Abstract
The 11 Rap proteins of Bacillus subtilis comprise a conserved family of tetratricopeptide (TPR)-containing regulatory proteins. Their activity is inhibited by specific Phr pentapeptides produced from the product of phr genes through an export-import maturation process. We found that one of the proteins, namely RapF, is involved in the regulation of competence to DNA transformation. The ComA response regulator and transcription factor for initiation of competence development is the target of RapF. Specific binding of RapF to the carboxy-terminal DNA-binding domain of ComA inhibits the response regulator's ability to bind its target DNA promoters. The PhrF C-terminal pentapeptide, QRGMI, inhibits RapF activity. The activity of RapF and PhrF in regulating competence development is analogous to the previously described activity of RapC and PhrC (L. J. Core and M. Perego, Mol. Microbiol. 49:1509-1522, 2003). In fact, the RapF and PhrF pair of proteins acts synergistically with RapC and PhrC in the overall regulation of the ComA transcription factor. Since the transcription of the RapC- and RapF-encoding genes is positively regulated by their own target ComA, an autoregulatory circuit must exist for the competence transcription factor in order to modulate its activity.
Signal transduction in prokaryotes is mainly carried out by the so-called two-component systems consisting of a histidine protein kinase and a response regulator. The kinase acts as a sensor of a specific signal which, upon binding, activates the kinase by inducing autophosphorylation of the protein kinase on a histidine residue. The phosphoryl group is subsequently transferred to a paired response regulator, thus activating its function, generally of transcription regulation, allowing the cells to respond and adapt to the specific signal (12).
A key issue for the proper functioning of a signal transduction system is its ability to balance the input signaling with the output response. This was thought to occur through regulation of the overall phosphorylation state of the system by means of protein phosphatases counteracting the protein kinases. This control mechanism was shown to be essential not only in eukaryotic signal transduction (17, 26) but also in some of the best understood bacterial two-component systems, i.e., sporulation in Bacillus subtilis and chemotaxis in Escherichia coli (30, 49). The B. subtilis sporulation system, in particular, was shown to be regulated by the opposing activity of five histidine kinases and six aspartyl phosphate phosphatases (15, 16, 31, 33). Among the latter, three proteins belong to the Spo0E family of phosphatases targeting the Spo0A response regulator and transcription factor of the phosphorelay (31). The other three (RapA, -B, and -E) belong to the Rap family of proteins and specifically dephosphorylate the Spo0F response regulator intermediate of the sporulation pathway (15, 33). The Rap family of proteins comprises 11 members in B. subtilis, all sharing high levels of amino acid sequence homology and a common structural organization characterized by six tetratricopeptide repeats (TPR) (19, 32, 40).
Seven of the 11 Rap proteins (RapA, -C, -E, -F, -G, -I, and -K) are regulated by specific pentapeptide inhibitors which result from an export-import pathway followed by the pre-pro precursor product of the rap-associated phr genes (29). Recently, a fourth member of the Rap family, RapC, was found to affect the B. subtilis two-component signal transduction system ComA-ComP, which controls the initiation of competence development to DNA transformation (6, 8). RapC was found to affect this system not through a dephosphorylation mechanism but by inhibition of the DNA-binding activity of the ComA transcription factor. Similarly, Ogura et al. reported that the RapG protein inhibited DNA binding activity of the DegU response regulator which controls many cellular processes, including exprotease production and siderophore formation (27). These results indicated that Rap proteins have not only evolved to carry out different specific mechanisms of regulation based on protein-protein interaction but also have introduced the concept that regulation of the level of phosphorylation of a signal transduction system is only one of the mechanisms developed by the cell to regulate the output of two-component systems.
In this report we show that another protein of the Rap family, RapF, and its associated PhrF peptide contribute to regulation of competence development synergistically with RapC-PhrC by inhibiting the DNA binding function of the ComA DNA-binding domain.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The B. subtilis strains used in this study are listed in Table 1. Strains were grown in Schaeffer's sporulation medium (38) in the presence of the appropriate antibiotic at the following concentrations: chloramphenicol, 5 μg/ml; kanamycin, 2 μg/ml; spectinomycin, 100 μg/ml; erythromycin, 5 μg/ml; lincomycin, 25 μg/ml. Competent cells were prepared by the method of Anagnostopoulos and Spizizen (1).
TABLE 1.
Bacillus subtilis strains used in this study
| Strain | Relevant genotypea | Originb |
|---|---|---|
| JH642 | Parental | Laboratory stock |
| JH646 | spo0A12 | Laboratory stock |
| JH651 | spo0H81 | Laboratory stock |
| JH12547 | spo0A12, abrB::Tn917mls | Laboratory stock (36) |
| JH12575 | abrB::Tn917mls | Laboratory stock (36) |
| JH12981 | amyE::rapA-lacZ aph3A | Laboratory stock |
| JH23059 | amyE::rapA-lacZ aph3A rapF::cat | pRapF37→JH12981 |
| JH23065 | amyE::rapA-lacZ aph3A rapC::spc rapF::cat | JH11196→JH23059 |
| JH11030 | amyE::rapA-lacZ aph3A rapC::cat | JH12981→JH12939 |
| JH11204 | amyE::rapF-lacZcat | pRapF32B→JH642 |
| JH11208 | amyE::rapF-lacZspc | pCm::Spc→JH11204 |
| JH11223 | comA::cat amyE::rapF-lacZspc | BD1626→JH11208 (47) |
| JH11224 | comP::cat amyE::rapF-lacZspc | BD1658→JH11208 (47) |
| JH11423 | amyE::rapA-lacZ aph3A phrC::spc | JH12981→JH11303 |
| JH11500 | amyE::rapA-lacZ aph3A phrF::cat | JH11478→JH12981 |
| JH11507 | amyE::phrF-lacZcat | pRapF38→JH642 |
| JH11508 | amyE::rapA-lacZ aph3A phrC::spc phrF::cat | JH11478→JH11423 |
| JH19163 | amyE::rapFphrF-lacZcat | pRapF39→JH642 |
| JH23060 | amyE::rapA-lacZ aph3A pSS8 | pSS8→JH12981 |
| JH23061 | amyE::rapA-lacZ aph3A pSS9 | pSS9→JH12981 |
| JH23062 | amyE::rapA-lacZ aph3A pSS10 | pSS10→JH12981 |
| JH23063 | amyE::rapA-lacZ aph3A pSS11 | pSS11→JH12981 |
| JH23064 | amyE::rapA-lacZaph3A pHT315 | pHT315→JH12981 |
| JH23066 | spo0A12 amyE::phrF-lacZcat | JH11507→JH646 |
| JH23067 | spo0A12 abrB::Tn917mls amyE::phrF-lacZcat | JH11507→JH12547 |
| JH23068 | abrB::Tn917mls amyE::phrF-lacZcat | JH11507→JH12575 |
| JH23069 | spo0H81 amyE::phrF-lacZcat | JH11507→JH651 |
| JH11196 | rapC::spc | pCm::Spc→JH12939 |
| JH12939 | rapC::cat | p0M47→JH642 |
| JH11298 | phrC::cat | p0M88→JH642 |
| JH11303 | phrC::spc | pCm::Spc→JH11298 |
| JH11478 | phrF::cat | pRapF35→JH642 |
All strains are derivatives of JH642 and thus carry the trpC2 phe-1 auxothrophic markers. Antibiotic resistance genes are expressed as follows: mls, erythromycin and lincomycin; aph3A, kamamycin; cat, chloramphenicol; spc, spectinomycin.
Arrows indicate construction by transformation using plasmid DNA or chromosomal DNA.
E. coli DH5α was used for plasmid construction and propagation. Cells were grown in Luria-Bertani (LB) medium containing antibiotics at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 20 μg/ml; spectinomycin, 200 μg/ml.
Assays for sporulation efficiency were carried out in Schaeffer's sporulation medium. Cells were grown for 28 h at 37°C, and then serial dilutions were plated before and after chloroform treatment.
β-Galactosidase assay.
B. subtilis cultures were grown in Schaeffer's sporulation medium. Samples were taken at hourly intervals and processed according to Miller (24). Activity was measured in Miller units (24).
Plasmid constructions.
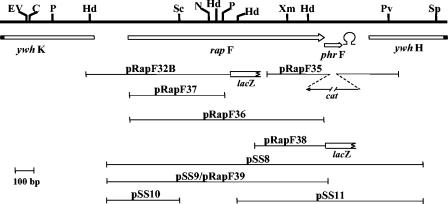
The plasmids used in this study are schematically represented in Fig. 1. The oligonucleotides used are listed in supplemental Table S1. A plasmid named p0F30, generously provided by Philippe Glaser (Institut Pasteur), contained a 3.3-kb chromosomal region carrying the rapF-phrF loci. Plasmid pRapF32B was constructed in the lacZ transcriptional fusion vector pJM116 (a derivative of pDH32 [9] carrying an extended multiple cloning site; M. Perego, unpublished data). An approximately 700-bp fragment was recovered by HindIII digestion of plasmid p0F30 and ligated into the HindIII site of pJM116. The correct orientation was determined by EcoRI-NdeI digestion.
FIG. 1.
Restriction map of the chromosomal region containing the rapF-phrF genes. Arrows indicate open reading frames and their direction. The position of a putative transcription terminator downstream of phrF is indicated. Fragments cloned in plasmids used in this study are indicated by lines. Restriction sites relevant to this study are indicated with the following symbols: C, ClaI; EV, EcoRV; Hd, HindIII; N, NdeI; P, PstI; Pv, PvuII; Sc, ScaI; Sp, SphI; Xm, XmnI. The ywhK and ywhH open reading frames are shown truncated.
Plasmid pRapF35 was constructed in the pJM105A vector (28) by cloning a 360-bp BamHI-EcoRI fragment on the left of the cat gene and a 450-bp SalI-KpnI on the right side. The left-side fragment was generated with oligonucleotides phrFΔEcoRI and phrFΔBamHI. The right-side fragment was amplified using oligonucleotides phrFΔSalI and phrFΔKpnI.
Plasmid pRapF37 was constructed in the integrative plasmid pJM103 (28) by cloning a 540-bp BamHI-PstI fragment obtained from plasmid pRapF36.
Plasmid pRapF36 was constructed in the pET16b expression vector (Novagen) by cloning a 1,170-bp BamHI fragment obtained by PCR amplification using ologonucleotides RapF5′BamHI and RapF3′BamHI.
Plasmid pRapF38 was constructed in the lacZ transcriptional fusion vector pDH32 (9) by cloning a 300-bp EcoRI-BamHI fragment generated by PCR amplification using oligonucleotides PhrF lac5′EcoRI and RapF3′BamHI.
Plasmid pRapF39 is a derivative of pDH32 (9) carrying the same fragment of plasmid pSS9. Plasmids pSS8, pSS9, pSS10, and pSS11 were constructed in the B. subtilis multicopy vector pHT315 (2). The fragment carried by pSS8 was generated by PCR amplification using oligonucleotides RapF5′EcoRI and PhrFΔKpnI. The fragment carried by pSS9 was generated by PCR amplification using oligonucleotides RapF5′EcoRI and RapF3′BamHI. The 440-bp fragment of pSS10 was obtained by digestion of pSS8 with EcoRI and ScaI. The fragment was then cloned in pHT315 digested with EcoRI and SmaI. The 654-bp fragment of plasmid pSS11 was obtained by digestion of pSS8 with XmnI and KpnI. The fragment was then cloned in pHT315 digested with SmaI and KpnI. All fragments derived from PCR amplification reactions were subject to sequence analysis to verify the fidelity of amplification.
The vector used for the expression of ComA, ComA-N, and ComA-C was pET28a (Novagen). The full-length comA gene was amplified using oligonucleotides ComA5′BspHI and ComA3′XhoI. After digestion with BspHI and XhoI, the fragment was ligated into pET28a digested with NcoI and XhoI, thus generating a fusion at the 3′ of the gene with six histidine codons. The plasmid expressing ComA-N (residues 1 to 126) was obtained by amplifying the 5′ domain of the comA gene using oligonucleotides ComA5′BspHI and ComAN′XhoI. Cloning of this fragment in pET28a digested with NcoI and XhoI generated a fusion of six histidine codons to the 3′ end of the gene. The 3′ portion of the comA gene encoding ComA-C (residues 146 to 214) was PCR amplified using oligonucleotides ComA-C′NdeI and ComA3′BamHI. Cloning of the fragment in pET28a-digested NdeI-BamHI resulted in the fusion of a tag comprising 6 histidine codons and 10 additional codons to the 5′ end of the comA gene. All PCR amplifications were carried out on genomic DNA extracted from strain JH642.
Protein expression and purification. His6-RapF.
One single colony of E. coli BL21(DE3) pLysS (Novagen) transformed with plasmid pET16bRapF(pRapF36)was grown overnight at 37°C in 20 ml of LB medium (ampicillin, 100 μg/ml). This culture was used to inoculate 2 liters of LB medium containing ampicillin (100 μg/ml) and grown at 37°C until the cells reached an optical density at 600 nm (OD600) of 0.5. The expression of RapF was induced with isopropyl-β-d-thiogalactopyranoside (IPTG) at 0.5 mM, and the culture was incubated for another 2 h. Cells were then harvested by centrifugation at 4°C. The pellet was resuspended in lysis buffer (50 mM HEPES, pH 7, 100 mM NaCl, 10 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 mM imidazole). The cells were lysed by sonication on ice and with one passage through a French press under 1,000 lb/in2 of pressure. After removal of the cell debris by ultracentrifugation at 143,000 × g for 1 h at 4°C, the supernatant was loaded onto a nickel nitrilotriacetic acid (NTA) agarose column (QIAGEN) and rinsed with 10 ml of washing buffer (50 mM HEPES, pH 7, 100 mM NaCl, 10 mM β-mercaptoethanol). The His6-RapF protein was eluted in washing buffer containing an increasing amount of imidazole (20 to 200 mM). The fractions containing the His6-RapF protein were dialyzed against the storage buffer (50 mM HEPES, pH 7, 100 mM NaCl, 10 mM dithiothreitol [DTT]) and concentrated with an Amicon Centriprep-30. The protein was stored in aliquots at −80°C after the addition of glycerol to 20% final concentration. The His6-RapF protein was analyzed by matrix assisted laser desorption-ionization time-of-flight (MALDI-TOF)-mass spectometry to ensure its correct size.
His6-ComA.
An overnight culture of E. coli BL21(DE3)pLysS carrying the plasmid pET16b-ComA was used to inoculate 1 liter of LB medium containing ampicillin (100 μg/ml). When the OD600 of the culture reached 0.7, IPTG was added to a final concentration of 1 mM; after 3 h of incubation at 37°C, the cells were collected by centrifugation, resuspended in 40 ml of ComA buffer (50 mM Tris-HCl, pH 7.5, 50 mM KCl, 2 mM EDTA, 5 mM imidazole, and 1 mM PMSF), and lysed with a French press. The cell lysate was centrifuged at 26,800 × g for 1 h at 4°C; the supernatant was applied to a nickel-NTA agarose column (QIAGEN) that was washed with 10 column volumes of washing buffer (50 mM Tris-HCl, pH 7.5, 50 mM KCl, 2 mM EDTA, 20 mM imidazole). His6-ComA was eluted with a gradient of imidazole, and the fractions containing the protein were pooled, dialyzed in ComA buffer (without imidazole) and concentrated with an Amicon centriprep-10. Glycerol was added to a final concentration of 20%, and the fractions were stored at −80°C.
His6-ComA C terminal and His6-ComA N terminal.
The N-terminal and the C-terminal domains of ComA were purified following an identical protocol. A 10-ml overnight culture from a single colony of E. coli BL21(DE3)pLysS containing plasmid pET28a-ComA-N or pET28-ComA-C was used to inoculate 1 liter of LB medium containing 30 μg/ml kanamycin and incubated with shaking at 37°C until the OD600 reached 0.6. IPTG was added to a 1 mM final concentration and incubation was continued for 2 h. The cells were harvested by centrifugation at 6,080 × g for 20 min at 4°C, resuspended in 40 ml of ComA-C-N buffer (50 mM Tris-HCl, pH 8, 200 mM KCl, 5 mM β-mercaptoethanol, 5 mM imidazole, and 1 mM PMSF), and lysed with a French press. The cell lysate was centrifuged at 30,590 × g for 30 min at 4°C, and the supernatant was applied to a nickel-NTA agarose column (QIAGEN) previously washed with 10 column volumes of ComA-C-N buffer. The column was washed with washing buffer (50 mM Tris-HCl, pH 8, 200 mM KCl, 5 mM β-mercaptoethanol, 10 mM imidazole). The protein was eluted with ComA-C-N buffer (without PMSF) containing increasing concentrations of imidazole (20 to 300 mM). The fractions eluted with 100 mM imidazole containing the ComA-C terminal protein were pooled and dialyzed against the storage buffer (50 mM EPPS [4-(2-hydroxyethyl)piperazine-1-propanesulfonic acid], pH 8, 200 mM KCl, 1 mM DTT), concentrated with an Amicon Centriprep-3, and stored at −80°C after the addition of glycerol to 20% (vol/vol). The fractions containing the ComA-N-terminal protein were pooled and dialyzed against the storage buffer (50 mM Tris-HCl, pH 8, 200 mM KCl, 10 mM β-mercaptoethanol). Proteins were concentrated using an Amicon Centriprep-10. Glycerol was added to 20% (vol/vol) final concentration, and the proteins were stored at −80°C. The concentration of each protein was measured by Bio-Rad's Protein Assay.
Native polyacrylamide gel electrophoresis (PAGE).
Native-PAGE was carried out according to Schägger and Von Jangow with some adjustments as previously described (13, 39). The gels were prepared with 10% acrylamide (29:1) in buffer 1 M Tris-HCl, pH 8.45, and the running buffer was 0.1 M Tris-HCl and 0.1 M Tricine (pH 8.45). The stacking gel was prepared with the same buffer using 4% acrylamide.
Yeast two-hybrid analysis.
Yeast strains used in this study were PJ69-4A (MATa, trp1-901, leu2-3, 112ura3-52, his-200, gal4Δ, gal80Δ, LYS2::GAL1-HIS3, GAL2-ADE2, met2::GAL7-lacZ) and PJ69-4α (MATa, trp1-901, leu2-3, 112ura3-52, his-200, gal4Δ, gal80Δ, LYS2::GAL1-HIS3, GAL2-ADE2, met2::GAL7-lacZ), kindly provided by Philip James (14). Plasmids pGBT9 and pGAD424, DNA-binding and activation domain fusion vectors, respectively, were obtained from Clontech.
The entire coding sequence of rapA, rapB, rapC, rapE, rapF, comA, and spo0F as well as the comA 5′ and 3′ ends (1 to 411 bp and 412 to 620 bp of the coding sequence) were amplified by PCR from B. subtilis 168 genomic DNA using the primers listed in Table S1. The amplified fragments were cloned in pGBT9 or pGAD424 and the inserts were verified by sequence analysis. The pGBT9 derivatives (TRP1) and the pGAD derivatives (LEU2) were used to transform the mating type yeast strains PJ69-4A and PJ69-4α, respectively, using essentially the method described by Yoshimura et al. (48). Transformants were mated in the appropriate liquid media using flat-bottom 96-well plates. After mating, the cultures were collected, washed with sterilized water, and then spotted on synthetic complete (SC) agar plates lacking leucine and tryptophane (SC-LW) for selection of LEU2 and TRP1 diploid cells. These cells were cultured in liquid SC-LW for 1 day and then were replicated on selection agar medium lacking histidine (SC-LEH) and supplemented with 1 mM 3-aminotrizaole (3-AT) to inhibit autoactivation of the HIS3 reporter gene.
Electrophoresis mobility shift assay.
The 313-bp NsiI-EcoRI fragment from pOM48 carrying the rapC promoter (6) was labeled with [α-32P]dATP using Klenow polymerase (New England Biolabs). The labeled fragment was purified from a 1% agarose gel using the QIAGEN gel extraction kit. Binding of ComA to the DNA fragment was performed at room temperature in half-strength ComA buffer (without imidazole) and half strength RapF buffer with DTT at 20 mM final concentration. Double-stranded poly[dA-dT] and poly[dG-dC] were added to 150 μg/ml final concentration. ComA was first incubated with the DNA for 5 min in the presence of poly deoxynucleoside triphosphates. Rap proteins were then added and the reactions allowed to proceed for an additional 5 min. The Phr peptides were then added and the reactions continued for an additional 5 min before stopping them by addition of loading dye. The samples were immediately loaded on a 5% prewarmed polyacrylamide gel running at 150 constant voltage in Tris-acetate-EDTA buffer. The gel was dried and exposed to a PhosphorImager screen (Amersham Molecular Dynamics).
RESULTS
rapF and phrF mutants affect ComA-dependent transcription.
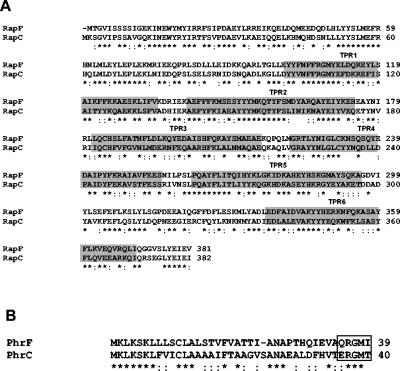
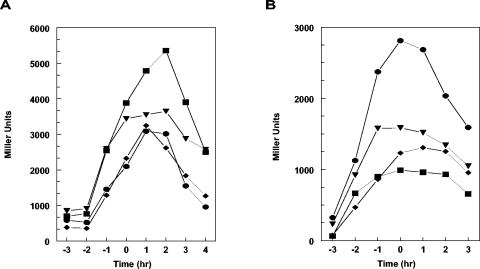
Within the B. subtilis Rap family of proteins, the RapC-RapF pair shows the highest level of conservation, with 57% of identical residues and 22.7% of conserved substitutions (Fig. 2A). Correspondingly, the 39-amino-acid product of the phrF gene is mostly related to the 40-amino-acid product of the phrC gene (48% of identity). The PhrF C-terminal pentapeptide (QRGMI) also shares three identical residues with the PhrC active pentapeptide (ERGMT) (Fig. 2B). Thus, it was not surprising that a deletion analysis carried out on the rap and phr genes of B. subtilis revealed that the RapF-PhrF pair of proteins affected competence development at its initial stages in a manner similar to the one described for the RapC-PhrC system (44, 6) (M. Perego, unpublished). In fact, it was observed that a deletion of the phrF gene significantly decreased the transcription of the ComA-dependent promoter of the rapA gene (Fig. 3B). Surprisingly, however, the deletion of the rapF gene did not significantly change rapA transcription (Fig. 3A). Similarly, a deletion of phrF resulted in a fourfold reduction of activity of a srfA-lacZ fusion construct while the deletion of rapF did not have any significant effect (data not shown).
FIG. 2.
Amino acid sequence alignment of RapF and RapC (A) or PhrF and PhrC (B). Alignments were obtained by the ClustalW program. Shaded areas in panel A identify the TPR domains characterizing Rap proteins. The box in panel B identifies the sequence of the active pentapeptide inhibitors. Asterisks indicate identical residues, while colons denote conserved residues.
FIG. 3.
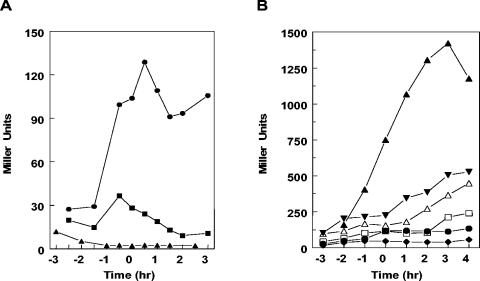
Transcription regulation of rapA in rapF, rapC, phrF, and phrC mutants. Strains carrying rapA-lacZ transcriptional fusion constructs were grown in Schaffer's sporulation medium. Time points were taken at hourly intervals before and after the transition (To) from exponential growth to stationary phase. A. •-, JH12981 wild type; -♦-, JH23059 rapF; -▾-, JH11030 rapC; -▪-, JH23065 rapC rapF. B. •-, JH12981 wild type; -♦-, JH11500 phrF; -▾-, JH11423 phrC; -▪-, JH11508 phrC phrF.
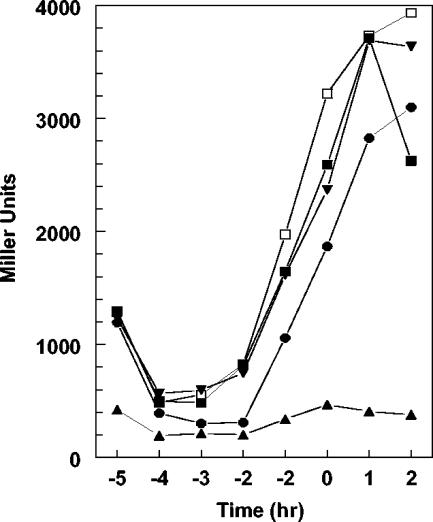
Since the effect of a negative regulator is often more evident when analyzed in an overproducing mutant, the rapF gene was cloned on the multicopy plasmid pHT315 (2) and its effect on rapA transcription was analyzed. As shown in Fig. 4, overproduction of RapF from plasmid pSS9 severely reduced β-galactosidase activity from the rapA-lacZ fusion promoter compared to the control strain carrying the rapF promoter alone (pSS10). The presence of phrF together with rapF in plasmid pSS8 restored a transcription level comparable to the control strain, while the multicopy phrF alone (pSS11) did not significantly affect the transcription of rapA.
FIG. 4.
Transcriptional regulation of rapA in RapF-PhrF overexpressing strains. Cultures for β-galactosidase analysis were grown in Schaeffer's sporulation medium containing erythromycin 5 μg/ml and lincomycin 25 μg/ml. Strains and symbols: -▴-, JH23061 (pSS9); -•-, JH23060 (pSS8); -▾-, JH23061 (pSS11); -▪-, JH23062 (pSS10); -□-, JH23064 (pHT315).
These results allowed us to conclude that RapF inhibited ComA-dependent transcription and PhrF counteracted this effect in a manner similar to the one described for RapC and PhrC.
Synergistic effect of RapC and RapF.
In an attempt to understand the relative contribution of RapC and RapF in regulating ComA-dependent transcription, β-galactosidase analyses were carried out on a strain containing the rapA-lacZ fusion in the presence of rapC and rapF single and double mutants. The analysis in the single-mutant strains showed that the absence of RapC resulted in a visible increase of rapA transcription while the effect of the absence of RapF, as mentioned above, was basically undetectable (Fig. 3A). The double deletion of rapC and rapF, however, had a synergistic effect as it increased the level of rapA transcription of approximately 25% compared to the wild-type strain (Fig. 3A).
When the effect of phr peptides was analyzed, we observed that the deletion of phrF had a stronger effect on rapA transcription than the deletion of phrC, and a synergistic effect of the double mutant phrC phrF was also detected (Fig. 3B). These results confirmed that RapC and RapF synergistically affect ComA activity.
Transcription regulation of rapF and phrF.
A chromosomal fragment presumably containing the rapF promoter region was cloned in the pJM115 transcriptional fusion vector, thus creating a fusion to the E. coli lacZ gene. This fragment contains an imperfect inverted repeat similar to the ComA boxes identified upstream of the srfA, degQ, and rapA promoter regions (25). After integration in the amyE chromosomal region, the transcription driven from this promoter was analyzed in various backgrounds. We observed that this rapF-lacZ fusion was affected when analyzed in strains mutated in the comA or comP genes encoding the two-component response regulator and histidine kinase, respectively, for the initiation of competence development (Fig. 5A). As previously described for the rapA and rapC promoters, the deletion of the comA response regulator had a more drastic effect than the deletion of comP. Thus, as previously shown for the rapA, rapC, and rapE genes, rapF is also under transcriptional control of the ComA competence factor (6, 15, 25).
FIG. 5.
Transcription analysis of the rapF (A) and phrF (B) promoters. Strains carrying the rapF-lacZ fusion construct pRapF32B or the phrF-lacZ plasmid pRapF38 were grown in Schaeffer's sporulation medium and assayed as described in Materials and Methods. A. -•-, JH12981 wild type; -▪-, JH11224 comP; -▴-, JH11223 comA. B. -□-, JH11507 wild type; -♦-, JH23066, spo0A; -•-, JH23069 spo0H; -▵, JH23068 abrB; -▾-, JH23067 spo0A abrB, -▴-, JH19163 (rapFphrF-lacZ).
The phrF gene is in an operon with rapF, as shown by the β-galactosidase analysis carried out on the JH19163 strain containing the lacZ fusion construct of plasmid pRapF39 (Fig. 5B). However, it was reported to have an independent promoter controlled by the sigma H sigma factor (23). Our analysis of transcription driven by the fragment cloned in plasmid pRapF38 (Fig. 1) and fused to the E. coli lacZ gene confirmed that the events controlling the initiation of the sporulation process have an impact on phrF transcription. In fact, we observed that an spo0A mutant severely impaired phrF transcription, and this effect was overcome by a mutation in the abrB transition phase regulator gene. In a sigH mutant, transcription of phrF did not reach the same level observed for the wild-type strain at 4 h past the transition phase (Fig. 5B), confirming that phrF induction during the stationary phase requires SigH.
RapF inhibits ComA DNA-binding activity and PhrF counteracts this effect.
Our previous work on the characterization of the mechanism of ComA regulation by the RapC-PhrC system showed that RapC binds the response regulator and inhibits its DNA-binding capability (6). The PhrC peptide was shown to bind to and inhibit RapC, thus restoring ComA-DNA binding in an in vitro assay using purified components.
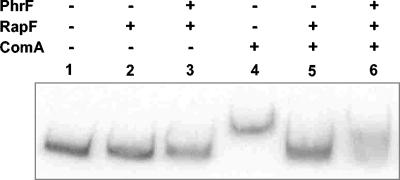
Because the genetic analysis showed that RapF and PhrF also affected the transcriptional activity of ComA, and because of the strong conservation between the RapF-RapC and the PhrF-PhrC proteins (Fig. 2), we tested whether a purified RapF protein could inhibit ComA binding to one of its target promoters, i.e., the rapC promoter. As shown in Fig. 6, inhibition of ComA binding to the rapC promoter was inhibited by RapF at equimolar concentration. The addition of a synthetic PhrF carboxy-terminal pentapeptide (QRGMI) partially restored ComA binding to the DNA promoter. The observation that a 10-fold excess of PhrF pentapeptide only partially inhibited RapF activity is most likely due to the fact that the synthesis of PhrF resulted in two products as shown by mass spectrometry analysis (data not shown). One product was of the expected mass (603 Da), and the other was 16 Da smaller (587 Da). The smaller product was most likely the result of a cyclization reaction occurring at the initial glutamine residue to give pyroglutamate with the loss of an NH2 group (Tony Wilkinson, Arthur Moir, and Andy Parsons, personal communication). Since this smaller size peptide accounted for approximately 40% of the total synthesis product, this contamination may explain the low efficiency of RapF inhibition by PhrF that we observed in vitro.
FIG. 6.
Electrophoresis mobility shift assay of ComA binding to the rapC promoter. The labeled 313-bp rapC promoter fragment was prepared as described in Materials and Methods and used at 1.5 nM final concentration per lane. ComA and RapF were used at 5 μM while the PhrF peptide was added at 50 μM final concentration.
RapF interacts with ComA and PhrF dissociates this complex.
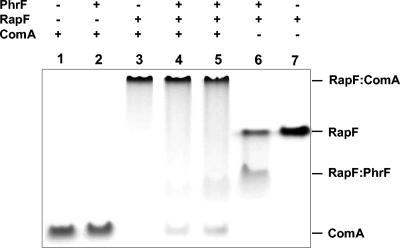
We previously showed that RapC formed a stable complex with ComA, and this complex was dissociated by the binding of PhrC to RapC (6). Since RapF and PhrF behaved like RapC and PhrC in inhibiting ComA binding to its target DNA promoter, we carried out an in vitro protein binding assay in native condition to demonstrate complex formation between RapF and ComA as well as RapF and PhrF. As shown in Fig. 7, the native gel-binding assay confirmed that RapF formed a stable complex with ComA, and this complex was dissociated when PhrF was added to the reaction mixture. Furthermore, the RapF:PhrF complex showed a faster mobility than the RapF dimer alone.
FIG. 7.
Interaction of RapF with ComA and PhrF. ComA (10 μM), RapF (10 μM), and PhrF (50 μM) were analyzed on a 10% Tris-Tricine-EDTA native gel stained with Coomassie blue. The gel was run at 4°C for 24 h at constant voltage (63 V). In lane 4, RapF was preincubated with ComA for 5 min before the addition of PhrF followed by a further 5-min incubation at room temperature. In lane 5, RapF was preincubated with PhrF and then ComA was added to the reaction mixture for a further 5-min incubation.
RapC and RapF interact with the carboxy-terminal DNA-binding domain of ComA.
The results described above raised the question of whether RapC and RapF inhibited ComA ability to bind to its target DNA by interacting with the amino terminal or the carboxy-terminal domain of the response regulator or both. In order to answer this question, a yeast two-hybrid system assay was devised for the identification of Rap protein targets. The rapA, rapB, rapC, rapE, and rapF genes were expressed as “bait” from plasmid pGBT9 while the spo0F, comA, comAN-terminal domain and comAC-terminal domain coding sequences were expressed as fusions to the GAL4 activation domain of the prey plasmid pGAD424 (Clontech Laboratories, Inc.)
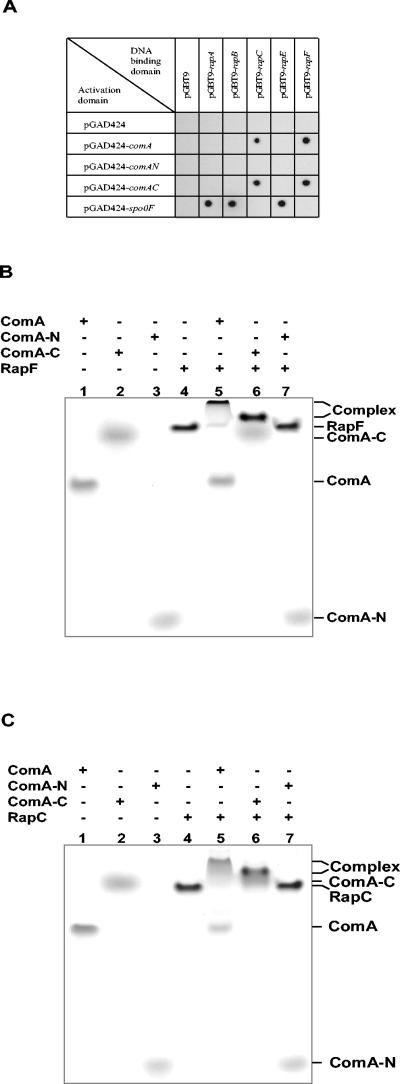
When baits and preys were cross analyzed, the control constructs expressing RapA, RapB, and RapE showed interaction with the plasmid expressing Spo0F, as expected. Instead, RapC and RapF interaction was observed with the construct expressing the full-length ComA as well as with the construct expressing the carboxy-terminal DNA binding domain of ComA. The construct expressing the amino-terminal response regulator domain of ComA did not interact with any of the constructs tested (Fig. 8A).
FIG. 8.
RapC and RapF interaction with ComA. A. Two-hybrid system analysis. Diploid strains were obtained by mating PJ69-4A (containing pGBT9 derivatives) with PJ69-4α (containing pGAD424 derivatives). Interaction between proteins was detected on selection medium (SC-LWH) supplemented with 1mM 3-AT 4 days after inoculation from liquid SC-LW medium. B. Native gel analysis of RapF interaction with ComA. Proteins (40 μM) were run on 10% acrylamide Tris-Tricine native gel for 40 h at constant voltage (63 V) at 4°C. C. Native gel analysis of RapC interaction with ComA. The gel was run as described in B.
This experiment provided strong evidence that the RapC and RapF proteins specifically interacted with the carboxy-terminal domain of ComA, thus inhibiting its DNA-binding activity.
RapC and RapF bind to the ComA carboxy-terminal domain in the native gel-binding assay.
The results of the DNA retardation assay and the yeast two-hybrid system prompted us to express and purify the amino-terminal and the carboxy-terminal domains of ComA as separate proteins in order to prove, in the native gel binding assay, the domain specificity of RapC and RapF. The full-length ComA protein and the ComA-N or ComA-C fragments were purified as described in Materials and Methods and used in the native gel binding assay previously described (13). As shown in Fig. 8B and C, RapF and RapC, respectively, formed a stable complex with the full-length ComA and the ComA carboxy-terminal domain but they did not interact with the ComA amino-terminal response regulator domain.
Thus, a second member of the Rap family of proteins acts in modulating the activity of a response regulator through a protein-protein interaction mechanism that does not involve a dephosphorylation reaction but rather acts by specifically inhibiting the DNA-binding capability of the transcription factor.
DISCUSSION
Competence in DNA transformation in B. subtilis is a complex developmental event that requires a sophisticated coordination of several physiological pathways in order to fully develop (8). Since competence is antagonistic to the terminal developmental pathway of sporulation, cells have devised cross check mechanisms that ensure a correct chain of events at any given time and/or environmental, metabolic, cell cycle condition. Although not all of these mechanisms are clearly understood at this time, we have developed a significant knowledge of how B. subtilis Rap proteins play a critical role in pathway cross regulation.
The 11 members of the B. subtilis Rap family share a highly conserved structural organization that places them in the widely spread group of proteins containing the structural domains called tetratricopeptide repeats (TPR) (32). TPR repeats are structurally conserved helical domains known to promote protein-protein or protein-ligand interactions (3, 4, 7, 10, 11). In previous studies we have demonstrated how the TPR domains of three Rap proteins, RapA, RapB, and RapE, are involved in the interaction of these proteins with either their target substrate or their specific inhibitor peptides (13). In fact, RapA, RapB, and RapE are negative regulators of the sporulation process by means of their ability to target the phosphorylated form of the Spo0F response regulator of the sporulation phosphorelay and induce its dephosphorylation (5, 15, 33). Rap activity is inhibited by specific pentapeptides (Phr) whose production follows an export-import pathway essential for their maturation from a precursor protein (29). Dephosphorylation of Spo0F∼P results in inhibition of the sporulation pathway, thus providing the cell with the possibility of remaining in vegetative growth or exploiting alternative developmental pathways known to be available at the transition phase of growth. These alternative pathways could be competence to DNA transformation, degradative enzyme production, motility, and perhaps others (42).
Competence development is well known for being a pathway antagonistic to sporulation in part because, during competence, the transcription of two of the Rap proteins, RapA and RapE, is induced by ComA (15, 25, 33). ComA also induces the transcription of the RapG coding gene thus affecting degradative enzyme production through the RapG regulation of the DegU response regulator transcription activity, as recently shown by Ogura et al. (27). Competence development, by means of the ComA response regulator and transcription factor, regulates the transcription of two additional Rap proteins, RapC and RapF, involved in regulation of ComA itself. RapC was shown to affect ComA activity, independently of the phosphorylation state of the response regulator, by inhibiting its ability to bind the DNA of target promoters (6). Here we showed that RapF acts similarly to RapC. We further analyzed this mechanism of DNA-binding inhibition by demonstrating that both, RapC and RapF, act on ComA by specifically binding to its DNA-binding domain in the C-terminal half portion of the protein. The binding of RapC or RapF to ComA is inhibited by the PhrC and PhrF pentapeptides, respectively. Both peptides are transcriptionally coupled to their corresponding rap gene, but they are also independently transcribed from a sigma H-dependent promoter located within the coding region of the preceding rap gene (23).
The transcriptional regulation of the rapC-phrC and rapF-phrF systems, although experimentally defined, creates a paradoxical situation in the bacterial physiology. In fact, transcription of the RapF- and RapC-encoding genes is dependent upon their own target ComA, perhaps as a part of an autoregulatory circuit. However, the peptides that inhibit the Rap proteins, PhrF and PhrC, are also induced at relatively higher levels when sporulation initiates and sigma H reaches its maximal activity. Then the question arises of why the negative regulators of competence development (RapC and RapF) would be inhibited by the peptides at a time when sporulation initiates, and thus competence should be repressed. Perhaps the induction of PhrC and PhrF by SigH is not effective on the corresponding Rap proteins due to limited reinportation of the peptides through the Opp transport system (34, 35, 37, 43, 45). If competition for OppA binding solely regulates the intracellular concentration of Phr peptides, as inferred from the structural characteristics of these transporters, then at the transition phase of growth the level of expression of phrA, the gene encoding the inhibitor of the sporulation regulator RapA, is approximately 15-fold higher than the expression of phrF and approximately 150-fold higher than the expression of phrC (18). This could result in a much higher concentration of exported PhrA peptide than PhrF or PhrC, and, consequently, higher reinportation of the former than the latter two, thus favoring sporulation development. Lack of inhibition of RapC and RapF would then eventually result in inhibition of ComA activity as seen by the decrease in rapA, rapC, or rapF transcription at approximately 2 h after the transition to stationary phase.
The existence of an autoregulatory circuit on ComA activity can also explain the observation that a deletion of rapF did not affect the transcription of rapA while the deletion of rapC did. In fact, deletion of one Rap regulator of ComA must impact on the overall level of transcription of the other. However, the increased activity of ComA that results from rapF deletion may be counterbalanced by the increased transcription of its negative regulator RapC, thus leaving the transcription of rapA unaffected. The additive effect of the double mutant rapC rapF and the inhibition of ComA-dependent transcription carried out by the overexpression of RapF clearly confirm the negative role of this protein in competence transcription regulation.
The PhrC peptide, also known as CSF for competence-stimulating factor (22, 44), was found to have three independent effects on B. subtilis physiology, each dependent on the concentration of the exogenously provided peptide (20). At low concentration it stimulated competence gene expression as a result of its inhibitory activity on RapC. At high concentration PhrC stimulated sporulation initiation most likely because of its inhibitory activity on the RapB sporulation phosphatase (13). Additionally, at high concentration, PhrC also inhibited competence gene expression, a phenotype that has never been explained experimentally. Our findings that RapF and PhrF are part of the autoregulatory circuit that controls competence gene expression synergistically with RapC/PhrC could provide an explanation for the unaccounted third phenotype of PhrC. The high level of homology between RapC and RapF or PhrC and PhrF suggested at first the possibility of peptide cross inhibition (RapC/PhrF or RapF/PhrC) or interference of one peptide with the activity of the other against its own target. Extensive native gel binding assay analyses, however, indicated that PhrC did not interact with RapF and it did not interfere in the RapF interaction with PhrF. Similarly, we did not observe any interaction of PhrF with RapC or interference in RapC-PhrC interaction (data not shown). Thus, the extreme level of specificity previously observed in Rap/Phr systems was confirmed.
One possibility we propose to explain the inhibitory effect on ComA-dependent transcription by high concentrations of PhrC is based again on the nonspecific mechanism of peptide reimportation by the Opp system. Extracellular addition of PhrC at high concentration may in fact reduce the overall reimportation of the PhrF peptide, with the consequence that RapF would not be inhibited and thus competence gene expression would decrease. This hypothesis could also explain the observation of Lazazzera et al. (20) that a mutated PhrC peptide (ARGMT) did not induce sporulation, most likely because it is unable to inhibit RapB, but could still inhibit competence gene expression because of its ability to compete with PhrF and other peptides for OppA. Furthermore, this mutant peptide could interfere with the RapF:PhrF interaction; single amino acid changes within a pentapeptide are known to generate very disparate effects (29). Other PhrC mutant peptides, EAGMT and ERGAT, were shown to be ineffective against both sporulation and competence, casting doubt on our theory of competition for OppA. However, a peculiar instability of these peptides in the medium cannot be ruled out. Curiously, none of the known active peptides in B. subtilis, B. thuringiensis (41), and B. anthracis (C. Bongiorni and M. Perego, unpublished data) contains an alanine residue in position 2 or 4 of the pentapeptide, suggesting perhaps a bias against this amino acid at least in these positions. Also, it should be pointed out that the tetrapeptide and nonapeptide ligands that cocrystallized with the Salmonella enterica serovar Typhimurium OppA and B. subtilis AppA transport proteins did not contain any alanine residues (21, 45). Perhaps the neutral nature of alanine and its lack of side chains reduces the affinity of the OppA-like transporter for peptides that contain this residue. Clearly a reevaluation of the effects of PhrC (CSF) and its mutants is necessary, in view of the existence of the RapF/PhrF system, in order to assign to them a correct function.
The finding that RapC and RapF bind to a DNA-binding domain of a transcription factor confirms our view of the Rap proteins as regulatory molecules that act by specifically interacting with their target, either a substrate or an inhibitor Phr peptide, thus affecting its activity. Protein-binding characteristics are provided by the structural organization of Rap proteins in the helical domains of the TPR type. The α-helices of the DNA-binding domain of ComA are likely to provide the surface of interaction with RapC and RapF. Molecular and structural studies are now under way to identify this interaction surface.
Supplementary Material
Acknowledgments
This research was supported, in part, by Public Health Service grant GM55594 from the National Institute of General Medical Sciences, National Institutes of Health. The Stein Beneficial Trust supported, in part, oligonucleotide synthesis and DNA sequencing.
Footnotes
Manuscript number 16655-MEM from The Scripps Research Institute.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arantes, O., and D. Lereclus. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108:115-119. [DOI] [PubMed] [Google Scholar]
- 3.Blatch, G. L., and M. Lassle. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932-939. [DOI] [PubMed] [Google Scholar]
- 4.Brinker, A., C. Scheufler, F. von der Mülbe, B. Bleckenstein, C. Herrmann, G. Jung, I. Moarefi, and F. U. Hartl. 2002. Ligand discrimination by TPR domains. J. Biol. Chem. 277:19265-19275. [DOI] [PubMed] [Google Scholar]
- 5.Burbulys, D., K. A. Trach, and J. A. Hoch. 1991. The initiation of sporulation in Bacillus subtilis is controlled by a multicomponent phosphorelay. Cell 64:545-552. [DOI] [PubMed] [Google Scholar]
- 6.Core, L. J., and M. Perego. 2003. TPR-mediated interaction of RapC with ComA inhibits response regulator-DNA binding for competence development in Bacillus subtilis. Mol. Microbiol. 49:1509-1522. [DOI] [PubMed] [Google Scholar]
- 7.Das, A. K., P. T. W. Cohen, and D. Barford. 1998. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 17:1192-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubnau, D., J. Hahn, M. Roggiani, F. Piazza, and Y. Weinrauch. 1994. Two-component regulators and genetic competence in Bacillus subtilis. Res. Microbiol. 145:403-411. [DOI] [PubMed] [Google Scholar]
- 9.Ferrari, E., D. J. Henner, M. Perego, and J. A. Hoch. 1988. Transcription of Bacillus subtilis subtilisin and expression of subtilisin in sporulation mutants. J. Bacteriol. 170:289-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatto, G. J., Jr., B. V. Geisbrecht, S. J. Gould, and J. M. Berg. 2000. Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat. Struct. Biol. 7:1091-1095. [DOI] [PubMed] [Google Scholar]
- 11.Groves, M. R., and D. Barford. 1999. Topological characteristics of helical repeat proteins. Curr. Opin. Struct. Biol. 9:383-389. [DOI] [PubMed] [Google Scholar]
- 12.Hoch, J. A., and T. J. Silhavy. 1995. Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 13.Ishikawa, S., L. J. Core, and M. Perego. 2002. Biochemical characterization of aspartyl phosphate phosphatase interaction with a phosphorylated response regulator and its inhibition by a pentapeptide. J. Biol. Chem. 277:20483-20489. [DOI] [PubMed] [Google Scholar]
- 14.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang, M., R. Grau, and M. Perego. 2000. Differential processing of propeptide inhibitors of Rap phosphatases in Bacillus subtilis. J. Bacteriol. 182:303-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang, M., W. Shao, M. Perego, and J. A. Hoch. 2000. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 38:535-542. [DOI] [PubMed] [Google Scholar]
- 17.Kurosawa, M. 1994. Phosphorylation and dephosphorylation of protein in regulating cellular function. J. Pharmacol. Toxicol. Methods 31:135-139. [DOI] [PubMed] [Google Scholar]
- 18.Lazazzera, B. A., I. G. Kurster, R. S. McQuade, and A. D. Grossman. 1999. An autoregulatory circuit affecting peptide signaling in Bacillus subtilis. J. Bacteriol. 181:5193-5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazazzera, B. A., T. Palmer, J. Quisel, and A. D. Grossman. 1999. Cell density control of gene expression and development in Bacillus subtilis, p. 27-46. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. American Society for Microbiology, Washington, D.C.
- 20.Lazazzera, B. A., J. M. Solomon, and A. D. Grossman. 1997. An exported peptide functions intracellularly to contribute to cell density signaling in B. subtilis. Cell 89:917-925. [DOI] [PubMed] [Google Scholar]
- 21.Levdikov, V. M., E. V. Blagova, J. A. Brannigan, L. Cladiere, A. A. Antson, M. N. Isupov, S. J. Seror, and A. J. Wilkinson. 2004. The crystal structure of YloQ, a circularly permuted GTPase essential for Bacillus subtilis viability. J. Mol. Biol. 340:767-782. [DOI] [PubMed] [Google Scholar]
- 22.Magnuson, R., J. Solomon, and A. D. Grossman. 1994. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell 77:207-216. [DOI] [PubMed] [Google Scholar]
- 23.McQuade, R. S., N. Comella, and A. D. Grossman. 2001. Control of a family of phosphatase regulatory genes (phr) by the alternate sigma factor sigma-H of Bacillus subtilis. J. Bacteriol. 183:4905-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Mueller, J. P., G. Bukusoglu, and A. L. Sonenshein. 1992. Transcriptional regulation of Bacillus subtilis glucose starvation-inducible genes: control of gsiA by the ComP-ComA signal transduction system. J. Bacteriol. 174:4361-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mustelin, T., and K. Tasken. 2003. Positive and negative regulation of T-cell activation through kinases and phosphatases. Biochem. J. 371:15-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogura, M., K. Shimane, K. Asai, N. Ogasawara, and T. Tanaka. 2003. Binding of response regulator DegU to the aprE promoter is inhibited by RapG, which is counteracted by extracellular PhrG in Bacillus subtilis. Mol. Microbiol. 49:1685-1697. [DOI] [PubMed] [Google Scholar]
- 28.Perego, M. 1993. Integrational vectors for genetic manipulation in Bacillus subtilis, p. 615-624. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 29.Perego, M. 1997. A peptide export-import control circuit modulating bacterial development regulates protein phosphatases of the phosphorelay. Proc. Natl. Acad. Sci. USA 94:8612-8617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perego, M. 1998. Kinase-phosphatase competition regulates Bacillus subtilis development. Trends Microbiol. 6:366-370. [DOI] [PubMed] [Google Scholar]
- 31.Perego, M. 2001. A new family of aspartyl-phosphate phosphatases targeting the sporulation transcription factor Spo0A of Bacillus subtilis. Mol. Microbiol. 42:133-144. [DOI] [PubMed] [Google Scholar]
- 32.Perego, M., and J. A. Brannigan. 2001. Pentapeptide regulation of aspartyl-phosphate phosphatases. Peptides 22:1541-1547. [DOI] [PubMed] [Google Scholar]
- 33.Perego, M., C. G. Hanstein, K. M. Welsh, T. Djavakhishvili, P. Glaser, and J. A. Hoch. 1994. Multiple protein aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in Bacillus subtilis. Cell 79:1047-1055. [DOI] [PubMed] [Google Scholar]
- 34.Perego, M., C. F. Higgins, S. R. Pearce, M. P. Gallagher, and J. A. Hoch. 1991. The oligopeptide transport system of Bacillus subtilis plays a role in the initiation of sporulation. Mol. Microbiol. 5:173-185. [DOI] [PubMed] [Google Scholar]
- 35.Perego, M., and J. A. Hoch. 1996. Cell-cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 93:1549-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perego, M., G. B. Spiegelman, and J. A. Hoch. 1988. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol. Microbiol. 2:689-699. [DOI] [PubMed] [Google Scholar]
- 37.Rudner, D. Z., J. R. LeDeaux, K. Ireton, and A. D. Grossman. 1991. The spo0K locus of Bacillus subtilis is homologous to the oligopeptide permease locus and is required for sporulation and competence. J. Bacteriol. 173:1388-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaeffer, P., J. Millet, and J. P. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 40.Scheufler, C., A. Brinker, G. Bourenkov, S. Pegoraro, L. Moroder, H. Bartunik, F. U. Hartl, and I. Moarefi. 2000. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101:199-210. [DOI] [PubMed] [Google Scholar]
- 41.Slamti, L., and D. Lereclus. 2002. A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. EMBO J. 21:4550-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith, I. 1993. Regulatory proteins that control late growth development, p. 785-800. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 43.Solomon, J., L. Su, S. Shyn, and A. D. Grossman. 2003. Isolation and characterization of mutants of the Bacillus subtilis oligopeptide permease with altered specificity of oligopeptide transport. J. Bacteriol. 185:6425-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Solomon, J. M., B. A. Lazazzera, and A. D. Grossman. 1996. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev. 10:2014-2024. [DOI] [PubMed] [Google Scholar]
- 45.Tame, J. R. H., G. N. Murshudov, E. J. Dodson, T. K. Neil, G. G. Dodson, C. F. Higgins, and A. J. Wilkinson. 1994. The structural basis of sequence-independent peptide binding by OppA protein. Science 264:1578-1581. [DOI] [PubMed] [Google Scholar]
- 46.Ward, B. K., R. K. Allan, D. Mok, S. E. Temple, P. Taylor, J. Dornan, P. J. Mark, D. J. Shaw, P. Kumar, M. D. Walkinshaw, and T. Ratajczak. 2002. A structure-based mutational analysis of cyclophilin 40 identifies key residues in the core tetratricopeptide repeat domain that mediate binding to Hsp90. J. Biol. Chem. 277:40799-70809. [DOI] [PubMed] [Google Scholar]
- 47.Weinrauch, Y., R. Penchev, E. Dubnau, I. Smith, and D. Dubnau. 1990. A Bacillus subtilis regulatory gene product for genetic competence and sporulation resembles sensor protein members of the bacterial two-component signal-transduction systems. Genes Dev. 4:860-872. [DOI] [PubMed] [Google Scholar]
- 48.Yoshimura, M., K. Asai, Y. Sadaie, and H. Yoshikawa. 2004. Interaction of Bacillus subtilis extracytoplasmic function (ECF) sigma factors with the N-terminal regions of their potential anti-sigma factors. Microbiology 150:591-599. [DOI] [PubMed] [Google Scholar]
- 49.Zhao, R., E. J. Collins, R. B. Bourret, and R. E. Silversmith. 2002. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nat. Struct. Biol. 9:570-575. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.