Abstract
Here we describe that the Helicobacter pylori sensor kinase produced by HP1364 and the response regulator produced by HP1365 and designated CrdS and CrdR, respectively, are both required for transcriptional induction of the H. pylori copper resistance determinant CrdA by copper ions. CrdRS-deficient mutants lacked copper induction of crdA expression and were copper sensitive. A direct role of CrdR in transcriptional regulation of crdA was confirmed by in vitro binding of CrdR to the crdA upstream region. A 21-nucleotide sequence located near the crdA promoter was shown to be required for CrdR binding.
Helicobacter pylori colonizes the human gastric mucosa and can cause severe gastric diseases (2, 10). In the hostile ecological niche, maintaining proper metal ion metabolism (38) is of critical importance for the pathogen. This has previously been shown for the homeostasis of iron, which turned out to be required for effective gastric colonization in animal models (38, 39). Copper ions play an important role in bacterial metabolism, because they function as cofactors for electron transport, oxidases, and hydroxylases (18, 19). On the other hand, copper catalyzes the generation of toxic hydroxyl radicals via Fenton-like reactions (20), and this necessitates mechanisms to keep the concentration of cytoplasmic copper ions below toxic levels. Whereas copper import occurs nonspecifically (13, 38), H. pylori controls the cytoplasmic copper concentration by efflux via the P-type ATPase CopA (14), which transports copper ions from the cytoplasm into the periplasmic space. Accumulating copper ions are detoxified via the copper resistance determinants CrdA (from HP1326), CrdB (from HP1327), and CzcB (from HP1328), which form together with the CzcA homolog from HP1329 a Czc-like metal export system, which was shown to contribute substantially to H. pylori copper resistance (40). The genetic organization of the H. pylori Crd system is orthologous to the Escherichia coli four-component copper export system Cus, which is proposed to transport Cu(I) ions from the periplasm across the outer membrane (12). Genome-wide RNA profiling revealed that H. pylori responds actively to changes in the environmental copper concentration and that the copper resistance determinant CrdA is strongly induced by copper at the transcriptional level (40). However, the underlying regulatory mechanisms were not investigated (40). Homologs of E. coli (24, 33), Pseudomonas (21, 22), or Ralstonia (35) copper regulators are absent in the H. pylori genome (1, 34), and the H. pylori metal regulator proteins Fur (6, 7, 38) and NikR (36, 37) are also not involved in crdA regulation (B. Waidner, F. N. Stähler, S. Bereswill, A. H. M. van Vliet, unpublished results). However, the transcriptional copper induction of E. coli Cus by CusRS, a two-component regulatory system (24) composed of a histidine kinase sensor protein and a cognate response regulator (23, 32), supported the idea that copper regulation of H. pylori crdA might be mediated by a similar type of regulator. The H. pylori genome contains only a small set of two-component signal transduction systems (1, 29, 34), and to date the regulated target genes have been defined for two of them (5, 9, 11). In the present study, we used mutational analysis and in vitro DNA/protein binding experiments to show that the H. pylori two-component regulatory system HP1364 (sensor homolog)/HP1365 (regulator homolog) is essential for transcriptional copper induction of crdA and for H. pylori copper resistance. Thus, we designated the HP1364 and HP1365 proteins as CrdS and CrdR, respectively. The putative sensor CrdS (from HP1364) was classified earlier as a member of an orthodox histidine kinase family (5, 9), and CrdR (from HP1365) was grouped into the OmpR family of response regulators (1, 5, 9, 29, 34). The CrdRS system was chosen because genes for bacterial copper regulators are often linked to their target genes and because the coding HP1364/HP1365 genes are located nearby the crdA locus in the H. pylori chromosome (1, 34, 40). Furthermore, the HP1364/HP1365 system was shown to be required for gastric colonization in a mouse infection model, but its target genes have not been determined so far (25).
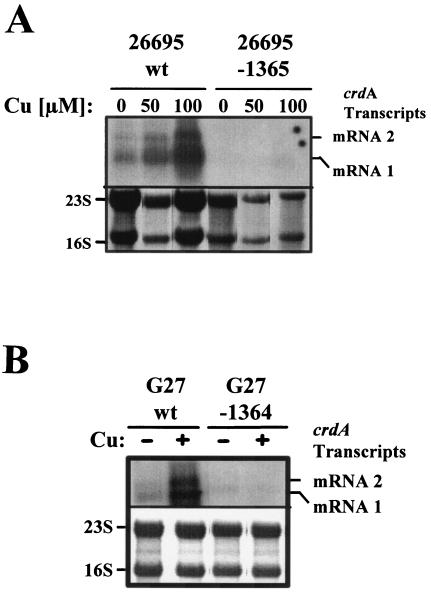
To investigate possible functions of HP1364 and HP1365 in crdA regulation, we inactivated both genes separately in H. pylori strain 26695 (Fig. 1) and analyzed copper-mediated induction of crdA transcription in the resulting crdS and crdR mutants by Northern blot hybridization. Strains and plasmids used are listed in Table 1. Cloning was performed in E. coli according to standard protocols (3). The cat gene with its own promoter (Pcat) was fused to upstream and downstream DNA regions of mutagenized genes by using a modified megaprimer PCR protocol (26, 28). DNA regions flanking Pcat were amplified by PCR from DNA of H. pylori strain 26695 using primers carrying 5′ extensions complementary to the 5′ and 3′ ends, respectively, of the Pcat cassettes (Table 2). Plasmids carrying mutagenized versions of both genes (listed in Table 1) were used for the mutagenesis of the corresponding genes in the H. pylori chromosome. Marker exchange mutagenesis in H. pylori strain 26695 was performed by electroporation according to standard procedures (15). H. pylori mutants 26695-1364 and 26695-1365 carrying Pcat inserted into the chromosomal crdS and crdR genes, respectively, were selected on Dent blood agar with 20 mg/liter chloramphenicol. Analysis of copper-induced crdA transcription by Northern blot hybridization (performed according to standard procedures as described in references 3, 6, and 39) revealed that mutant strains 26695-1365 (Fig. 2A) and 26695-1364 (not shown) both completely lacked the copper-mediated increase of the small crdA mRNAs observed in the wild-type (wt) strain 26695 (Fig. 2A), indicating that CrdRS acts as a copper regulator of CrdA. Previous studies reported pronounced differences in the transcriptional regulation of two-component systems in different H. pylori strains (9, 11). Thus, we studied copper induction of crdA in the well-characterized H. pylori mutant strains G27/HP1364::km and G27/HP1365::km (5, 41; kindly provided by Dagmar Beier, Würzburg, Germany). Kinetic analyses revealed that copper-induced crdA transcription (40) occurred within minutes in the H. pylori wt strain G27 and was completely abolished in the crdS (Fig. 2B) and crdR (not shown) mutants.
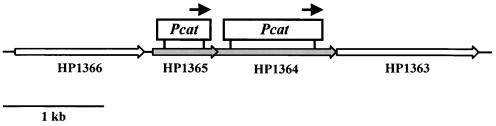
FIG. 1.
Schematic overview and mutational analysis of HP1364 and HP1365. Genes are numbered according to the annotated genome sequence of H. pylori strain 26695 (32). HP1364 and HP1365 are indicated by grey arrows, and neighboring genes are indicated by white arrows. The insertion sites of the Pcat resistance cassettes are marked with white boxes. The directions of resistance genes are displayed by black arrows.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Plasmids | ||
| pZERO-2 | Cloning vector, MCS in lacZ′, neo, Kmr | Invitrogen |
| p1364-PCAT | pZERO-2, Δ HP1364::Pcat, Cmr, Kmr | This study |
| p1365-PCAT | pZERO-2, HP1365::Pcat, Cmr, Kmr | This study |
| pASK-IBA3 | Expression vector, tetR, Ptet, bla, Apr | IBA |
| pIBA3-1365 | pASK-IBA3 carrying the HP1365 coding sequence under the control of the tet promoter cloned in the BsaI site | This study |
| H. pylori strains | ||
| 26695 | wt, containing the entire cag PAI | 32 |
| 26695-1364 | 26695, ΔHP1364::Pcat, Cmr | This study |
| 26695-1365 | 26695, ΔHP1365::Pcat, Cmr | This study |
| G27 | Clinical isolate | 41 |
| G27/HP1364::km | G27, ΔHP1364::km, Kmr | 5 |
| G27/HP1365::km | G27, ΔHP1365::km, Kmr | 5 |
MCS, multiple cloning site; PAI, pathogenicity island.
TABLE 2.
Oligonucleotides used in this study
| Genea | Application | Primer | Sequence (5′→3′) | Primer | Sequence (5′→3′)b |
|---|---|---|---|---|---|
| HP1326 | Antisense RNA probe | 1326-L1 | GGGTTATGGGTTTAAACGCA | 1326-R1T7 | T7-TTATAAATCCAGGCTTGTTT |
| Promoter HP1326 | P1326-L1 | CTTGTTATCTTAATGTAAAG | 1326-R2 | TTCACTTCCAAGTCATTAGC | |
| P1326-L2 | TATTATCCGTTCGCAACAAG | P1326-DIG | GGTTTGCTCCCATGCGTTTA | ||
| P1326-L3 | AAACACCATTTCCACAATTT | ||||
| HP1364 | Mutagenesis, upstream | 1364-L1 | GGCAATTAGCGCTACAATAC | PCAT1364-R1 | 1-CTAATCACTAGCATCAACAC |
| HP1365 | Mutagenesis, downstream | CAT1364-L1 | 2-ACAAGATCACAGAATTAAGC | 1364-R1 | CCTATCACGCAGTCTATTAA |
| Mutagenesis, upstream | 1365-L1 | AGAGAGATTATCATAATTGG | PCAT1365-R1 | 1-CTCCTTAACGCTCTCGCTTA | |
| Mutagenesis, downstream | CAT1365-L1 | 2-TTCCACCTTGCGCACTTATA | 1365-R1 | GGCGGTGTTGTTGGTTGTCT | |
| Expression in E. coli | 1365-ASK3-L1 | TGGTAGGTCTCAAATGAATGCAAAAAA | 1365-ASK3-R1 | ATGGTAGGTCTCAGCGCTTAGTGGGTTAAAGCG | |
| AGATTTTTTTACTAGAAGACG | ATAGCCAAC | ||||
| Pcat | cat gene with promoter | CATS1 | TCCGGTTTTTGTTAATCCGCC | CATAS1 | TTACGCCCCGCCCTGCCA |
Gene numbers refer to the H. pylori 26695 genome sequence (32).
The 5′ extensions used for fusion of PCR products to the cat gene by megaprimer PCR are labeled as follows: 1, (5′-GGCGGATTAACAAAAACCGGA), complementary to the 5′ region of the cat gene with promoter; 2, (5′-TGGCAGGGCGGGGCGTAA), complementary to the 3′ end of the cat gene; T7, (5′-CTAATACGACTCACTATAGGGAGA) adds a T7 promoter sequence for creation of digoxigenin-labeled antisense RNA. The BsaI restriction site used for the HP1365 expression via the IBA system is underlined.
FIG. 2.
The role of CrdR (HP1365) and CrdS (HP1364) in copper-mediated induction of crdA transcription. Copper-induced crdA transcripts, marked on the right (mRNAs 1 and 2), were visualized by hybridization of total RNA (15 μg), isolated from H. pylori strains that were grown in BBF supplemented with copper with a specific antisense RNA probe. (A) Induction of crdA transcription in the H. pylori wt strain 26695 and in its isogenic HP1365 mutant (26695-1365) grown for 48 h in BBF (0) and in BBF supplemented with 50 and 100 μM copper chloride. The lower panel shows methylene blue stains of the same RNA samples after blotting to the nylon membrane prior to hybridization. (B) Copper-induced crdA transcription in H. pylori strain G27-2 and its isogenic HP1364 mutant. The upper panel shows total RNA (15 μg) isolated from bacteria incubated without (−) or with (+) 1 mM copper chloride for 1 h. The lower panel shows methylene blue stains of the same RNA samples after blotting to the nylon membrane prior to hybridization. Sizes of crdA mRNAs were determined by using the digoxigenylated RNA length standards from Roche Diagnostics (Set I, No. 1526529).
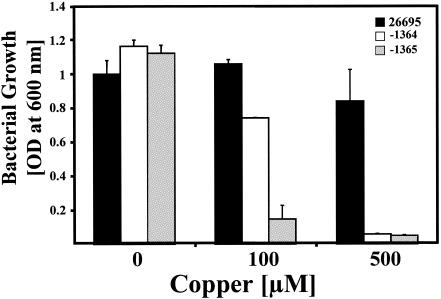
Subsequently, we investigated by growth inhibition experiments whether the CrdRS-mediated copper induction of crdA transcription is required for H. pylori copper resistance. To this end, H. pylori wt strain 26695 and mutants that were normally cultured on Dent blood agar (36) were grown in brucella broth with 5% fetal calf serum (BBF) (total copper content, 0.5 μM [6]). At an optical density at 600 nm (OD600) of 1.0, these precultures were diluted to a ratio of 1:150 in BBF supplemented with copper chloride (CuCl2) (C6641; Sigma). Bacterial growth was determined by reading the OD600 after 48 h. Experiments were performed in triplicate and were repeated at least three times. Results from growth experiments demonstrated that crdS and crdR mutations generally do not limit bacterial fitness but that both genes are required for copper resistance (Fig. 3). Similar results were obtained for crdS (see above) and crdR (not shown) mutants of H. pylori strain G27.
FIG. 3.
The role of the HP1364 and HP1365 genes in copper resistance. The H. pylori wt strain 26695 (black) and the isogenic mutants 26695-1364 (white) and -1365 (grey) were cultivated for 48 h in BBF medium, and growth inhibition was determined by measuring the OD600s. The medium was supplemented with copper at increasing concentrations as indicated on the x axis. The data represent mean values of three independent determinations. Standard deviations are indicated.
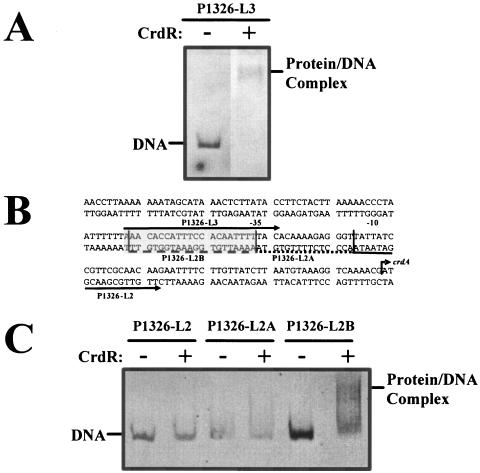
To test whether CrdR can directly interact with the crdA promoter, we performed DNA-binding experiments with a purified recombinant version of the H. pylori CrdR protein produced in E. coli using the Strep-Tag (30) protein expression system from IBA (Göttingen, Germany) according to the manufacturer's instructions. The HP1365 coding sequence of H. pylori strain 26695 was amplified using the primer pair 1365ASK3-L1/1365ASK3-R1 (Table 2) and cloned via BsaI restriction sites added as 5′ extensions into the plasmidpASKIBA-3 (IBA-Göttingen). Solubilization and purification of the resulting inclusion bodies were performed according to the method of Sambrook et al. (27). Various parts of the DNA region upstream of crdA were amplified by PCR from genomic DNA of H. pylori strain 26695 with primers P1326-L1, -L2, and -L3 (Fig. 4A; Table 2) in combination with primers 1326-R2 or P1326-DIG (Table 2). The formation of HP1365 protein-DNA complexes with the crdA DNA was analyzed by electrophoretic mobility shift assays (EMSA). Recombinant HP1365 protein (2,000 nM) was incubated for 20 min in 20 μl fivefold-concentrated binding buffer (10 mM Tris-HCl, pH 8.0, 5 mM dithiothreitol, 5% glycerol, 50 mM acetyl phosphate). Then, 0.14 pM of the target DNA, 30 μl glycerin (50%), and distilled water were added. After 30 min, the samples were electrophoresed on a 7% nondenaturing polyacrylamide gel, and protein-DNA complexes were visualized by staining with ethidium bromide or were blotted to a membrane and detected with the digoxigenin detection kit (Roche) if primer 1326-DIG was used for amplification. The results showed that the mobility of the PCR product generated with primer P1326-L3 was strongly retarded in the presence of CrdR (Fig. 4A), indicating that CrdR binds to crdA DNA. The addition of acetyl phosphate (50 mM), which can act as a phosphoryl donor for many response regulators (17), did not influence CrdR binding to the crdA DNA probe (not shown). The fact that CrdR binding was not observed with a shorter PCR product (Fig. 4B) generated with primer P1326-L2 and lacking the DNA region between primers P1326-L3 and -L2 (Fig. 4B and C) indicated that CrdR binding is sequence specific and that the target sequence is located in a short stretch of nucleotides directly upstream of the crdA coding sequence (Fig. 4B). Further analysis of the binding site by using PCR products generated with the prolongated primers P1326-L2A and -L2B (Fig. 4B) revealed that the binding of CrdR to the crdA promoter region depends on the presence of a 21-nucleotide region located directly upstream of the −35 RNA polymerase binding site (Fig. 4C).
FIG. 4.
Binding of the recombinant CrdR protein to the crdA promoter region. Different regions in front of the crdA gene were analyzed for binding of the recombinant CrdR (rCrdR) protein by EMSA. (A) EMSA with crdA upstream DNA generated by PCR with primers P1326-L3 and 1326-R2. The positions of the crdA DNA probe (DNA) and the protein complex with CrdR are indicated. PCR products were incubated without (−) and with (+) CrdR. The picture represents a black-and-white image of the ethidium bromide-stained agarose gel visualized under UV light. (B) Schematic overview of the sequence in front of the crdA gene. Primers P1326-L3 and -L2, depicted by arrows, and primers P1326-L2A and -L2B, depicted by dotted and dashed lines, respectively, were used for the generation of PCR products for determination of the CrdR binding site. Putative −35 and −10 binding sites for RNA polymerase are indicated. The ATG start codon is marked by a black arrowhead. The DNA region required for binding of CrdR is shown in grey. (C) Determination of the HP1365 binding site in the crdA promoter using PCR products generated with primers P1326-L2, -L2A, and -L2B (localizations and sequences are marked in panel B). The positions of the crdA DNA probe (DNA) and the protein complex with CrdR are indicated. PCR products were incubated without (−) and with (+) rHP1365 protein. The picture represents a black-and-white image of the ethidium bromide-stained agarose gel visualized under UV light.
We conclude that the transcriptional induction of the H. pylori CrdA copper resistance determinant by copper ions is directly mediated by the CrdRS two-component system encoded by the HP1365/1364 genes. The average daily copper intake, which is in the range of 1 mg (4), and the copper content of up to 200 mg/kg of body weight in copper-rich foods allow the estimate that H. pylori is exposed to copper ions in the micromolar range. Together with the finding that H. pylori CrdS and CrdR mutants are not able to colonize the stomach in mice (25), this indicates that H. pylori copper homeostasis plays a crucial role in the adaptation to the gastric environment. In this context, the first description of an H. pylori copper regulator and an environmental stimulus sensed by an H. pylori two-component regulatory system supports the importance of these regulation systems in gastric adaptation. The CrdR binding region is located directly upstream of the crdA promoter and contains a sequence signature in the form of a mirror repeat (AACACC-ATTT-CCACAA) (Fig. 4B). The CrdR binding region is not homologous to copper regulator binding sites in other bacteria and was not detected in other H. pylori promoters by a genome-wide screen, indicating that CrdR function might be limited to crdA regulation. The fact that in vitro binding of CrdR was not influenced by acetyl phosphate leads to the assumption that CrdR, like other response regulators (8, 16, 31), is able to interact with the crdA promoter even in the unphosphorylated form. However, the role of phosphorylation in CrdS and CrdR regulation remains to be analyzed in more detail. Earlier studies showed that the deletion of the input domain of HP1364 did not result in autophosphorylation and that HP1365 could be phosphorylated neither by HP1364 nor by the other H. pylori histidine kinases (5). In summary, the results presented here clearly support a direct functional relation between CrdR and CrdS (5) and demonstrate that CrdS most likely acts as a sensor for environmental copper ions. This is further supported by the fact that copper added at a concentration of 1 mM to CrdR DNA binding reactions did not influence the interaction of CrdR with its binding site (data not shown). The discovery of a copper regulator in H. pylori will support further studies on the role of metal ion homeostasis in gastric adaptation.
Acknowledgments
This study was supported by a grant from ALTANA Pharma AG, Konstanz, Germany, to S.B. and by grant KI 201/9-3 from the Deutsche Forschungsgemeinschaft to M.K.
We thank Dagmar Beier (Würzburg, Germany) for providing the H. pylori strain G27 and its isogenic HP1364 and HP1365 mutants and Christian Bogdan (Department of Medical Microbiology and Hygiene, Freiburg) for helpful comments on the manuscript.
REFERENCES
- 1.Alm, R. A., L.-S. L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Asaka, M., M. Kudo, M. Kato, T. Sugiyama, and H. Takeda. 1998. Long-term Helicobacter pylori infection—from gastritis to gastric cancer. Aliment. Pharmacol. Ther. 12(Suppl. 1):9-15. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Short protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 4.Barceloux, D. G. 1999. Copper. J. Toxicol. Clin. Toxicol. 37:217-230. [DOI] [PubMed] [Google Scholar]
- 5.Beier, D., and R. Frank. 2000. Molecular characterization of two-component systems of Helicobacter pylori. J. Bacteriol. 182:2068-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bereswill, S., S. Greiner, A. H. M. van Vliet, B. Waidner, F. Fassbinder, E. Schiltz, J. G. Kusters, and M. Kist. 2000. Regulation of ferritin-mediated cytoplasmic iron storage by the ferric uptake regulator homolog (Fur) of Helicobacter pylori. J. Bacteriol. 182:5948-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bijlsma, J. J. E., B. Waidner, A. H. M. van Vliet, N. J. Hughes, S. Häg, S. Bereswill, D. J. Kelly, C. M. J. E. Vandenbroucke-Grauls, M. Kist, and J. G. Kusters. 2002. The Helicobacter pylori homologue of the ferric uptake regulator is involved in acid resistance. Infect. Immun. 70:606-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boucher, P. E., K. Murakami, A. Ishihama, and S. Stibitz. 1997. Nature of DNA binding and RNA polymerase interaction of the Bordetella pertussis BvgA transcriptional activator at the fha promoter. J. Bacteriol. 179:1755-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dietz, P., G. Gerlach, and D. Beier. 2002. Identification of target genes regulated by the two-component system HP166-HP165 of Helicobacter pylori. J. Bacteriol. 184:350-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forsyth, M. H., P. Cao, P. P. Garcia, J. D. Hall, and T. L. Cover. 2002. Genome-wide transcriptional profiling in a histidine kinase mutant of Helicobacter pylori identifies members of a regulon. J. Bacteriol. 184:4630-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franke, S., G. Grass, C. Rensing, and D. H. Nies. 2003. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 185:3804-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fulkerson, J. F. J., R. M. Garner, and H. L. Mobley. 1998. Conserved residues and motifs in the NixA protein of Helicobacter pylori are critical for the high affinity transport of nickel ions. J. Biol. Chem. 273:235-241. [DOI] [PubMed] [Google Scholar]
- 14.Ge, Z., and D. E. Taylor. 1996. Helicobacter pylori genes hpcopA and hpcopP constitute a cop operon involved in copper export. FEMS Microbiol. Lett. 145:181-188. [DOI] [PubMed] [Google Scholar]
- 15.Ge, Z., and D. E. Taylor. 1997. Helicobacter pylori DNA transformation by natural competence and electroporation, p. 145-152. In C. L. Clayton and H. L. Mobley (ed.), Helicobacter pylori protocols. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 16.Liu, W., and F. M. Hulett. 1997. Bacillus subtilis PhoP binds to the phoB tandem promoter exclusively within the phosphate starvation-inducible promoter. J. Bacteriol. 179:6302-6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukat, G. S., W. R. McCleary, A. M. Stock, and J. B. Stock. 1992. Phosphorylation of bacterial response regulator proteins by low molecular weight phosphor-donors. Proc. Natl. Acad. Sci. USA 89:718-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malmstrom, B. G., and J. Leckner. 1998. The chemical biology of copper. Curr. Opin. Chem. Biol. 2:286-292. [DOI] [PubMed] [Google Scholar]
- 19.McGuirl, M. A., and D. M. Dooley. 1999. Copper-containing oxidases. Curr. Opin. Chem. Biol. 3:138-144. [DOI] [PubMed] [Google Scholar]
- 20.Miller, R. A., and B. E. Britigan. 1997. Role of oxidants in microbial pathophysiology. Clin. Microbiol. Rev. 10:1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mills, S. D., C. K. Lim, and D. A. Cooksey. 1994. Purification and characterization of CopR, a transcriptional activator protein that binds to a conserved domain (cop box) in copper-inducible promoters of Pseudomonas syringae. Mol. Gen. Genet. 244:341-351. [DOI] [PubMed] [Google Scholar]
- 22.Mills, S. D., C. A. Jasalavich, and D. A. Cooksey. 1993. A two-component regulatory system required for copper-inducible expression of the copper resistance operon of Pseudomonas syringae. J. Bacteriol. 175:1656-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizuno, T. 1998. His-Asp phosphotransfer signal transduction. J. Biochem. (Tokyo) 123:555-563. [DOI] [PubMed] [Google Scholar]
- 24.Munson, G. P., D. L. Lam, F. W. Outten, and T. V. O'Halloran. 2000. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J. Bacteriol. 182:5864-5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panthel, K., P. Dietz, R. Haas, and D. Beier. 2003. Two-component systems of Helicobacter pylori contribute to virulence in a mouse infection model. Infect. Immun. 71:5381-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfeiffer, J., J. Guhl, B. Waidner, M. Kist, and S. Bereswill. 2002. Magnesium uptake by CorA is essential for viability of the gastric pathogen Helicobacter pylori. Infect. Immun. 70:3930-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Sarkar, G., and S. S. Sommer. 1990. The “megaprimer” method of site-directed mutagenesis. BioTechniques 8:404-407. [PubMed] [Google Scholar]
- 29.Scarlato, V., I. Delany, G. Spohn, and D. Beier. 2001. Regulation of transcription in Helicobacter pylori: simple systems or complex circuits? Int. J. Med. Microbiol. 291:107-117. [DOI] [PubMed] [Google Scholar]
- 30.Skerra, A., and T. G. Schmidt. 2000. Use of the Strep-Tag and streptavidin for detection and purification of recombinant proteins. Methods Enzymol. 326:271-304. [DOI] [PubMed] [Google Scholar]
- 31.Spohn, G., and V. Scarlato. 1999. Motility of Helicobacter pylori is coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J. Bacteriol. 181:593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 33.Stoyanov, J. V., J. L. Hobman, and N. L. Brown. 2001. CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA. Mol. Microbiol. 39:502-511. [DOI] [PubMed] [Google Scholar]
- 34.Tomb, J.-F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 35.van der Lelie, D., T. Schwuchow, U. Schwidetzky, S. Wuertz, W. Baeyens, M. Mergeay, and D. H. Nies. 1997. Two-component regulatory system involved in transcriptional control of heavy-metal homoeostasis in Alcaligenes eutrophus. Mol. Microbiol. 23:493-503. [DOI] [PubMed] [Google Scholar]
- 36.van Vliet, A. H. M., E. J. Kuipers, B. Waidner, B. J. Davies, N. de Vries, C. W. Penn, C. M. J. E. Vandenbroucke-Grauls, M. Kist, S. Bereswill, and J. G. Kusters. 2001. Nickel-responsive induction of urease expression in Helicobacter pylori is mediated at the transcriptional level. Infect. Immun. 69:4891-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Vliet, A. H. M., S. W. Poppelaars, B. J. Davies, J. Stoof, S. Bereswill, M. Kist, C. W. Penn, E. J. Kuipers, and J. G. Kusters. 2002. NikR mediates nickel-responsive transcriptional induction of urease expression in Helicobacter pylori. Infect. Immun. 70:2846-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Vliet, A. H. M., S. Bereswill, and J. G. Kusters. 2001. Ion metabolism and transport, p. 193-206. In H. L. T. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, D.C. [PubMed]
- 39.Waidner, B., S. Greiner, S. Odenbreit, H. Kavermann, J. Velayudhan, F. Stähler, J. Guhl, E. Bissé, A. H. M. van Vliet, S. C. Andrews, J. G. Kusters, D. J. Kelly, R. Haas, M. Kist, and S. Bereswill. 2002. Essential role of ferritin Pfr in Helicobacter pylori iron metabolism and gastric colonization. Infect. Immun. 70:3923-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waidner, B., K. Melchers, I. Ivanov, H. Loferer, K. W. Bensch, M. Kist, and S. Bereswill. 2002. Identification by RNA profiling and mutational analysis of the novel copper resistance determinants CrdA (HP1326), CrdB (HP1327), and CzcB (HP1328) in Helicobacter pylori. J. Bacteriol. 184:6700-6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiang, Z., S. Censini, P. F. Bayeli, J. L. Telford, N. Figura, R. Rappuoli, and A. Covacci. 1995. Analysis of expression of CagA and VacA virulence factors in 43 strains of Helicobacter pylori reveals that clinical isolates can be divided into two major types and that CagA is not necessary for expression of the vacuolating cytotoxin. Infect. Immun. 63:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]