Abstract
Pseudomonas aeruginosa is an opportunistic pathogen that causes chronic lung infections in cystic fibrosis patients and is a major source of nosocomial infections. This bacterium controls many virulence factors by using two quorum-sensing systems, las and rhl. The las system is composed of the LasR regulator protein and its cell-to-cell signal, N-(3-oxododecanoyl) homoserine lactone, and the rhl system is composed of RhlR and the signal N-butyryl homoserine lactone. A third intercellular signal, the Pseudomonas quinolone signal (PQS; 2-heptyl-3-hydroxy-4-quinolone), also regulates numerous virulence factors. PQS synthesis requires the expression of multiple operons, one of which is pqsABCDE. Previous experiments showed that the transcription of this operon, and therefore PQS production, is negatively regulated by the rhl quorum-sensing system and positively regulated by the las quorum-sensing system and PqsR (also known as MvfR), a LysR-type transcriptional regulator protein. With the use of DNA mobility shift assays and β-galactosidase reporter fusions, we have studied the regulation of pqsR and its relationship to pqsA, lasR, and rhlR. We show that PqsR binds the promoter of pqsA and that this binding increases dramatically in the presence of PQS, implying that PQS acts as a coinducer for PqsR. We have also mapped the transcriptional start site for pqsR and found that the transcription of pqsR is positively regulated by lasR and negatively regulated by rhlR. These results suggest that a regulatory chain occurs where pqsR is under the control of LasR and RhlR and where PqsR in turn controls pqsABCDE, which is required for the production of PQS.
Pseudomonas aeruginosa is a ubiquitous gram-negative bacterium that can infect insects, plants, and animals. As an opportunistic pathogen of humans, P. aeruginosa causes acute infections in immunocompromised individuals and chronic lung infections in cystic fibrosis patients. Such infections are made possible through the production of an arsenal of virulence factors, many of which are regulated by cell-to-cell signals (see reference 36 for a review). P. aeruginosa produces at least three small compounds that function as intercellular communication signals. The acyl homoserine lactone signals, N-(3-oxododecanoyl) homoserine lactone (3-oxo-C12-HSL) and N-butyryl homoserine lactone (C4-HSL), have been well studied and function in combination with the LuxR homologs LasR and RhlR, respectively (16, 30, 31, 33, 34). Together, these quorum-sensing signals control 6 to 11% of the P. aeruginosa genome (44, 46, 48). The third P. aeruginosa intercellular signal is a quinolone compound that was identified as 2-heptyl-3-hydroxy-4-quinolone (the Pseudomonas quinolone signal [PQS]) (37). This signal controls multiple virulence factors and is intertwined in the quorum-sensing cascade, where it appears to be a regulatory link between the las and rhl quorum-sensing systems (14, 27). PQS is produced in the lungs of cystic fibrosis patients infected with P. aeruginosa (9) and is required for virulence in nematodes, plants, and mice (5, 15, 23, 25, 40). PQS also induces apoptosis and decreases viability in eukaryotic cells (4).
The synthesis of PQS requires several putative enzymes encoded by the pqsABCDE, phnAB, and pqsH operons (10, 15). In addition, PqsR (also known as MvfR [5]) is a LysR-type regulator required for the synthesis of PQS (10, 15) and at least 55 related 4-quinolone compounds (24). A recent genomics study indicated that 143 genes were differentially regulated in a pqsR mutant (12), demonstrating the global regulatory nature of the PQS system. Initial studies on the expression of the PQS synthetic genes indicated that the genes are governed by a complex regulatory scheme. PqsR was found to have a positiveeffect on the transcription of the pqsABCDE and phnAB operons (5, 26). In addition, the transcription of the pqsABCDE promoter was positively regulated by LasR-3-oxo-C12-HSL and negatively regulated by RhlR-C4-HSL (26). Whether the LasR, RhlR, and PqsR transcriptional regulators were acting in a direct or indirect manner to control pqsABCDE expression was not determined. In this study, the interactions that occur at the pqsABCDE promoter were investigated. We demonstrate that the effects of LasR and RhlR on the pqsABCDE operon occur indirectly through PqsR. We also show that PqsR, but not LasR or RhlR, binds directly to the pqsABCDE promoter and that this binding is augmented by the presence of PQS.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids used in this study are listed in Table 1. P. aeruginosa strains were maintained at −70°C in 10% skim milk (Becton Dickinson, Sparks, MD). Freshly plated cells from skim milk stocks were used to begin all experiments. P. aeruginosa cultures were grown in peptone tryptic soy broth (32). When necessary to maintain plasmids, cultures were supplemented with 200 μg/ml carbenicillin. Escherichia coli strain DH5α was cultured in Luria-Bertani broth (42) and supplemented with 100 μg/ml ampicillin and/or 30 μg/ml chloramphenicol to maintain plasmids. Liquid cultures were grown at 37°C and shaken at 250 rpm.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or phenotype | Reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F′/endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF) U169 deoR [φ80 dlac Δ(lacZ)M15 recA1] | 49 |
| P. aeruginosa strains | ||
| PAO1 | Wild type | 19 |
| MP551 | pqsR::TnIS phoA/hah::Tc; derived from strain PAO1 | 15 |
| PAO-R1 | lasR::Tc; derived from strain PAO1 | 17 |
| PDO111 | rhlR::Tn501; derived from strain PAO1 | 2 |
| Plasmids | ||
| pACYC184 | General purpose cloning vector | 6 |
| pPCS11 | tacp-lasR on pACYC184 | 38 |
| pECP8 | tacp-lasR on pEX1.8 | This study |
| pJPP8 | tacp-rhlR on pEX1.8 | 35 |
| pDSW8 | tacp-pqsR on pEX1.8 | This study |
| pMTP58 | Minimum tiling pathway cosmid 58 which contains pqsR; Tetr | 20 |
| pLP170 | lacZ transcriptional fusion vector | 39 |
| pMWC1003 | pqsR′-lacZ transcriptional fusion | This study |
| pEX1.8 | P. aeruginosa expression vector | 35 |
Construction of plasmids.
To obtain a PqsR expression plasmid, PCR was used to generate a 1,039-bp DNA fragment that began at the pqsR start codon (ATG) and ended 36 bp downstream from the stop codon. The oligonucleotide primers for PCR were engineered to contain a blunt end at the start codon and a single HindIII site downstream from the stop codon. Plasmid pEX1.8, a tacp expression vector with both an E. coli and a P. aeruginosa origin of replication, was digested with EcoRI, treated with Klenow fragment to fill in the 5′ overhang, and then digested with HindIII. After digestion with HindIII, the pqsR-containing fragment was ligated into pEX1.8 to yield pDSW8. Plasmid pECP8 was constructed using the same strategy by ligating a PCR-derived DNA fragment that contains the coding region of lasR into pEX1.8. Plasmids pDSW8 and pECP8 contain a tacp-pqsR and a tacp-lasR fusion, respectively, in which there is optimal spacing between the ribosome binding site from pEX1.8 and the start codon of pqsR or lasR. To construct a pqsR′-lacZ reporter plasmid, a 924-bp fragment corresponding to bp −618 to +210 relative to the pqsR translational start site was amplified by PCR with oligonucleotide primers and P. aeruginosa strain PAO1 chromosomal DNA. The amplified product was digested with BamHI to yield a 698-bp fragment that was ligated into the BamHI-digested lacZ fusion vector pLP170. The resulting plasmid, pMWC1003, harbors a pqsR′-lacZ transcriptional fusion. All gene fusions were sequenced to ensure cloning integrity.
Preparation of E. coli cell lysates containing PqsR, LasR, or RhlR.
Overnight cultures of E. coli strain DH5α containing expression vectors (pDSW8, pECP8, or pJPP8) were subcultured to an absorbance of 0.1 at 600 nm. When desired, a specific cell-to-cell signal or an organic extract of a wild-type P. aeruginosa culture was dried in flasks before the subculture was added. Final concentrations of signals were as follows: 10 μM 3-oxo-C12-HSL, 10 μM C4-HSL, and 20 μM PQS. The organic extract was prepared by extracting a 24-h P. aeruginosa culture with acidified ethyl acetate (37). The final amount of organic extract resuspended in the subculture was equivalent to twice the subculture volume (e.g., an entire extract from a 20-ml culture extraction was resuspended in a 10-ml subculture). Subcultures were then grown for 2 h, and IPTG (isopropyl-β-d-thiogalactoside) was added at a concentration of 1 mM to induce the tacp-pqsR, tacp-lasR, or tacp-rhlR fusion. After two additional hours of growth, cells were harvested and passed through a French pressure cell at 735 lb/in2 to yield a whole-cell lysate. Protein assays (Bio-Rad, Hercules, CA) were performed on cell lysates to determine protein concentrations for DNA mobility shift assays.
DNA mobility shift assays.
PCR was used to generate DNA fragments containing the pqsA (253 bp), lasB (255 bp), and rhlA (216 bp) promoter regions. DNA fragments were labeled with [γ-32P]ATP (Perkin-Elmer, Wellesley, MA) by using T4 polynucleotide kinase (Invitrogen, Carlsbad, CA). The binding assays were carried out in buffer containing 10 mM Tris-HCl (pH 8.0), 60 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, and 10% glycerol (28). Each reaction mixture contained 0.3 μg of salmon sperm DNA, 104-cpm-radiolabeled DNA, and 0 to 60 μg of protein. Reaction mixtures were incubated at room temperature for 20 min and separated by electrophoresis at 4°C on a native 6% polyacrylamide gel in 0.5× Tris-borate-EDTA buffer. Radiolabeled bands were visualized by autoradiography. X-ray film was exposed for either a short period (approximately 2 to 3 h) or a longer period (24 or 72 h) as noted in Results.
Primer extension analysis of the pqsR transcript.
RNA was purified from P. aeruginosa strain PAO1(pMTP58) by a CsCl density gradient separation technique as described previously (26). Cosmid pMTP58 was included to increase the number of copies of pqsR mRNA. Two primers were used to locate the pqsR transcriptional start site. The primers were as follows: 5′-CACGTGATTCAGGTTATGAATAGGCA-3′, which corresponds to nucleotides +27 to +2 relative to the pqsR start codon, and 5′-AGCGGAGGAAATCGAACCGGAGGCGA-3′, which corresponds to nucleotides +72 to +46 relative to the pqsR start codon. Primers were radiolabeled using [γ-32P]ATP and T4 polynucleotide kinase (Invitrogen), and extensions were performed as described previously (26) using 40 μg of RNA and Superscript II RNase H− reverse transcriptase (Invitrogen). Primer extension reaction mixtures were electrophoresed on a sequencing gel along with mixtures for DNA sequencing reactions completed using a T7 Sequenase version 2.0 DNA sequencing kit (USB) and pMTP58 as a template.
Monitoring pqsR′-lacZ expression.
Freshly plated cells of P. aeruginosa strains PAO1, PAO-R1, PDO111, and MP551, all containing pMWC1003, were used to inoculate 10-ml overnight cultures. Overnight cultures were washed with fresh media and used to inoculate 10-ml subcultures to an absorbance of 0.05 at 660 nm. Subcultures were grown to mid-logarithmic phase and washed with fresh media, and 1.0-ml aliquots (starting at an A660 of 0.05) were added to 13-ml culture tubes. After 24 h of growth, β-galactosidase (β-Gal) activity was measured in duplicate samples. All data are reported in Miller units (29) as the means ± the standard deviations (σn−1) of results from three separate experiments.
Monitoring pqsR′-lacZ expression in E. coli.
Freshly plated cells of E. coli strain DH5α harboring plasmid pMWC1003 and either pPCS11 or pACYC184 were used to inoculate overnight cultures in A medium (42) supplemented with 25 mM glucose and 1 mM MgSO4. Overnight cultures were used to inoculate subcultures to an absorbance of 0.08 at 600 nm and grown for 3 h in the presence of 1 mM IPTG. Cultures were then transferred into tubes containing evaporated 3-oxo-C12-HSL (or no 3-oxo-C12-HSL as a control) and were grown for an additional 90 min (the final 3-oxo-C12-HSL concentration was 100 nM). β-Gal activity was then measured in duplicate samples. All data are reported in Miller units as the means ± the standard deviations (σn−1) of results from three separate experiments.
PQS analysis.
Freshly plated P. aeruginosa cultures were used to inoculate 1-ml cultures for overnight growth. Cultures were extracted with acidified ethyl acetate as previously described (3). Extracts were dried, resuspended in a small volume of 1:1 ethyl acetate-acetonitrile, and separated by thin-layer chromatography (TLC) as described previously (37). Resolved TLC plates were photographed under long-wave UV light.
RESULTS
LasR and RhlR do not interact with the pqsA promoter.
Previously, it was shown that the transcription of pqsA is positively controlled by the las quorum-sensing system and negatively controlled by the rhl quorum-sensing system and that these two systems appear to compete for regulatory effects (26). These results, along with the discovery of two putative quorum-sensing operator sequences upstream from the pqsA promoter (26), suggested that regulation by and competition between the two systems may be occurring in this region. To determine whether LasR and/or RhlR binds the pqsA promoter, DNA mobility shift assays were performed in both the presence and absence of each protein's respective cell-to-cell signal. Surprisingly, neither LasR nor RhlR interacted with the pqsA promoter. In the presence or absence of 3-oxo-C12-HSL, LasR caused no shift in DNA mobility for the pqsA promoter (Fig. 1A). To ensure that our LasR-containing lysate would interact with a specific DNA fragment, we showed that the lysate caused an interaction with the lasB promoter (Fig. 1C). The two different shifted complexes seen in this control assay were similar to that seen by Schuster et al. (45) and indicated that our cell lysate contained active LasR. Similarly, in the presence or absence of C4-HSL, RhlR did not produce a shift in DNA mobility for the pqsA promoter (Fig. 1B). In a control experiment, the RhlR-containing cell lysate caused a mobility shift of the rhlA promoter in the absence (Fig. 1D) and presence (data not shown) of C4-HSL. While this shift was small, it was comparable to that reported by Medina et al. (28) and indicated that the cell lysate contained active RhlR.
FIG. 1.
Neither LasR nor RhlR binds the pqsA promoter. (A) Radiolabeled DNA containing the pqsA promoter region was added to a LasR-containing cell lysate that was prepared in the absence (lanes 1 to 5) or presence (lanes 6 to 10) of 10 μM 3-oxo-C12-HSL (C12). (B) Radiolabeled DNA from the pqsA promoter region was added to an RhlR-containing cell lysate prepared in the absence (lanes 1 to 5) or presence (lanes 6 to 10) of 10 μM C4-HSL (C4). (C) Radiolabeled DNA from the lasB promoter region was added to a LasR-containing cell lysate prepared in the presence of 10 μM 3-oxo-C12-HSL. (D) Radiolabeled DNA from the rhlA promoter region was added to an RhlR-containing cell lysate prepared in the absence of C4-HSL. The total protein concentration of each lysate was determined, and the following amounts of protein were added per reaction: lanes 1 and 6, 0 μg; lanes 2 and 7, 10 μg; lanes 3 and 8, 20 μg; lanes 4 and 9, 40 μg; and lanes 5 and 10, 60 μg. Total binding reaction mixtures were then electrophoresed on nondenaturing 6% polyacrylamide gels. Gels were dried, and overlaid X-ray film was exposed for approximately 2 or 24 h before development. Results presented are from film exposed for 2 h, which showed the same number of bands as film exposed for at least 24 h (data not shown).
PqsR interacts with the pqsA promoter.
It was shown previously that pqsA promoter activity requires the presence of pqsR (26). This gene, which is required for PQS synthesis (10, 15), is homologous to members of the LysR-type transcriptional regulator family. Many LysR-type transcriptional activators contain a conserved DNA binding domain and a variable coinducer binding domain (see reference 43 for a review). These transcription factors typically bind a promoter region in the absence of a coinducer and bind a nearby site in the presence of a coinducer (43). Others have shown that PqsR interacts with the phnAB promoter in the absence of a coinducer and that pqsR positively regulates the expression of phnAB (5). Because pqsA expression requires pqsR, it seemed likely that PqsR was interacting with the pqsA promoter. To test this hypothesis, E. coli cell lysates containing PqsR were used in DNA mobility shift assays with the pqsA promoter region as described above. As we suspected, PqsR interacted with the pqsA promoter, thereby causing a shift in DNA mobility (Fig. 2A). This interaction occurred in the absence of any P. aeruginosa-produced factors which presumably would mean that the PqsR coinducer was not present. To determine whether the interaction with the pqsA promoter could be augmented by a coinducer, E. coli strain DH5α(pDSW8) was grown in the presence of an ethyl acetate extract of culture supernatant from wild-type P. aeruginosa (strain PAO1). Most interestingly, the PqsR-containing lysate from these cells caused a much stronger DNA shift than that from cells which were grown in the absence of the organic extract (Fig. 2B). This shift was clearly visible when the gel had been exposed to X-ray film for only 3 h, while the shift with PqsR alone was not visible (Fig. 2B). (The autorad shown in Fig. 2A was exposed for 72 h in order to detect the mobility shift.) These data indicated that the PqsR coinducer was contained in the ethyl acetate extract of the P. aeruginosa culture supernatant. Since PQS is present in such an extract (along with many other compounds) and PqsR controls PQS production, we speculated that PQS would be a likely candidate for a coinducer of PqsR. To determine if this was correct, E. coli strain DH5α(pDSW8) was grown in the presence of synthetic PQS in order to prepare a PqsR-containing cell lysate for the DNA mobility shift assay. Most excitingly, we found that in the presence of PQS, the interaction of PqsR with the pqsA promoter region was greatly enhanced (Fig. 2C). As in Fig. 2B, the autorad presented in Fig. 2C was exposed to X-ray film for only 3 h, and the majority of labeled DNA had shifted to a higher molecular weight. Extracts prepared in the absence of PQS did not produce a detectable mobility shift when X-ray film was exposed for 3 h (Fig. 2C). This result showed that PQS greatly enhanced the interaction between PqsR and a target promoter (pqsA), thereby implying that PQS acts as a coinducer for PqsR. Control experiments also showed that the addition of excess unlabeled competitor DNA (a P. aeruginosa promoter that is not controlled by PqsR) at various concentrations that were up to 200-fold higher than the concentration of the radiolabeled pqsA promoter had no effect on the mobility of pqsA in the presence of PqsR and PQS (data not shown). This indicated that the interaction between the PqsR-PQS complex and the pqsA promoter was specific. Finally, it can be noted that a minor shifted band was observed above the major shifted complex on autorads exposed for longer times when PqsR was prepared in the presence of P. aeruginosa culture extract or synthetic PQS (data not shown). Such a result suggests that, as seen with another LysR homolog (7), PqsR and the PqsR-PQS complex bind to different locations in the pqsA promoter region.
FIG. 2.
Binding of PqsR to the pqsA promoter. Radiolabeled DNA containing the pqsA promoter was incubated with E. coli cell lysates containing PqsR. PqsR-containing cell lysate was prepared in the absence (lanes 1 to 5 of panels A, B, and C) or the presence of an ethyl acetate extract of a P. aeruginosa culture (lanes 6 to 10 of panel B) or 20 μM PQS (lanes 6 to 10 of panel C). The total protein concentration of each lysate was determined, and the following amounts of protein were added per reaction: lanes 1 and 6, 0 μg; lanes 2 and 7, 10 μg; lanes 3 and 8, 20 μg; lanes 4 and 9, 40 μg; and lanes 5 and 10, 60 μg. Total binding reaction mixtures were then electrophoresed on nondenaturing 6% polyacrylamide gels. Gels were dried, and overlaid X-ray film was exposed for approximately 3 (B and C) or 72 (A) h.
pqsR is controlled by the las and rhl quorum-sensing systems.
The data presented in Fig. 1 and 2 indicate that pqsA is controlled indirectly by LasR and RhlR and directly by PqsR. We suspected that the indirect effects of LasR and RhlR on pqsA expression may be occurring through PqsR. Before constructing a pqsR′-lacZ transcriptional fusion to study pqsR regulation, we mapped the pqsR transcriptional start site through primer extension analysis. These experiments showed that two primary transcriptional start sites existed for pqsR (Fig. 3A and B). The two sites were 190 and 278 bp upstream from the pqsR ATG start codon, respectively. The results of repeat experiments with the same primer are presented in Fig. 3. A second primer was used in a separate primer extension analysis, and this analysis resulted in the identification of the same start sites (data not shown). A minor primer extension product also appeared between the mapped sites in only one experiment (Fig. 3B), but the significance of this product is not known. The transcriptional start site that is farther upstream contains an appropriately located region that matched five out of six bases of the consensus sequences for both the −35 and −10 regions of a σ70-type promoter (Fig. 3C). No sequences similar to known promoter consensus sequences could be identified for the transcriptional start site that is closer to the pqsR start codon.
FIG. 3.
Mapping the pqsR start of transcription. (A) Primer extension analysis of the pqsR transcript. Sequencing reaction mixtures are labeled according to nucleotide (A, C, G, or T), and lane P contains the mixture for the primer extension reaction performed with RNA isolated from P. aeruginosa strain PAO1(pMTP58). Extension products are labeled TS1 and TS2. (B) Repeat of primer extension analysis as described for panel A. Panel B is presented to show that TS1 and TS2 are at the same locations as in panel A. (C) Promoter region of pqsR. The pqsR transcriptional start sites are indicated by bent arrows at TS1 and TS2, and the pqsR ATG start codon is underlined. The −35 and −10 regions of a potential σ70-type promoter upstream from TS1 are boxed and labeled. A putative quorum-sensing operator sequence (47) is also boxed, with highly conserved nucleotides in bold.
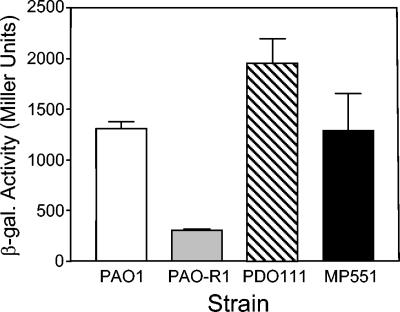
To determine the relationship between LasR, RhlR, and PqsR, we monitored the expression of a pqsR′-lacZ transcriptional fusion in strains PAO1 (wild type), PAO-R1 (lasR), PDO111 (rhlR), and MP551 (pqsR). Interestingly, the data showed that the wild-type strain PAO1 produced 1,308 ± 68 Miller units of activity from the pqsR′-lacZ fusion and that the lasR mutant produced only 303 ± 14 Miller units of activity (Fig. 4). This indicated that lasR controlled pqsR transcription in a positive manner. To confirm that this control was happening in a direct manner, we examined the effect of LasR and 3-oxo-C12-HSL on pqsR expression in E. coli. The resulting data showed that in the presence of only LasR, pqsR′-lacZ was expressed at a level similar to that of the background β-Gal activity produced by the parent vector control (Fig. 5). However, when 3-oxo-C12-HSL was added in the presence of LasR, pqsR′-lacZ expression greatly increased (Fig. 5). This showed that LasR and 3-oxo-C12-HSL were required and sufficient for pqsR induction in E. coli, which indicated that their effect on pqsR is direct.
FIG. 4.
Transcriptional regulation of pqsR in P. aeruginosa. P. aeruginosa strains PAO1 (wild type), PAO-R1 (lasR), PDO111 (rhlR), and MP551 (pqsR) containing plasmid pMWC1003 (pqsR′-lacZ) were cultured as described in Materials and Methods and assayed for β-Gal activity. Data are presented in Miller units as the means ± σn−1 of results from duplicate assays from three separate experiments.
FIG. 5.
LasR and 3-oxo-C12-HSL induce pqsR expression in E. coli. Subcultures were grown for 90 min in the presence or absence of 3-oxo-C12-HSL, and duplicate β-Gal activity assays were performed. Data are presented in Miller units as the means ± σn−1 of results from three separate experiments. Lane 1, E. coli strain DH5α(pMWC1003, pPCS11) without 3-oxo-C12-HSL; lane 2, E. coli strain DH5α(pMWC1003, pPCS11) with 3-oxo-C12-HSL; lane 3, E. coli strain DH5α(pMWC1003, pACYC184) (control).
The data presented in Fig. 4 also show that the expression of pqsR′-lacZ in an rhlR mutant, strain PDO111, produced 1,958 ± 238 Miller units, which is an increase (approximately 1.5 times higher) in expression over that seen in the wild-type strain PAO1. Although the increase in expression seen in strain PDO111 was minor, it nevertheless demonstrated that rhlR can have a negative regulatory effect on pqsR transcription. This supports previous data which showed that the rhl quorum-sensing system represses pqsA transcription (26). These data could not be confirmed as described above in an E. coli bioassay due to the high level of background β-Gal activity produced by the expression vector in E. coli (Fig. 5 and data not shown). Finally, we found that pqsR is not autoregulated at the transcriptional level under the conditions tested. The expression of pqsR′-lacZ in the pqsR mutant, strain MP551, was very similar to that seen in the parent strain, PAO1 (Fig. 4). Overall, our results indicated that pqsR is positively regulated by lasR and may be regulated by rhlR in a negative manner. Taken together with the previous report on the regulation of the pqsA promoter by lasR, rhlR, and pqsR, these data suggest that a regulatory chain occurs where pqsR is under the control of lasR and rhlR and, in turn, pqsR controls the pqsABCDE operon, which is required for the production of PQS.
PqsR complements PQS production in a lasR mutant.
The data discussed above imply that the decreased production of PQS in a lasR mutant should be complemented by the expression of pqsR. To investigate this hypothesis and connect our findings to a phenotype, we constructed a pqsR expression plasmid (pDSW8) that contains pqsR controlled by the tacp promoter (see Materials and Methods). This plasmid was used to transform P. aeruginosa strain PAO-R1, and PQS production was monitored by TLC analysis of ethyl acetate extracts of the P. aeruginosa cultures. As shown previously (37), the lasR mutant (containing a control plasmid) does not make a detectable amount of PQS under these conditions (Fig. 6, lane 3). However, when strain PAO-R1 contained pDSW8, PQS was produced at a level comparable to that in the wild-type strain PAO1 (Fig. 6, lanes 2 and 4). These data agree with those of Diggle et al. (14), who showed that PQS production is dependent on factors in addition to LasR. The data presented in Fig. 6 demonstrate that the expression of PqsR complemented the production of PQS in a lasR mutant and provide additional evidence to support our conclusion that pqsR is controlled by LasR in P. aeruginosa.
FIG. 6.
PqsR complements PQS production in a lasR mutant. Ethyl acetate extracts of the indicated cultures were separated by TLC and visualized under UV light. Lanes 1 and 5, synthetic PQS; lane 2, extract from strain PAO1; lane 3, extract from strain PAO-R1(pEX1.8); lane 4, extract from strain PAO-R1(pDSW8).
DISCUSSION
This research began with a desire to learn more about the regulation of PQS production, with a specific focus on the control of the pqsABCDE operon. Our data quickly led us into a study that included multiple transcriptional regulators that were found to be working in a regulatory chain to control PQS production. Previous data had indicated that LasR and RhlR were having a positive and a negative effect, respectively, on the pqsA promoter (26). The pqsA promoter region had been identified previously (26), and we used these data to develop a DNA mobility shift assay to monitor interactions that occurred in the pqsA regulatory region. Our initial data showed that neither LasR nor RhlR, with or without their specific activating signals, would interact with the pqsA promoter (Fig. 1). This finding suggested that these regulators may instead be controlling another regulator that affects pqsA. We hypothesized that PqsR would be the most likely intermediate regulator in this chain. PqsR is a LysR homolog that has been shown to control both the pqsABCDE and phnAB operons (5, 26). LysR homologs usually function in conjunction with a coinducer and interact with a DNA region of dyad symmetry near the −35 region of a promoter (43). In the absence of a coinducer, LysR-type regulators also interact with a DNA sequence approximately 30 bp upstream from the −35 region of a promoter (43). Our data first showed that PqsR alone would interact with the pqsA promoter region in a relatively inefficient manner (Fig. 2). However, the presence of PQS greatly enhanced the interaction of PqsR with the pqsA promoter region (Fig. 2). This exciting result amounts to the discovery of a target protein for PQS and implies that PQS is a coinducer for PqsR. Such a finding is an important step in the elucidation of how PQS affects P. aeruginosa.
A search of the pqsA promoter region revealed two appropriately located sequences that resembled LysR-type protein binding sites (43). The first sequence begins at −65 relative to the pqsA transcriptional start site and has A and T nucleotides that are 11 bp apart. This region consists of a 5-bp sequence that shares dyad symmetry with a sequence 10 bp downstream. The second potential PqsR binding site sequence extends from −51 to −39 and again has a T and an A that are 11 bp apart and a 5-bp region that shares dyad symmetry with a sequence that begins 2 bp downstream. While these sites are similar to LysR-type protein binding sites in both sequence and location, additional analysis of the pqsA promoter region is required to determine exactly which sequences are bound by PqsR and the PqsR-PQS complex.
The fact that pqsA was controlled by PqsR, LasR, and RhlR but that only PqsR interacted with the pqsA promoter region logically led to the hypothesis that pqsR must be controlled by LasR and RhlR. With this in mind, our studies were then directed toward the control of pqsR. The pqsR gene is located adjacent to the PQS synthetic operons, pqsABCDE and phnAB. The data presented in Fig. 4 show that pqsR is positively regulated by the las quorum-sensing system and negatively regulated by the rhl quorum-sensing system. The ability of LasR and 3-oxo-C12-HSL to activate pqsR was also demonstrated in E. coli, thereby indicating that LasR and 3-oxo-C12-HSL are required and sufficient for pqsR activation (Fig. 5). These data are in agreement with data from two different mRNA microarray analysis experiments in studies of quorum sensing-controlled genes in P. aeruginosa (18, 44). However, the results in Fig. 4 and 5 conflict with those of Cao et al. (5), who suggested that there is no regulatory effect of either lasR or rhlR on pqsR. The most likely cause for these conflicting data is that our pqsR′-lacZ reporter fusion contains 171 bp of additional upstream DNA compared to the fusion used by Cao et al. (5). While their fusion should contain both promoter regions that were identified in Fig. 3, it does not contain a potential quorum-sensing operator centered 248 bp upstream from the transcriptional start site of extension product TS1 (Fig. 3C). Déziel et al. (12) also allude to a potential quorum sensing-controlled operator sequence that was absent from the pqsR reporter fusion used by Cao et al. (5). This sequence contains the highly conserved (47) CT and AG nucleotides at positions 3 and 4 and 17 and 18, respectively. In addition, it matches 14 out of 20 bp of the quorum-sensing operator of the rsaL promoter, which is regulated by the las quorum-sensing system (11). Our pqsR′-lacZ fusion contains 85 bp beyond this putative operator, which could explain why the data in Fig. 4 and 5 show that LasR controls pqsR while Cao et al. (5) reported that it does not. It must be pointed out here that the potential operator sequence is quite distant from the pqsR transcriptional start sites so its presence may be purely coincidental. The quorum-sensing control of pqsR may be through this site or a less-conserved operator that cannot be identified by sequence comparisons.
With regard to the negative regulation of PQS production, our data derived from P. aeruginosa experiments indicated that rhlR had a negative effect on pqsR transcription (Fig. 4). The rhl quorum-sensing system did not have as great a negative effect on pqsR as it did on pqsA, which leads us to speculate that a small effect on the expression of this transcriptional regulator could be amplified in a gene that it controls (e.g., pqsA).
Taken together, our data show that PQS is an important part of the cell-to-cell signaling hierarchy of P. aeruginosa. The production of all three P. aeruginosa cell-to-cell signals appears to be finely regulated and interconnected. To try to help understand this complex regulatory scheme, we have included a model that shows how each signal affects the others (Fig. 7). In this model, the cell-to-cell signaling cascade starts with the induction of lasR and lasI. LasR is regulated by multiple factors, including Vfr and GacA (1, 41), and lasI is tightly controlled by positive autoregulation (in conjunction with LasR) and two negative regulators (RsaL and QscR) (8, 11). LasR-3-oxo-C12-HSL then positively influences rhlR and rhlI (22, 38) and pqsH (48), which encodes a putative monooxygenase that is proposed to catalyze the final step of PQS synthesis (13). A competitive regulatory event occurs at the pqsR promoter, with LasR-3-oxo-C12-HSL and RhlR-C4-HSL inducing and repressing pqsR (Fig. 4 and 5), respectively. PqsR then interacts with PQS, and the PqsR-PQS complex regulates the pqsA promoter (Fig. 2) (26) (and probably the phnA promoter [5]) to cause an increased amount of PQS to be produced. The positive feedback on PQS production that is implied by PQS's acting as a coinducer for PqsR has been seen in a similar regulatory cascade in which ToxR (a LysR homolog) controls toxoflavin production in Burkholderia glumae (21). PqsR-PQS then has a positive effect on rhlI (27), which could lead to negative feedback on PQS production. The effect of PqsR-PQS on rhlI production can be demonstrated only in a lasR mutant (15), which suggests that the effect occurs at a time when lasR is not being expressed. All three cell-to-cell systems also control numerous cell functions, including many virulence factors (44, 46).
FIG. 7.
Model of the P. aeruginosa cell-to-cell signaling hierarchy. The details of this model are deliberated in Discussion. Plus and minus symbols indicate positive and negative effects, respectively. The LasI and RhlI signal synthase proteins were left out due to space limitations. Virulence factors include the many virulence determinants and other cell factors that are regulated by the transcriptional activator-signal complexes.
In summary, we have begun to unravel the complex regulatory cascade that governs PQS synthesis in P. aeruginosa. The discovery that PQS appears to be a coinducer for PqsR is important for our understanding of PQS bioactivity and will lead to future studies on the function of PQS. The ability of PQS to alter gene regulation and virulence in P. aeruginosa is an indicator of a promising target for therapeutics aimed at decreasing the pathogenesis of this opportunistic pathogen.
Acknowledgments
This work was supported by a research grant from the National Institute of Allergy and Infectious Diseases (grant R01-AI46682).
We thank J. Farrow and C. Pesci for help in manuscript preparation and thoughtful insight.
REFERENCES
- 1.Albus, A. M., E. C. Pesci, L. J. Runyen-Janecky, S. E. H. West, and B. H. Iglewski. 1997. Vfr controls quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brint, J. M., and D. E. Ohman. 1995. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J. Bacteriol. 177:7155-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calfee, M. W., J. P. Coleman, and E. C. Pesci. 2001. Interference with Pseudomonas quinolone signal synthesis inhibits virulence factor expression by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 98:11633-11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calfee, M. W., J. G. Shelton, J. A. McCubrey, and E. C. Pesci. 2005. Solubility and bioactivity of the Pseudomonas quinolone signal are increased by a Pseudomonas aeruginosa-produced surfactant. Infect. Immun. 73:878-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao, H., G. Krishnan, B. Goumnerov, J. Tsongalis, R. Tompkins, and L. G. Rahme. 2001. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc. Natl. Acad. Sci. USA 98:14613-14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, M., and I. P. Crawford. 1991. In vitro determination of the effect of indoleglycerol phosphate on the interaction of purified TrpI protein with its DNA-binding sites. J. Bacteriol. 173:1590-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chugani, S. A., M. Whiteley, K. M. Lee, D. D'Argenio, C. Manoil, and E. P. Greenberg. 2001. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 98:2752-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collier, D. N., L. Anderson, S. L. McKnight, T. L. Noah, M. Knowles, R. Boucher, U. Schwab, P. Gilligan, and E. C. Pesci. 2002. A bacterial cell to cell signal in the lungs of cystic fibrosis patients. FEMS Microbiol. Lett. 215:41-46. [DOI] [PubMed] [Google Scholar]
- 10.D'Argenio, D. A., M. W. Calfee, P. B. Rainey, and E. C. Pesci. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 184:6481-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Kievit, T., P. C. Seed, J. Nezezon, L. Passador, and B. H. Iglewski. 1999. RsaL, a novel repressor of virulence gene expression in Pseudomonas aeruginosa. J. Bacteriol. 181:2175-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Déziel, E., S. Gopalan, A. P. Tampakaki, F. Lépine, K. E. Padfield, M. Saucier, G. Xiao, and L. G. Rahme. 2005. The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-l-homoserine lactones. Mol. Microbiol. 55:998-1014. [DOI] [PubMed] [Google Scholar]
- 13.Déziel, E., F. Lépine, S. Milot, J. He, M. N. Mindrinos, R. G. Tompkins, and L. G. Rahme. 2004. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. USA 101:1339-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diggle, S. P., K. Winzer, S. R. Chhabra, K. E. Worrall, M. Cámara, and P. Williams. 2003. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol. Microbiol. 50:29-43. [DOI] [PubMed] [Google Scholar]
- 15.Gallagher, L. A., S. L. McKnight, M. S. Kuznetsova, E. C. Pesci, and C. Manoil. 2002. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 184:6472-6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gambello, M. J., and B. H. Iglewski. 1991. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 173:3000-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gambello, M. J., S. Kaye, and B. H. Iglewski. 1993. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect. Immun. 61:1180-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hentzer, M., H. Wu, J. B. Andersen, K. Riedel, T. B. Rasmussen, N. Bagge, N. Kumar, M. A. Schembri, Z. Song, P. Kristoffersen, M. Manefield, J. W. Costerton, S. Molin, L. Eberl, P. Steinberg, S. Kjelleberg, N. Hoiby, and M. Givskov. 2003. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 22:3803-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holloway, B. W., V. Krishnapillai, and A. F. Morgan. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43:73-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, B., C. B. Whitchurch, L. Croft, S. A. Beatson, and J. S. Mattick. 2000. A minimal tiling path cosmid library for functional analysis of the Pseudomonas aeruginosa PAO1 genome. Microb. Comp. Genomics 5:189-203. [DOI] [PubMed] [Google Scholar]
- 21.Kim, J., J.-G. Kim, Y. Kang, J. Y. Jang, G. J. Jog, J. Y. Lim, S. Kim, H. Suga, T. Nagamatsu, and I. Hwang. 2004. Quorum sensing and the LysR-type transcriptional activator ToxR regulate toxoflavin biosynthesis and transport in Burkholderia glumae. Mol. Microbiol. 54:921-934. [DOI] [PubMed] [Google Scholar]
- 22.Latifi, A., M. Foglino, K. Tanaka, P. Williams, and A. Lazdunski. 1996. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 21:1137-1146. [DOI] [PubMed] [Google Scholar]
- 23.Lau, G. W., H. Ran, F. Kong, D. J. Hassett, and D. Mavrodi. 2004. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect. Immun. 72:4275-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lepine, F., S. Milot, E. Deziel, J. He, and L. G. Rahme. 2004. Electrospray/mass spectrometric identification and analysis of 4-hydroxy-2-alkylquinolines (HAQs) produced by Pseudomonas aeruginosa. J. Am. Soc. Mass Spectrom. 15:862-869. [DOI] [PubMed] [Google Scholar]
- 25.Mahajan-Miklos, S., M. W. Tan, L. G. Rahme, and F. M. Ausubel. 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96:47-56. [DOI] [PubMed] [Google Scholar]
- 26.McGrath, S., D. S. Wade, and E. C. Pesci. 2004. Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS). FEMS Microbiol. Lett. 230:27-34. [DOI] [PubMed] [Google Scholar]
- 27.McKnight, S. L., B. H. Iglewski, and E. C. Pesci. 2000. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 182:2702-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medina, G., K. Juárez, B. Valderrama, and G. Soberón-Chávez. 2003. Mechanism of Pseudomonas aeruginosa RhlR transcriptional regulation of the rhlAB promoter. J. Bacteriol. 185:5976-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 30.Ochsner, U. A., A. K. Koch, A. Fiechter, and J. Reiser. 1994. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J. Bacteriol. 176:2044-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ochsner, U. A., and J. Reiser. 1995. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:6424-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohman, D. E., S. J. Cryz, and B. H. Iglewski. 1980. Isolation and characterization of a Pseudomonas aeruginosa PAO mutant that produces altered elastase. J. Bacteriol. 142:836-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearson, J. P., K. M. Gray, L. Passador, K. D. Tucker, A. Eberhard, B. H. Iglewski, and E. P. Greenberg. 1994. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 91:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearson, J. P., L. Passador, B. H. Iglewski, and E. P. Greenberg. 1995. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:1490-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearson, J. P., E. C. Pesci, and B. H. Iglewski. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pesci, E. C., and B. H. Iglewski. 2003. Quorum sensing, p. 55-65. In D. L. Burns, J. T. Barbieri, B. H. Iglewski, and R. Rappuoli (ed.), Bacterial protein toxins. ASM Press, Washington, D.C.
- 37.Pesci, E. C., J. B. Milbank, J. P. Pearson, S. McKnight, A. S. Kende, E. P. Greenberg, and B. H. Iglewski. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:11229-11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pesci, E. C., J. P. Pearson, P. C. Seed, and B. H. Iglewski. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3127-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Preston, M. J., P. C. Seed, D. S. Toder, B. H. Iglewski, D. E. Ohman, J. K. Gustin, J. B. Goldberg, and G. B. Pier. 1997. Contribution of proteases and LasR to the virulence of Pseudomonas aeruginosa during corneal infections. Infect. Immun. 65:3086-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rahme, L. G., M. W. Tan, L. Le, S. M. Wong, R. G. Tompkins, S. B. Calderwood, and F. M. Ausubel. 1997. Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 94:13245-13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reimmann, C., M. Beyeler, A. Latifi, H. Winteler, M. Foglino, A. Lazdunski, and D. Haas. 1997. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24:309-319. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 44.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuster, M., M. L. Urbanowski, and E. P. Greenberg. 2004. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl. Acad. Sci. USA 101:15833-15839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whiteley, M., and E. P. Greenberg. 2001. Promoter specificity elements in Pseudomonas aeruginosa quorum-sensing-controlled genes. J. Bacteriol. 183:5529-5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]