. Abbreviations: Asp, aspartic acid; IA, iminoaspartic acid; Qa, quinolinic acid; Na, nicotinic acid; NaAD, nicotinic acid adenine dinucleotide. The known genes are indicated for the appropriate reactions. Question marks indicate activities that have been assayed or are likely to exist but for which no gene or protein has been identified. Proteins in circles are transport proteins, and an asterisk indicates that a protein has been mutationally altered to transport either NmR or NMN.
A simple nadA, nadB, or nadC auxotroph can obtain pyridines from exogenous nicotinamide mononucleotide (NMN) by several routes. In route 1, the nicotinamide (Nm) moiety is removed from NMN by an unknown periplasmic glycohydrolase and is imported for use as pyridine source; this route allows growth on 10 μM NMN (2). Additional routes of pyridine assimilation become apparent when route 1 is blocked by a mutation (e.g., pncA, encoding Nm deamidase). Use of the alternative pathway (route 2) requires 100 μM NMN (10, 48). All strains studied here carry an nadA or nadB mutation (to block de novo pyridine synthesis) and a pncA mutation (to block assimilation via route 1). Here we describe use of NMN by routes other than route 1.
In cells lacking route 1, NMN assimilation was shown to depend on the PnuC transport protein and on a function of the NadR regulatory protein (37, 41, 48). A role for PnuC in transport is consistent with the multiple membrane-spanning domains of this molecule (34) and the presence of a functional signal sequence (48). It was initially thought that PnuC transports intact NMN, based on very convincing double-labeling experiments that demonstrated cotransport of the nucleotide phosphate and pyridine ring (21). The role of NadR in transport was initially attributed to a posited regulatory interaction between internal NadR and the PnuC transporter (11, 47). This contrasted with the finding that in Haemophilus either of two periplasmic phosphatases converts NMN to NmR prior to transport (17). The situation in Haemophilus did not initially seem relevant to Salmonella since the PnuC protein and the relevant phosphatases have sequences that are substantially different from those of their Salmonella counterparts. Here we provide evidence that Salmonella and Haemophilus actually use NMN in similar ways and both convert it to NmR prior to uptake.
The Salmonella NadR protein is now known to have two enzymatic activities in addition to serving as a transcriptional repressor (12). The NadR(R) function represses transcription of the nadB and pncB genes and the nadA-pnuC operon when NAD levels are high (7, 37). When NAD levels are low, the repressor activity is lost and NadR expresses two enzyme activities, NmR kinase activity (20) and NMN adenylyltransferase activity (30), both of which are feedback inhibited by NAD (12). The NmR kinase, NadR(T), contributes to transport of pyridine by trapping NmR inside the cells as the charged pyridine compound NMN (Fig. 1). The NMN adenylyltransferase activity is contributed by a domain that also mediates feedback regulation of all three activities by NAD (12).
The key observation in reinterpreting NMN utilization was that there is a class of mutants in which route 2 is blocked and NMN deamidase is apparently eliminated (6). Here we present evidence that these mutants actually lack the periplasmic acid phosphatase AphA (18, 19, 33, 40, 42), which removes phosphate from NMN in the periplasm and produces NmR for transport by PnuC. Evidence reported here supports reinterpretation of four previous conclusions regarding NMN assimilation in Salmonella.
MATERIALS AND METHODS
Strains.
The strains were derived from S. enterica serovar Typhimurium LT2 (Table 1). The transposable elements Tn10dTc and T-POP are transposition-defective derivatives of Tn10 (31, 44). The MudJ element is a transposition-defective derivative of phage Mu (4). Allele designations with the suffix (sw) indicate mutations (swap) in which the coding sequence was replaced with a drug resistance cassette by linear transformation (29). Transduction crosses were mediated by the generalized transducing phage P22(HT105, int) (5, 36).
TABLE 1.
Strains
| Strain | Genotype | Source or reference |
|---|---|---|
| TR5987 | nadB51 pncA15 trpA49 | Lab collection |
| TR6601 | nadB51 pncA15 trpA49 pnuC290 | 21 |
| TR7253 | nadB51 pncA15 trpA49 pnuC269 | This study |
| TR7254 | nadB51 pncA15 trpA49 pnuC270 | This study |
| TR7255 | nadB51 pncA15 trpA49 pnuC271 | This study |
| TR7256 | nadB51 pncA15 trpA49 pnuC272 | This study |
| TR7257 | nadB51 pncA15 trpA49 pnuC273 | This study |
| TR7258 | nadB51 pncA15 trpA49 pnuC274 | This study |
| TR7259 | nadB51 pncA15 trpA49 pnuC275 | This study |
| TR7260 | nadB51 pncA15 trpA49 pnuC276 | This study |
| TR7261 | nadB51 pncA15 trpA49 pnuC277 | This study |
| TR7262 | nadB51 pncA15 trpA49 aphA2* | This study |
| TR7263 | nadB51 pncA15 trpA49 aphA3* | This study |
| TR7264 | nadB51 pncA15 trpA49 aphA4* | This study |
| TR7466 | nadD157 zbe-1028::Tn10 | Lab collection |
| TR7467 | nadD158 zbe-1028::Tn10 | Lab collection |
| TR7468 | nadD159 zbe-1028::Tn10 | Lab collection |
| TR7469 | nadD187 zbe-1028::Tn10 | Lab collection |
| TR7470 | nadD188 zbe-1028::Tn10 | Lab collection |
| TT10740 | nadE381(Ts) nadB499::MudJ leu-401(del:leuA-ppsB) ara-9 gal-205 | Lab collection |
| TT13007 | nadB499::MudJ pncA278::Tn10dCm | Lab collection |
| TT13160 | pnuC103::MudJ | Lab collection |
| TT13498 | nadB499::MudJ pncA180::Tn10 | Lab collection |
| TT13499 | nadB499::MudJ pncA180::Tn10 nadE381(Ts) | Lab collection |
| TT14890 | pncA278::Tn10dCm nadA219::MudJ[Lac+ del1052(KnspnuC aroG zbh-3652::Tn10d-tet)] | Lab collection |
| TT14946 | nadB499::MudJ pncA278::Tn10dCm nadR312 | Lab collection |
| TT15483 | nadB499::MudJ nadR511spncA278::Tn10dCm pnuC130* del1085(serB-pnuA) | Lab collection |
| TT15540 | nadR511spncA278::Tn10dCm nadA532(C) zbh-3652::Tn10dTc nadB499::MudJ pnuC133* del1085(serB-pnuA) | Lab collection |
| TT15541 | nadR511spncA278::Tn10dCm nadA532(C) zbh-3652::Tn10dTc nadB499::MudJ pnuC134* del1085(serB-pnuA) | Lab collection |
| TT15564 | pncA278::Tn10dCm pnuD135* nadA219::MudJ[Lac+ del1052(KnspnuC aroG zbh-3652::Tn10dTc)] | Lab collection |
| TT15608 | nadR511spncA278::Tn10dCm nadA532(C) zbh-3652::Tn10dTc nadB499::MudJ pnuC152* del1085(serB-pnuA) | Lab collection |
| TT15609 | nadR511spncA278::Tn10dCm nadA532(C) zbh-3652::Tn10dTc nadB499::MudJ pnuC153 del1085(serB-pnuA) | Lab collection |
| TT15620 | pncA278::Tn10dCm pnuD136* serB1463::Tn10 nadA219::MudJ[Lac+ del1052(KnspnuC aroG zbh-3652::Tn10dTc)] | Lab collection |
| TT15621 | pncA278::Tn10dCm pnuD136* serB1463::Tn10 nadR312 nadA219::MudJ[Lac+ del1052(KnspnuC aroG zbh-3652::Tn10dTc)] | Lab collection |
| TT15622 | pncA278::Tn10dCm pnuD136* serB1463::Tn10 nadR331 pnuD136* nadA219::MudJ[Lac+ del1052(KnspnuC aroG zbh-3652::Tn10dTc)] | Lab collection |
| TT15623 | pncA278::Tn10dCm pnuD136* serB1463::Tn10 nadR322 pnuD136* nadA219::MudJ[Lac+ del1052(KnspnuC aroG zbh-3652::Tn10dTc)] | Lab collection |
| TT15870 | nadB499::MudJ pncA278::Tn10dCm nadR260 | Lab collection |
| TT15871 | nadB499::MudJ pncA278::Tn10dCm nadR275 | Lab collection |
| TT15872 | nadB499::MudJ pncA278::Tn10dCm nadR312 | Lab collection |
| TT15878 | nadB499::MudJ pncA278::Tn10dCm pnuC127* nadR511 | Lab collection |
| TT18391 | nadB103 pncA278::Tn10dCm lig2::MudJ/pBR313/598/8/1b (T4 lig+ Ampr) | 6 |
| TT20738 | nadB51 pncA278::Tn10dCm qor::Tn10dTc aphA1 | 6 |
| TT20191 | pncA278::Tn10dCm DEL1052(nadA-pnuC) del1890(pnuD136*-glyA) pnuD262::MudJ | Lab collection |
| TT22416 | nadA219::MudJ pncA278::Tn10dCm qor::Tn10dTc aphA1 | This study |
| TT22458 | nadB499::MudJ pncA278::Tn10dCm qor::Tn10dTc aphA1/pBAD HisB AphA | This study |
| TT22638 | nadB103 pncA278::Tn10dCm lig2::MudJ aphA5/pBR313/598/8/1b(T4lig+ Ampr) | This study |
| TT22639 | nadB103 pncA278::Tn10dCm lig2::MudJ aphA6/pBR313/598/8/1b(T4 lig+ Ampr) | This study |
| TT22640 | nadB103 pncA278::Tn10dCm lig2::MudJ aphA7 | This study |
| /pBR313/598/8/1b(T4 lig+ Ampr) | ||
| TT22641 | nadB103 pncA278::Tn10dCm lig2::MudJ aphA8/pBR313/598/8/1b(T4 lig+ Ampr) | This study |
| TT22642 | nadB103 pncA278::Tn10dCm lig2::MudJ aphA9/pBR313/598/8/1b(T4 lig+ Ampr) | This study |
| TT22643 | nadB103 pncA278::Tn10dCm lig2::MudJ aphA10/pBR313/598/8/1b(T4 lig+ Ampr) | This study |
| TT22644 | nadB103 pncA278::Tn10dCm lig2::MudJ aphA11/pBR313/598/8/1b(T4 lig+ Ampr) | This study |
| TT22645 | nadB103 pncA278::Tn10dCm lig2::MudJ aphA12/pBR313/598/8/1b(T4 lig+ Ampr) | This study |
| TT22852 | nadB499::MudJ pncA180::Tn10 aphA13::Cm(sw) | This study |
| TT22855 | nadB51 pncA15 pnuC278 zbe-1028::Tn10 | This study |
| TT22856 | nadB51 pncA15 pnuC278 zbe-1028::Tn10 nadD188 | Lab collection |
| TT22857 | nadB51 pncA15 pnuC278 zbe-1028::Tn10 nadD157 | Lab collection |
| TT22858 | nadB51 pncA15 pnuC278 zbe-1028::Tn10 nadD158 | Lab collection |
| TT22859 | nadB51 pncA15 pnuC278 zbe-1028::Tn10 nadD159 | Lab collection |
| TT22890 | nadB499::MudJ pncA180::Tn10 aphA15::Cm(sw) | This study |
| TT22897 | nadB51 pncA15 trpA49 pnuC269 nadI609::Tn10dTc | This study |
| TT22898 | nadB51 pncA15 trpA49 pnuC270 nadI609::Tn10dTc | This study |
| TT22899 | nadB51 pncA15 trpA49 pnuC272 nadI609::Tn10dTc | This study |
| TT22901 | nadB51 pncA15 trpA49 pnuC273 nadI609::Tn10dTc | This study |
| TT22903 | nadB51 pncA15 trpA49 pnuC274 nadI609::Tn10dTc | This study |
| TT22905 | nadB51 pncA15 trpA49 pnuC275 nadI609::Tn10dTc | This study |
| TT22907 | nadB51 pncA15 trpA49 pnuC276 nadI609::Tn10dTc | This study |
| TT22909 | nadB51 pncA15 trpA49 pnuC277 nadI609::Tn10dTc | This study |
| TT22911 | nadB51 pncA15 trpA49 aphA2 nadI609::Tn10dTc | This study |
| TT22913 | nadB51 pncA15 trpA49 aphA3 nadI609::Tn10dTc | This study |
| TT22915 | nadB51 pncA15 trpA49 aphA4 nadI609::Tn10dTc | This study |
| TT22943 | pncA278::Tn10dCm pnuD135* nadI609::Tn10dTc nadA219::MudJ[Lac+ del1052(KnspnuC aroG zbh-3652::Tn10dTc)] | This study |
| TT22947 | pncA278::Tn10dCm pnuD135* aphA1 nadA219::MudJ[Lac+ DEL1052(KnspnuC aroG zbh-3652::Tn10dTc)] | This study |
| TT24215 | nadB499::MudJ pncA286::Tn10dTc pnuC103::MudCm | This study |
| TT24636 | nadB51 pncA15 aphA13::Cm(sw) | This study |
| TT24637 | nadB51 pncA15 phoN52::Tn10dTc | This study |
| TT24638 | nadB51 pncA15 aphA13::Cm(sw) phoN52::Tn10dTc | This study |
| TT24639 | nadB51 pncA15 trpA49 pnuC269 aphA13::Cm(sw) | This study |
| TT24640 | nadB51 pncA15 aphA1 deoR1::Cm(sw) | This study |
| TT24641 | E. coli TOP10 (Invitrogen)/pTrcHis2-TOPO aphA | This study |
| TT24642 | nadB51 pncA15 trpA49 pnuC103::MudJ aphA3* | This study |
Media.
The rich medium was either LB medium or nutrient broth (Difco) supplemented with 5g/liter NaCl. Minimal E medium was supplemented with 0.2% glucose (43). All media were solidified with 1.5% agar (BBL). The final concentrations of antibiotics (Sigma) in rich media were as follows: tetracycline, 20 μg/ml; kanamycin, 40 μg/ml; ampicillin, 100 μg/ml; and chloramphenicol, 20 μg/ml. In minimal media, the antibiotic concentrations were 10 μg/ml tetracycline, 100 μg/ml kanamycin, 100 μg/ml ampicillin, and 5 μg/ml chloramphenicol. The amino acid concentrations used have been described previously (9). Calf intestinal alkaline phosphatase and shrimp alkaline phosphatase were purchased from Sigma. Nicotinamide ribonucleoside (NmR) was prepared from NMN using calf intestinal alkaline phosphatase as described previously (17).
PCR and DNA sequencing.
Insertions of Tn10dTc and T-POP were localized by sequencing a single-primer PCR product that included the junction of the element and the adjacent chromosomal sequence, as described previously (14). Primer TP93 (ACCTTTGGTCACCAACGCTTTTCC) primed outward-directed synthesis from the ends of the Tn10dTc and T-POP elements; the same primer initiated synthesis at low stringency in the opposite direction from random sites. PCR products containing the junction between the transposon and the chromosome were sequenced with the nested Tn10 primer TP91 (ATCATTAGGGGATTCATCAG).
To characterize pnuC mutations, the locus was amplified by PCR with primers TP681 (ATACAGGTTGATGCGGCGCTAC), which binds 108 bases upstream of pnuC, and TP682 (GCGTCAACGAATTGCTGGAAGG), which binds 60 bases downstream of pnuC. The aphA locus was sequenced following amplification using primers TP756 (GATAACAGTGCCCTCCGGCCTGAC) and TP757 (GTCTCCTGAATAGTGTGCAGCAAG).
To clone aphA in plasmid pBAD/HisB (Invitrogen), primers TP742 (GAAGATCTCGATAACAGTGCCCTCCGCTGACAAT) and TP743 (CCCAAGCTTACGTCTCCTGAATAGTGTGCAGCAAG) were used. To clone AphA into the histidine-tagging vector TrcHis2-TOPO (Invitrogen), primers TP1592 (TAAACCATGGCCAAGGAGGAATAATAAATGAAAAAAATAACCCTGGCG) and TP1593 (TTCGAATTCGTACTCCGAGTTGACAATGA) were used. All products were sequenced at the University of Utah Health Sciences DNA Sequence Facility.
Mapping transposon insertions by PFGE.
The DNA of insertion mutants was cut with the restriction enzyme XbaI as described previously (3), and fragments were separated by pulsed-field gel electrophoresis (PFGE) with a custom-built apparatus. The Tn10 and T-POP elements included an XbaI site, which is rare in the chromosome of S. enterica. The approximate chromosomal positions of insertions were inferred by comparing the set XbaI restriction fragments (separated by PFGE) observed for wild-type and insertion mutants (22).
Cloning the aphA gene.
The aphA coding sequence was amplified by PCR from strain TR10000 using primers that included BglII and HindIII restriction sites and, following cleavage with these enzymes, was cloned into a pBAD/HisB plasmid (Invitrogen) for use in complementation tests. A C-terminal histidine-tagged AphA protein was produced by cloning the aphA coding sequence into pTrcHis2-TOPO (Invitrogen) at the NcoI and BamHI restriction sites. The constructs were verified by sequencing using primers provided by Invitrogen.
Assay of AphA activity.
AphA protein tagged with six histidine residues was purified on a ProBond column used according to manufacturer's instructions (Invitrogen). The enzyme was approximately 10% pure as judged by sodium dodecyl sulfate gel electrophoresis. Overexpression was difficult because high levels of the enzyme were lethal to cells.
The NMN and NADP phosphatase activities were assessed by the method of Ames (1). Purified protein (or 25 μl suspended cells for the whole-cell assay) was mixed with 90 μl of reaction mixture (0.1 M MgCl2, 0.5 M sodium acetate, 50 mM NMN or NADP) in a 200-μl (total volume) reaction mixture, and the mixture was incubated for 20 min at 37°C. The reaction was stopped by adding 0.7 ml of a solution consisting of 1 part 10% ascorbic acid and 6 parts 0.42% ammonium molybdate in 1 N H2SO4 and incubating the mixture at 45°C for 20 min. The cells were then pelleted, and the absorbance at 820 nm of the supernatant solution was determined and compared to a standard curve made with known PO4 concentrations (1). The same procedure was used to assay AphA activity in whole cells. Cells were grown in 100-ml LB cultures at 37°C to an optical density at 600 nm of 0.6 and resuspended in 2 ml of saline.
The NMN phosphatase activity was confirmed using [carbonyl-14C]NMN that was prepared by pyrophosphorolysis of [carbonyl-14C]NAD (Amersham Pharmacia) with nucleotide pyrophosphatase (Sigma) according to the method of Zhu et al. (47). These reactions were performed at 37°C in 0.04-ml reaction mixtures containing 4.0 mM [carbonyl-14C]NMN, 5 mM MgCl2, and 100 mM sodium acetate (pH 5.6). Each reaction was stopped by heating the mixture (98°C for 90 s), and 10 μl of the mixture was spotted onto cellulose-F plates (Merck, Darmstadt, Germany) for identification and quantification of the products. Chromatography was performed by the method of Kasarov and Moat (16) using a 1 M ammonium acetate (pH 5)-ethanol (30:70) solvent system. Reactants and products were identified by comparing Rf values with the Rf values of unlabeled standards visualized using a UV lamp and quantified using a PhosphorImager SI system (Molecular Dynamics).
Isolation of point mutants unable to assimilate NMN.
Mutants were isolated from the nadB pncA strain TT18391 by mutagenizing cells with diethyl sulfate (Eastman), plating single cells on minimal medium plus nicotinic acid, and replica plating the resulting colony arrays onto minimal medium containing either NMN or Nm. Isolated mutants could use nicotinic acid but not NMN as a pyridine source.
Selection and mapping of mutants that exhibited improved growth on a low concentration of NMN.
Cells from 20 independent 1-ml overnight cultures of strain TR5987 (trpA nadB pncA) were washed and plated on minimal medium with a low concentration of NMN (10−5 M). One mutant was isolated from each culture as a fast-growing colony that appeared above a lawn of weak confluent growth.
Mutations in the pnuC region were identified by their cotransducibility with a Tn10 insertion (zbb-3652::Tn10dTc) near pnuC. Mutations not linked to pnuC were mapped by isolating a cotransducible Tn10 insertion and sequencing PCR fragments that included the junction between this element and the chromosome.
Construction of deletions.
Deletions of aphA and deoR were constructed by replacing the coding sequences of these genes with a chloramphenicol resistance cassette (8, 29, 46). The chloramphenicol resistance (Cmr) gene of plasmid pACYC184 was amplified using primer pairs with homology to the targeted gene (aphA or deoR) at the 5′ end and homology to the Cmr gene at the 3′ end. The resulting PCR products were used to transform strains TR6579 and TT22889, which carried a plasmid (pPT223) encoding the lam, bet, and exo genes of phage lambda expressed from a lac promoter (28).
Cross-feeding tests.
Various NAD pathway mutants were stabbed into a top agar layer containing a lawn of TT20738 (nadB pncA aphA) cells, which required pyridine for growth. The diameter of the lawn of growth surrounding the inoculum was measured after 16 and 24 h of incubation. To demonstrate the feeding of cells by exogenous NmR, commercial calf intestinal phosphatase or shrimp alkaline phosphatase (10 U) was spotted onto NMN-containing solid medium seeded with cells embedded in a top agar layer. Minimal agar was supplemented with 200 μM NMN, 0.2% glucose, and 1% tryptophan.
RESULTS
Role of AphA phosphatase in NMN assimilation.
A mutant (TT20738) blocked in NMN assimilation by route 2 was reported to lack NMN deamidase (6). However, an improved assay showed that it had normal NMN deamidase activity. A Tn10dTc insertion that was 50% cotransducible with the mutation causing the growth defect was mapped by PFGE and by sequencing the junction between Tn10 and the chromosome. The insertion is within the qor gene at min 92. Sequencing of the region between qor and uvrA revealed a base substitution in the gene for acid phosphatase (aphA1; bp 637; G → A).
A plasmid carrying the wild-type aphA gene restored the ability to use NMN (Table 2). Deletion of the aphA gene (constructed by linear transformation) also prevented use of NMN as a source of pyridine (strains TT22852 and TT22890) (Table 2). Evidence described below suggests that periplasmic AphA converts exogenous NMN to NmR, which is then imported and used as a pyridine source. It seems likely that the previous conclusion that there was a deamidase defect (6) was reached because the assay method included thin-layer chromatography methods that did not resolve NmR (produced by AphA) from nicotinic acid mononucleotide (NaMN) (produced by deamidation).
TABLE 2.
Growth phenotypes with standard and low concentrations of NMN
| Strain(s) | Relevant genotypea | Growth with:
|
|
|---|---|---|---|
| 100 μM NMN | 10 μM NMN | ||
| TT13007 | nadB pncA | + | − |
| TT24215 | nadB pncA pnuC | − | − |
| TT14946 | nadB pncA nadR(T−) | − | − |
| TT20738, TT22890 | nadB pncA aphA | − | − |
| TT22458 | nadB pncA aphA/pBAD(aphA) | + | − |
| TR7262-TR7264 | nadB pncA aphA* | + | + |
| TT22911 | nadB pncA aphA* nadR(T−) | − | − |
| TT24642 | nadB pncA aphA* pnuC | − | − |
| TR7253-TR7261 | nadB pncA pnuC* | + | + |
| TT24639 | nadB pncA pnuC* aphA | + | + |
| TT22897 | nadB pncA pnuC* nadR(T−) | + | + |
| TT15564 | nadA pncA pnuC pnuD* | + | − |
| TT22947 | nadA pncA pnuC pnuD* aphA | − | − |
| TT22943 | nadA pncA pnuC pnuD* nadR(T−) | − | − |
Full genotypes are shown in Table 1. All pnuC mutations are all null types except pnuC* which allows uptake of intact NMN as well as NmR. Phenotypes indicated in parentheses are as follows: nadR(T−), deficient in use of NMN; aphA*, increases the level of AphA; pnuD*, allows an unrelated transporter to import NmR.
Isolation of additional mutants unable to use NMN.
Ninety-five new mutants unable to assimilate NMN (see Materials and Methods) were isolated. Thirty-five of these mutants displayed a simple failure to use NMN; 14 mutations affected aphA, and 12 mutations affected nadR. Mapping attempts failed for nine additional mutations, suggesting that there was a more complex genotype. All of the eight aphA mutations sequenced were C/G-to-T/A transitions, as expected for mutations induced by diethyl sulfate (see Materials and Methods). The inferred amino acid changes were as follows: aphA5, S77F; aphA8, R139C; aphA7, D194N; aphA10, D196N; aphA9 and aphA11, A213T; aphA12, Q221UAG; and aphA6, A224T. It is not clear why this analysis yielded no pnuC mutants, which were common in previous analyses and clearly prevent growth on NMN (6, 48).
Mutants with improved ability to assimilate NMN (aphA* and pnuC*).
A parent nadB pncA strain (TR5987) was plated on minimal glucose medium containing a level of NMN (10 μM) lower than that required for growth (100 μM). Six of 18 independent spontaneous mutants that were able to grow on 10 μM NMN carried revertants of the parent nadB mutation (and required no exogenous pyridine). Three carried mutations that increased expression of the periplasmic AphA phosphatase, and nine mutations caused a qualitative change in the PnuC protein (below).
The three mutations that upregulated AphA were the result of a G-to-A transition at a single site 68 bases upstream of the aphA coding sequence and were designated aphA*. The affected sequence was perfectly conserved in Escherichia coli and S. enterica and shares features with known DeoR repressor binding sites in the E. coli genome. Because the AphA enzyme removes phosphate from nucleotides and is known to be important for nucleotide assimilation (24, 45), it seemed reasonable that aphA might be repressed by DeoR, a regulator of genes involved in use of deoxynucleotides as carbon sources. However, a constructed deoR deletion mutation caused no discernible improvement in the ability to grow on NMN, suggesting that the AphA* mutations relieve aphA repression by some other regulatory protein.
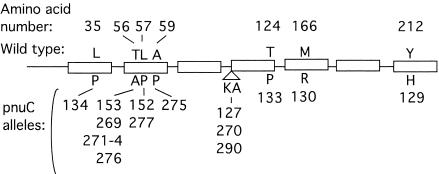
The nine mutations (pnuC*269 to pnuC*277) that affected the pnuC gene all caused amino acid substitutions at highly conserved positions in one of the seven inferred membrane-spanning regions of the protein (Fig. 2). Evidence that these regions actually span the membrane has been reported recently (34). Mutants with this phenotype were reported previously but not sequenced (10, 21, 37). One of the previously isolated mutations (used to demonstrate transport of intact NMN) also affected pnuC (pnuC*290 in Fig. 2). The ability of these mutants to grow on lower levels of NMN (a PnuC* phenotype) was attributed to changes that allowed NMN transport by PnuC (which normally transports only NmR).
FIG. 2.
Location and description of pnuC* mutations. All pnuC* mutations were isolated based on the ability of mutants to grow on 10 μM NMN (see Materials and Methods). The amino substitutions and positions in predicted transmembrane segments (boxes) are indicated. The base changes for the mutations are as follows. In allele pnuC129 bp 631 is changed from T to C. In allele pnuC130 bp 497 is changed from T to G. In allele pnuC133 bp 370 is changed from A to C. In allele pnuC275 bp 175 is changed from G to C. In alleles pnuC134 and pnuC143 bp 104 is changed from T to C. In alleles pnuC152 and pnuC277 bp 170 is changed from T to C. In alleles pnuC153, pnuC269, pnuC271 to pnuC274, and pnuC276 bp 166 is changed from A to C. Alleles pnuC127, pnuC270, and pnuC290 have a duplication of the six-base sequence AAAAGC encoding amino acids 108 (K) and 109 (A) which adds K and A residues just before the fourth membrane-spanning region. Mutation pnuC*-290 was originally designated pnuB (21).
Model for NMN assimilation.
The results described above suggest a model for NMN assimilation by route 2. According to this model (Fig. 1), periplasmic AphA removes phosphate from NMN to form NmR. Transport of NmR from the periplasm into the cell via wild-type PnuC is driven by internal phosphorylation of NmR by the NmR kinase activity of the trifunctional NadR protein. This kinase reaction adds a charge to the internal pyridine, thereby trapping it within the cell, driving uptake, and forming internal NMN (which can be converted to NAD). According to this model, AphA* mutants (see above) improve NMN assimilation by producing more periplasmic AphA (and therefore more NmR), while PnuC* mutants are able to transport intact NMN (in addition to NmR) and thereby circumvent the need for both the AphA periplasmic phosphatase and the internal NmR kinase activity of NadR.
This model predicts that uptake of the labeled pyridine moiety from NMN should depend on AphA (to remove phosphate), on PnuC (to transport NmR), and on the internal NmR kinase of NadR (to produce internal NMN). A dependence on PnuC and NadR kinase for transport of pyridine from NMN has been shown previously (47). Furthermore, the model predicts that uptake of the labeled pyridine ring from NMN via PnuC should show preference for NmR over NMN (which requires dephosphorylation prior to transport); this was recently demonstrated for the similar PnuC transporter of E. coli (34). Other aspects of the model were tested as described below.
Assays of NMN phosphatase in whole cells.
Total periplasmic phosphatase activity in suspended whole cells was estimated by using NMN as the substrate and detecting released phosphate (see Materials and Methods). Table 3 shows the measured phosphate release data, and these data suggest that an aphA mutation reduces phosphate release only about twofold. However, this assay is likely to seriously underestimate NMN-specific phosphatase activity because most exogenous NMN is cleaved by a periplasmic glycohydrolase (route 1) to Nm plus ribose 5-phosphate. Phosphatases that do not act on NMN may release phosphate from NMN-derived ribose 5-phosphate and thus contribute a high background of AphA-independent phosphate release. The acid phosphatase PhoN is a likely candidate since it makes a significant contribution to the background release of phosphate. PhoN is unlikely to act directly on NMN because phoN mutations do not impair growth on NMN, while aphA mutations reduce this growth severely.
TABLE 3.
NMN phosphatase activity in whole cells
| Strain | Genotypea | Total phosphate released (SD)b | Release corrected for PhoN activityc |
|---|---|---|---|
| TR5958 | nadB pncA | 2.0 (0.12) | 1.2 |
| TT24636 | nadB pncA aphA | 0.9 (0.05) | 0.1 |
| TT24637 | nadB pncA phoN | 1.2 (0.08) | 1.2 |
| TT24638 | nadB pncA aphA phoN | 0.08 (0.01) | 0.08 |
| TT22855 | nadB pncA pnuC* | 2.0 (0.09) | 1.2 |
| TR7262 | nadB pncA aphA* | 4.0 (0.11) | 3.2 |
Complete genotypes are shown in Table 1.
Assays were performed with whole cells in duplicate as described in Materials and Methods. The activity is expressed in nmol PO4/min per 25 μl cell suspension (optical density at 600 nm, 0.6).
Values were corrected for the contribution of PhoN to total phosphate release, which was inferred to come from derived ribose 5-phosphate rather than NMN itself (see text). The PhoN contribution (2.0 − 1.2 = 0.8 nmol PO4/min per 25 μl) was subtracted from the total phosphate release seen in all phoN+ cells.
To better estimate the AphA contribution to total phosphate release, values were corrected for the contribution of PhoN. The PhoN contribution was taken to be the deficit in total phosphate release caused by a phoN mutation (2.0 − 1.2 = 0.8 nmol PO4/min/25 μl cell suspension). When the PhoN contribution was subtracted, aphA mutations resulted in a 10-fold reduction in NMN phosphatase activity and aphA* mutations resulted in a threefold increase. The validity of this correction could be directly tested using an NMN glycohydrolase (NMN → Nm) mutation to prevent formation of ribose 5-phosphate from NMN; however, such mutants are not available, and the gene(s) encoding this activity is not known.
Purified AphA has NMN and NADP phosphatase activity.
Histidine-tagged AphA protein was overproduced and purified 48-fold (200-fold compared with uninduced levels) as described in Materials and Methods. The partially purified enzyme removed 5′-phosphate from NMN with a specific activity of 4 μmol/min/mg. This activity was demonstrated in two ways (see Materials and Methods): first, by measuring released phosphate and second, by measuring released NmR by thin-layer chromatography. The 48-fold purification of this NMN phosphatase activity from the starting extract was accompanied by a 42-fold increase in NADP 2′-phosphatase activity (to 10 μmol/min/mg). The latter reaction was shown to generate free phosphate and NAD; the detection methods used are described in Materials and Methods. This suggests that AphA catalyzes both removal of the 5′-phosphate from NMN and removal of the 2′-phosphate from NADP. The Haemophilus AphA homolog [e(P4)] is also known to dephosphorylate both NMN and NADP, and e(P4)-deficient mutants are unable to grow on NADP (32).
Unlike the situation in Haemophilus, Salmonella does not require the NADP phosphatase activity of AphA for use of NADP. While AphA is essential for use of NADP or NAD as a pyridine source in Salmonella, this could reflect only the need to remove phosphate from derived NMN, as described above. Consistent with this, pnuC* strains (which used NMN intact) used both NADP and NAD even in the absence of AphA. The nadB pncA pnuC* strain TT22897 grew on NAD or NADP as a pyridine source, and this ability was not impaired by an added aphA null mutation (tested in strain TT24639 on E medium containing glucose with both 0.1 mM and 0.2 mM NADP). The difference between Haemophilus and Salmonella may be explained by the Salmonella PnuE pyrophosphatase (13, 27), which cleaves periplasmic NAD to NMN. If PnuE pyrophosphatase cleaves NADP as well as NAD, then AphA would be required only for converting the NMN produced to NmR, explaining the dispensability of AphA for NADP use by Pnu* mutants.
Evidence that AphA acts in the periplasm.
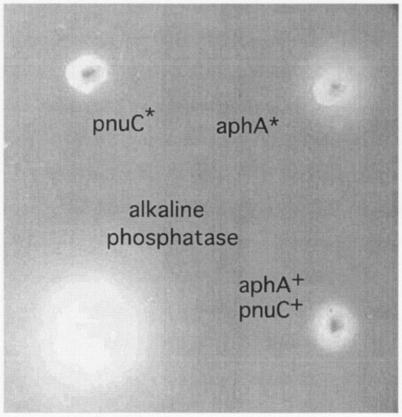
The inability of aphA mutants to use NMN is corrected by exogenous alkaline phosphatase. Furthermore, on NMN medium, colonies of an nadB pncA aphA* mutant excrete a compound (presumably NmR) that allows growth of an nadB pncA aphA mutant (which cannot use NMN directly). Figure 3 shows the surface of a plate containing minimal medium with NMN (100 μM) seeded with a lawn of nadB pncA aphA (TT20738) cells. Growth of the lawn was stimulated by a spot of an alkaline phosphatase solution or an aphA* cell culture. In contrast, a spot of a pnuC* culture, which grew faster than the lawn because of improved import of intact NMN, did not feed the surrounding lawn, and an aphA+ strain fed the lawn only minimally. These results are consistent with the idea that aphA* mutants overproduce AphA and therefore convert more periplasmic NMN to NmR, which can diffuse away and stimulate the growth of nadB pncA aphA cells, circumventing their lack of AphA phosphatase.
FIG. 3.
Effects of external phosphatase and cross-feeding on an NAD-limited lawn. The plate contained nadB pncA aphA (TT20738) cells embedded in minimal top agar on top of minimal agar (the agar contained 200 μM NMN, 0.2%glucose, and 1% tryptophan, as described in Materials and Methods). Growth of the lawn was stimulated by added alkaline phosphatase (bottom left) and by a spotted culture of an aphA* mutant (TR7264) and was stimulated slightly by a spotted culture of the wild type (TR5987) or a PnuC* mutant (TR7253).
For the growth stimulation described above (attributed to NmR), cells in the lawn had to possess both a PnuC+ transporter and the internal NadR(T) kinase function (see below). Thus, aphA* mutants were inferred to overproduce periplasmic NmR, whose use by a lawn required the PnuC and NadR(T) functions but not AphA. The compound excreted by AphA* cells or produced by alkaline phosphatase could not be either Nm or nicotinic acid. Use of Nm by the lawn was prevented by the pncA mutation, and use of either Nm or nicotinic acid did not depend on NadR. The results of these cross-feeding experiments are summarized in Table 4.
TABLE 4.
Cross-feeding of pyridine by aphA* cells
| Strain in lawn | Relevant genotype of lawna | Fed by aphA* |
|---|---|---|
| TT13007 | nadB pncA | Yes |
| TT14890 | nadA pncA pnuC | No |
| TT15620 | nadA pncA pnuC pnuD* | Yes |
| TT15621 | nadA pncA pnuC pnuD* nadR(R+ T−) | No |
| TT15622 | nadA pncA pnuC pnuD* nadR(R− T+) | Yes |
| TT15623 | nadA pncA pnuC pnuD* nadR(R− T−) | No |
| TT15870 | nadB pncA nadR(R− T+) | Yes |
| TT15871 | nadB pncA nadR(R− T−) | No |
| TT15872 | nadB pncA nadR(R+ T−) | No |
| TT20191 | nadA pncA pnuC pnuD | No |
| TT15564 | nadA pncA pnuC pnuD* | Yes |
Complete genotypes are shown in Table 1.
AphA-dependent use of NMN (route 2) depends on both NadR and PnuC functions.
If aphA* mutations improve growth on NMN by increasing periplasmic NmR production, as suggested by the model, then the growth of aphA* mutants on NMN should depend on both PnuC and the NmR kinase function of NadR. A pnuC103::MudJ insertion (from strain TT13160) eliminated the ability of aphA* mutants to grow on NMN (at both 10 and 100 μM). All nadR mutations that eliminated growth of an aphA+ strain on NMN [nadR(T−) mutations] also eliminated growth of aphA* mutants on NMN. Evidence has been presented elsewhere (12) that nadR(T−) mutants lack NmR kinase activity, suggesting that this kinase is critical for use of NMN by Route 2, as predicted by the model.
NMN assimilation requires AphA and NadR(T) even when another transporter replaces PnuC.
In the model suggested here, AphA, PnuC, and NadR kinase produce, transport, and phosphorylate NmR with no required protein-protein interactions. Thus, growth on NMN would be expected even if PnuC were replaced by some other transporter of NmR. The previous model for route 2 suggested that NadR interacts directly with PnuC to stimulate its activity when NAD levels are low (47). An unrelated transporter of NmR would be unlikely to interact directly with NadR or require regulatory stimulation by it or any other component of the pyridine metabolic pathway.
Growth on NMN can be restored to strains with pnuC deleted by point mutations that recruit an unrelated substitute transporter. One class of such mutations (pnuD*) alters a transporter whose normal substrate is unknown but which shows homology with known nucleoside transporters (15). The ability of a pnuD* pnuC deletion double mutant to grow on NMN was eliminated by an nadR kinase mutation or by an aphA mutation (Table 2). Growth of a pnuD* pnuC double mutant could be stimulated by material (presumably NmR) cross-fed from aphA* mutants (Table 4). Thus, cells that use a foreign, recruited transporter still require both NadR kinase activity and AphA phosphatase to grow on NMN, and they require only the kinase activity for growth on NmR. Their growth is independent of PnuC. A foreign transport protein would be unlikely to form or need critical regulatory protein-protein interactions between NadR and PnuC. This argues against our initial model (in which PnuC activity is stimulated by direct interaction with NadR).
In PnuC* mutants, NMN can be assimilated without AphA or NadR (NmR kinase).
The arguments described above suggest that the normal PnuC transporter acts on NmR, but not NMN. The previous report (21) that PnuC transports intact NMN was based on strains with improved use of NMN due to a pnuC mutation, now designated pnuC*290 (Fig. 2). Thus, the conclusion that NMN can be transported intact is correct, but it applies only to strains with a pnuC* mutation.
If strains with a pnuC* mutation can transport intact NMN, they should use exogenous NMN without the AphA or NadR(kinase) functions described above (i.e., route 3 rather than route 2). This hypothesis was tested by adding aphA and nadR mutations to a strain carrying a pnuC* mutation. As predicted by the model, the resulting strain retained the ability to use NMN. Thus, a pnuC* mutation opens a third pathway (route 3) for NMN use and restores growth on NMN to an aphA mutant or an nadR (kinase) mutant, both of which are defective in route 2. (Route 1 is blocked by a pncA mutation.)
Wild-type PnuC prefers NmR to NMN.
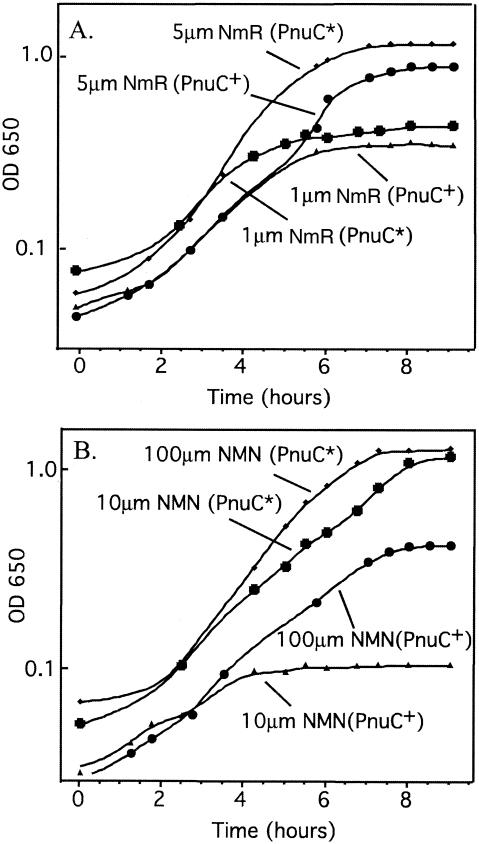
Growth on NMN by route 2 requires 100 μM NMN, while route 1 requires only 10 μM NMN. This behavior supports the model in which the normal substrate of PnuC is NmR (not NMN) and suggests that route 2 requires a high concentration of NMN because it depends on formation of NmR by a nonspecific phosphatase (AphA). Consistent with this model, 1 μM NmR (but 100 μM NMN) is sufficient to support the growth of an nadB pncA mutant (Fig. 4). Furthermore, pnuC* mutations, which allow transport of intact NMN, have little effect on the ability of cells to use NmR. The main role of PnuC may be to facilitate influx and efflux of NmR (see below).
FIG. 4.
Comparison of growth of PnuC* and PnuC+ strains on NMN and NmR. The growth of an nadB pncA pnuC+ strain (TR5987, parent) was compared to the growth of an isogenic strain with pnuC*-269 (TR7253) on 1 μM and 5 μM NmR (A) and on 10 μM and 100 μM NMN (B). OD 650, optical density at 650 nm.
Role of PnuC in pyridine efflux.
The findings described above suggest that PnuC facilitates diffusion of NmR and that the driving force for uptake is addition of the charged phosphate, which traps internal NMN. As a diffusion facilitator, PnuC might contribute to excretion when the internal concentration of NmR is high. The effect of pnuC mutations on NmR excretion was tested directly.
A lawn of nadC cells was plated on medium with 10 mM quinolinic acid, which the lawn could not use as a pyridine source (Fig. 1). Cells blocked for nadB and pncB were spotted on this lawn and found to excrete a diffusible compound that supported lawn growth (data not shown). The donor cells took up quinolinic acid and excreted some other pyridine that could be used by the lawn (which could not use Nm or nicotinic acid). Excretion was strongly reduced by a pnuC mutation in the feeding cells, suggesting that PnuC has a role in export. The excreted compound was inferred to be NmR in view of evidence presented here that NmR is the normal substrate of PnuC. The potential importance of pyridine excretion is discussed below.
DISCUSSION
Wild-type S. enterica assimilates NMN primarily by route 1, which involves periplasmic removal of Nm from the ribose ring. Route 2 dephosphorylates NMN to form periplasmic NmR, which is then imported by PnuC. The internal NmR is phosphorylated by the NadR kinase activity, which contributes to NmR transport by adding a phosphate, trapping the internal pyridine, and initiating its conversion to NAD. Intact NMN is imported (route 3) only by certain mutant strains (pnuC*) with an altered PnuC transporter. This suggests that the normal role of the PnuC transporter is uptake of NmR rather than NMN. The four points below reinterpret previous conclusions regarding NMN assimilation by Salmonella.
NMN is not normally transported intact.
Direct transport of NMN by S. enterica was demonstrated very clearly (21) using NMN isotopically labeled in both the phosphate and the pyridine ring. There is evidence that this conclusion applies only to the strains used, which carried a mutant (pnuC*) transporter. PnuC+ cells transport only NmR.
PnuC* mutations broaden the specificity of transport rather than increasing total activity.
The mutants for which transport of intact NMN was demonstrated (called pnuB mutants) were originally thought to have elevated expression or activity of PnuC (21, 37). Evidence in this study shows that the mutations actually allow PnuC to transport intact NMN in addition to NmR (its normal substrate).
Mutants previously thought to lack NMN deamidase actually lack the periplasmic phosphatase (AphA).
The previous conclusion was based on an enzyme assay that did not distinguish between produced NaMN (inferred) and NmR (the actual product). Mapping may have been complicated by multiple mutations (as observed for some of the new mutants isolated here). However, an aphA mutation extracted genetically from the original strains was found in this study to prevent NMN use by limiting conversion of periplasmic NMN to the transportable NmR.
NadR protein does not regulate activity of an NMN transporter but participates directly in uptake of NmR by adding a phosphate and thereby trapping NMN within the cell.
The rate at which the activity of the transport system allows pyridine-labeled NMN to enter cells is controlled by internal NAD levels, and this regulation requires the NadR(T) function (47). This observation was initially attributed to activation of PnuC by the NadR protein when NAD levels are low. Evidence obtained in this study suggests instead that NadR contributes enzymatically to NmR transport by converting cytoplasmic NmR to NMN. Internal NAD reduces uptake of label from NMN (47) by feedback inhibiting the NmR kinase activity of NadR (12).
The assimilation of NMN in Salmonella (by route 2) is very similar to the assimilation in Haemophilus, in which either of two different external phosphatases is required to convert NMN to transportable NmR to satisfy the natural requirement of this organism for pyridines (17, 34). Haemophilus converts transported NmR to NAD by two sequential activities provided internally by the Haemophilus NadR protein (NmR kinase and NMN adenylyl transferase) (20). In Salmonella the adenylyltransferase activity is vestigial (12, 20, 30), and NadR converts NmR only to NMN, which is then deamidated to NaMN before conversion to NAD by the NadD and NadE activities (Fig. 1).
The AphA protein used by Salmonella is a nonspecific periplasmic acid phosphatase (40) that contributes to assimilation of various purine and pyrimidine nucleotides by removing phosphate (24, 45) and thus does not appear to be dedicated to uptake of pyridines. Wild-type S. enterica appears to obtain pyridines by uptake of either free nicotinic acid, Nm, or (via PnuC) NmR (all of which are used at external concentrations below 1 μM). Assimilation of NMN by route 1 is also efficient (requiring 10 μM NMN), perhaps because it generates perplasmic Nm, one of Salmonella's preferred pyridines. Assimilation of NMN by route 2 is less efficient (requiring 100 μM NMN) because it relies on a nonspecific phosphatase to generate NmR, some of which may be lost by diffusion. It seems likely that the primary natural function of PnuC is not in NMN assimilation but rather in import and excretion of NmR.
Loss of excess pyridines may be critical under some growth conditions. The essential DNA ligase is strongly inhibited by NMN (25, 49). During highly aerobic growth, the pyridine nucleotide cycle is activated, which converts NAD(P) to NMN with the potential to inhibit ligase (26). Following UV irradiation of E. coli, pyridines are known to be lost from the cell (35, 39). We suspect that these observations are related in that excess NMN may be converted to NmR and allowed to escape from the cell via the PnuC facilitator to avoid toxic inhibition of ligase.
Acknowledgments
This work was supported in part by NIH grant GM23408.
We thank Peter Anderson, Marian Price-Carter, Eric Kofoid, Hotcherl Jeong, Chris Mace, Toto Olivera, David Sheppard, Tiffany Tsang, and Sid Velick for helpful suggestions. We are grateful to Janet Shaw, in whose lab some of the experiments were done.
REFERENCES
- 1.Ames, B. N. 1966. Assay of inorganic phosphate. Methods Enzymol. 8:115-118. [Google Scholar]
- 2.Andreoli, A. J., T. W. Okita, R. Bloom, and T. A. Grover. 1972. The pyridine nucleotide cycle: presence of a nicotinamide mononucleotide-specific glycohydrolase in Escherichia coli. Biochem. Biophys. Res. Commun. 49:264-269. [DOI] [PubMed] [Google Scholar]
- 3.Bergthorsson, U., and H. Ochman. 1995. Heterogeneity of genome sizes among natural isolates of Escherichia coli. J. Bacteriol. 177:5784-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castilho, B. A., P. Olfson, and M. J. Casadaban. 1984. Plasmid insertion mutagenesis and lac gene fusion with mini-mu bacteriophage transposons. J. Bacteriol. 158:488-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, R. K., D. Botstein, T. Watanabe, and Y. Ogata. 1972. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high-frequency-transducing lysate. Virology 50:883-898. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, W., and J. Roth. 1995. Isolation of NAD cycle mutants defective in nicotinamide mononucleotide deamidase in Salmonella typhimurium. J. Bacteriol. 177:6711-6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cookson, B. T., B. M. Olivera, and J. R. Roth. 1987. Genetic characterization and regulation of the nadB locus of Salmonella typhimurium. J. Bacteriol. 169:4285-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 10.Foster, J. W., D. M. Kinney, and A. G. Moat. 1979. Pyridine nucleotide cycle of Salmonella typhimurium: isolation and characterization of pncA, pncB, and pncC mutants and utilization of exogenous nicotinamide adenine dinucleotide. J. Bacteriol. 137:1165-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster, J. W., Y. K. Park, T. Penfound, T. Fenger, and M. P. Spector. 1990. Regulation of NAD metabolism in Salmonella typhimurium: molecular sequence analysis of the bifunctional nadR regulator and the nadA-pnuC operon. J. Bacteriol. 172:4187-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grose, J. H., U. Bergthorsson, and J. R. Roth. 2005. Regulation of NAD synthesis by the trifunctional NadR protein of Salmonella enterica. J Bacteriol 187:2774-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grose, J. H., and J. Schalheim. Unpublished results.
- 14.Hermann, S. R., J. A. Miller, S. O'Neill, T. T. Tsao, R. M. Harding, and J. L. Dale. 2000. Single-primer amplification of flanking sequences. BioTechniques 298:1176-1180. [DOI] [PubMed] [Google Scholar]
- 15.Jeong, H., and N. Zhu. Unpublished results.
- 16.Kasarov, L. B., and A. G. Moat. 1980. Convenient method for enzymic synthesis of [14C]nicotinamide riboside. Methods Enzymol. 66:120-122. [DOI] [PubMed] [Google Scholar]
- 17.Kemmer, G., T. J. Reilly, J. Schmidt-Brauns, G. W. Zlotnik, B. A. Green, M. J. Fiske, M. Herbert, A. Kraiss, S. Schlor, A. Smith, and J. Reidl. 2001. NadN and e(P4) are essential for utilization of NAD and nicotinamide mononucleotide but not nicotinamide riboside in Haemophilus influenzae. J. Bacteriol. 183:3974-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kier, L. D., R. Weppelman, and B. N. Ames. 1977. Resolution and purification of three periplasmic phosphatases of Salmonella typhimurium. J. Bacteriol. 130:399-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kier, L. D., R. M. Weppelman, and B. N. Ames. 1979. Regulation of nonspecific acid phosphatase in Salmonella: phoN and phoP genes. J. Bacteriol. 138:155-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurnasov, O. V., B. M. Polanuyer, S. Ananta, R. Sloutsky, A. Tam, S. Y. Gerdes, and A. L. Osterman. 2002. Ribosylnicotinamaide kinase domain of NadR protein: identification and implications in NAD biosynthesis. J. Bacteriol. 184:6906-6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, G., J. Foster, P. Manlapaz-Ramos, and B. M. Olivera. 1982. Nucleoside salvage pathway for NAD biosynthesis in Salmonella typhimurium. J. Bacteriol. 152:1111-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, S. L., A. Hessel, and K. E. Sanderson. 1993. The XbaI-BlnI-CeuI genomic cleavage map of Salmonella enteritidis shows an inversion relative to Salmonella typhimurium LT2. Mol. Microbiol. 10:655-664. [DOI] [PubMed] [Google Scholar]
- 23.Maggio-Hall, L. A., and J. C. Escalante-Semerena. 2003. Alpha-5,6-dimethylbenzimidazole adenine dinucleotide (alpha-DAD), a putative new intermediate of coenzyme B12 biosynthesis in Salmonella typhimurium. Microbiology 149:983-990. [DOI] [PubMed] [Google Scholar]
- 24.Makde, R. D., V. Kumar, G. D. Gupta, J. Jasti, T. P. Singh, and S. K. Mahajan. 2003. Expression, purification, crystallization and preliminary X-ray diffraction studies of recombinant class B non-specific acid phosphatase of Salmonella typhimurium. Acta Crystallogr. Sect. D 59:1849-1852. [DOI] [PubMed] [Google Scholar]
- 25.Olivera, B. M., Z. W. Hall, Y. Anraku, J. R. Chien, and I. R. Lehman. 1968. On the mechanism of the polynucleotide joining reaction. Cold Spring Harbor Symp. Quant. Biol. 33:27-34. [DOI] [PubMed] [Google Scholar]
- 26.Park, U. E., B. M. Olivera, K. T. Hughes, J. R. Roth, and D. R. Hillyard. 1989. DNA ligase and the pyridine nucleotide cycle in Salmonella typhimurium. J. Bacteriol. 171:2173-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park, U. E., J. R. Roth, and B. M. Olivera. 1988. Salmonella typhimurium mutants lacking NAD pyrophosphatase. J. Bacteriol. 170:3725-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poteete, A., and A. Fenton. 1984. Lambda rec-dependent growth and recombination of phage P22. Virology 134:161-167. [DOI] [PubMed] [Google Scholar]
- 29.Price-Carter, M., J. Tingey, T. A. Bobik, and J. R. Roth. 2001. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J. Bacteriol. 183:2463-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raffaelli, N., T. Lorenzi, P. L. Mariani, M. Emanuelli, A. Amici, S. Ruggieri, and G. Magni. 1999. The Escherichia coli NadR regulator is endowed with nicotinamide mononucleotide adenylyltransferase activity. J. Bacteriol. 181:5509-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rappleye, C. A., and J. R. Roth. 1997. A Tn10 derivative (“T-POP”) for isolation of insertions with conditional (tetracycline-dependent) phenotypes. J. Bacteriol. 179:5827-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reidl, J., S. Schlor, A. Kraiss, J. Schmidt-Brauns, G. Kemmer, and E. Soleva. 2000. NADP and NAD utilization in Haemophilus influenzae. Mol. Microbiol. 35:1573-1581. [DOI] [PubMed] [Google Scholar]
- 33.Rossolini, G. M., M. C. Thaller, R. Pezzi, and G. Satta. 1994. Identification of an Escherichia coli periplasmic acid phosphatase containing a 27 kDa-polypeptide component. FEMS Microbiol. Lett. 118:167-173. [DOI] [PubMed] [Google Scholar]
- 34.Sauer, E., M. Merdanovic, A. Mortimer, G. Bringmann, and J. Reidl. 2004. PnuC and the utilization of the nicotinamide riboside analog 3-aminopyridine in Haemophilus influenzae. Antimicrob. Agents Chemother. 48:4532-4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schenley, R. L., P. A. Swenson, and J. G. Joshi. 1979. Cessation of respiration after far-ultraviolet irradiation of Escherichia coli B/r: loss of unaltered pyridine nucleotides to the medium. Radiat. Res. 79:611-621. [PubMed] [Google Scholar]
- 36.Schmieger, H. 1971. A method for detection of phage mutants with altered transducing ability. Mol. Gen. Genet. 110:378-381. [DOI] [PubMed] [Google Scholar]
- 37.Spector, M. P., J. M. Hill, E. A. Holley, and J. W. Foster. 1985. Genetic characterization of pyridine nucleotide uptake mutants of Salmonella typhimurium. J. Gen Microbiol. 131:1313-1322. [DOI] [PubMed] [Google Scholar]
- 38.Starai, V. J., I. Celic, R. N. Cole, J. D. Boeke, and J. C. Escalante-Semerena. 2002. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298:2390-2392. [DOI] [PubMed] [Google Scholar]
- 39.Swenson, P. A., and R. L. Schenley. 1970. Role of pyridine nucleotides in the control of respiration in ultraviolet-irradiated Escherichia coli B/r cells. J. Bacteriol. 104:1230-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thaller, M. C., S. Schippa, A. Bonci, S. Cresti, and G. M. Rossolini. 1997. Identification of the gene (aphA) encoding the class B acid phosphatase/phosphotransferase of Escherichia coli MG1655 and characterization of its product. FEMS Microbiol. Lett. 146:191-198. [DOI] [PubMed] [Google Scholar]
- 41.Tirgari, S., M. P. Spector, and J. W. Foster. 1986. Genetics of NAD metabolism in Salmonella typhimurium and cloning of the nadA and pnuC loci. J. Bacteriol. 167:1086-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uerkvitz, W., and C. F. Beck. 1981. Periplasmic phosphatases in Salmonella typhimurium LT2. A biochemical, physiological, and partial genetic analysis of three nucleoside monophosphate dephosphorylating enzymes. J. Biol. Chem. 256:382-389. [PubMed] [Google Scholar]
- 43.Vogel, H. J., and D. M. Bonner. 1956. Acethylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 44.Way, J. C., M. A. Davis, D. Morisato, D. E. Roberts, and N. Kleckner. 1984. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene 32:369-379. [DOI] [PubMed] [Google Scholar]
- 45.Weppelman, R., L. D. Kier, and B. N. Ames. 1977. Properties of two phosphatases and a cyclic phosphodiesterase of Salmonella typhimurium. J. Bacteriol. 130:411-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, N., B. M. Olivera, and J. R. Roth. 1991. Activity of the nicotinamide mononucleotide transport system is regulated in Salmonella typhimurium. J. Bacteriol. 173:1311-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu, N., B. M. Olivera, and J. R. Roth. 1989. Genetic characterization of the pnuC gene, which encodes a component of the nicotinamide mononucleotide transport system in Salmonella typhimurium. J. Bacteriol. 171:4402-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zimmerman, S. B., J. W. Little, C. K. Oshinsky, and M. Gellert. 1967. Enzymatic joining of DNA strands: a novel reaction of diphosphopyridine nucleotide. Proc. Natl. Acad. Sci. USA 57:1841-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]