Abstract
In this study, the occurrence and chromosomal clustering of genes encoding C1 transfer reactions linked to tetrahydromethanopterin (H4MPT) were analyzed in a variety of proteobacteria and in representatives of the Planctomycetes via genomic analysis or via partial sequencing by cosmid walking. Although a tendency for clustering was found common for the genes of interest, significant variations in gene order and the degree of clustering were uncovered both between and within different groups of Proteobacteria and between Proteobacteria and Planctomycetes. Phylogenetic analyses suggested that the evolution of genes encoding H4MPT-linked reactions in Proteobacteria involved lateral transfers within Proteobacteria and possibly between Proteobacteria and other phyla. Gene cluster comparisons revealed a number of novel genes potentially involved in the C1 transfer reactions, and these were analyzed by mutation and expression analyses. Four genes, a homolog of pabB, and three genes conserved between methanogenic Archaea and Bacteria possessing H4MPT-linked functions, orfY, orf1, and afpA were shown to be involved in formaldehyde oxidation/detoxification, as judged by specific mutant phenotypes. In particular, pabB contributes to the biosynthesis of para-aminobenzoic acid, a precursor of both tetrahydrofolate and H4MPT, and afpA apparently encodes a novel dihydromethanopterin reductase, based on mutant complementation experiments.
One of the major breakthroughs in the understanding of methylotrophy in Bacteria during the recent decade, the recognition of the tetrahydromethanopterin (H4MPT)-linked pathway for formaldehyde oxidation as a major C1 oxidation pathway, was due to a serendipitous discovery of a cluster of genes in Methylobacterium extorquens AM1 that are homologous to the genes involved in methanogenesis in Archaea (7). It has since been demonstrated that this pathway is nearly ubiquitous in gram-negative methylotrophs (26). More recently, the pathway's presence has been expanded beyond methylotrophs (20), and even beyond Proteobacteria into the Planctomycetes (5, 10). Although phylogenetic analysis has argued against recent lateral transfer of the genes in question between methanogenic Archaea and Proteobacteria (5), the history of these genes in Bacteria remains poorly understood. Although some congruence has been observed between the phylogenetic positions of the respective species within Proteobacteria and phylogenies of genes involved in H4MPT-linked reactions (13, 15), the relationships of the latter are not always well resolved and sometimes are complicated by the presence of multiple gene copies. In the present study we attempted a more comprehensive analysis of gene islands in Bacteria involved in H4MPT-linked C1 transfers via analysis of available genomic sequences, via expanding gene databases by cosmid walking, and via mutagenesis and phylogenetic analysis. The following major objectives were pursued: (i) to determine whether gene clustering patterns are conserved within specific groups of Bacteria, (ii) to determine whether such patterns reflect phylogenies of the respective genes, (iii) to determine whether these patterns reflect organismal phylogenies, and (iv) to attempt identification of novel C1 transfer genes based on the clustering patterns.
MATERIALS AND METHODS
Sequence analysis.
The genome of Methylobacterium extorquens AM1 was analyzed as described in reference 3. The genome of Methylobacillus flagellatus KT was analyzed as described in reference 14. The genomes of Methylococcus capsulatus Bath, Rhodopirellula baltica, Gemmata obscuriglobus, and Burkholderia xenovorans LB400 were analyzed as described in reference 5). The sequences of interest were retrieved from the genome of Methylobium petroleophilum PM1 (http://genome.jgi-psf.org/draft_microbes/metpe/metpe.home.html) via BLAST analyses using the sequences of B. xenovorans as queries as described previously (5). Partial sequences of gene islands encoding H4MPT-linked C1 transfer reactions from Xanthobacter autotrophicus, Hyphomicrobium zavarzinii, Methylosinus sp. strain LW2, Methylomonas sp. strain LW13, and Methylomicrobium sp. strain AMO were obtained via cosmid walking as described in reference 5. Environmental sequences of interest were detected as described in reference 14.
Phylogenetic analysis.
For phylogenetic analyses, the PHYLIP package (9) was used. Distance and parsimony methods were used, and 100 bootstrap analyses were performed. Concatenated polypeptide sequences were generated as follows. The respective polypeptide sequences were translated from the respective cosmid sequences or retrieved from the respective genomic databases as described in reference 5. Separate polypeptide sequences (or truncated sequences) were aligned by using the CLUSTAL W program (23), and all of the sequences were truncated to be of the same length. The truncated polypeptide sequences of each organism were then fused together. To clarify the position of M. flagellatus sequences, the following polypeptides were concatenated in the following order (ranging in length from 1,410 to 1,544 amino acid residues): OrfY-Mch-Orf5-Orf7-Fae-Orf17. To clarify the position of X. autotrophicus sequences, the following polypeptides were concatenated in the following order (ranging in length from 2,333 to 2,401 amino acid residues): Mch-Orf5-Orf9-FhcC-FhcD-FhcA-FhcB. In the case of G. obscuriglobus, two homologs of fhcD are found in the genome (53 and 39% identity with the fhcD from M. extorquens, respectively, at the amino acid level). The one with the higher identity to the proteobacterial FhcDs was used in the analysis. The concatenated sequences were realigned by using the CLUSTAL W program, and the alignments were manually curated.
Mutant generation and mutant complementation.
Mutations in the following genes of M. extorquens were generated essentially as described in reference 18: mxaE1, mxaD2, mxaD3, pabB, pcbD, pts, fae2, and fae3 (Table 1). A double fae2 fae3 mutant was generated essentially as described in reference 18. Mutants in the following genes of M. flagellatus were generated by using the pCM184 suicide vector (18): orf5, orfY, mtdB, mptG, orf1, and afpA (Table 1). For mutant complementation, the previously described broad-host-range expression vector pCM80 (17) was used.
TABLE 1.
Genes mutated in this study
| Genea | Known or predicted function | C1 growth defectb |
|---|---|---|
| mxaE1* | Unknown function in methanol oxidation | No* |
| mxaD1* | Methanol oxidation, interaction between methanol dehydrogenase and cytochrome cL | No* |
| mxaD2* | Methanol oxidation, interaction between methanol dehydrogenase and cytochrome cL | No* |
| pabB* | Component 1 of PABA synthase, biosynthesis of H4F and H4MPT | Yes* |
| pcbD* | Pterin-4a-carbinolamine dehydratase, regeneration of BH4 | No* |
| pts* | 6-Pyruvoyl tetrahydropterin synthase, biosynthesis of BH4 | No* |
| fae2* | Formaldehyde-activating enzyme | No* |
| fae3* | Formaldehyde-activating enzyme | No* |
| fae2 fae3 | No* | |
| orf1* | ATP-utilizing protein, ATP grasp family | Yes† |
| afpA* | Archaeal flavoprotein, FMN-binding electron transfer protein | Yes† |
| orf5† | Unknown function in biosynthesis of H4MPT | Yes† |
| mtdB† | Methylene-H4MPT dehydrogenase | Yes† |
| mptG† | Ribofuranosylaminobenzene 5-phosphate synthase, biosynthesis of H4MPT | Yes† |
| orfY* | Predicted ATP-dependent carboligase | Yes† |
*, Function is predicted based on sequence similarity; †, function has been characterized in M. extorquens. See text for references.
*, Mutated in M. extorquens; †, mutated in M. flagellatus.
Mutant characterization.
Growth characteristics of the M. extorquens mutants were tested on methanol-supplemented or succinate-supplemented media or on succinate-supplemented media in the presence of methanol vapors, as described earlier (6). To complement the pabB mutation, a range of concentrations (1 nM to 1 mM) of para-aminobenzoic acid (PABA) was used as a supplement. To characterize the phenotypes of the M. flagellatus mutants, a standard medium with added formaldehyde was used as described previously (4).
RESULTS
Gene islands encoding H4MPT-linked C1 transfer reactions in Bacteria.
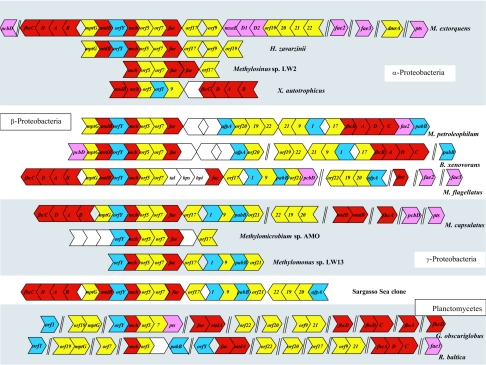
A cluster of genes with homologs in methanogenic archaea (the “archaeal-like” gene island) has been characterized in M. extorquens, and most of the “archaeal-like” genes have been shown to be involved in oxidation of formaldehyde to formate via H4MPT-linked derivatives (6, 7, 27). Analysis of the yet-incomplete genome of M. extorquens revealed no other “archaeal” gene islands or gene homologs in the chromosome, with the exceptions of two distant homologs of fae, fae2 and fae3, of unknown function (5). In the present study, we expanded analysis of genes involved in H4MPT-linked reactions to include other proteobacterial species, as well as representatives of Planctomycetes. Genomic sequences are now available for three methylotrophic proteobacteria other than M. extorquens: Methylococcus capsulatus Bath (28), Methylobium petroleophilum PM1 (http://genome.jgi-psf.org/draft_microbes/metpe/metpe.home.html), and Methylobacillus flagellatus KT (http://genome.jgi-psf.org/draft_microbes/metfl/metfl.home.html). In addition, genomic sequences of some nonmethylotrophic bacteria were shown to contain genes involved in H4MPT-linked C1 transfers, i.e., the genome of Burkholderia xenovorans LB400 and the genomes of the three planctomycetes: Gemmata obscuriglobus, Gemmata Wa1-1, and Rhodopirellula baltica (5, 10, 20). We also expanded “archaeal” gene analysis in Proteobacteria by partially sequencing these chromosomal regions from Methylomicrobium sp. strain AMO, Methylomonas sp. strain LW13, Hyphomicrobium zavarzinii, Xanthobacter autotrophicus, and Methylosinus sp. strain LW2 (5). In addition, clusters of genes homologous to the “archaeal” genes in M. extorquens have been identified in the environmental shotgun sequence database generated from the Sargasso Sea DNA (14). The newly identified gene clusters were aligned with the gene cluster in M. extorquens and analyzed in terms of gene order conservation (Fig. 1).
FIG. 1.
Clusters of genes encoding reactions of the H4MPT-linked C1 transfer pathway in Bacteria. Genes for C1 transfer reactions are shown in red; previously characterized genes for cofactor biosynthesis or not-yet-identified functions are shown in yellow. Genes with representatives mutated in the present study are shown in blue (C1-negative phenotype) or purple (C1-positive phenotype). Genes not involved in this study are left blank. Genes not contiguous on chromosomes are separated by double lines.
In general, we observed a high degree of gene order conservation in methylotrophic proteobacteria. However, various signs of chromosomal rearrangements particular to specific lineages were observed, such as insertions, deletions, and inversions. Analysis of the genomic sequence of M. capsulatus, a γ-proteobacterial methanotroph, revealed that the genes in question were not clustered together on the chromosome but formed three different gene islands separated by large distances on the chromosome. An additional homolog of fhcA was also present, one not linked to other C1 genes (28). Gene order in the main island was highly conserved relative to the order of genes in M. extorquens between fhcC and orf9. However, mtdB was not found in the main island in M. capsulatus. Instead, two copies of mtdB were found as a separate island on the chromosome. There is an insertion of an open reading frame (orf1) between orf17 and orf9 of M. capsulatus not found in the M. extorquens island or anywhere in the almost complete genome of M. extorquens. This gene has homologs in archaeal genomes, and its translated product possesses kinase motifs, based on COG (for clusters of orthologous genes) analysis (http://www.ncbi.nlm.nih.gov/COG/), but nothing is known about its function. Although in M. extorquens three genes with no archaeal homologs (homologous to mxaE and mxaD genes) were found as part of a methylotrophy gene island, separating orf9 from orf19 to -22 in the M. capsulatus island, a homolog of pabB, a gene predicted to encode component I of PABA synthase, was found downstream of orf9, together with orf21. orf22, orf19, and orf20 formed a separate gene island in the M. capsulatus chromosome. The partially sequenced gene islands from two other γ-proteobacterial methanotrophs, Methylomicrobium sp. strain AMO and Methylomonas sp. strain LW13 revealed a gene order similar to the one in the main island in M. capsulatus (Fig. 1); however, in Methylomicrobium sp. strain AMO, insertions of genes not conserved in other clusters were found (Fig. 1).
Analysis of the recently generated genomic sequence of a β-proteobacterial methylotroph, M. flagellatus revealed the presence of two “archaeal-like” gene islands. The main island aligned very well with the main island of M. capsulatus; however, three insertions are present in the M. flagellatus island. The gene for mtdB is located between mptG and orfY, as in M. extorquens. Three genes are inserted between orf7 and fae, encoding, respectively, transaldolase (tal), hexulosephosphate synthase (hps), and hexulosephosphate isomerase (hpi), enzymes of the ribulose monophosphate cycle for formaldehyde assimilation (1). Another novel gene, tentatively designated pcbD, is found downstream of orf21 in M. flagellatus, which is predicted to encode pterin-4a-carbinolamine dehydratase (an enzyme involved in regeneration of the cofactor tetrahydrobiopterin, BH4 [24]), based on COG analysis. However, no nucleotide or amino acid conservation exists between characterized eukaryotic (24) and putative bacterial pcbD genes (National Center for Biotechnology Information). A pcbD homolog is present in a larger C1 island of M. extorquens, separated by 16.5 kb from fhcC and by ca. 7 kb from fch, which encodes methenyl tetrahydrofolate (H4F) cyclohydrolase, an enzyme involved in methylotrophy (3). A pcbD homolog in M. capsulatus is found ca. 7 kb apart from fch-mtdA pair, but no other C1 genes are present in the vicinity. MtdA encodes methylene tetrahydrofolate dehydrogenase, which is also involved in methylotrophy in M. extorquens AM1 (3).
The second gene island of M. flagellatus, containing orf19, -20, and -22, aligns with the respective island in M. capsulatus. However, one additional gene is present upstream of orf20, having homologs in Archaea, encoding a conserved archaeal flavoprotein (afpA) (8). In addition to the two gene islands, three other archaeal gene homologs were found in the genome. One of them was highly similar to fae in the main gene island, whereas two others were similar to the fae homologs, fae2 and fae3, previously identified in M. extorquens.
We have recently identified gene islands involved in H4MPT-linked C1 transfers in the shotgun sequence library of environmental DNA from the Sargasso Sea that belong to as-yet-unidentified bacteria (14). One of these clusters in shown in Fig. 1. The gene order in this cluster shares many similarities with that for M. capsulatus and M. flagellatus.
Three partial cluster sequences from α-proteobacteria representing Hyphomicrobium, Methylosinus, and Xanthobacter were aligned with the cluster in M. extorquens (Fig. 1). A partial cluster of nine genes has been sequenced from another Methylobacterium species closely related to M. extorquens (16), showing identical gene order and a high degree of sequence conservation at the DNA level, and thus is not shown in Fig. 1. The partial gene islands from H. zavarzinii and Methylosinus sp. strain LW2 revealed a high degree of gene order conservation with the Methylobacterium species, with some exceptions. For example, the cluster from H. zavarzinii contained no insert of mxa gene homologs found in M. extorquens, whereas the cluster in Methylosinus sp. strain LW2 contained two fae homologs. In contrast, the gene order in the partial cluster from X. autotrophicus was strikingly different from all other known clusters: no mptG was present upstream of mtdB (a region of 5 kb has been sequenced [data not shown]), no orfY was present between mtdB and mch, and no genes were conserved in their order downstream of orf5. Instead, orf5 was followed by orf1 and orf9, downstream of which an inverted cluster of the fhc genes was found. Additional sequence of 15 kb downstream of fhcB did not reveal the presence of any other genes of interest.
The genomes of two other β-proteobacteria were involved in our analyses, those of closely related organisms belonging to Burkholderiaceae, M. petroleophilum PM1 and B. xenovorans LB400. Although M. petroleophilum can grow on some C1 compounds (K. M. Scow and K. R. Hristova, unpublished data), B. xenovorans is not known to be a methylotroph (20). Analysis of the genome of M. petroleophilum revealed that most of the genes of interest were located in a single gene island. The gene order between mptG and fae was conserved with relation to the order in most of the islands discussed above, whereas the rest of the island was represented by an inversion of the cluster of genes represented by afpA-orf17 in the environmental clone, in which pabB was missing. The fhc gene cluster was located downstream of orf17 and was inverted relative to its position in other methylotrophs belonging to α-, β-, and γ-proteobacterial groups. A homolog of fae2 from M. extorquens and M. flagellatus was found downstream of fhcC, followed by pabB. The C1 transfer gene island in B. xenovorans had a very similar structure with the following exceptions. A cluster of cbb genes (encoding genes of the Calvin-Benson-Bassham cycle) was inserted between orf20 and orf19 (20; data not shown in Fig. 1), no fae2 homolog was present in the cluster or anywhere in the genome, a pabB hmolog was present elsewhere in the genome, and a pcbD homolog was present upstream of mptG.
As is seen in Fig. 1, C1 transfer gene clustering in Planctomycete genomes is much looser compared to proteobacterial genomes. However, the mch-orf5 order was conserved in the genome of R. baltica, and the order of orfY-mch-orf5-orf7 was conserved in G. obscuriglobus. Besides this, clusters of three (fhcADC) and two (fhcDC) genes are found in the genomes of R. baltica and G. obscuriglobus, respectively (Fig. 1). However, no homolog of fhcB was detected in R. baltica (10), whereas in G. obscuriglobus two homologs of fhcD were identified that greatly diverged in their sequences (46% identity between the translated amino acid sequences). The low coverage of the genome of Gemmata sp. strain Wa1-1 (5) allowed detection of only a few gene clusters, and these were the same as in G. obscuriglobus (data not shown). In all Planctomycete genomes, orf19 was linked to mptG and orf9 was linked to orf21, showing novel clustering types. In R. baltica, a pabB homolog was found linked to the mch-orf5 cluster, whereas in G. obscuriglobus, a homolog of pts, a gene potentially encoding an enzyme involved in BH4 biosynthesis, 6-pyruvoyl-tetrahydropterin synthase, was found downstream of orf7. fae genes in Planctomycetes, unlike those in Proteobacteria, were found linked to genes revealing homology to genes encoding either MtdA or MtdB enzymes (44 to 49% and 28 to 30% identity at the amino acid level, respectively). MtdA and MtdB have related sequences (ca. 30% identity), MtdA being a NADP-specific dehydrogenase that utilizes both methylene-H4F and methylene-H4MPT, whereas MtdB uses both NAD and NADP but is specific to H4MPT (3, 12).
Phylogenetic analysis.
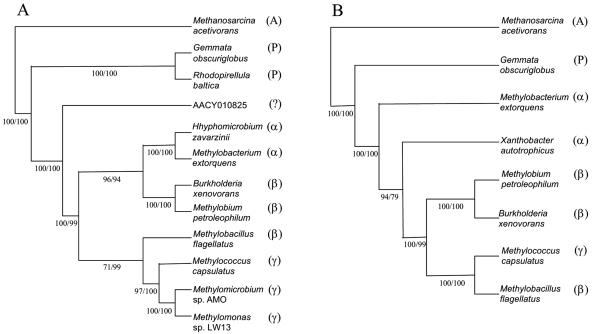
Analysis of these gene clusters has revealed a great deal of divergence in gene order within the groups of the α-proteobacterial and β-proteobacterial species. The gene order in the M. flagellatus cluster was more similar to that in γ-proteobacterial clusters than to that in clusters of other β-proteobacteria, whereas the gene order in X. autotrophicus was unique within the bacterial set sampled. These distinct organizations may indicate that the C1 clusters in these strains have a separate evolutionary history relative to other methylotrophs within their own respective classes of proteobacteria. To test this hypothesis, we performed phylogenetic analyses with the subsets of sequences available. In accordance with our previous experience, due to the divergence of the genes in question, analysis of single polypeptide sequences often results in unresolved branching patterns, reflected by low bootstrap values. However, using concatenated polypeptide sequences results in increased phylogenetic signal and thus a better resolution in phylogenetic analyses (14). To assess the phylogenetic position of the “archaeal” genes in M. flagellatus relative to other bacterial species, we used concatenates of six polypeptide sequences as described in Materials and Methods. Both distance and parsimony analyses resulted in identical branching patterns, placing M. flagellatus sequences with the sequences of γ-proteobacterial methylotrophs, with high bootstrap values, while the sequences of other β-proteobacteria, B. xenovorans and M. petroleophilum, branched together (Fig. 2A). Such a branching pattern suggests separate evolutionary histories for the two types of C1 clusters found in β-proteobacteria. Similar analyses were performed with a subset of sequences to obtain insights into the position of the X. autotrophicus sequences within the group tested. Concatenates of seven polypeptide sequences were used as described in Materials and Methods. Distance and parsimony analyses produced identical branching patterns in phylogenetic trees (Fig. 2B). In these trees, the sequences of X. autotrophicus clearly separated from the sequences of M. extorquens and from all other sequences, suggesting that the genes in question have distinct evolutionary histories within α-proteobacteria as well.
FIG. 2.
Phylogenetic analysis of concatenated polypeptides representing subsets of H4MPT-linked C1 transfer genes (see Materials and Methods). Bootstrap values are shown for distance/parsimony analyses. (A) Tree resolving the position of the M. flagellatus sequences using six sequences in the concatenates. (B) Tree resolving the position of the X. autotrophicus sequences using seven sequences in the concatenates. A, P, α, β, and γ indicate Archaea, Planctomycetes, and α-, β-, and γ-Proteobacteria, respectively. AACY010825 is the sequence recovered from the Sargasso Sea metagenome.
Functional analysis of novel genes conserved in C1 transfer clusters. (i) Mutagenesis in M. extorquens.
An insert of three nonarchaeal genes is present in the gene cluster of M. extorquens (Fig. 1). One of these genes is homologous to mxaE, a gene of unknown function in the gene cluster (mox cluster) involved in methanol oxidation (3), while two others are both homologous to mxaD, another gene in the mox cluster that has been shown to be involved in interaction between methanol dehydrogenase and cytochrome cL (25). We mutated all three mox gene homologs to test their involvement in C1 metabolism. The resulting mutants did not reveal any visible defects in growth on methanol (Table 1); thus, the roles of these mxa gene homologs remain unknown.
Another gene persistently present in methylotroph genomes and in some cases in the C1 transfer clusters is the gene tentatively designated pcbD, based on COG analysis. Known pcbD genes encode pterin-4a-carbinolamine dehydratase (24), an enzyme involved in regeneration of tetrahydrobiopterin (BH4). This enzyme also possesses a second function, as a regulatory protein (24). A gene encoding a homolog of an enzyme involved in the BH4 biosynthesis pathway, 6-pyruvoyl tetrahydropterin synthase (pts [24]), is clustered with the C1 transfer genes in G. obscuriglobus, and this gene is recognizable in many of the sequenced genomes. We used M. extorquens as a model to test for possible involvement of these genes in C1 metabolism but did not observe any deficiency for growth on methanol for mutants lacking either pcbD or pts (Table 1).
We have previously identified two distant homologs of fae in the genome of M. extorquens, tentatively designated fae2 and fae3, which were not part of any C1 clusters (5). Homologs of both fae2 and fae3 were identified in the genome of M. flagellatus, a close homolog of fae3 was identified in R. baltica (10), and a homolog of fae2 was identified in the genome of M. petroleophilum. In the latter case, fae2 was part of the C1 transfer cluster (Fig. 1). We mutated orf2 and orf3 in M. extorquens, and we also obtained a double fae2 fae3 mutant. All three mutants grew with wild-type characteristics on both succinate and methanol (Table 1). Overexpression of either fae2 or fae3 in the background of a fae mutant of M. extorquens (27) did not lead to complementation of the phenotype of the mutant (14, the present study).
Analysis of H4MPT-linked C1 transfer gene islands also revealed the presence of a nonarchaeal gene, a homolog of pabB. This gene encodes component 1 of PABA synthase, the enzyme involved in biosynthesis of PABA, the precursor in biosyntheses of both H4F and H4MPT (11). To test if mutants in pabB would reveal a specific phenotype, we mutated a pabB homolog in M. extorquens. This gene is not a part of the C1 island shown in Fig. 1, and it was identified in the genomic database as the highest hit with pabB homologs present in other C1 islands. Null mutants in pabB were selected on succinate-supplemented medium with or without added PABA (1 mM). pabB mutants grew on succinate or methanol plates supplemented with 1 mM PABA but not on either substrate in the absence of PABA supplementation. The presence of wild-type on the same plates as pabB mutants permitted slow growth of the mutants in a manner consistent with cross-feeding. This phenomenon likely explains our success in isolating pabB mutants in the absence of PABA: release of PABA from lysed cells and/or single-crossover exconjugants may have obviated the need for PABA supplementation. We determined the minimal PABA supplement concentration to allow growth of the mutants on succinate to be ca. 10 nM. We then tested the influence of added methanol on growth of pabB mutants in the presence of the minimal supplementary concentration of PABA. The addition of ca. 1 mM methanol caused significant growth inhibition of the pabB mutant (Table 1). Methanol sensitivity has been shown to be a trait of mutants defective in H4MPT-linked formaldehyde oxidation (6, 12, 19, 27). Increasing the concentration of PABA supplement gradually alleviated methanol sensitivity, and the mutants could grow on methanol with wild-type rate in the presence of 1 mM PABA.
(ii) Mutagenesis in M. flagellatus.
Two novel genes were found conserved in a number of proteobacterial gene clusters that were not identifiable in the genome of M. extorquens (3) but have homologs in Archaea: orf1 and afpA. No function has been proposed for homologs of orf1 in Archaea; however, AfpA in Archaeoglobus fulgidus has been proposed to be involved in electron transfer between ferredoxin and the CO dehydrogenase/acetyl coenzyme A synthase (CODH/ACS) complex (8). The possible involvement of orf1 and afpA in the C1 transfer pathway was tested via mutagenesis in M. flagellatus. Previously, mch from this organism has been mutated, and the phenotype of this mutant suggested that the pathway was not essential for methylotrophy in this organism but played an auxiliary function in formaldehyde detoxification (4). However, in the main model organism for studying H4MPT-linked C1 transfer reactions, M. extorquens, we demonstrated that mutations in different genes involved in the pathway resulted in phenotypes that differed in the degree of formaldehyde sensitivity (6, 19). To provide a reference database for characterization of the mutants in genes of unknown functions in M. flagellatus, we also mutated genes previously characterized in M. extorquens, i.e., mptG, mtdB, and orf5. We also tested the function of orfY by mutation. The function of this gene remained unknown in M. extorquens. We had originally reported obtaining knockout mutants in this gene in M. extorquens (7), but later tests have shown these mutants were single-crossover recombinants (unpublished data). Our recent attempts to generate orfY-null mutants in M. extorquens were unsuccessful, suggesting that this gene is essential in this organism.
Mutants of M. flagellatus were generated via allelic exchange as described in Materials and Methods. Double-crossover mutations were generated in all of the genes tested, as judged by diagnostic PCR (data not shown). Their sensitivity to formaldehyde was investigated as previously described (4) and compared to the phenotypes of the wild-type strain and the previously characterized mch mutant, respectively. All of the mutants investigated showed increased formaldehyde sensitivity similar to the previously described mutant in mch; however, the new mutants showed slightly lower formaldehyde sensitivity (between 2.5 and 3 mM versus 2 mM for the mch mutant) (Table 1). The functions of mtdB and mptG have been determined (12, 22), and a function has been proposed for orf5 in H4MPT biosynthesis (6). Based on the phenotypes of the new mutants and also on gene location within the formaldehyde oxidation islands, orfY, orf1, and afpA are likely to be involved in the H4MPT-linked pathway as well. To test whether orfY from M. extorquens could complement orfY mutants of M. flagellatus, we expressed orfY from M. extorquens in the orfY mutant of M. flagellatus and tested the recombinants for formaldehyde sensitivity. The recombinants carrying the copy of the M. extorquens gene in trans showed wild-type sensitivity toward formaldehyde.
Expression of orf1 and afpA in the dmrA mutant of M. extorquens.
Although orf1 and afpA genes are present in many of the “archaeal” gene islands characterized here, neither of these genes is identifiable in the almost-complete genome of M. extorquens. Conversely, a gene has been identified in M. extorquens that is involved in the last step of the H4MPT biosynthetic pathway, a gene that encodes dihydromethanopterin reductase (dmrA [2, 21, 22]), that is not recognizable in the genomes of M. capsulatus, M. flagellatus, B. xenovorans, and M. petroleophilum or in the planctomycete genomes. Mutagenesis of orf1 and afpA in M. flagellatus suggests their involvement in some step of the H4MPT-linked C1 transfer pathway (see above). To test whether orf1 or afpA could fulfill the function of dmrA, we expressed the orf1 genes from M. capsulatus and Methylomonas sp. strain LW13, and the afpA genes from M. flagellatus and B. xenovorans in the dmrA mutant of M. extorquens and tested the transconjugants for growth on methanol and methanol sensitivity. Although expression of orf1 genes had no effect on the phenotype of the dmrA mutant, expression of afpA genes resulted in reversal of the dmrA mutant phenotype to the wild-type phenotype, i.e., AfpA could fulfill the function of DmrA. Since the substrate for the DmrA, reaction, dihydromethanopterin, is not commercially available, we were not able to confirm the function of AfpA via enzyme activity measurements.
DISCUSSION
In this study we investigated the structure of clusters of genes encoding reactions of H4MPT-linked C1 transfer and correlated these with specific bacterial groups. We demonstrated that, whereas tight clustering of the genes in question is characteristic of Proteobacteria and a great deal of gene order conservation occurs, the cluster structure and the degree of clustering vary greatly both within the specific groups and between the groups. We have previously reported that gene sequences of Planctomycetes involved in H4MPT-linked reactions diverge significantly from their proteobacterial counterparts and occupy an intermediate phylogenetic position between Archaea and Proteobacteria (5). Here we demonstrate that the degree of gene clustering in Planctomycetes is also intermediate between Archaea and Proteobacteria: whereas almost no clustering for the genes in question occurs in Archaea, a certain degree of clustering takes place in Planctomycetes. However, only two instances of a cluster structure common to both Planctomycetes and Proteobacteria were found (mch-orf5 and fhcDC). The results of this analysis point toward long and separate histories of the genes in question in Archaea, Proteobacteria, and Planctomycetes.
In a previous study, we argued against a single recent event of lateral transfer of genes for H4MPT-linked reactions between Archaea and Bacteria (5). However, the data presented here suggest that lateral transfers within groups—specifically, within Proteobacteria—have likely occurred. The difference in gene cluster structure and in the phylogenetic positions of β-proteobacterial genes represented by M. flagellatus on one hand and by B. xenovorans and M. petroleophilum on the other may have two alternative explanations, both involving lateral transfers. First, an ancestor of M. flagellatus may have lost the gene cluster typical of β-proteobacteria and subsequently acquired a new cluster via a lateral transfer from a γ-proteobacterial methanotroph. Second, the genes found in modern representatives of Burkholderiaceaea are a result of lateral transfer into the ancestor of both Burkholderia and Methylobium from a source not known at this time. Similarly, the history of the genes for H4MPT-linked reactions in X. autotrophicus may involve a lateral transfer from an unknown source since they diverge significantly in both sequence and clustering patterns from genes in other α-proteobacteria. It is interesting that, whereas the cluster structure found in environmental clones from the Sargasso Sea is similar to that from γ-proteobacteria and M. flagellatus, gene sequences diverge significantly from both and form a separate branch on phylogenetic trees (14). It is possible that the clusters found in the Sargasso Sea group and γ- and β-proteobacterial methylotrophs have descended from a single ancestral cluster but were exposed to different types of selective pressures, resulting in great sequence divergence.
Clustering of H4MPT-linked C1 transfer genes on the chromosome of M. extorquens has been advantageous for gaining insights into details of C1 metabolic pathways, including gene discovery and identification. Involving more species in gene cluster analyses as described here led to the discovery of additional genes potentially involved in the H4MPT-linked C1 transfer pathway (Table 1), at least in some of the organisms producing methanopterin or its derivatives. orfY is predicted to encode an ATP-grasp family protein, possibly a carboxylase, in accordance with COG analysis. Its role in H4MPT-linked C1 transfer remains unknown. Since all of the genes for the catalytic reactions in the pathway are known, orfY may be involved in either cofactor biosynthesis or regulation of the pathway. The presence of orf1 in a number of C1 transfer gene clusters (Fig. 1) and the phenotype of the orf1 mutant in M. flagellatus suggest this gene is also involved in the pathway. However, orf1 is not present in the almost complete genome of M. extorquens. This may imply either that orf1 is involved in a reaction not essential in M. extorquens (for example, a cofactor modification) or that a nonhomologous substitution for this gene is present in M. extorquens. Our data point toward pabB being involved in the biosynthesis of PABA, a precursor of both H4F and H4MPT. Based on mutant analysis in M. extorquens, higher PabB activity is required during growth on C1 compounds compared to multicarbon compounds, a finding in agreement with previous data on the major role of the H4MPT-linked formaldehyde oxidation pathway in C1 oxidation. afpA appears to encode a novel dihydromethanopterin reductase. Ding and Ferry have recently purified an AfpA homolog from A. fulgidus and demonstrated that it is an FMN-binding electron carrier protein (8). These authors also analyzed the distribution of AfpAs in prokaryotes based on the genomic sequences available at the time and concluded that these were restricted to methanogenic archaea. Ding and Ferry proposed a function for AfpA in an electron transport chain in A. fulgidus in which electrons originating from the CODH/ACS complex are transferred to AfpA mediated by ferredoxin, thus assigning a role for AfpA in energy generation. We demonstrated here that afpA homologs are present in the C1 gene islands of most of the bacteria synthesizing H4MPT or its derivatives. Our data on complementation of the dmrA mutant, as well as the absence of CODH/ASC in the bacteria in question, suggest that afpA homologs perform an alternative physiological function in the biosynthesis of H4MPT. In methanogenic Archaea not relying on CODH/ACS complex for energy generation (such as M. thermoautotrophicus or M. jannaschii), afpA homologs may also encode DMR enzymes. M. extorquens possesses a DMR that is a homolog of dihydrofolate reductase, encoded by dmrA (2, 21, 22). However, thus far, dmrA is unique to M. extorquens and not found in the genomes of other Bacteria and Archaea synthesizing H4MPT. Interestingly, the genome of M. capsulatus lacks both dmrA and afpA, suggesting that yet another nonhomologous DMR may exist.
Acknowledgments
This study was supported by the NSF Microbial Observatories program (MCB-0131957) and in part by a grant from the NIH (GM36296).
The Joint Genome Institute is acknowledged for sequencing the genomes of M. flagellatus and M. petroleophilum, and The Institute for Genomic Research is acknowledged for early release of the genome sequence of G. obscuriglobus (all three projects are funded by the DOE).
REFERENCES
- 1.Anthony, C. 1982. Biochemistry of methylotrophs. Academic Press, London, England.
- 2.Caccamo, M. A., C. M. Malone, and M. E. Rasche. 2004. Biochemical characterization of a dihydromethanopterin reductase involved in tetrahydromethanopterin biosynthesis in Methylobacterium extorquens AM1. J. Bacteriol. 186:2068-2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chistoserdova, L., S.-W. Chen, A. Lapidus, and M. E. Lidstrom. 2003. Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J. Bacteriol. 185:2980-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chistoserdova, L., L. Gomelsky, J. A. Vorholt, M. Gomelsky, Y. D. Tsygankov, and M. E. Lidstrom. 2000. Analysis of two formaldehyde oxidation pathways in Methylobacillus flagellatus KT, a ribulose monophosphate cycle methylotroph. Microbiology 146:233-238. [DOI] [PubMed] [Google Scholar]
- 5.Chistoserdova, L., C. Jenkins, M. G. Kalyuzhnaya, C. J. Marx, A. Lapidus, J. A. Vorholt, J. T. Staley, and M. E. Lidstrom. 2004. The enigmatic planctomycetes may hold a key to the origins of methanogenesis and methylotrophy. Mol. Biol. Evol. 21:1234-1241. [DOI] [PubMed] [Google Scholar]
- 6.Chistoserdova, L., M. E. Rasche, and M. E. Lidstrom. 2005. Novel dephosphotetrahydromethanopterin biosynthesis genes discovered via mutagenesis in Methylobacterium extorquens AM1. J. Bacteriol. 187:2508-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chistoserdova, L., J. A. Vorholt, R. K. Thauer, and M. E. Lidstrom. 1998. C1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic archaea. Science 281:99-102. [DOI] [PubMed] [Google Scholar]
- 8.Ding, Y. H., and J. G. Ferry. 2004. Flavin mononucleotide-binding flavoprotein family in the domain Archaea. J. Bacteriol. 186:90-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felsenstein, J. 2003. Inferring phylogenies. Sinauer Associates, Inc., Sunderland, Mass.
- 10.Glöckner, F. O., M. Kube, M. Bauer, H. Teeling, T. Lombardot, W. Ludwig, D. Gade, A. Beck, K. Borzym, K. Heitmann, R. Rabus, H. Schlesner, R. Amann, and R. Reinhardt. 2003. Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc. Natl. Acad. Sci. USA 100:8298-8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham, D. E., and R. H. White. 2002. Elucidation of methanogenic coenzyme biosyntheses: from spectroscopy to genomics. Nat. Prod. Rep. 19:133-147. [DOI] [PubMed] [Google Scholar]
- 12.Hagemeier, C. H., L. Chistoserdova, M. E. Lidstrom, R. K. Thauer, and J. A. Vorholt. 2000. Characterization of a second methylene tetrahydromethanopterin dehydrogenase from Methylobacterium extorquens AM1. Eur. J. Biochem. 267:3762-3769. [DOI] [PubMed] [Google Scholar]
- 13.Kalyuzhnaya, M. G., M. E. Lidstrom, and L. Chistoserdova. 2004. Utility of environmental probes targeting ancient enzymes: methylotroph detection in Lake Washington. Microb. Ecol. 48:463-472. [DOI] [PubMed] [Google Scholar]
- 14.Kalyuzhnaya, M. G., O. Nercessian, A. Lapidus, and L. Chistoserdova. 8 April 2005, posting date. Fishing for biodiversity: novel methanopterin-linked C1 transfer genes deduced from the Sargasso Sea metagenome. Environ. Microbiol. doi: 10.1111/j.1462-2920.2005.00798.x. [DOI] [PubMed]
- 15.Kalyuzhnaya, M. G., O. Nercessian, M. E. Lidstrom, and L. Chistoserdova. Development and application of polymerase chain reaction primers based on fhcD for environmental detection of methanopterin-linked C1-metabolism in bacteria. Environ. Microbiol., in press. [DOI] [PubMed]
- 16.Kayser, M. F., Z. Ucurum, and S. Vuilleumier. 2002. Dichloromethane metabolism and C1 utilization genes in Methylobacterium strains. Microbiology 148:1915-1922. [DOI] [PubMed] [Google Scholar]
- 17.Marx, C. J., and M. E. Lidstrom. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other gram-negative bacteria. Microbiology 147:2065-2075. [DOI] [PubMed] [Google Scholar]
- 18.Marx, C. J., and M. E. Lidstrom. 2002. A broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. BioTechniques 33:1062-1067. [DOI] [PubMed] [Google Scholar]
- 19.Marx, C. J., L. Chistoserdova, and M. E. Lidstrom. 2003. Formaldehyde-detoxifying role of the tetrahydromethanopterin-linked pathway in Methylobacterium extorquens AM1. J. Bacteriol. 185:7160-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marx, C. J., J. A. Miller, L. Chistoserdova, and M. E. Lidstrom. 2004. Multiple formaldehyde oxidation/detoxification pathways in Burkholderia fungorum LB400. J. Bacteriol. 186:2173-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marx, C. J., B. N. O'Brien, J. Breezee, and M. E. Lidstrom. 2003. Novel methylotrophy genes of Methylobacterium extorquens AM1 identified by using transposon mutagenesis including a putative dihydromethanopterin reductase. J. Bacteriol. 185:669-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rasche, M. E., S. A. Havemann, and M. Rosenzvaig. 2004. Characterization of two methanopterin biosynthesis mutants of Methylobacterium extorquens AM1 by use of a tetrahydromethanopterin bioassay. J. Bacteriol. 186:1565-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight Matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thöny, B., G. Auerbach, and N. Blau. 2000. Tetrahydrobiopterin biosynthesis, regulation and functions. Biochem. J. 347:1-16. [PMC free article] [PubMed] [Google Scholar]
- 25.Toyama, H., H. Inagaki, K. Matsushita, C. Anthony, and O. Adachi. 2003. The role of the MxaD protein in the respiratory chain of Methylobacterium extorquens during growth on methanol. Biochim. Biophys. Acta 1647:372-375. [DOI] [PubMed] [Google Scholar]
- 26.Vorholt, J. A., L. Chistoserdova, S. M. Stolyar, R. K. Thauer, and M. E. Lidstrom. 1999. Distribution of tetrahydromethanopterin-dependent enzymes in methylotrophic bacteria and phylogeny of methenyl tetrahydromethanopterin cyclohydrolases. J. Bacteriol. 181:5750-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vorholt, J. A., C. J. Marx, M. E. Lidstrom, and R. K. Thauer. 2000. Novel formaldehyde-activating enzyme in Methylobacterium extorquens AM1 required for growth on methanol. J. Bacteriol. 182:6645-6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward, N., (swsl)O. Larsen, J. Sakwa, L. Bruseth, H. Khouri, A. S. Durkin, G. Dimitrov, L. Jiang, D. Scanlan, K. H. Kang, M. Lewis, K. E. Nelson, B. Methe, M. Wu, J. F. Heidelberg, I. T. Paulsen, D. Fouts, J. Ravel, H. Tettelin, Q. Ren, T. Read, R. T. DeBoy, R. Seshadri, S. L. Salzberg, H. B. Jensen, N. K. Birkeland, W. C. Nelson, R. J. Dodson, S. H. Grindhaug, I. Holt, I. Eidhammer, I. Jonasen, S. Vanaken, T. Utterback, T. V. Feldblyum, C. M. Fraser, J. R. Lillehaug, and J. A. Eisen. 2004. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol. 2:e303. [DOI] [PMC free article] [PubMed] [Google Scholar]