Abstract
Certain marine unicellular cyanobacteria of the genus Synechococcus exhibit a unique type of swimming motility characterized by the absence of flagella or any other obvious organelles of motility. While the abundant cell surface-associated 130-kDa glycoprotein SwmA is known to be required for the generation of thrust, identification of other components of the motility apparatus has, until recently, been unsuccessful. Here we report on the development of a transposon mutagenesis system for use with marine Synechococcus sp. strain WH8102, a model organism for which the genome has been sequenced. Utilizing this mutagenesis technique, we have isolated 17 independent mutants impaired in swimming motility. These 17 transposon insertions are located in nine open reading frames, which cluster in three separate regions of the genome. Included within these clusters are several multicomponent transport systems as well as a number of glycosyltransferases.
The unicellular marine cyanobacteria of the genus Synechococcus are ubiquitous in the world's oceans and comprise a major fraction of the photosynthetic marine picoplankton (29-31). As such, they are important contributors to global primary production and may generate as much as 30% of total oceanic primary production (29). A genetic manipulation system has been described previously (5, 6) which has allowed molecular investigation into the adaptations of this globally important organism. Here we describe an addition to the molecular tools available for use with marine Synechococcus spp. with the development of a transposon mutagenesis technique. We have applied this method of mutagenesis to investigate the unique swimming motility exhibited by marine Synechococcus spp.
Among the adaptations exhibited by marine Synechococcus spp. is the ability of certain strains to swim through their liquid environment without the use of flagella (33). Transmission electron microscopy techniques, such as negative staining and quick-freeze fixation, fracture, and etching as well as high-intensity dark-field microscopy, have been employed in attempts to visualize structures, without success (35). Additionally, shearing experiments and motility-dependent amplitude spectra confirm the lack of flagella or other extracellular appendages used for motility (35). Both jet propulsion and self-electrophoresis have been ruled out as possible mechanisms for motility (24). This leaves the cell surface itself as the only remaining structure with potential for generating a propulsive force. Ehlers et al. have proposed a model by which cells could move at the observed speeds by propagating longitudinal waves across their surfaces (9).
Nonflagellar motility is not limited to marine Synechococcus spp. A diverse array of bacteria (16), including other cyanobacteria (12), exhibit nonflagellar motility, but in virtually every case this movement is not swimming but rather a movement associated with surfaces. Marine Synechococcus spp. are different in that in spite of their cells' clear lack of flagella, they swim through their liquid environment and do not move along surfaces. With the exception of the helical, wall-less bacteria of the genus Spiroplasma, which swim by means of conformational deformations of each cell's helical cytoskeletal filament (11, 36), Synechococcus spp. are the only bacteria known to swim without flagella. For a review of swimming motility in marine Synechococcus spp., see the work of Brahamsha (7).
While the basic tools required for genetic manipulations in Synechococcus spp. have been described previously (5, 6), a functional transposon mutagenesis system would be a powerful additional tool for molecular and genetic studies. The integration of a transposon into the host chromosome creates an insertional mutation, which can be easily cloned and sequenced to determine the site of insertion. We used the delivery vector pRL27 (15), which has a mini-Tn5 derivative engineered to enhance transposition frequency, for transposon mutagenesis in Synechococcus sp. strain WH8102, and our results have shown that this tool is useful in identifying new genetic loci involved in swimming motility. As this was the first use of transposon mutagenesis in a marine Synechococcus sp., preliminary experiments were conducted to characterize the use of this delivery vector and transposon. When combined with an appropriate screen or selection, the method described here will be applicable to the study of various aspects of Synechococcus physiology. We have developed a screen to identify mutants impaired in the ability to swim, and this screen in combination with transposon mutagenesis has identified several regions of the chromosome that contain genes necessary for nonflagellar swimming motility.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains MC1061(pRK24, pRL528), DH5α, BW20767 (15), and Transformax EC100D pir+ (Epicentre, Madison, WI) were grown in Luria-Bertani medium (28). When appropriate, ampicillin (100 μg/ml), kanamycin (50 μg/ml), and chloramphenicol (10 μg/ml) were used for the selection and maintenance of plasmids in E. coli. Cyanobacterial strains were grown either in SN medium (32) made with seawater obtained from the Scripps Pier (Scripps Institution of Oceanography, La Jolla, CA) or in SN medium prepared with synthetic ocean water (25). Cyanobacterial cultures were incubated at 25°C with a constant illumination of 25 microeinsteins m−2 s−1 and were maintained as either 4-ml cultures in 17- by 100-mm polystyrene tubes (Becton Dickinson, Franklin Lakes, NJ) or as 50-ml cultures in 125-ml glass flasks without shaking. SN pour plates for obtaining isolated colonies were prepared as previously described (5), with the single modification of reducing the agar concentration to 0.2% (wt/vol), which aids in screening for nonmotile colonies. Kanamycin was added to a final concentration of 25 μg/ml for pour plates and 20 μg/ml for liquid cultures, where appropriate, to maintain selection for insertions.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Sourceb or reference |
|---|---|---|
| Strains | ||
| Synechococcus sp. strain WH8102 | Motile strain, recipient in conjugations with pRL27 and pMUT100 constructions | J. Waterbury |
| E. coli | ||
| MC1061 | Host for pRK24, pRL528; used as a donor in pJM construction conjugations | 8 |
| DH5α | Recipient in transformations | BRL |
| BW20767 | Used as donor for pRL27 conjugations | 21 |
| Transformax EC100D pir+ | Expresses pir gene product for propagation of vectors containing the R6Kγ origin of replication | Epicentre |
| Plasmids | ||
| pMUT100 | Kanr, Tetr; suicide vector | 5 |
| pRL27 | Kanr; mini-Tn5 plasposon (oriR6K) delivery vector | 15 |
| pRK24 | Tcr, Ampr; conjugal plasmid, RK2 derivative | 5, 22 |
| pRL528 | Cmr; helper plasmid, carries mob | 5, 10 |
| pCR2.1-TOPO | Kanr, Ampr; PCR product cloning vector | Invitrogen |
| pJM37 | pMUT100 containing SYNW0079 fragment of nt 78600 to 78939 | This work |
| pBB1000 | pMUT100 containing swmA fragment of nt 84507 to 84926 | This work |
| pJM62 | pMUT100 containing SYNW0087 fragment of nt 88793 to 89004 | This work |
| pJM18 | pMUT100 containing SYNW0088 fragment of nt 89414 to 89635 | This work |
| pJM59 | pMUT100 containing SYNW0192 fragment of nt 193443 to 193661 | This work |
| pJM40 | pMUT100 containing SYNW0193 fragment of nt 194020 to 194287 | This work |
| pJM20 | pMUT100 containing swmB fragment of nt 913153 to 913396 | This work |
| pJM35 | pMUT100 containing SYNW0957 fragment of nt 947811 to 948110 | This work |
| pJM34 | pMUT100 containing SYNW0960 fragment of nt 953811 to 954171 | This work |
nt, nucleotide. Numbering according to reference 23.
BRL, Bethesda Research Laboratories, Gaithersburg, MD; Invitrogen, Carlsbad, CA; Epicentre, Madison, WI.
Transposon mutagenesis.
Biparental matings of E. coli and the Synechococcus sp. were carried out as previously described (5). E. coli strain BW20767, which harbors the plasposon delivery vector pRL27 (15), was used as the donor in conjugations with Synechococcus sp. strain WH8102. Following a 48-h incubation, cells were resuspended and plated in SN pour plates containing 0.2% agar and kanamycin (25 μg/ml) to obtain isolated colonies. Following the appearance of colonies, which usually took from 2 to 3 weeks, the plates were screened visually for putative nonmotile mutants displaying a compact, dense colony morphology. These colonies were transferred into and grown in liquid SN medium and examined by phase-contrast microscopy to confirm the loss of swimming motility in liquid. The absence of the E. coli donor strain was verified as described previously (5).
Plasmid rescue.
The plasposon pRL27 allows for one-step cloning of a transposon insertion and its flanking DNA (15). Chromosomal DNA was prepared from a 50-ml culture of a Synechococcus transconjugant as described previously (5). This purified DNA was digested with BamHI, which does not cut within the transposon sequence. The resulting fragments were ligated using T4 DNA ligase (Roche Applied Science, Indianapolis, IN) to generate a transposon junction plasmid consisting of the transposon and the flanking chromosomal DNA. The material from the ligation reaction was then electroporated into E. coli strain Transformax EC100D pir+ (Epicentre, Madison, WI) following the manufacturer's recommendations. Following electroporation, the E. coli isolate was plated on Luria-Bertani plates containing kanamycin. Plasmid DNA was isolated from the transformants using a QIAprep spin miniprep kit (QIAGEN, Valencia, CA). The site of insertion of the transposon was determined by sequencing, utilizing the outward-directed transposon-specific primer tpnRL 17-1 (5′-AACAAGCCAGGGATGTAACG-3′) (15). DNA sequencing was performed with Megabace reagents (Amersham, Piscataway, NJ) on a Megabace 500 sequencer.
Directed mutagenesis.
Directed inactivations were accomplished by cloning a completely internal fragment of a gene into the suicide vector pMUT100 as previously described (5). Twenty-mer oligonucleotide primers (Integrated DNA Technologies, Inc., Coralville, IA) were used to amplify DNA fragments (details given in Table 1) for cloning into pMUT100. These constructions were introduced into Synechococcus sp. strain WH8102 by conjugation with E. coli followed by subsequent selection of exconjugants on solidified media containing kanamycin. Clonal isolates were grown in liquid medium to confirm the mutant phenotype of the original transposon mutant. Complete segregation of mutant chromosomes was confirmed by Southern blotting and by PCR. For the PCR test, chromosomal DNA from the cyanobacterial mutant strain was isolated using a DNeasy tissue kit (QIAGEN, Valencia, CA) with the following modification: prior to proteinase K treatment, cells were incubated in a solution containing 20 mM Tris-Cl (pH 8.0), 2 mM Na2EDTA, 1.2% Triton X-100, and 50 mg/ml lysozyme for 30 min at 37°C. This DNA was then used as a template for PCR analysis utilizing primers flanking the fragment used for inactivation. Primer pairs used in these tests were confirmed to amplify a fragment of the expected size from wild-type DNA. Failure to amplify a fragment of the wild-type size from a mutant strain's DNA confirms the absence of the intact gene among the clonal population of mutant cells. This same DNA sample was used as a template in another PCR, utilizing primers directed to another open reading frame (ORF) as a positive control to confirm that the DNA was of sufficient quality for PCR amplification.
Genome information.
Complete genomic sequence information for Synechococcus sp. strain WH8102 is available at http://genome.ornl.gov/microbial/syn_wh/ (23).
RESULTS
Initial characterization.
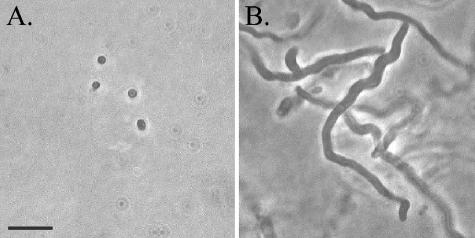
After an initial conjugation of Synechococcus sp. strain WH8102 with E. coli strain BW20767 (containing the plasposon delivery vector pRL27), 11 kanamycin-resistant transconjugants were selected randomly without application of a specific phenotypic screen. Following plasmid rescue, the site of transposon integration was determined for each strain by sequencing outwards from the transposon into flanking chromosomal DNA. The transposon insertion site was unique for each isolate. Genomic DNA isolated from these strains was also subjected to digestion with SmaI and to Southern analysis utilizing a probe designed against a portion of the kanamycin resistance gene present within the transposon. For each strain assayed, the probe hybridized with a single band consistent in size with that predicted by the genome sequence (data not shown). The majority of these strains did not have a phenotype that was obviously different from that of the wild-type strain, with a few exceptions. Two nonmotile strains were isolated and are described in greater detail below. The cells of another isolate, while still motile, were extremely elongated, ranging from 10 to 100 times the typical cell length (Fig. 1). In this strain, transposon insertion was into a putative septum site-determining gene, minD, which is present on the chromosome in the typical operon arrangement minCDE. The proteins of the minCDE system act in concert as a negative regulator of septation site formation in E. coli (26), and the phenotype of the Synechococcus minD mutant is consistent with a similar role for the Min proteins in Synechococcus. Directed mutagenesis at this locus confirmed the filamentous phenotype observed in the transposon mutant; nevertheless, these elongated cells retain the ability to swim.
FIG. 1.
Phase-contrast micrographs of wild-type WH8102 (A) and minD mutant (B) cells. Scale bar, 10 μm (both panels).
Nonmotile mutants.
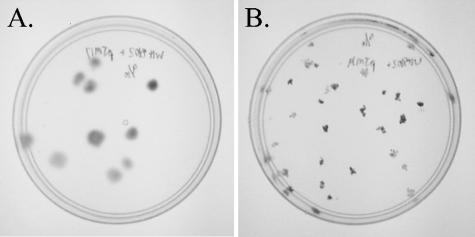
To screen for mutants with impaired ability to swim, a screen was developed to distinguish between motile and nonmotile isolates based on colony morphology. Solid SN medium prepared as described previously (5), with the single modification of reducing the agar concentration to 0.2%, allowed for the identification of nonmotile isolates. Nonmotile cells produce small, dense colonies while motile cells produce larger, more diffuse colonies (Fig. 2). Nonmotile isolates were grown for multiple transfers in liquid medium to confirm total loss of swimming motility.
FIG. 2.
Pour plates of Synechococcus sp. strain WH8102 transconjugants for isolation and screening of mutants. Shown are both a motile transconjugant (A) and a nonmotile transconjugant (B).
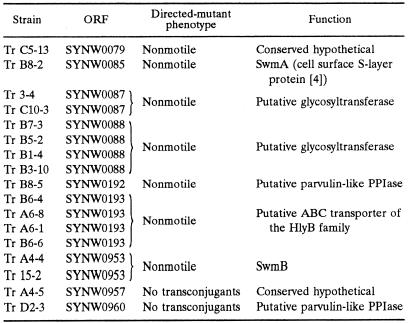
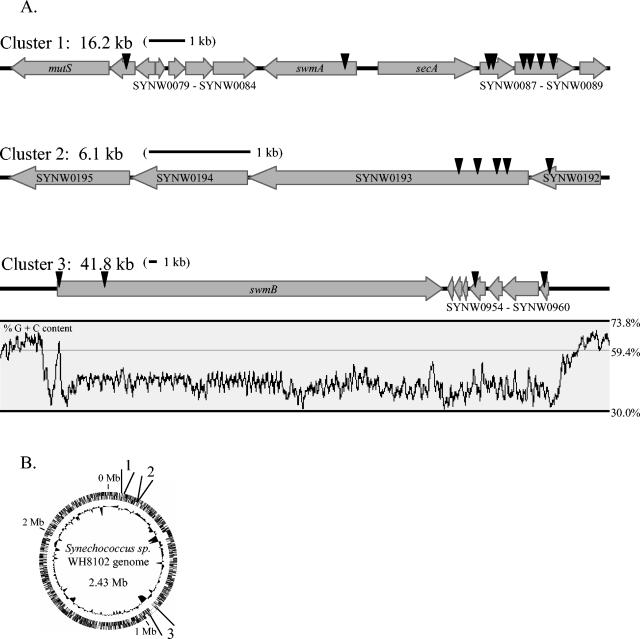
Utilizing this screen, 17 nonmotile transposon mutant strains were obtained (Table 2). These 17 independent transposon insertions are located in nine ORFs, which cluster into three separate regions of the genome (Fig. 3). Eight transposon insertions are located in a 12-kb region (cluster 1) that includes the previously identified motility gene swmA (5). Another five transposon insertions are tightly grouped into a 1.4-kb region that appears to be the beginning of an operon containing four genes (cluster 2). The last four insertions are spaced out over a 41-kb region that contains, among other ORFs, one large ORF of 32.4 kb (cluster 3).
TABLE 2.
Transposon insertions yielding nonmotile mutants
FIG. 3.
Chromosomal regions containing genes involved in swimming motility as identified by transposon mutagenesis. (A) Gene arrangement and location of each transposon insertion (arrowheads) are shown. A 1-kb scale bar is included for each cluster. Percent G+C content (window = 200 bp) is included for the chromosomal region encompassing cluster 3. (B) Location of clusters 1 to 3 on a circular chromosome. The outer ring contains predicted ORFs for both strands, and the inner ring shows associated G+C content (deviation from average).
To confirm that the phenotype observed for these mutants was the result of transposon insertion, directed mutations were constructed for each ORF identified by transposon mutagenesis as required for swimming motility. These directed mutations confirmed the motility phenotype of the original mutations. None of the mutants described here exhibits the attached rotating behavior of the swmA mutant. Two ORFs (SYNW0957 and SYNW0960) interrupted by transposon insertion were resistant to inactivation by directed mutagenesis. Multiple attempts were made to create a directed mutation near the site of transposon insertion of these problematic ORFs, and in no instance were we able to obtain transconjugants. DNA from the original transposon mutant isolates (strains Tr A4-5 and Tr D2-3) was analyzed by PCR, utilizing primers flanking the site of transposon insertion. Positive PCR amplification of the wild-type-sized product from these isolates confirmed that the original transposon mutants were incomplete segregants which still possessed a nondisrupted intact gene.
On the basis of their proximity to one another, some genes in which insertions occurred are likely to be arranged in operons, and hence polar effects on downstream genes are possible. The start codon of SYNW0088 is 5 bp from the stop codon of SYNW0087. Likewise, in cluster 2, the stop codon of SYNW0192 overlaps the start codon of SYNW0193, and the stop and start codons of the next two downstream ORFs are 2 and 5 bp apart, respectively. On the other hand, neither swmA (SYNW0085) nor SYNW0953 is transcribed with other genes, while the intergenic distances between SYNW0079, SYNW0957, SYNW0960, and SYNW0088 and their downstream genes (159 bp, 59 bp, 118 bp, and 123 bp, respectively) indicate that they are unlikely to be cotranscribed. Determining which, if not all, of ORFs SYNW0087, SYNW0192, SYNW0193, SYNW0194, and SYNW0195 are needed for motility will require complementation experiments. Unfortunately, these experiments are currently not possible with Synechococcus sp. strain WH8102, as kanamycin is the only selectable marker available. We are working to remedy this.
DISCUSSION
The transposon delivery vector pRL27 has been mobilized into a diverse array of bacterial species (15). As this was its first use in a marine Synechococcus sp., we report here on preliminary characterization of the behavior of this transposon delivery vector in this host. Sequence analysis of the first 11 isolated mutant strains indicates that each transposon insertion is unique, and Southern analysis of DNA isolated from these same mutant strains shows that for each strain, transposon insertion occurred only once, indicating that insertion of multiple transposons is likely to be rare.
Motile cells grown embedded in solidified media produce large, diffuse colonies among which nonmotile isolates can be identified by their small, dense colony morphology. Using this simple visual screen, we have isolated 17 nonmotile transposon mutants and characterized the site of transposon insertion for each. All motility genes identified to date cluster into three discrete regions on the chromosome. Various transporters and transport-related genes are present within these clusters. Both cluster 2 and cluster 3 contain a set of three genes that appear to encode a multicomponent transport apparatus. Moreover, these three genes are arranged in the same order in both clusters. Each set contains a gene for an ABC transporter of the protein-1 exporter (HlyB) family (SYNW0193 and SYNW0959) (27), a gene for a membrane fusion protein (SYNW0194 and SYNW0958) which is an auxiliary component possibly spanning the periplasm to connect the ABC transporter with an outer membrane component, and a gene whose product has limited similarity to peptidyl-prolyl isomerases (PPIases) (SYNW0192 and SYNW0960). Such PPIases have been shown to exhibit chaperone-like activity involved in the maturation of outer membrane porins in E. coli (2). Additionally, the SYNW0957 product shows very weak similarity to the MotA/TolQ/ExbB proton channel family (PFAM accession no. PF01618), which is also involved in protein translocation across both membranes of the gram-negative bacterial cell wall. In addition to multicomponent transporters, the products of several ORFs in these clusters (SYNW0087, SYNW0088, SYNW0084, and SYNW0195) show similarity to glycosyltransferases. Interestingly, the only previously known component of the motility apparatus, SwmA, is glycosylated (4) and has been shown to form an S-layer on the surface of these cells outside of the outer membrane (20). Both of these characteristics of SwmA correspond well with the types of genes found in the motility clusters. The presence of multiple transporters and multiple glycosyltransferases suggests that other components of the motility apparatus may be located outside the outer membrane and may be posttranslationally modified by glycosylation.
One exceptional gene present in cluster 3 (SYNW0953), hereinafter called swmB, is more than 32 kb long and comprises more than 1% of the total genome of this organism. While the function of SwmB is still unclear, preliminary experiments have shown that this protein is associated with the outer membrane (J. McCarren and B. Brahamsha, unpublished). More than half of the sequence of swmB is composed of regions of large tandem repeats. Database searches against the SwmB sequence indicate limited similarity to bacterial RTX proteins (named for repeats in toxin, which are secreted toxins that possess tandem copies of a nine-amino-acid motif) (34) and that this similarity is restricted to the repetitive portions of SwmB. While SwmB shares similarity with RTX proteins, SwmB does not actually contain the RTX repeat. Cluster 3 is also exceptional in its percent G+C content compared to that of the rest of the genome (Fig. 2). Synechococcus sp. strain WH8102 has a genome average G+C content of 59.4% (23) while the G+C content of cluster 3 is 42.2% and exhibits an abrupt departure from the genome average immediately before swmB and after SYNW0961. The dramatically different percent G+C contents suggest that these ORFs may have been acquired by a recent horizontal gene transfer event. While the ORFs in cluster 3 may be a recent acquisition, clusters 1 and 2 have percent G+C contents typical of Synechococcus sp. strain WH8102, suggesting that all of the components required for motility were not acquired in a single horizontal transfer event.
While swimming motility in marine Synechococcus spp. is distinct from the surface-associated gliding motility observed in other bacteria, it does share characteristics with the gliding motility observed in the Cytophaga-Flavobacteria-Bacteroides group, which occurs without slime extrusion or apparent appendages (17). Following chance attachment, both types of cells are observed to rotate about their point of attachment at a rate of 1 to 2 rotations per s (14, 35). Additionally, several parallels can be drawn between the motility genes of Flavobacterium johnsoniae identified in reports from the laboratory of McBride and coworkers (1, 13, 18, 19) and the types of genes identified in the data presented here. The first is the importance of transporters to both mechanisms of motility. Two sets of putative multicomponent ABC transporters have been identified in our current results. Similarly, several genes homologous to multicomponent ABC transporters (gldA, gldF, and gldG) and other transport-related genes (secDF) are all required for F. johnsoniae motility. A second similarity shared by both mechanisms of motility is that they employ PPIase-like proteins. While the PPIase-like ORF products required for Synechococcus and F. johnsoniae motility are from separate and evolutionarily unrelated families, the implication of functionally related proteins is of interest. Lastly, some genes with limited similarity to swmB have been implicated in Cytophaga-Flavobacteria-Bacteroides-type gliding motility. Recently, both a gene encoding an RTX autotransporter protein (T. Braun, S. Nelson, M. Uppal, and M. McBride, Abstr. Gen. Meet. Am. Soc. Microbiol. 2004, abstr. 104, 2004) and an exceptionally large and repetitive ORF (S. Nelson, personal communication) have been found to be involved in F. johnsoniae gliding motility. Also worthy of note in light of the low G+C content of cluster 3 genes, bacteria of the family Flavobacteriaceae have a characteristically low G+C content, ranging from 28 to 44% (3). How surface-associated gliding motility and nonflagellar swimming motility are related is difficult to envision, but the identification of similar types of genes involved in both phenomena suggests some possible relationship.
How the products of these recently identified genes function has yet to be determined. Exporting components of the motility apparatus to the cell surface appears to be important. In addition to allowing for the correct localization of the motility apparatus, perhaps these transporters play a more active role in generating swimming motility. Furthermore, a number of glycosyltransferases appear to be implicated. Whether these function to glycosylate protein components of the motility apparatus or whether they are involved in some other aspect of polysaccharide biosynthesis important for motility is unclear. Although how Synechococcus spp. are able to swim is still not understood, these findings have begun to identify the genes required for swimming motility.
Acknowledgments
We thank Rachel Larsen and William Metcalf for the gift of pRL27 and, along with Eric Webb, for helpful discussions.
This work was supported by grants NSF MCB97-27759 and DOE DE-FG03-O1ER63148.
REFERENCES
- 1.Agarwal, S., D. W. Hunnicutt, and M. J. McBride. 1997. Cloning and characterization of the Flavobacterium johnsoniae (Cytophaga johnsonae) gliding motility gene, gldA. Proc. Natl. Acad. Sci. USA 94:12139-12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behrens, S., R. Maier, H. de Cock, F. X. Schmid, and C. A. Gross. 2001. The SurA periplasmic PPIase lacking its parvulin domains functions in vivo and has chaperone activity. EMBO J. 20:285-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernardet, J. F., Y. Nakagawa, B. Holmes, et al. 2002. Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int. J. Syst. Evol. Microbiol. 52:1049-1070. [DOI] [PubMed] [Google Scholar]
- 4.Brahamsha, B. 1996. An abundant cell-surface polypeptide is required for swimming by the nonflagellated marine cyanobacterium Synechococcus. Proc. Natl. Acad. Sci. USA 93:6504-6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brahamsha, B. 1996. A genetic manipulation system for oceanic cyanobacteria of the genus Synechococcus. Appl. Environ. Microbiol. 62:1747-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brahamsha, B. 1999. Genetic manipulations in Synechococcus spp. of marine cluster A. Bull. Inst. Oceanogr. (Monaco) 19:517-527. [Google Scholar]
- 7.Brahamsha, B. 1999. Non-flagellar swimming in marine Synechococcus. J. Mol. Microbiol. Biotechnol. 1:59-62. [PubMed] [Google Scholar]
- 8.Casadaban, M., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 9.Ehlers, K. M., A. D. T. Samuel, H. C. Berg, and R. Montgomery. 1996. Do cyanobacteria swim using traveling surface waves? Proc. Natl. Acad. Sci. USA 93:8340-8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elhai, J., and C. P. Wolk. 1988. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 167:747-754. [DOI] [PubMed] [Google Scholar]
- 11.Gilad, R., A. Porat, and S. Trachtenberg. 2003. Motility modes of Spiroplasma melliferum. Mol. Microbiol. 47:657-669. [DOI] [PubMed] [Google Scholar]
- 12.Hoiczyk, E. 2000. Gliding motility in cyanobacteria: observations and possible explanations. Arch. Microbiol. 174:11-17. [DOI] [PubMed] [Google Scholar]
- 13.Hunnicutt, D. W., M. J. Kempf, and M. J. McBride. 2002. Mutations in Flavobacterium johnsoniae gldF and gldG disrupt gliding motility and interfere with membrane localization of GldA. J. Bacteriol. 184:2370-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lapidus, I. R., and H. C. Berg. 1982. Gliding motility of Cytophaga sp. strain U67. J. Bacteriol. 151:384-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsen, R. A., M. M. Wilson, A. M. Guss, and W. W. Metcalf. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178:193-201. [DOI] [PubMed] [Google Scholar]
- 16.McBride, M. J. 2001. Bacterial gliding motility: Multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 55:49-75. [DOI] [PubMed] [Google Scholar]
- 17.McBride, M. J. 2004. Cytophaga-flavobacterium gliding motility. J. Mol. Microbiol. Biotechnol. 7:63-71. [DOI] [PubMed] [Google Scholar]
- 18.McBride, M. J., and T. F. Braun. 2004. GldI is a lipoprotein that is required for Flavobacterium johnsoniae gliding motility and chitin utilization. J. Bacteriol. 186:2295-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McBride, M. J., T. F. Braun, and J. L. Brust. 2003. Flavobacterium johnsoniae GldH is a lipoprotein that is required for gliding motility and chitin utilization. J. Bacteriol. 185:6648-6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCarren, J., J. Heuser, R. Roth, N. Yamada, M. Martone, and B. Brahamsha. 2005. Inactivation of swmA results in the loss of an outer cell layer in a swimming Synechococcus strain. J. Bacteriol. 187:224-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metcalf, W. W., W. Jiang, L. L. Daniels, S.-K. Kim, A. Haldimann, and B. L. Wanner. 1996. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1-13. [DOI] [PubMed] [Google Scholar]
- 22.Meyer, R., D. Figurski, and D. R. Helinski. 1977. Physical and genetic studies with restriction endonucleases on the broad host range plasmid RK2. Mol. Gen. Genet. 152:129-135. [DOI] [PubMed] [Google Scholar]
- 23.Palenik, B., B. Brahamsha, F. W. Larimer, M. Land, L. Hauser, P. Chain, J. Lamerdin, W. Regala, E. E. Allen, J. McCarren, I. Paulsen, A. Dufresne, F. Partensky, E. A. Webb, and J. Waterbury. The genome of a motile marine Synechococcus. Nature 424:1037-1042, 2003. [DOI] [PubMed]
- 24.Pitta, T. P., and H. C. Berg. 1995. Self-electrophoresis is not the mechanism for motility in swimming cyanobacteria. J. Bacteriol. 177:5701-5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price, N. M., G. I. Harrison, J. G. Hering, R. J. Hudson, P. M. V. Nirel, B. Palenik, and F. M. M. Morel. 1989 1988. Preparation and chemistry of the artificial algal culture medium Aquil. Biol. Oceanogr. 6:443-461. [Google Scholar]
- 26.Rothfield, L. I., Y. L. Shih, and G. King. 2001. Polar explorers: membrane proteins that determine division site placement. Cell 106:13-16. [DOI] [PubMed] [Google Scholar]
- 27.Saier, M. H., Jr. 1999. A functional-phylogenetic system for the classification of transport proteins. J. Cell. Biochem. 1999(Suppl. 32-33):84-94. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Scanlan, D. J., and N. J. West. 2002. Molecular ecology of the marine cyanobacterial genera Prochlorococcus and Synechococcus. FEMS Microbiol. Ecol. 40:1-12. [DOI] [PubMed] [Google Scholar]
- 30.Waterbury, J. B., S. W. Watson, R. R. L. Guillard, and L. E. Brand. 1979. Widespread occurrence of a unicellular, marine, planktonic, cyanobacterium. Nature 277:293-294.16069038 [Google Scholar]
- 31.Waterbury, J. B., S. W. Watson, R. L. Guillard, and L. E. Brand. 1986. Biological and ecological characterization of the marine unicellular cyanobacterium Synechococcus. Can. Bull. Fish. Aquat. Sci. 214:71-120. [Google Scholar]
- 32.Waterbury, J. B., and J. M. Willey. 1988. Isolation and growth of marine planktonic cyanobacteria. Methods Enzymol. 167:100-105. [Google Scholar]
- 33.Waterbury, J. B., J. M. Willey, D. G. Franks, F. W. Valois, and S. W. Watson. 1985. A cyanobacterium capable of swimming motility. Science 230:74-76. [DOI] [PubMed] [Google Scholar]
- 34.Welch, R. A., C. Forestier, A. Lobo, S. Pellett, W. Thomas, Jr., and G. Rowe. 1992. The synthesis and function of the Escherichia coli hemolysin and related RTX exotoxins. FEMS Microbiol. Immunol. 5:29-36. [DOI] [PubMed] [Google Scholar]
- 35.Willey, J. M. 1988. Characterization of swimming motility in a marine cyanobacterium. Ph.D. thesis. Woods Hole Oceanographic Institution and Massachusetts Institute of Technology, Cambridge, Mass.
- 36.Wolgemuth, C. W., O. Igoshin, and G. Oster. 2003. The motility of mollicutes. Biophys. J. 85:828-842. [DOI] [PMC free article] [PubMed] [Google Scholar]