Abstract
In Drosophila, the S3 ribosomal protein has been shown to act as a DNA glycosylase/AP lyase capable of releasing 8-hydroxyguanine (8-OH-Gua) in damaged DNA. Here we describe a second Drosophila protein (dOgg1) with 8-OH-Gua and abasic (AP) site DNA repair activities. The Drosophila OGG1 gene codes for a protein of 327 amino acids, which shows 33 and 37% identity with the yeast and human Ogg1 proteins, respectively. The DNA glycosylase activity of purified dOgg1 was investigated using γ-irradiated DNA and gas chromatography/isotope dilution mass spectrometry (GC/IDMS). The dOgg1 protein excises 8-OH-Gua and 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyGua) from γ-irradiated DNA. with kcat/KM values of 21.0 × 10–5 and 11.2 × 10–5 (min–1 nM–1), respectively. Enzymatic assays using oligodeoxyribonucleotides containing a single lesion show that dOgg1 displays a marked preference for DNA duplexes containing 8-OH-Gua, 8-OH-Ade or an AP site placed opposite a cytosine. The cleavage of the 8-OH-Gua-containing strand results from the excision of the damaged base followed by a β-elimination reaction at the 3′-side of the resulting AP site. Cleavage of 8-OH-Gua.C duplex involves the formation of a reaction intermediate that is converted into a stable covalent adduct in the presence of sodium borohydre. dOgg1 complements the mutator phenotype of fpg mutY mutants of Escherichia coli. Whole-mount in situ hybridizations on tissues at different stages of Drosophila development reveal that the dOGG1 messenger is expressed uniformly at a low level in cells in which mitotic division occurs. Therefore, Drosophila possesses two DNA glycosylase activities that can excise 8-OH-Gua and formamidopyrimidines from DNA, dOgg1 and the ribosomal protein S3.
INTRODUCTION
Reactive oxygen species (ROS) induce DNA damage that has been implicated to play a role in mutagenesis, carcinogenesis and aging (1–5). ROS produce a variety of damages in DNA including base and sugar damage, sites of base loss, strand breaks and DNA–protein crosslinks (6,7). Oxidatively damaged DNA bases are thought to be primarily repaired by the base excision repair (BER) pathway in prokaryotes and eukaryotes (8–11). 8-Hydroxyguanine (8-OH-Gua) is a prevalent DNA lesion caused by oxidizing agents and ionizing radiation and is highly mutagenic (12–15). In Escherichia coli, two DNA glycosylases prevent mutagenesis by 8-OH-Gua: the Fpg protein which excises 8-OH-Gua in damaged DNA and the MutY protein which excises the adenine residues incorporated by DNA polymerases opposite 8-OH-Gua. Inactivation of both the fpg (mutM) and mutY(micA) genes of E.coli results in a strong G:C to T:A mutator phenotype (12–15). In Saccharomyces cerevisiae, a DNA glycosylase that is encoded by the OGG1 gene and named yOgg1, catalyses the removal of 8-OH-Gua and formamidopyrimidines from damaged DNA (16–19). Furthermore, Ogg1-deficient strains of S.cerevisiae exhibit a G:C to T:A mutator phenotype (20,21). In mammalian cells, 8-OH-Gua is released by DNA glycosylases showing strong sequence homology with the yeast Ogg1 and named mOgg1 and hOgg1 for the mouse and human enzymes, respectively (22–28). To investigate the biological role of the Ogg1 protein in mammalian cells, ogg1–/– mice have been generated (29,30). Null animals are viable and show no marked pathological changes up to 12 months. However, Ogg1-deficient mice show an abnormal accumulation of 8-OH-Gua in their genome and exhibit a significantly higher spontaneous mutation rate in non-proliferative tissues compared to the isogenic wild-type (29,30). These results strongly suggest that excision of 8-OH-Gua by DNA glycosylases such as the bacterial Fpg and the eukaryotic Ogg1, prevent mutations induced by endogenous ROS. Although they have similar biological functions, the Fpg and Ogg1 proteins do not exhibit significant sequence homology (19). Indeed, Ogg1 belongs to a family of DNA glycosylases/AP lyases, the signature of which is the α-helix–hairpin–α-helix-Gly/Pro-Asp motif together with a conserved catalytic lysine, which we refer to as the HhH-GPD/K family (18,31), the prototype of this family being the Nth (Endo III) protein of E.coli (18,32).
Yeast and mammalian Ogg1 proteins have very similar, if not identical, substrate specificities and catalytic properties. The Ogg1 proteins are monomers of ~40 kDa endowed with both DNA glycosylase and AP lyase activities (reviewed in 12). They excise 8-OH-Gua, 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine (Me-FapyGua) and 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyGua) from damaged DNA (16–19,22–28). In addition, the Ogg1 proteins efficiently remove 8-hydroxyadenine (8-OH-Ade) placed opposite a cytosine in oligodeoxyribonucleotides (33,34). Yeast and human Ogg1 proteins are also endowed with an AP lyase activity that incises DNA at AP sites placed opposite a cytosine via a β-elimination reaction (16,24). The catalytic mechanism of the Ogg1 proteins involves the formation of a transient covalent imino enzyme–DNA intermediate presumably between the ɛ-NH2 group of a catalytic lysine residue and the C1’ of the abasic sugar moiety (16,24,25,31).
In Drosophila, the ribosomal protein S3 has been shown to possess an 8-OH-Gua DNA glycosylase/AP lyase activity (35–37). The glutathione S-transferase (GST) fusion of the Drosophila S3 (GST-S3) efficiently releases 8-OH-Gua and FapyGua from γ-irradiated DNA (35). These results suggested that S3 could play a role in DNA repair in addition to its involvement in protein synthesis. In the present study, we show that Drosophila also possesses an OGG1 homolog, dOGG1. The data presented show that this gene expresses a functional DNA glycosylase/AP lyase, capable of excising 8-OH-Gua and FapyGua from DNA exposed to γ-irradiation. Our results also show that dOgg1 efficiently incises 34mer DNA duplexes containing 8-OH-Gua, 8-OH-Ade and AP sites placed opposite to a cytosine. The biochemical properties of the dOgg1 protein were compared to those of S3. The expression pattern of the dOGG1 mRNA in Drosophila larvae and ovaries is also presented. Therefore, Drosophila is the first eukaryotic organism to be found having two functional 8-OH-Gua DNA glycosylase activities.
MATERIALS AND METHODS
Materials
Modified DNA bases, their stable isotope-labeled analogs and other materials for GC/IDMS were obtained as described previously (38). Calf thymus DNA and poly(dG.dC).poly(dG.dC) were purchased from Sigma and Boehringer, respectively. Restriction endonucleases, DNA polymerases, T4 DNA kinase and T4 DNA ligase were from New England Biolabs. DNA repair proteins from E.coli (Ung, Endo III, Endo IV or Fpg) and yeast Ogg1 were purified from overproducing strains of E.coli according to standard protocols (our laboratory stocks).
Preparation of DNA substrates
DNA samples for GC/IDMS analysis were prepared as follows. Calf thymus DNA was dissolved in phosphate buffer (pH 7.4) at a concentration of 0.3 mg/ml. An aliquot of this solution was bubbled with N2O and irradiated with γ-rays in a 60Co γ-source at a dose of 80 Gy (dose rate 35.5 Gy/min). Subsequently, unirradiated and irradiated DNA solutions were dialyzed against 10 mM phosphate buffer (pH 7.4). The [3H]-Me-FapyGua-poly(dG.dC).poly(dG.dC) substrate was prepared as previously described (39). The oligodeoxyribonucleotides used in this study are 34mers with the following sequences: 5′-GGCTTCATCGTTGTC[X]CAGACCTGGTGGATACCG-3′ with [X] for 8-OH-Gua, 8-OH-Ade or uracil (OligoExpress, Grenoble), respectively. The complementary sequences with each of the four DNA bases placed opposite [X] in the duplex were from OligoExpress (Grenoble, France). To generate the AP sites, the 34mer DNA containing uracil was incubated in the presence of uracil DNA glycosylase as described (16).
Cloning, expression and purification of the Drosophila dOgg1 protein
An expressed sequence tag (EST) clone coding for dOgg1 was retrieved from the Berkeley Drosophila Genome Project database. The cDNA in a pBluescript vector (Stratagene) was sequenced using an ALFexpress automated sequencer (Amersham Pharmacia Biotech). The open reading frame identified was PCR amplified using the Pfu polymerase (Stratagene) with the following primers: 5′ CGGGATCCATGAAGGCTGTTTTAC and 5′ TAGATAAGATCACTTTTTAGG. The resulting fragment was digested with BamH1 and ligated to plasmid pGEX-4T1 (Amersham Pharmacia Biotech) digested with SmaI and BamH1. The plasmid obtained (pPR195) codes for a GST peptide with the Drosophila OGG1 cDNA fused at its C-terminus. The plasmid was introduced into the E.coli strain BH410 (fpg–). Escherichia coli BH410 harboring pPR195 was grown at 37°C in LB broth medium (2 l) containing 150 µg/ml ampicillin until the absorbance at 600 nm reached 0.3 and induced for 16 h at 20°C in the presence of IPTG (1 mM). Cells were collected and stored at –80°C. Cell pellets were resuspended in 10 ml/g of lysis buffer (20 mM Tris–HCl, pH 8.0, 1 mM Na2EDTA, 250 mM NaCl, 0.8 µg/ml antipain, 0.8 µg/ml leupeptine, 0.8 µg/ml aprotinin) and sonicated. After centrifugation of the cell lysate, the supernatant fraction was dialyzed against PBS buffer and applied to glutathione–Sepharose 4B (Pharmacia Biotech) equilibrated with PBS buffer. The Sepharose was washed with PBS and proteins eluted with a buffer containing 50 mM Tris–HCl pH 8.0 and 30 mM reduced glutathione. Fractions containing the enzyme activity were pooled and dialyzed overnight against a buffer containing 50 mM Tris–HCl pH 8.0 and 50 mM NaCl and subsequently 4 h against 50 mM Tris–HCl pH 8.0 with 150 mM NaCl and 2.5 mM CaCl2. Thrombin (ICN) was added (10 U/mg of GST–dOgg1 fusion protein) and the mixture incubated for 2 h at 25°C. Reactions were stopped by adding Na2EDTA to 5 mM final. Proteins were dialyzed against a buffer containing 20 mM Tris–HCl pH 8.0, 2 mM Na2EDTA, 50 mM NaCl and 2% glycerol and applied to a MonoS column (FPLC system, Pharmacia Biotech). Proteins were eluted with a linear salt gradient (50–800 mM NaCl). The active fractions were pooled and their protein concentration determined by the method of Bradford (40). Purification of dOgg1 was followed by SDS–PAGE and using the excision of [3H]-Me-FapyGua from [3H]-Me-FapyGua-poly(dG.dC).poly(dG.dC) as an activity assay (39).
Enzymatic assays and GC/IDMS analysis
The determination of the dependence of excision on the enzyme amount and on the incubation time, the measurement of excision kinetics and the GC/IDMS analysis were performed as described previously (41). The amount of dOgg1 used for the measurement of excision kinetics corresponded to an enzyme concentration of 267 nM.
Assays for cleavage of 34mer DNA duplexes
The DNA strand containing the lesion (8-OH-Gua, 8-OH-Ade or AP site) was 32P-labeled at the 5′-end and annealed with each of the four complementary sequences (16,33,34). The reaction mixtures (10 µl final volume) contained 25 mM Tris–HCl pH 7.6, 2 mM Na2EDTA, 50 mM NaCl, 50 fmol of 32P-labeled DNA duplex and dOgg1. Reactions were performed at 37°C for 15 min. Reactions were stopped by adding 6 µl of formamide dye and subjected to 7 M urea–20% PAGE (16,34). Gels were scanned and quantified using a PhosphorImager (Molecular Dynamics) (33).
Trapping assay
The reaction mixture (20 µl final volume) contained 25 mM Tris–HCl pH 7.6, 2 mM Na2EDTA, 100 fmol of labeled 8-OH-Gua.C duplex, 50 mM of NaBH4 and 50 ng of either dOgg1, yOgg1 or Fpg. The reaction was carried out at 37°C for 20 min. The reactions were stopped by addition of 10 µl of SDS–PAGE loading buffer/formamide blue dye and heating at 90°C for 3 min. The products of the reactions were separated onto 15% SDS–PAGE and analyzed as previously described (16).
Mutagenesis experiments
The complementation of the spontaneous mutator phenotype of an E.coli fpg mutY double mutant (PR195) by the expression of the different GST fusion proteins was analyzed by determining the frequency of rifampicin-resistant cells in 20 independent cultures (26).
Whole-mount in situ hybridization
Whole-mount RNA in situ hybridization on ovaries and larvae was performed with the complete dOgg1 cDNA. Digoxigenin-labeled sense and antisense RNA probes were synthesized using the RNA Genius kit (Boehringer Mannheim) according to manufacturer’s protocol. The hybridization procedures were carried out according to Tautz and Pfeifle (42) with minor modifications (43) and other modifications as follow: Dissected tissues were fixed in 4% paraformaldehyde in PBS–0.1% Tween-20 (PBT) for 20 min at room temperature (RT) and washed three times in PBT. Tissues were then incubated in boiling water for 5 min. Prehybridization was in hybridization solution (HS) for 1 h at 65°C, and hybridization was overnight at 65°C. After rinsing twice in HS for 20 min at 65°C, tissues were washed three times in PBT for 20 min at RT. After the incubation with anti-DIG antibody (1/2000 in PBT) for 1 h at RT, tissues were washed three times in PBT for 20 min at RT and brought into staining buffer. Tissues were stained for 1 h at RT, washed three times in PBT and mounted in 70% glycerol.
RESULTS
Isolation of a Drosophila cDNA coding for an Ogg1 protein homolog
A cDNA clone coding for a peptide showing a high degree of homology to the human and yeast Ogg1 proteins was identified in the EST database from the Berkeley Drosophila Genome Project. The dOGG1 cDNA is 1284 bp long and codes for a 327 amino acid peptide. Sequence alignments show that the protein coded, dOgg1, shares 33 and 37% identity with the yeast and human Ogg1, respectively. Furthermore, dOgg1 possesses all the hallmarks of the Ogg1 proteins. In particular, the highly conserved active site motif helix–hairpin–helix motif-PVD/K is present between residues 209 and 260 (Fig. 1A). Comparison of the cDNA with the complete Drosophila genome allowed the identification of the corresponding genomic sequence for the dOGG1 gene and its localization to chromosome 1. Comparison between the cDNA and the genomic sequences shows that dOGG1 has four exons separated by introns whose sizes are 58, 61 and 59 bp (Fig. 1B). The transcribed region spans 1.5 kb of genomic sequences.
Figure 1.
Drosophila OGG1 sequence analysis. (A) Sequence alignment of the dOgg1 protein with its yeast (Sc), plant (At) and human (Hs) homologs. Black and gray boxes identify identical and similar residues respectively. Asterisks in the consensus line indicate conservation of the amino acid in all four proteins. (B) Structure of the dOGG1 gene. Boxes indicate exons.
Expression and purification of the Drosophila Ogg1 protein
The Drosophila OGG1 cDNA was cloned into the bacterial vector pGEX-4T1 to express dOgg1 fused to the C-terminus of GST. The GST–dOgg1 protein was purified from bacterial lysates by affinity to glutathione–Sepharose. Treatment with thrombine allowed the cleavage of the GST tag. The dOgg1 protein was separated from GST and thrombin after chromatography on a MonoS column. The purity of the dOgg1 protein was assessed by the observation of a single protein band on a SDS–PAGE with a molecular mass of ~38 kDa, which agreed well with the expected mass (Fig. 2). The specificity of the site of cleavage was confirmed by a single N-terminal sequence (data not shown). The purification steps were monitored by measuring the excision of [3H]-Me-FapyGua by the protein fractions. For the purified dOgg1 protein, the specificity constant (kcat/KM) for the excision of Me-FapyGua is 214 × 10–5 (min–1 nM–1). This value is very similar to that measured for the human α-hOgg1 protein (33).
Figure 2.
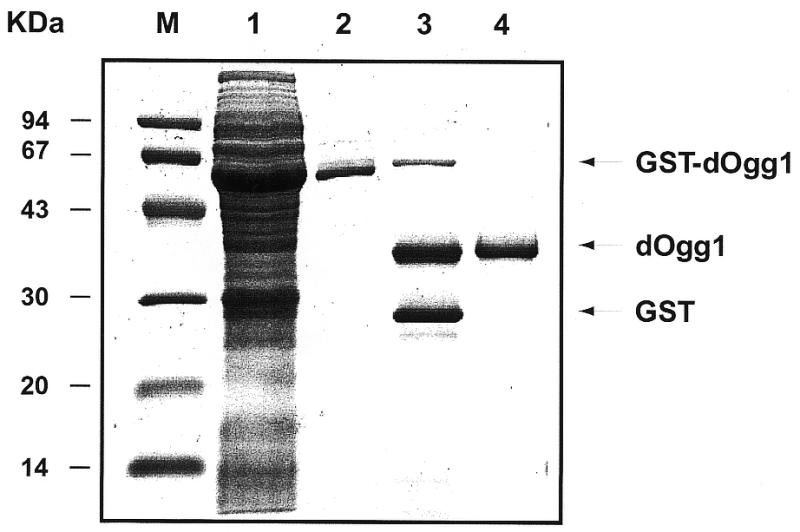
Gel analysis of the dOgg1 purification steps. After expression in E.coli the purification fractions were analyzed by SDS–PAGE. M, molecular weight markers; lane 1, total cell lysate; lane 2, eluate from the glutathione–Sepharose 4B; lane 3, products of the thrombin cleavage reaction; lane 4, eluate from the MonoS column.
Excision of modified bases by the Drosophila dOgg1 from γ-irradiated DNA: GC/IDMS analysis
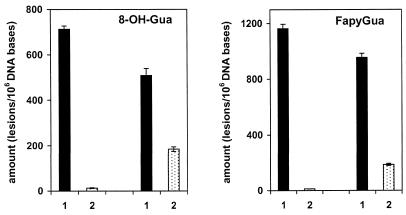
We investigated the ability of the dOgg1 protein to excise modified DNA bases from γ-irradiated DNA. Using GC/IDMS, 17 modified bases were identified and quantified in DNA γ-irradiated under N2O (44). Of these modified bases, dOgg1 protein efficiently excised FapyGua and 8-OH-Gua. No other modified base was excised significantly under the conditions used in this work. The excision of 8-OH-Gua and FapyGua was assessed by their appearance in supernatant fractions of DNA substrates incubated with active enzyme (Fig. 3). The amounts of 8-OH-Gua and FapyGua in DNA pellets after incubation of γ-irradiated DNA with active protein were significantly reduced when compared to those in DNA pellets incubated with heat-inactivated enzyme or without enzyme. The amounts found in the supernatant fractions of DNA substrates incubated with active enzyme were similar to those removed from the pellets of the same DNA substrate, demonstrating the excision of the lesions (Fig. 3). Kinetic parameters were determined by measurement of excision at six different concentrations of FapyGua and 8-OH-Gua with the total amount of DNA remaining constant in each sample. The excised amounts of these products in supernatant fractions were used for the determination of the initial velocity. Excision followed Michaelis–Menten kinetics (45). Kinetic constants and standard deviations (n = 6) were calculated using Lineweaver–Burk plots and a linear least-squares analysis of the data (data not shown). Kinetic parameters calculated from these plots are given in Table 1. The specificity constant (kcat/KM) for excision of 8-OH-Gua is 1.9-fold higher than that for FapyGua (Table 1). This result suggests that, for the dOgg1 protein, 8 -OH-Gua may be a better substrate than FapyGua in N2O γ-irradiated DNA.
Figure 3.
Excision of 8-OH-Gua and FapyGua by dOgg1 from DNA γ-irradiated under N2O. Dark columns (1), pellets. Light columns (2), supernatant fractions. (Left) 100 µg of γ-irradiated DNA were incubated with 4 µg of heat inactivated dOgg1. (Right) 100 µg of γ-irradiated DNA were incubated with 4 µg of active dOgg1. The identification and the quantification of the products were achieved by GC/IDMS.
Table 1. Kinetic constants for excision of 8-OH-Gua and FapyGua by Drosophila Ogg1 protein from DNA γ-irradiated under N2O.
| Lesion | Vmax (nM min–1)a | KM (nM)a | kcat × 103 (min–1)a | kcat/KM × 105 (min–1 nM–1)a |
|---|---|---|---|---|
| 8-OH-Gua | 15.0 ± 0.3 | 267 ± 16b | 56.2 ± 1.1 | 21.0 ± 0.4b |
| FapyGua | 12.7 ± 0.2 | 422 ± 22 | 47.5 ± 0.7 | 11.2 ± 0.2 |
aValues represent the mean ± standard deviation (n = 6). kcat = Vmax/[enzyme]: ([enzyme] = 267 nM). The concentration ranges of the FapyGua and 8-OH-Gua were 0.82 – 3.8 µM, and 0.41 – 2.3 µM, respectively.
bStatistically different from the value in line 2 (P < 0.05).
Cleavage of 34mer DNA duplexes containing 8-OH-Gua, 8-OH-Ade or an AP site placed opposite each of the four DNA bases
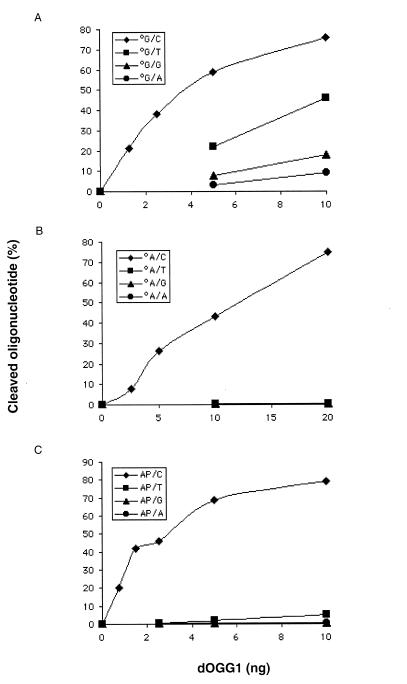
The specificity of the dOgg1 protein was also investigated using DNA lesions embedded in 34mer DNA duplexes as substrates. These substrates were also used to analyze the opposite base dependence of the dOgg1 activity on 8-OH-Gua, 8-OH-Ade and AP site. For this purpose, 34mer harboring one of these lesions was hybridized to complementary sequences harboring each of the four normal DNA bases opposite the lesion. The cleavage activity of the dOgg1 protein on the strand harboring the lesion was analyzed for each of the four DNA duplexes (Fig. 4). In the case of 8-OH-Gua, dOgg1 efficiently cleaves 8-OH-Gua.C and 8-OH-Gua.T duplexes whereas it cleaves 8-OH-Gua.G and 8-OH-Gua.A duplexes at a very slow rate (Fig. 4A). In the case of 8-OH-Ade and AP site, dOgg1 efficiently cleaves 8-OH-Ade.C and AP.C duplexes (Fig. 4B and C). In contrast, dOgg1 does not cleave 34mer DNA duplexes containing an 8-OH-Ade or an AP site placed opposite a thymine, a guanine or an adenine (Fig. 4B and C). This last result explains why 8-OH-Ade is not released from γ-irradiated DNA substrates which contain 8-OH-Ade.T base pairs.
Figure 4.
Cleavage activity of dOgg1 on DNA duplexes harboring single lesions. (A) Cleavage of 34mer oligodeoxyribonucleotides carrying a single 8-OH-Gua paired to each of the four normal DNA bases. (B) Cleavage of 34mer oligodeoxyribonucleotides carrying a single 8-OH-Ade paired to each of the four normal DNA bases. (C) Cleavage of 34mer oligodeoxyribonucleotides carrying a single AP site paired to each of the four normal DNA bases. The DNA duplexes were 32P-labeled at the 5′-end of the strand carrying the lesion. Incubations were performed at 37°C for 15 min. The products were separated on 20% PAGE containing 7 M urea and quantified using a PhosphorImager.
Catalytic mechanism of the dOgg1 protein
To further investigate the catalytic mechanism of dOgg1, we compared the products generated by dOgg1, yOgg1 and Fpg after cleavage of an 8-OH-Gua.C duplex. Figure 5A shows that dOgg1 and yOgg1 generate the same product (P1) corresponding to the cleavage of the phosphodiester bond at the 3′-side of the AP site resulting from the excision of the 8-OH-Gua [18]. It is widely accepted that this product results from a β-elimination reaction catalyzed by the AP lyase activity of this class of enzymes (46). In contrast, the Fpg protein generates a product that migrates faster (P3) and results from successive β- and δ-elimination reactions at the AP site generated by the glycosylase activity (16,46). Control experiments show that the dOgg1 product P1 is not modified by post-treatment with Endo III (Fig. 5A). In contrast, the addition of Endo IV after cleavage by the dOgg1 protein results in a nearly quantitative conversion of the dOgg1 product P1 into another product (P2) which is probably generated by the release of the trans-4-hydroxy-2-pentenal-5-phosphate at the 3′-end of the P1 fragment (16,36,46). These results strongly suggest that the mechanism of strand cleavage by the dOgg1 protein involves the release of the 8-OH-Gua residue followed by a β-elimination reaction at the 3′-side of the resulting AP site.
Figure 5.
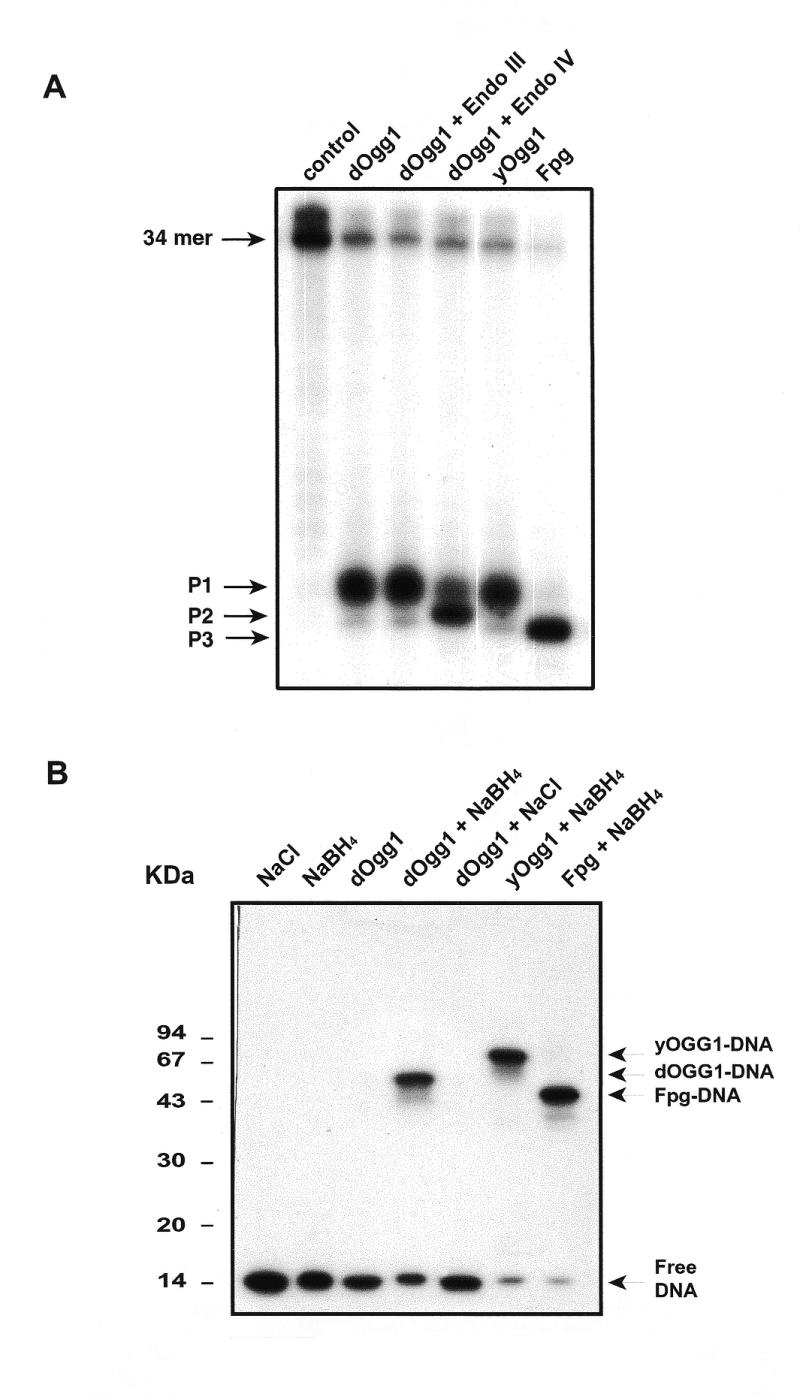
Analysis of the products of the dOgg1 repair reaction. (A) Mechanism of strand cleavage. The 34mer carrying a single 8-OH-Gua residue was 32P-labeled at its 5′ end and hybridized with a complementary sequence carrying a cytosine opposite the lesion. Incubation were performed with 10 ng of dOgg1, yOgg1 or Fpg protein for 15 min at 37°C. When indicated, reaction mixtures were then incubated with 10 ng of either endonuclease III (Nth) or endonuclease IV (Nfo). The products were separated by denaturing 20% PAGE. P1, P2 and P3 are the products described in the Results. (B) NaBH4-mediated trapping assay. The 34mer carrying a single 8-OH-Gua residue was labeled at its 5′-end and hybridized with a complementary sequence carrying a cytosine opposite the lesion. Fifty nanograms of either Fpg, yOgg1 or dOgg1 proteins were allowed to react for 20 min at 37°C with the 32P-labeled 34mer duplex DNA, containing a single 8-OH-Gua.C, in the presence of 50 mM of either NaCl or NaBH4. The products of the reactions were separated using 15% SDS–PAGE.
The mechanism of DNA strand cleavage after excision of 8-OH-Gua by DNA glycosylases/AP lyases such as the bacterial Fpg or the yeast and human Ogg1 involves the nucleophilic attack at the C1’ of the sugar moiety leading to the formation of an enzyme–DNA Schiff base intermediate (46,47). To probe for such an intermediate, the Ogg1 protein was allowed to react with a labeled 8-OH-Gua.C substrate in the presence of the reducing agent NaBH4 and the products of the reactions were analyzed by SDS–PAGE. Figure 5B shows that the presence of NaBH4 in the assay mixture results both in the inhibition of the cleavage activity of the dOgg1 protein and in the formation of a shifted band (trapped complex) with an apparent molecular weight of 55–60 kDa (Fig. 5B). The dOgg1–DNA complex can be digested with proteinase K to yield a product that co-migrates with free DNA (data not shown). Control experiments show that the dOgg1-DNA complex is not formed when NaCl is present instead of NaBH4 (Fig. 5B). Figure 5B also shows the formation of a Fpg–DNA and yeast Ogg1–DNA complexes in the presence of NaBH4. These results suggest that the Ogg1 protein forms a transient Schiff base intermediate, which is converted into a covalent protein–DNA adduct in the presence of NaBH4.
Expression of the dOgg1 proteins in E.coli fpg mutY complements the mutator phenotype
Plasmid PR195 expressing GST–dOgg1 protein was transformed into E.coli strain PR195 in which the mutY and fpg genes are disrupted. This strain displays a strong spontaneous mutator phenotype due to its incapacity to eliminate errors induced by the presence of 8-OH-Gua in its DNA. The rates of mutation to rifampicin resistance were determined (26). Table 2 shows that the expression of the Drosophila protein in this strain reduces the mutation frequencies to rifampicin resistance as effectively as the human Ogg1 protein expression, partially complementing the spontaneous mutator phenotype.
Table 2. Frequencies of spontaneous mutation to rifampicin resistance in E.coli PR195 (fpg, mutY) expressing GST, GST–hOGG1 or GST–dOGG1.
| Protein expressed | Rifampicin resistant cells/108 cells |
|---|---|
| GST | 257 ± 35 |
| GST–hOGG1 | 64 ± 17 |
| GST–dOGG1 | 75 ± 18 |
Expression pattern of dOGG1 in Drosophila ovaries and larvae
We examined the distribution of the dOGG1 transcripts in ovaries and larvae by whole-mount RNA in situ hybridization. In all the tested tissues, a specific signal has been observed after a long time of staining compared to a control probe (Fig. 6). This result suggests that the dOGG1 mRNA might be expressed at a low level in these tissues. Ovaries are composed of about 16 ovarioles, each with a germarium at its anterior tip and progressively older egg chambers toward the posterior. An egg chamber consists of three cell types: the oocyte connected to 15 polyploide nurse cells, both surrounded by a monolayer of somatic follicle cells. The subsequent development of the egg chamber is divided into 14 stages. From stage 10B onwards, the nurse cells expel their entire content into the oocyte. At the end of the oogenesis, the columnar follicle cells synthesize the vitelline membrane and the chorion of the egg and then both nurse cells and follicle cells degenerate. Hybridization on ovaries with an antisense dOGG1 probe revealed a signal in the cytoplasm of the nurse cells from stage 3 (data not shown) and in the oocyte’s cytoplasm from stage 10B onwards (Fig. 6A and B). Hybridization on third instar larvae with an antisense dOGG1 probe revealed a specific signal in the imaginal discs which are the precursors of the epidermal structures of the head, thorax and external genitalia of the adult fly (Fig. 6C and D). In late third instar larvae, they appear as sac-like epithelial organs.
Figure 6.
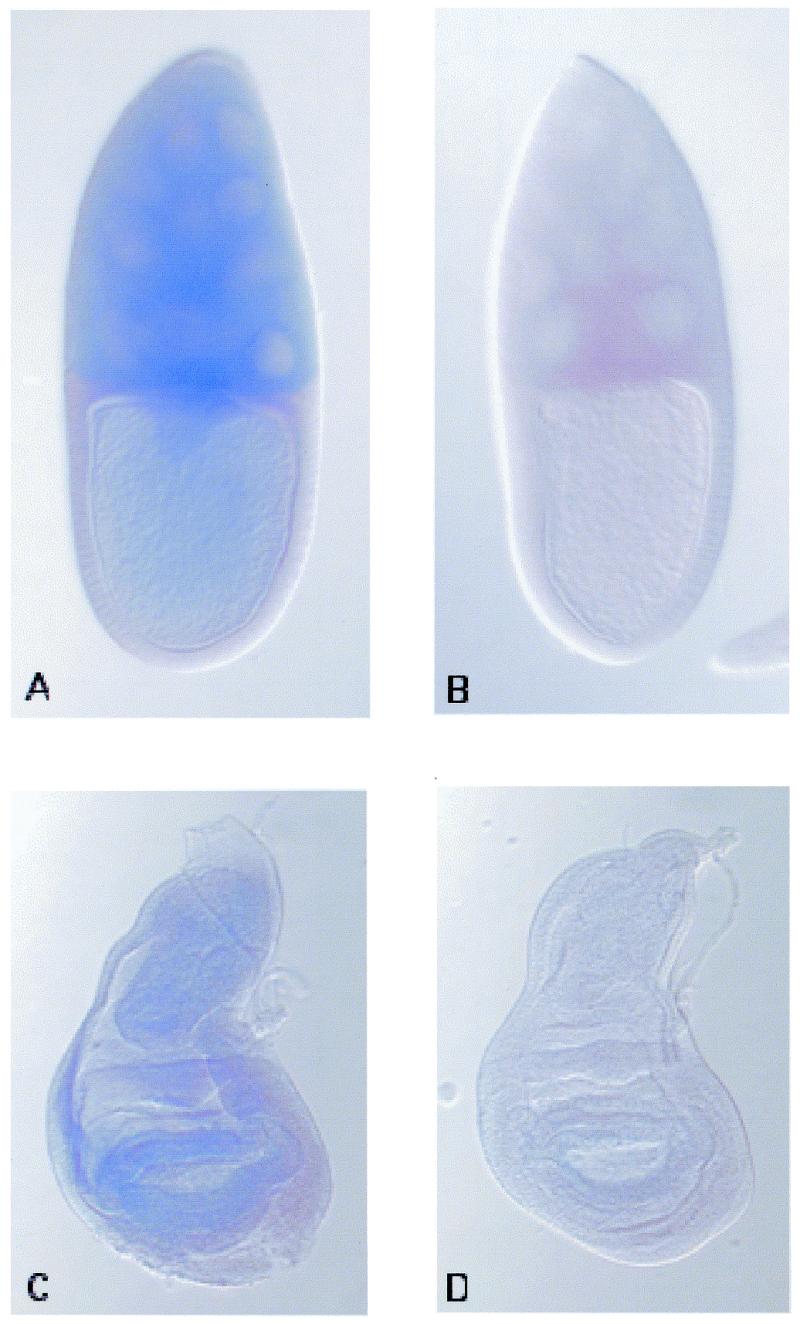
Whole-mount in situ hybridization on stage 10B egg chambers (A and B) and third instar larvae imaginal discs (C and D). The antisense probe (A and C) reveals an expression of the dOGG1 transcript in germ cells in the egg chamber and a uniform expression in the imaginal disc, as the control sense probe (B and D) exhibits no staining.
DISCUSSION
The unavoidable but potentially mutagenic DNA base alterations resulting from endogenous ROS attack are primarily removed by the BER pathway (8–11). The first step in this ubiquitous repair pathway is the recognition and removal of the altered base by a DNA glycosylase catalyzing the cleavage of the glycosylic bond between the modified base and the sugar moiety, leaving an AP site in DNA. Subsequently, the resulting AP site is incised and the repair is completed by the successive actions of a phosphodiesterase, a DNA polymerase and a DNA ligase (8–11). In yeast three DNA glycosylases are involved in the removal of oxidatively-damaged DNA bases, namely Ntg1, Ntg2 and Ogg1 (48). Structural and/or functional homologs of these DNA glycosylases have been found in higher eukaryotes (2). The biological function of this class of enzymes is thought to prevent genetic instability due to endogenous DNA base damage such as 8-OH-Gua. Assessments of 8-OH-Gua formation and mutagenic potential are strongly supported by the G:C to T:A mutator phenotype of E.coli, S.cerevisiae and mouse strains deficient in 8-OH-Gua DNA glycosylase activity (11–15, 18,19,29,30).
Three classes of 8-OH-Gua DNA glycosylases have been identified in living organisms: (i) The Fpg family in Bacteria and Arabidopsis thaliana (49); (ii) the Ogg1 family in Archae (50), yeast and mammals (2); and (iii) The S3 ribosomal protein in Drosophila (35–37). In the present study, we report sequence alignments that indicate the presence of Ogg1 homologs in A.thaliana and Drosophila (Fig. 1A). The high degree of sequence homology and the conservation of critical blocks of amino acids strongly suggest that these are functional 8-OH-Gua DNA glycosylases from the Ogg1 family (2). The results from the characterization of the purified dOgg1 protein from Drosophila presented in this paper show that the dOgg1 protein is indeed a DNA glycosylase that removes Me-FapyGua, FapyGua and 8-OH-Gua from damaged DNA as do the yeast and human Ogg1 (16–19,22–28). Previous experiments had suggested that there is no Fapy glycosylase activity in Drosophila cells (51). Consistently, experiments carried out in cell extracts from a Drosophila cell line, although positive for 8-oxoG DNA glycosylase activity, failed to detect an activity capable of removing Me-FapyGua from DNA (data not shown). Moreover, it was suggested that no BER system was present in Drosophila (52,53). These contradictory results suggest that such an activity in crude extracts might be subject to some inhibitory factor. Alternatively, the Fapy glycosylase activity could be too low to be detected in the extracts. The kinetic constants for excision of the various lesions are very similar in the different organisms studied. For example, kcat values for the excision of 8-OH-Gua are 0.084, 0.038 and 0.056 (min–1) for yeast, human and Drosophila Ogg1 proteins, respectively. The dOgg1 protein also shows a marked preference for lesions placed opposite a cytosine as do the yeast and human Ogg1 (16,24). The GC/IDMS experiments fail to show a DNA glycosylase activity of the dOgg1 protein on pyrimidines. This makes it an unlikely candidate for the activity reported in cell extracts (51). Analysis of the genomic sequences suggests that the activity on fragmented pyrimidines could be coded by the putative Endonuclease III open reading frame. Finally, the dOgg1 protein possesses an AP lyase activity which generates the same product as do the yeast and human Ogg1. In conclusion, the substrate specificity and catalytic properties of the dOgg1 protein are, to the best of our knowledge, identical to that of the yeast and human Ogg1 (2). Therefore sequence homologies reveal functional Ogg1 proteins in evolutionary distant organisms such as S.cerevisiae, Homo sapiens and Drosophila which in turn strongly suggests that the A.thaliana sequence also encodes a functional Ogg1 (Fig. 1A).
These results lead us to conclude that Drosophila has two 8-OH-Gua DNA glycosylase activities, S3 and dOgg1, because of this apparent redundancy, it was important to analyze in detail the properties and the expression of the dOgg1 protein. Indeed, dOgg1 and S3 have very similar properties. Both dOgg1 and S3 are DNA glycosylases that excise 8-OH-Gua and FapyGua from γ-irradiated DNA. Specificity constants for the excision of 8-OH-Gua and FapyGua by dOgg1 (Table 1) and S3 (35) have been measured using the same experimental conditions. The results show that kcat/KM values for excision of 8-OH-Gua and FapyGua by dOgg1 were 8.1- and 4.5-fold higher than that by S3, respectively. The better kcat/KM values of dOgg1, compared to S3, are due to lower KM values. Furthermore, both dOgg1 and S3 have a marked preference for 8-OH-Gua placed opposite a cytosine. Finally, dOgg1 and S3 are AP lyases whose reaction mechanisms involve the formation of a covalent imino enzyme–DNA intermediate. dOgg1 and S3 have then very similar, if not identical, substrate specificities and potentially the same biological function.
In the present study, we also show that dOGG1 mRNA is ubiquitously expressed in Drosophila tissues. The experiments presented suggest that the nurse cells synthesize the dOGG1 transcript and then expel it in the oocyte. The zygote is transcriptionaly inactive during the first 14 division of embryogenesis. Therefore, the accumulation of the dOGG1 transcript in the oocyte might be important for the mitotic divisions that occur during early embryogenesis, in order to avoid oxidative damage-induced mutations. No dOGG1 expression is detected in follicle cells. These cells will not divide any more as they degenerated at the end of the oogenesis. Then, we could expect that they do not need an important DNA repair machinery to keep their DNA without damage. The presence at a detectable level of the dOGG1 transcript in the imaginal discs could be related to the rapid divisions of the disc’s cells during larval development. Since S3 is essential, both S3 and Ogg1 are likely to be present in most Drosophila cells. The dOgg1 possesses a cluster of basic amino acids at its C-terminal end which is probably used as a nuclear localization signal as previously reported for the human Ogg1 (24). Similarly, the S3 ribosomal protein has been suggested to possess a nuclear localization signal and to bind the nuclear matrix (37). Therefore, both dOgg1 and S3 may have access to the nuclear DNA. All the data point then to the fact both dOgg1 and S3 are functional in vivo. Although dOgg1 seems to have somehow better kinetic properties than S3 the relative importance of their activities in the Drosophila cells remains to be determined. The transcription-coupled repair of 8-OH-Gua in Ogg1-deficient rodent cells reveals the existence of an unidentified Ogg1-independent repair pathway in eukaryotes (54). If it exists in Drosophila, this could be a third repair system for 8-OH-Gua.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Drs Jean Cadet and André Guy for the kind gift of oligodeoxyribonucleotides containing 8-OH-Gua or 8-OH-Ade used in this study, and Dr A. Lepesant for his advice. They also thank Marc Audebert for the sequencing of the plasmids used in this work. This work was supported by the Centre National de la Recherche Scientifique (CNRS), the Commissariat à l’Energie Atomique (CEA) and the Comité de Radioprotection of Electricité de France (EDF). Certain commercial equipment or materials are identified in this paper in order to specify adequately the experimental procedure. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment identified are necessarily the best available for the purpose.
REFERENCES
- 1.Beckman K.B. and Ames,B.N. (1997) Oxidative decay of DNA. J. Biol. Chem., 272, 19633–19636. [DOI] [PubMed] [Google Scholar]
- 2.Boiteux S. and Radicella,J.P. (2000) The human OGG1 gene: structure, functions and its implication in the process of carcinogenesis. Arch. Biochem. Biophys., 377, 1–8. [DOI] [PubMed] [Google Scholar]
- 3.Breimer L.H. (1990) Molecular mechanisms of oxygen radical carcinogenesis and mutagenesis: the role of DNA base damage. Mol. Carcinog., 3, 188–197. [DOI] [PubMed]
- 4.Feig D.I., Reid,T.M. and Loeb,L.A. (1994) Reactive oxygen species in tumorigenesis. Cancer Res., 54, 1890s–1894s. [PubMed] [Google Scholar]
- 5.Wiseman H. and Halliwell,B. (1996) Damage to DNA by reactive oxygen and nitrogen species: role in inflammatory disease and progression to cancer. Biochem. J., 313, 17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cadet J., Berger,M., Douki,T. and Ravanat,J.L. (1997) Oxidative damage to DNA: formation, measurement and biological significance. Rev. Physiol. Biochem. Pharmacol., 131, 1–87. [DOI] [PubMed] [Google Scholar]
- 7.Dizdaroglu M. (1992) Oxidative damage to DNA in mammalian chromatin. Mutat. Res., 275, 331–342. [DOI] [PubMed] [Google Scholar]
- 8.Boiteux S. and Laval,J. (1997) Repair of oxidized purines in DNA. In Hickson,I.D. (ed.), Base Excision Repair of DNA damage. Springer, Austin, TX, pp. 31–44.
- 9.Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.
- 10.Krokan H.E., Standal,R. and Slupphaug,G. (1997) DNA glycosylases in the base excision repair of DNA. Biochem. J., 325, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindahl T. and Wood,R.D. (1999) Quality Control by DNA Repair. Science, 286, 1897–1905. [DOI] [PubMed] [Google Scholar]
- 12.Boiteux S. and Radicella,J.P. (1999) Excision repair of 8-Oxoguanine in eukaryotes. The Ogg1 proteins. In Dizdaroglu,K. (ed.), Advances in DNA damage and repair. Kluwer Academic, New York, pp. 35–45.
- 13.Grollman A.P. and Moriya,M. (1993) Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet., 9, 246–249. [DOI] [PubMed]
- 14.Michaels M.L., Tchou,J., Grollman,A.P. and Miller,J.H. (1992) A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry, 31, 10964–10968. [DOI] [PubMed] [Google Scholar]
- 15.Tajiri T., Maki,H. and Sekiguchi,M. (1995) Functional cooperation of MutT, MutM and MutY proteins in preventing mutations caused by spontaneous oxidation of guanine nucleotide in Escherichia coli. Mutat. Res., 336, 257–267. [DOI] [PubMed]
- 16.Girard P.M., Guibourt,N. and Boiteux,S. (1997) The Ogg1 protein of Saccharomyces cerevisiae: a 7,8-dihydro-8- oxoguanine DNA glycosylase/AP lyase whose lysine 241 is a critical residue for catalytic activity. Nucleic Acids Res., 25, 3204–3211. [DOI] [PMC free article] [PubMed]
- 17.Karahalil B., Girard,P.M., Boiteux,S. and Dizdaroglu,M. (1998) Substrate specificity of the Ogg1 protein of Saccharomyces cerevisiae: excision of guanine lesions produced in DNA by ionizing radiation- or hydrogen peroxide/metal ion-generated free radicals. Nucleic Acids Res., 26, 1228–1233. [DOI] [PMC free article] [PubMed]
- 18.Nash H.M., Bruner,S.D., Scharer,O.D., Kawate,T., Addona,T.A., Spooner,E., Lane,W.S. and Verdine,G.L. (1996) Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr. Biol., 6, 968–980. [DOI] [PubMed] [Google Scholar]
- 19.van der Kemp P.A., Thomas,D., Barbey,R., de Oliveira,R. and Boiteux,S. (1996) Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N- methylformamidopyrimidine. Proc. Natl Acad. Sci. USA, 93, 5197–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruner S.D., Nash,H.M., Lane,W.S. and Verdine,G.L. (1998) Repair of oxidatively damaged guanine in Saccharomyces cerevisiae by an alternative pathway. Curr. Biol., 8, 393–403. [DOI] [PubMed] [Google Scholar]
- 21.Thomas D., Scot,A.D., Barbey,R., Padula,M. and Boiteux,S. (1997) Inactivation of OGG1 increases the incidence of G. C–>T. A transversions in Saccharomyces cerevisiae: evidence for endogenous oxidative damage to DNA in eukaryotic cells. Mol. Gen. Genet., 254, 171–178. [DOI] [PubMed] [Google Scholar]
- 22.Aburatani H., Hippo,Y., Ishida,T., Takashima,R., Matsuba,C., Kodama,T., Takao,M., Yasui,A., Yamamoto,K. and Asano,M. (1997) Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res., 57, 2151–2156. [PubMed] [Google Scholar]
- 23.Arai K., Morishita,K., Shinmura,K., Kohno,T., Kim,S.R., Nohmi,T., Taniwaki,M., Ohwada,S. and Yokota,J. (1997) Cloning of a human homolog of the yeast OGG1 gene that is involved in the repair of oxidative DNA damage. Oncogene, 14, 2857–2861. [DOI] [PubMed] [Google Scholar]
- 24.Bjoras M., Luna,L., Johnsen,B., Hoff,E., Haug,T., Rognes,T. and Seeberg,E. (1997) Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. EMBO J., 16, 6314–6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu R., Nash,H.M. and Verdine,G.L. (1997) A mammalian DNA repair enzyme that excises oxidatively damaged guanines maps to a locus frequently lost in lung cancer. Curr. Biol., 7, 397–407. [DOI] [PubMed] [Google Scholar]
- 26.Radicella J.P., Dherin,C., Desmaze,C., Fox,M.S. and Boiteux,S. (1997) Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 94, 8010–8015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roldan-Arjona T., Wei,Y.F., Carter,K.C., Klungland,A., Anselmino,C., Wang,R.P., Augustus,M. and Lindahl,T. (1997) Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc. Natl Acad. Sci. USA, 94, 8016–8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenquist T.A., Zharkov,D.O. and Grollman,A.P. (1997) Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc. Natl Acad. Sci. USA, 94, 7429–7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klungland A., Rosewell,I., Hollenbach,S., Larsen,E., Daly,G., Epe,B., Seeberg,E., Lindahl,T. and Barnes,D.E. (1999) Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl Acad. Sci. USA, 96, 13300–13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minowa O., Arai,T., Hirano,M., Monden,Y., Nakai,S., Fukuda,M., Itoh,M., Takano,H., Hippou,Y., Aburatani,H., Masumura,K., Nohmi,T., Nishimura,S. and Noda,T. (2000) Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc. Natl Acad. Sci. USA, 97, 4156–4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guibourt N., Castaing,B., Van Der Kemp,P.A. and Boiteux,S. (2000) Catalytic and DNA binding properties of the ogg1 protein of Saccharomyces cerevisiae: comparison between the wild type and the K241R and K241Q active-site mutant proteins. Biochemistry, 39, 1716–1724. [DOI] [PubMed] [Google Scholar]
- 32.Thayer M.M., Ahern,H., Xing,D., Cunningham,R.P. and Tainer,J.A. (1995) Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J., 14, 4108–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dherin C., Radicella,J.P., Dizdaroglu,M. and Boiteux,S. (1999) Excision of oxidatively damaged DNA bases by the human alpha-hOgg1 protein and the polymorphic alpha-hOgg1(Ser326Cys) protein which is frequently found in human populations. Nucleic Acids Res., 27, 4001–4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Girard P.M., D’Ham,C., Cadet,J. and Boiteux,S. (1998) Opposite base-dependent excision of 7,8-dihydro-8-oxoadenine by the Ogg1 protein of Saccharomyces cerevisiae. Carcinogenesis, 19, 1299–1305. [DOI] [PubMed] [Google Scholar]
- 35.Deutsch W.A., Yacoub,A., Jaruga,P., Zastawny,T.H. and Dizdaroglu,M. (1997) Characterization and mechanism of action of Drosophila ribosomal protein S3 DNA glycosylase activity for the removal of oxidatively damaged DNA bases. J. Biol. Chem., 272, 32857–32860. [DOI] [PubMed] [Google Scholar]
- 36.Sandigursky M., Yacoub,A., Kelley,M.R., Deutsch,W.A. and Franklin,W.A. (1997) The Drosophila ribosomal protein S3 contains a DNA deoxyribophosphodiesterase (dRpase) activity. J. Biol. Chem., 272, 17480–17484. [DOI] [PubMed] [Google Scholar]
- 37.Yacoub A., Augeri,L., Kelley,M.R., Doetsch,P.W. and Deutsch,W.A. (1996) A Drosophila ribosomal protein contains 8-oxoguanine and abasic site DNA repair activities. EMBO J., 15, 2306–2312. [PMC free article] [PubMed] [Google Scholar]
- 38.Dizdaroglu M. (1994) Chemical determination of oxidative DNA damage by gas chromatography–mass spectrometry. Methods Enzymol., 234, 3–16. [DOI] [PubMed]
- 39.Boiteux S., Belleney,J., Roques,B.P. and Laval,J. (1984) Two rotameric forms of open ring 7-methylguanine are present in alkylated polynucleotides. Nucleic Acids Res., 12, 5429–5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 41.Audebert M., Radicella,J.P. and Dizdaroglu,M. (2000) Effect of single mutations in the OGG1 gene found in human tumors on the substrate specificity of the ogg1 protein. Nucleic Acids Res., 28, 2672–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tautz D. and Pfeifle,C. (1989) A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma, 98, 81–85. [DOI] [PubMed] [Google Scholar]
- 43.Suter B. and Steward,R. (1991) Requirement for phosphorylation and localization of the Bicaudal-D protein in Drosophila oocyte differentiation. Cell, 67, 917–926. [DOI] [PubMed] [Google Scholar]
- 44.Dizdaroglu M., Bauche,C., Rodriguez,H. and Laval,J. (2000) Novel substrates of Escherichia coli nth protein and its kinetics for excision of modified bases from DNA damaged by free radicals. Biochemistry, 39, 5586–5592. [DOI] [PubMed] [Google Scholar]
- 45.Gutfreund H. (1972) Enzymes: Physical principals. Wiley-Interscience, London, UK.
- 46.McCullough A.K., Dodson,M.L. and Lloyd,R.S. (1999) Initiation of base excision repair: glycosylase mechanisms and structures. Annu. Rev. Biochem., 68, 255–285. [DOI] [PubMed] [Google Scholar]
- 47.Dodson M.L., Michaels,M.L. and Lloyd,R.S. (1994) Unified catalytic mechanism for DNA glycosylases. J. Biol. Chem., 269, 32709–32712. [PubMed] [Google Scholar]
- 48.Girard P.M. and Boiteux,S. (1997) Repair of oxidized DNA bases in the yeast Saccharomyces cerevisiae. Biochimie, 79, 559–566. [DOI] [PubMed] [Google Scholar]
- 49.Ohtsubo T., Matsuda,O., Iba,K., Terashima,I., Sekiguchi,M. and Nakabeppu,Y. (1998) Molecular cloning of AtMMH, an Arabidopsis thaliana ortholog of the Escherichia coli mutM gene and analysis of functional domains of its product. Mol. Gen. Genet., 259, 577–590. [DOI] [PubMed] [Google Scholar]
- 50.Gogos A. and Clarke,N.D. (1999) Characterization of an 8-oxoguanine DNA glycosylase from Methanococcus jannaschii. J. Biol. Chem., 274, 30447–30450. [DOI] [PubMed] [Google Scholar]
- 51.Breimer L.H. (1986) A DNA glycosylase for oxidized thymine residues in Drosophila melanogaster. Biochem. Biophys. Res. Commun., 134, 201–204. [DOI] [PubMed] [Google Scholar]
- 52.Deutsch W.A. and Spiering,A.L. (1982) A new pathway expressed during a distinct stage of Drosophila development for the removal of dUMP residues in DNA. J. Biol. Chem., 257, 3366–3368. [PubMed] [Google Scholar]
- 53.Green D.A. and Deutsch,W.A. (1983) Repair of alkylated DNA: Drosophila have DNA methyltransferases but not DNA glycosylases. Mol. Gen. Genet., 192, 322–325. [DOI] [PubMed] [Google Scholar]
- 54.Le Page F., Klungland,A., Barnes,D.E., Sarasin,A. and Boiteux,S. (2000) Transcription coupled repair of 8-oxoguanine in murine cells: the ogg1 protein is required for repair in nontranscribed sequences but not in transcribed sequences. Proc. Natl Acad. Sci. USA, 97, 8397–8402. [DOI] [PMC free article] [PubMed] [Google Scholar]