Abstract
The type 3 synthase from Streptococcus pneumoniae is a processive β-glycosyltransferase that assembles the type 3 polysaccharide [3)-β-d-GlcUA-(1→4)-β-d-Glc-(1→] by a multicatalytic process. Polymer synthesis occurs via alternate additions of Glc and GlcUA onto the nonreducing end of the growing polysaccharide chain. In the presence of a single nucleotide sugar substrate, the type 3 synthase ejects its nascent polymer and also adds a single sugar onto a lipid acceptor. Following single sugar incorporation from either UDP-[14C]Glc or UDP-[14C]GlcUA, we found that phospholipase D digestion of the Glc-labeled lipid yielded a product larger than a monosaccharide, while digestion of the GlcUA-labeled lipid resulted in a product larger than a disaccharide. These data indicated that the lipid acceptor contained a headgroup and that the order of addition to the lipid acceptor was Glc followed by GlcUA. Higher-molecular-weight product synthesized in vitro was also sensitive to phospholipase D digestion, suggesting that the same lipid acceptor was being used for single sugar additions and for polymer formation. Mass spectral analysis of the anionic lipids of a type 3 S. pneumoniae strain demonstrated the presence of glycosylated phosphatidylglycerol. This lipid was also observed in Escherichia coli strains expressing the recombinant type 3 synthase. The presence of the lipid primer in S. pneumoniae membranes explained both the ability of the synthase to reinitiate polysaccharide synthesis following ejection of its nascent chain and the association of newly synthesized polymer with the membrane. Unlike most S. pneumoniae capsular polysaccharides, the type 3 capsule is not covalently linked to the cell wall. The present data indicate that phosphatidylglycerol may anchor the type 3 polysaccharide to the cell membrane.
The capsule of the gram-positive pathogen Streptococcus pneumoniae is essential for its virulence (4, 16, 18a). Capsule serves to prevent opsonophagocytosis by interfering with the recognition of complement bound to the bacterial surface by phagocytic receptors on immune cells (7, 40, 41) and, to a lesser extent, by reducing the amount of complement bound (1). Production of a capsule is also critical for colonization of the nasopharynx of mice (26). S. pneumoniae capsules demonstrate a high level of diversity, with 90 distinct serotypes that differ in their monosaccharide composition and/or their glycosidic linkages (19, 37). Thus far, two different mechanisms have been described for capsule synthesis in S. pneumoniae (10, 12, 17, 23). The majority of capsules are synthesized by a mechanism analogous to that of Wzy-dependent O-antigen synthesis in gram-negative bacteria, where the repeating-unit structure of the capsule is assembled on a lipid acceptor that is subsequently transported across the cytoplasmic membrane and polymerized into polysaccharide (39). At some point during polymerization, polysaccharide is transferred onto the cell wall (33). The transfer is size independent, and polymer ranging from very small to very large molecular size is detectable on both membrane and cell wall fractions (5). Generally, polysaccharides synthesized by this mechanism require a separate glycosyltransferase to form each glycosidic linkage of the repeat unit, as well as an enzyme to polymerize the repeat units.
The second mechanism of synthesis, utilized in the production of type 3 and 37 capsules, requires only a single enzyme for formation of all the glycosidic linkages and polymerization (2, 12, 25). The type 3 synthase alternately adds Glc and GlcUA onto the nonreducing end of the polysaccharide chain by a processive mechanism whereby the growing polysaccharide remains tightly associated with a carbohydrate binding site on the synthase (10). A similar mechanism of synthesis is used by the enzymes involved in forming hyaluronan, chitin, and cellulose (24, 28), although hyaluronan from Streptococcus pyogenes and Streptococcus equisimilis has recently been shown to grow from the reducing end (34). These enzymes are classified in glycosyltransferase family 2 (8). The type 3 synthase, encoded by cps3S, is one of two gene products encoded in the type 3 capsule locus that is required for type 3 polysaccharide synthesis. The other, Cps3D, is a UDP-Glc dehydrogenase that oxidizes UDP-Glc to form UDP-GlcUA, the second substrate for type 3 polysaccharide formation (12).
Both the type 3 synthase contained in S. pneumoniae membranes and the recombinant synthase contained in Escherichia coli membranes will rapidly assemble type 3 polysaccharide in vitro in the presence of both nucleotide sugar substrates (9, 10, 32). When one of these substrates becomes limiting or only a single nucleotide sugar substrate is present, synthesis is terminated. In E. coli membranes, the nascent polymer is ejected from the enzyme following termination and can no longer be extended, although it remains associated with the membrane. In addition, a single sugar is added onto a glycerophosphate lipid (9). In contrast, termination of synthesis in S. pneumoniae membranes results in only about 50% of the polymer being retained on the membrane and the remainder being released into solution (10, 15). In both systems, polysaccharide ejection is time, temperature, and substrate concentration dependent, indicating that it is an enzymatic process (9, 15).
Although it has been suggested that processive glycosyltransferases do not require a primer for synthesis of their polymers (28), characterization of the type 3 synthase expressed in E. coli and of the cellulose synthase CesA demonstrated that these enzymes initiate synthesis of their respective polymers on lipid acceptors (9, 27). The formation of novel glycolipid products is also observed when glycosyltransferases from other gram-positive bacteria are expressed in E. coli. Jorasch et al. found, however, that these products are not synthesized in their native bacterial membranes (21). Here, we demonstrate that the type 3 synthase contained in S. pneumoniae membranes is able to synthesize glycolipids with properties similar to those observed for synthase-containing E. coli membranes and that initiation of type 3 polysaccharide synthesis utilizes phosphatidylglycerol.
MATERIALS AND METHODS
Materials.
UDP-[14C]Glc (257 mCi/mmol) was obtained from Andotek, UDP-[14C]GlcUA (338 mCi/mmol) from ICN, and UDP-[3H]Glc (1 Ci/mmol) from Amersham Pharmacia Biotech. Econo-Safe scintillation cocktail was from Research Products International, Corp. Mutanolysin, Sephacryl S-500HR, Sephacryl S-300, UDP-Glc, and UDP-GlcUA were obtained from Sigma. Nonidet P-40 was from Calbiochem, and Todd-Hewitt broth, yeast extract, and tryptone were from Difco. Clostridium perfringens phospholipase C (66 units/mg of protein), Streptomyces chromofuscus phospholipase D (PLD) (4,980 units/mg solid), type VIII β-glucuronidase from E. coli, and β-glucosidase from Caldocellum saccharolyticum were from Sigma. Silica gel G thin-layer plates and 3MM chromatography paper were from Whatman.
Analytical methods.
Chromatography on Sephacryl S-500 and Sephacryl S-300 was carried out on 1.4- by 37-cm columns. Chromatography on Sephadex G-10 was carried out on a 1- by 37-cm column. All columns were eluted with a solution consisting of 0.1% Nonidet P-40, 0.02% sodium azide, and either 200 mM ammonium acetate (pH 6.5) or 5 mM Tris (pH 7.5) containing 200 mM NaCl, as described previously (15). Silica gel G plates were chromatographed in butanol:acetic acid:water (44:16:40) or chloroform:methanol:1 M ammonium acetate (pH 7.5) (65:25:4) followed by autoradiography. Samples spotted on 3MM paper were chromatographed in butanol:acetic acid:water (44:16:40) or ethanol:1 M ammonium acetate (pH 5.5) (65:35). Paper chromatograms were cut into 1-cm strips, and the amount of radioactivity was determined by liquid scintillation counting. Preparation of type 3 polysaccharide-specific depolymerase and digestion with the depolymerase were carried out as described previously (15). The assays for polysaccharide ejection and release were as described previously (15). Phosphorus analysis on extracted lipid was done as described previously (14). Hexose analysis was performed using a modified version of the phenol sulfuric assay (3). Briefly, lipid samples were dried under nitrogen and suspended in 400 μl water. Ten microliters of 90% (wt/wt) phenol was added to the sample and heated to 50°C. One milliliter of concentrated sulfuric acid was added, and the sample was vortexed immediately. Samples were heated at 95 to 100°C for 5 min and, after cooling to room temperature, the absorbance was read at 490 nm.
Growth conditions and membrane preparations.
The type 3 parent strain, WU2 (6), and its isogenic mutants JD614 (Cps3D mutant [12]) and JD908 (Cps3S− [13]) were grown in Todd-Hewitt broth supplemented with 0.5% yeast extract at 37°C. The cps3D mutation in JD614 results in the production of <6% of the parental levels of capsule as determined by reactivity with a type 3-specific monoclonal antibody (26). The mutation was localized to the 5′ end of the gene by repair with a cps3D fragment to yield the parental phenotype (12). Sequence analysis revealed that the mutation resulted in a T83I change (this study). Membranes from all three S. pneumoniae strains were isolated as described previously (10). E. coli strains JD424 (Cps3S+) and JD422 (vector control strain) (12) were grown in L broth (10 g tryptone, 5 g yeast extract, 5 g NaCl, 1 g glucose per liter) containing 100 μg/ml ampicillin. These strains are derivates of E. coli TG-1 that contain either the plasmid vector pKK223-3 (JD422) or cps3S cloned into pKK223-3 (JD424) (12).
Isolation and identification of Glc-containing glycerophosphate lipids from E. coli and S. pneumoniae membranes.
One-liter cultures of S. pneumoniae strain JD614 and E. coli strain JD424 were grown as described above. Bacteria were pelleted by centrifugation at 7,000 × g and washed with 500 ml of phosphate-buffered saline (PBS) (140 mM NaCl, 3 mM KCl, 5 mM Na2HPO4 [pH 7.4]), and the pellet was frozen at −80°C overnight. Cells were thawed at room temperature, suspended in 13 ml sterile water, and extracted with a single-phase Bligh and Dyer solvent mixture by addition of 25 ml of methanol, 12.5 ml of chloroform, and 0.13 ml of glacial acetic acid. After 60 min of stirring, the insoluble material was removed by centrifugation at 5,000 × g for 10 min and the supernatant saved. The insoluble material was suspended in 3 ml water and extracted again with a single-phase Bligh and Dyer mixture by adding 25 ml methanol, 12.5 ml chloroform, and 10 ml water. The mixture was stirred at room temperature for 60 min, and the insoluble material was removed by centrifugation. The two supernatants were combined and extracted with a two-phase Bligh and Dyer mixture by the addition of 32 ml chloroform and 16 ml water. The lower chloroform phase was removed, and the upper aqueous phase was extracted with 50 ml of the lower phase of a preequilibrated solution containing chloroform:methanol:water (1:1:0.8). The lower chloroform phase was combined with the previous lower phase and washed once with 80 ml of the upper phase of the preequilibrated solution. The lower chloroform phase was applied to a 7-ml diethyaminoethyl cellulose column prewashed with 20 ml of methanol. Lipids were eluted with 100 ml of a 0 to 0.1 M ammonium acetate gradient, and 3-ml fractions were collected. Total phosphorus was analyzed in the even-numbered fractions, and a single peak was detected in fractions 6 through 10. These fractions were combined and dried under a stream of nitrogen, suspended in 100 μl of chloroform:methanol (1:1), and chromatographed on silica gel G plates in chloroform:methanol:1 M ammonium acetate (pH 7.4) (65:25:4) for 2 h. Lipids were visualized by iodine staining. The lipid band that comigrated with the [14C]Glc-labeled lipid synthesized in vitro was scraped from the TLC plate and dissolved in 2 ml of methanol. Lipids were dried under a stream of nitrogen and suspended in 50 μl of methanol. Tandem mass spectrometry (MS/MS) analysis of the lipids was performed by the UAB Mass Spectrometry Facility using either a Micromass Q-TOF instrument in the electrospray ionization mode or a PE ScieX triple quadrupole in the negative-ion mode.
Synthesis of 3H-polysaccharide-synthase complex.
Type 3 polysaccharide-enzyme complexes were synthesized in reaction mixtures consisting of 100 mM HEPES (pH 8.0), 10 mM MnCl2, UDP-[3H]Glc (20 μCi/mmol), 25 μM UDP-Glc, 25 μM UDP-GlcUA, and S. pneumoniae membranes (400 to 1,600 μg of total protein). The reactions were incubated for 5 min at 35°C and stopped by placing on ice. The volumes of the reactions were brought to 2 ml with 100 mM HEPES (pH 7.5) containing 10% glycerol, and the membranes were washed five times, as described previously (15).
Synthesis of glycolipid products.
Glycolipids were synthesized in 200-μl reaction mixtures containing 100 mM HEPES (pH 8.0), 10 mM MnCl2, either 1 μM UDP-[14C]Glc or 1 μM UDP-[14C]GlcUA and S. pneumoniae membranes (300 μg of total protein). Following incubation at 35°C for 20 min, the reaction was terminated by the addition of 2 ml of an ice-cold solution containing 100 mM HEPES (pH 7.5) and 10% glycerol, and the membranes were sedimented by centrifugation at 100,000 × g for 30 min. The membranes were washed twice with 2.5 ml of wash solution, as described above. For chromatography on paper, the membranes were suspended in 100 μl of wash solution and spotted on paper. For separation by thin-layer chromatography (TLC) and gel filtration, the membranes were suspended in 5 mM EDTA (pH 7.5) and heated at 100°C for 5 min. The labeled glycolipids were extracted by adding Nonidet P-40 to a final concentration of 0.5%. The insoluble material was sedimented by centrifugation at 10,000 × g for 5 min, the supernatants were saved, and the pellets were extracted a second time. The supernatants containing the glycolipid fraction were combined for analyses. Apparent Km values for UDP-Glc and UDP-GlcUA in the formation of the Glc- and GlcUA-labeled lipids were determined by incubating WU2 membranes (30 μg of total protein) in 100-μl reaction mixtures containing 100 mM HEPES (pH 8.0), 10 mM MnCl2, and increasing concentrations of either UDP-[14C]Glc (1 μM to 100 μM) or UDP-[14C]GlcUA (0.2 μM to 20 μM) for 5 to 10 min at 35°C. Reactions were stopped and the membranes pelleted and washed as described above. The membranes were suspended in 50 μl of water, spotted on 3MM chromatography paper, and chromatographed for 2 h in butanol:acetic acid:water (44:16:40). The amount of radioactivity that migrated faster than Glc was determined, and Km values were calculated using the kinetics program Enzfitter (Elsevier-Biosoft).
Saponification, acid hydrolysis, β-glycosidase, and phospholipase digestion of glycolipid products.
Glycolipids were saponified at 35°C for 20 min in 0.4 ml of 80% methanol containing 0.1 N NaOH. The mixture was neutralized with acetic acid, and the methanol was removed under a stream of nitrogen. Dried samples were suspended in water and analyzed by either paper chromatography or TLC. Acid hydrolysis was carried out in either 1.0 N or 0.1 N HCl at 100°C for various amounts of time. The hydrolysates were neutralized with NaOH, and samples were analyzed by paper chromatography. Phospholipase digestions were conducted in reaction mixtures of the appropriate buffer, pH, and temperature as recommended by Sigma, with the addition of Nonidet P-40 and 10 mM CaCl2, which have been shown to enhance these activities (11, 20). A minimum of 10 units of phospholipase was added, except for 1 unit of C. perfringens phospholipase C. The reactions were terminated by heating to 100°C for 4 min, the mixtures were clarified by centrifugation at 13,000 × g for 5 min, and the products were analyzed by paper, thin-layer, or Sephacryl S-300 chromatography. β-Glucosidase (5 units for 16 h) and β-glucuronidase (376 units for 6 h) digestions were performed following PLD digestion, as described previously (10). Digested samples were analyzed by paper chromatography in ethanol:1 M ammonium acetate (pH 5.5) (65:35).
RESULTS
The type 3 synthase from S. pneumoniae membranes synthesizes glycolipids in vitro.
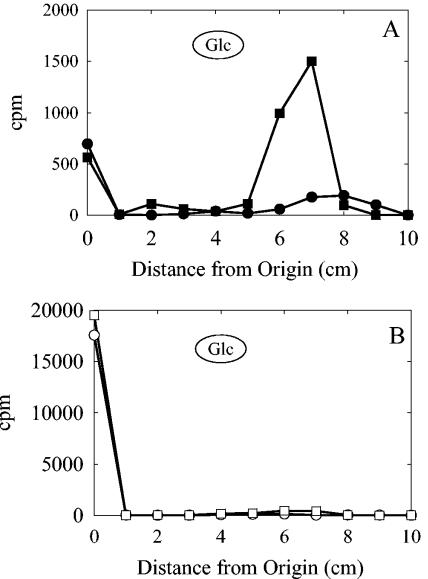
Membranes isolated from the S. pneumoniae type 3 strain WU2 were incubated with either 1 μM UDP-[14C]Glc or 1 μM UDP-[14C]GlcUA, and the products were analyzed by paper chromatography. In each case, the radioactivity was incorporated into a product that migrated faster than Glc and a second product that remained at the origin, consistent with the formation of a glycolipid and high-molecular-weight products, respectively (Fig. 1A). Approximately 10-fold more glycolipid was formed with UDP-GlcUA than with UDP-Glc. The apparent Km values for formation of the glycolipid products with UDP-GlcUA and UDP-Glc were 12 μM and 42 μM, respectively. The addition of 1 μM of the second nucleotide sugar to either reaction resulted in a shift from glycolipid formation to predominately polymer formation (Fig. 1B). No labeled products were synthesized using membranes from a type 3 strain in which the type 3 synthase had been insertionally inactivated (data not shown).
FIG. 1.
The type 3 synthase forms a hydrophobic product when incubated with a single nucleotide sugar. Membranes from type 3 S. pneumoniae strain WU2 were incubated for 20 min at 35°C in 200-μl reaction mixtures containing 100 mM HEPES (pH 8.0), 10 mM MnCl2, and either (A) 1 μM UDP-[14C]Glc (•) or 1 μM UDP-[14C]GlcUA (▪) or (B) 1 μM UDP [14C]Glc and 1 μM UDP-GlcUA (○) or 1 μM UDP-[14C]GlcUA and 1 μM UDP-Glc (□). Membranes were sedimented by centrifugation and washed twice with 100 mM HEPES (pH 7.5) containing 10% glycerol. Aliquots (50 μl) were spotted on paper and chromatographed for 2 h in butanol:acetic acid:water (44:16:40). The chromatogram was cut into 1-cm strips, and the amount of radioactivity was determined by liquid scintillation counting. The circled Glc indicates the migration of the Glc standard.
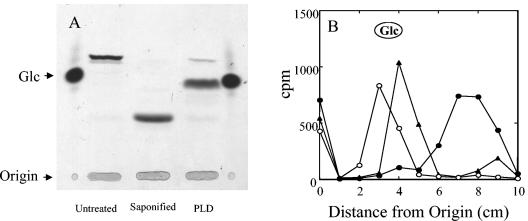
Following separation of the GlcUA-labeled lipid product by TLC in butanol:acetic acid:water (44:16:40), a single major band was observed (Fig. 2A, untreated). The Glc-labeled lipid also migrated as a single band when separated by TLC in the same solvent (data not shown). The migration of the GlcUA-labeled (Fig. 2A) and Glc-labeled lipid products (Fig. 2B) changed following either saponification with 0.1 N NaOH for 30 min or digestion with PLD, suggesting that the Glc- and GlcUA-labeled products were glycerophosphate-based lipids as opposed to polyprenylphosphate-based lipids. Neither the Glc- nor the GlcUA-labeled lipids were sensitive, however, to phospholipase C (data not shown). Acid hydrolysis of the GlcUA-labeled product in 0.1 N HCl for 10 min and 30 min at 100°C resulted in degradation of only 20% and 40% of the product, respectively, again consistent with a glycerophosphate lipid. Hydrolysis of the Glc- and GlcUA-labeled products in 1 N HCl at 100°C for 2 h resulted in complete degradation of the products into their respective monosaccharides (data not shown). Also consistent with a glycerophosphate lipid, the GlcUA-lipid was insensitive to hydrolysis in 50% phenol for 2 h at 70°C (data not shown), whereas undecaprenyl-diphosphate and -monophosphate sugars have respective half-lives of 4 to 10 min and 60 min under identical conditions (35).
FIG. 2.
Glycolipid products are sensitive to saponification and PLD treatment. Lipids were labeled by incubating WU2 membranes in reactions containing 100 mM HEPES (pH 7.5), 10 mM MnCl2, and either 1 μM UDP-[14C]Glc or 1 μM UDP-[14C]GlcUA for 20 to 30 min at 35°C. Membranes were sedimented and washed twice with 100 mM HEPES (pH 7.5) containing 10% glycerol and then solubilized with 0.5% Nonidet P-40 containing 0.05 M EDTA. Solubilized products were saponified with 0.1 N NaOH for 20 min at 35°C or treated with 5 units of PLD for 1 h at 30°C in 25 mM Tris-acetate (pH 8) containing 10 mM CaCl2. (A) Untreated, saponified, and PLD-treated [14C]GlcUA-labeled samples were subjected to TLC on silica plates in butanol:acetic acid:water (44:16:40). Products were visualized by autoradiography. (B) Untreated (•), saponified (○), and PLD-treated (▴) [14C]Glc-labeled samples were chromatographed on paper in butanol:acetic acid:water (44:16:40) for 2 h. The chromatogram was cut into 1-cm strips, and the amount of radioactivity was determined by liquid scintillation counting.
The lipid acceptor contains a head group.
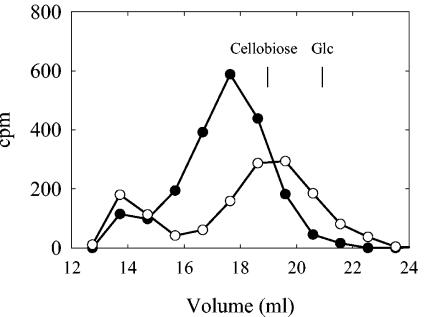
Following PLD digestion and chromatography on a Sephadex G-10 column, the GlcUA-labeled product eluted prior to the disaccharide cellobiose [β-d-Glc-(1→4)-d-Glc], while the Glc-labeled product coeluted with cellobiose (Fig. 3), indicating that the lipid acceptor may contain an additional residue attached to the phosphate. When the PLD-digested Glc- and GlcUA-labeled lipids were chromatographed on paper, they migrated faster than their respective monosaccharides (shown for Glc-labeled lipid in Fig. 2B), again indicating that the lipid acceptor contained a head group. The PLD-digested Glc-labeled product was sensitive to β-glucosidase digestion, liberating Glc, but was not sensitive to β-glucuronidase digestion (data not shown). Treatment of the GlcUA-labeled product with β-glucuronidase following PLD digestion resulted in degradation of approximately 50% of the product and yielded GlcUA (data not shown). The GlcUA-labeled product was not sensitive to β-glucosidase digestion. These data suggest that the Glc-labeled lipid serves as an acceptor for GlcUA addition.
FIG. 3.
Glc- and GlcUA-labeled PLD-liberated products are different sizes. [14C]GlcUA-labeled (•) and [14C]Glc-labeled (○) lipids synthesized as for Fig. 2A were solubilized and treated with PLD as described in the legend to Fig. 2. Samples were applied to a Sephadex G-10 column and eluted with 0.2 M NaCl containing 0.2 M ammonium acetate and 0.02% sodium azide at a flow rate of 0.3 ml/min. Samples (0.5 ml) were collected, and the amount of radioactivity present in the even-numbered fractions was determined.
Purification of a glycosylated lipid from S. pneumoniae and E. coli membranes expressing the type 3 synthase.
Total lipids were extracted from the type 3 S. pneumoniae strains JD614 and JD902 (Cps3S−) as well as from E. coli strains JD424 (Cps3S+) and JD422 (vector control). JD614 contains a point mutation in cps3D (12) that causes a single amino acid change (T83I; this study) in the UDP-Glc dehydrogenase, resulting in <6% of parental levels of capsule (12, 26). JD614 was used in place of the wild-type strain WU2 because the reduction in capsule facilitates lipid extraction. Membranes from JD614 exhibited 60% of the in vitro polymer synthesis activity of the parent strain, but the enzyme was able to catalyze synthesis of high-molecular-weight polysaccharide under the same conditions used for WU2 membranes (data not shown). Additionally, when JD614 membranes were incubated with single nucleotide sugars, glycolipids with characteristics identical to those described for WU2 were synthesized in nearly equivalent amounts (data not shown). Following separation of the anionic lipids by TLC, a major lipid product that comigrated with the [14C]Glc-labeled-lipid synthesized in vitro using JD614 membranes was observed in the lipid extract from JD614 and JD424 (data not shown). The ratio of hexose to phosphorous of the final lipid product from both strains was 1.2 (Table 1). This lipid represented approximately 4% of the chloroform-extractable phosphorous in S. pneumoniae and 1% in E. coli. No corresponding product was found in JD422, and only a minor band was found in JD902 (see below).
TABLE 1.
Phosphorous and hexose levels during lipid purification
| Fraction or process | Result for strain
|
|||||
|---|---|---|---|---|---|---|
|
S. pneumoniae JD614
|
E. coli JD424
|
|||||
| Amt of hexosea (μmol) | Amt of phosphorousa (μmol) | Hexose/phosphorous ratio | Amt of hexosea (μmol) | Amt of phosphorousa (μmol) | Hexose/phosphorous ratio | |
| Chloroform extract | 8 | 2 | 4 | 2.3 | 74.5 | 0.03 |
| Anionic lipids | 0.2 | 1.2 | 0.17 | 1.8 | 14.5 | 0.12 |
| TLCb | 0.086 | 0.076 | 1.2 | 1 | 0.8 | 1.25 |
Aliquots of the fractions indicated were measured for total hexose and phosphorous content as described in Materials and Methods. These values were used to determine the hexose and phosphorous content in the total fraction, which is indicated.
Lipid band purified by thin-layer chromatography.
Mass spectral analysis of anionic lipids of type 3 S. pneumoniae and E. coli strains expressing the type 3 synthase.
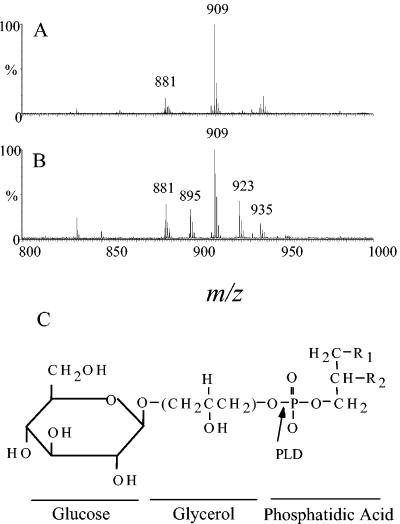
Mass spectral analysis of the lipid band purified from JD614 revealed mass ions at m/z 909 and m/z 881, which are compatible with a monoglycosylated phosphatidylglycerol with fatty acids 16:0 and 18:1 and fatty acids 16:0 and 16:1, respectively (Fig. 4A and Table 2). Phosphatidylglycerol isolated from the same anionic fraction as the glycosylated phosphatidylglycerol also showed mass ions that were compatible with the same fatty acid content (data not shown). These fatty acids are consistent with those previously reported to be the predominant fatty acids in S. pneumoniae glycerophosphate lipids (36). The mass ions at m/z 909 and m/z 881 were not detected in the lipid band from JD902, indicating that the presence of glycosylated phosphatidylglycerol in S. pneumoniae membranes is dependent on the presence of the type 3 synthase (data not shown). The level of phosphorous in the lipid band from JD902 was approximately one-third that for JD614 (data not shown), suggesting that the glycosylated phosphatidylglycerol made up a significant portion of the lipid band for JD614. Many of the mass ions observed in the purified lipid band from both JD902 and JD614 have not been conclusively identified. However, several of the mass ions were consistent with lysophosphatidylglycerol (data not shown).
FIG. 4.
Mass spectral analysis of purified anionic lipids from S. pneumoniae and E. coli expressing the type 3 synthase. Anionic lipids purified from (A) S. pneumoniae strain JD614 and (B) E. coli strain JD424 as described in Materials and Methods were analyzed by mass spectrometry. (C) Putative structure of lipid product identified by mass spectrometry. Arrow indicates site of PLD activity; R1 and R2 indicate positions of fatty acids. The linkage between phosphatidic acid and glycerol could be between either hydroxyl group on the glycerol headgroup.
TABLE 2.
Predicted compounds for mass ion peaks
| m/z | Compound |
|---|---|
| 253 | 16:1 fatty acid |
| 255 | 16:0 fatty acid |
| 281 | 18:1 fatty acid |
| 881 | Glycosylated phosphatidylglycerol with 16:0 and 16:1 fatty acids |
| 895 | Glycosylated phosphatidylglycerol with a 16:0 fatty acid and a C17 fatty acid with a cyclopropane ring |
| 909 | Glycosylated phosphatidylglycerol with 16:0 and 18:1 fatty acids |
| 923 | Glycosylated phosphatidylglycerol with a 16:0 fatty acid and a C19 fatty acid with a cyclopropane ring |
| 935 | Glycosylated phosphatidylglycerol with two 18:1 fatty acids |
Mass ion peaks at m/z 881 and m/z 909 were also observed with the comigrating lipid band from E. coli strain JD424, which expresses the recombinant type 3 synthase (Fig. 4B). In addition, mass ion peaks at m/z 895, m/z 923, and m/z 935 were also observed with the lipid from JD424. The peak at m/z 935 was consistent with glycosylated phosphatidylglycerol containing two 18:1 fatty acids, while the peaks at m/z 895 and m/z 923 were consistent with glycosylated phosphatidylglycerol containing a 16:0 fatty acid and either a C17 fatty acid carrying a cyclopropane ring or a C19 fatty acid carrying a cyclopropane ring, respectively. The presence of these fatty acids was confirmed by MS/MS of these mass ions (data not shown). The set of mass ion peaks observed with the lipid from JD424 was nearly identical to that observed for glycosylated phosphatidylglycerol synthesized when the glycosyltransferase Ugt106B1 from Staphylococcus aureus was expressed in E. coli (21). For E. coli strain JD422 (vector control), no lipid that comigrated with the [14C]Glc-labeled lipid synthesized in vitro using JD614 membranes was observed; however, two iodine-reactive lipid bands that migrated in the region were observed. Analysis of these two lipids by MS revealed no mass ions that corresponded to glycosylated phosphatidylglycerol (data not shown). The mass spectral data in combination with the size and migration of the products generated following treatment of the [14C]Glc-labeled lipid with PLD, mild alkali, and acid provide compelling evidence that the lipid acceptor is glucosylated phosphatidylglycerol (Fig. 4C).
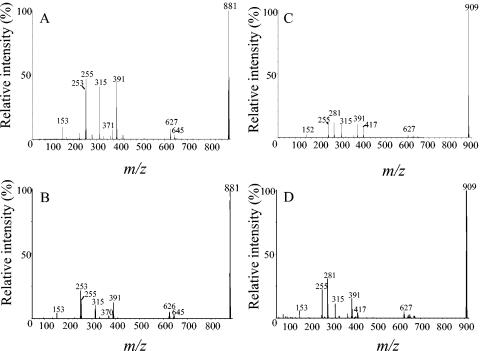
To further confirm that the lipids with mass ions at m/z 881 and m/z 909 from E. coli and S. pneumoniae were the same, they were subjected to MS/MS. As can be seen in Fig. 5A and B, the MS/MS spectra for the mass ion at m/z 881 were nearly identical for S. pneumoniae and E. coli, with peaks at m/z 253 and m/z 255, which correspond to the fatty acids 16:1 and 16:0, respectively. Similarly, the MS/MS spectra for the peak at m/z 909 were nearly identical between S. pneumoniae and E. coli (Fig. 5C and D). The peaks at m/z 255 and m/z 281 were consistent with the fatty acids 16:0 and 18:1, respectively. Besides the mass ion peaks that correspond to the fatty acids, the MS/MS spectra of the mass ions at m/z 881 and m/z 909 were nearly identical (compare Fig. 5A and 5C for S. pneumoniae and Fig. 5B and 5D for E. coli), confirming that they were the same lipid with different fatty acid chains.
FIG. 5.
MS/MS analysis of mass ions m/z 881 and m/z 909 from S. pneumoniae and E. coli anionic lipid fractions. The mass ions m/z 881 from (A) S. pneumoniae and (B) E. coli and m/z 909 from (C) S. pneumoniae and (D) E. coli, identified from the MS of an anionic lipid in Fig. 4, were analyzed by MS/MS. The S. pneumoniae mass ions were analyzed using a Micromass Q-TOF instrument in the electrospray ionization mode, while the E. coli mass ions were analyzed using a PE ScieX triple quadrupole in the negative-ion mode.
Polysaccharide synthesized using JD614 membranes mimics that of E. coli membranes in the polysaccharide ejection and release assay.
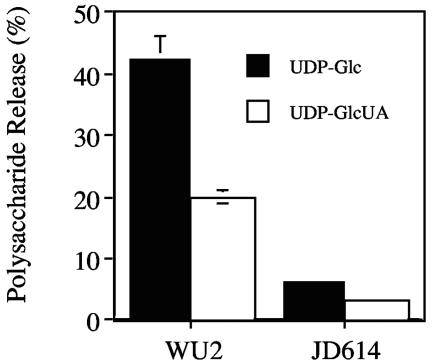
To examine polymer release, membrane-polysaccharide-enzyme complexes were first synthesized and then incubated with a 100 μM concentration of either UDP-Glc or UDP-GlcUA. As previously shown (15), membranes of the type 3 parent strain WU2 released approximately 40% and 20% of the bound polysaccharide in the presence of UDP-Glc and UDP-GlcUA, respectively (Fig. 6). In contrast, very little of the polysaccharide synthesized using JD614 membranes was released under identical conditions. Despite retention of the polysaccharide on the JD614 membranes, the polymer could not be extended into a higher molecular size, indicating it had been ejected from the enzyme (see below). These results were similar to those observed with E. coli membranes (9) and membranes from other S. pneumoniae type 3 strains that contain point mutations in cps3D or in pgm, which encodes a phosphoglucomutase (data not shown). These strains produce reduced amounts of capsule (0% to 25% of parental levels), and their mutations have been described previously (13, 18, 26). Since these S. pneumoniae and E. coli strains produce very little to no type 3 polysaccharide due to reduced UDP-GlcUA levels, they presumably lack the large amount of preformed, large-molecular-size polysaccharide that serves as an acceptor for sugar addition in WU2 (15). This makes strains like JD614 useful for examining type 3 polysaccharide initiation.
FIG. 6.
JD614 membranes release less polysaccharide than WU2 membranes. Polysaccharide-enzyme complexes were synthesized as described in Materials and Methods. Labeled complexes (10,000 cpm for WU2 and 15,000 cpm for JD614) were incubated with either 100 μM UDP-Glc (solid bars) or 100 μM UDP-GlcUA (open bars), and polysaccharide release from the membranes was assayed as described previously (15). The data presented are averages (± standard deviations) for duplicate release assays using the same polysaccharide-enzyme complex. The amount of release observed with no nucleotide sugar (5% for WU2 and 2% for JD614) was subtracted from each sample prior to averaging data.
Initiation of polymer synthesis on a glycerophosphate lipid.
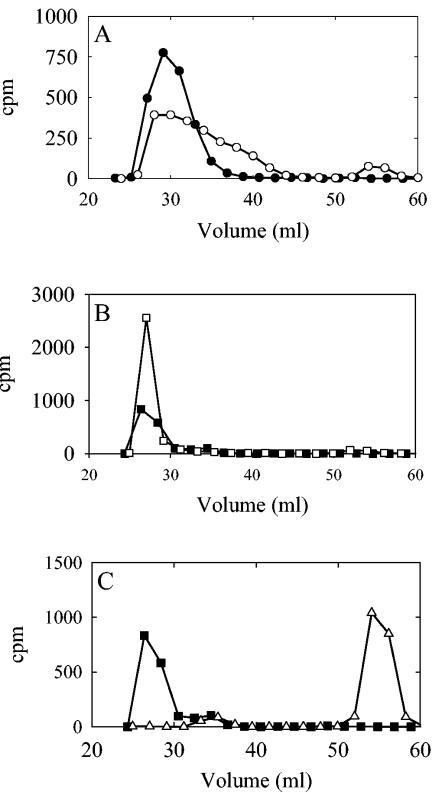
JD614 membranes were used to pulse label polysaccharide with [14C]Glc in a 1-min reaction with low substrate concentrations followed by a 5-min chase with 500 μM unlabeled substrate. The majority of the products in the pulse and chase samples remained at the origin following paper chromatography in butanol:acetic acid:water (44:16:40), indicating that mostly polymer or oligosaccharides were formed (data not shown). Following PLD treatment of the polysaccharide in the pulse reaction and analysis of the products by Sephacryl S-300 chromatography, a shift in the size of a portion of the polymer to a slightly smaller molecular size and the generation of what appeared to be small oligosaccharides were observed (Fig. 7A). Nearly all of the product labeled in the pulse was chased into a high-molecular-weight product, confirming that it was still engaged with the synthase. The size of the polymer in the chase sample, however, was not changed following PLD treatment (Fig. 7B). This result indicates that the portion of the polysaccharide whose migration changed following PLD digestion was chased into a higher molecular weight, where removal of the lipid moiety by PLD would not affect its elution on a Sephacryl S-300 column. All of the polysaccharide synthesized during the chase was sensitive to type 3 polysaccharide depolymerase from Bacillus palustris, indicating that all the product was type 3 polysaccharide (Fig. 7C).
FIG. 7.
Type 3 polysaccharide is synthesized on a glycerophosphate lipid. Type 3 polysaccharide was labeled in a 600-μl-volume pulse reaction containing 100 mM HEPES (pH 7.0), 10 mM MnCl2, 1 μM UDP-[14C]Glc, 1 μM UDP-GlcUA, and membranes from type 3 strain JD614 for 1 min at 35°C. A 300-μl aliquot was taken and placed on ice. The reaction was then chased for 5 min at 35°C with 500 μM of UDP-Glc and UDP-GlcUA. The membranes from the pulse and chase samples were sedimented by centrifugation, washed twice, and resuspended in 100 μl of wash buffer. Samples were solubilized and treated with PLD, as described in the legend to Fig. 2. (A) Pulse samples, untreated (•) and PLD digested (○), and (B) chase samples, untreated (▪) and PLD digested (□), were chromatographed on a Sephacryl S-300 column and eluted as described above. (C) A portion of the untreated chase sample (▪) was digested with type 3 polysaccharide-specific depolymerase (▵) in a 200-μl-volume reaction containing 0.1 M morpholineethanesulfonic acid (pH 6.0) for 15 min at 35°C. For all samples, 1-ml fractions were collected and the amount of radioactivity present in the even-numbered fractions was determined.
The type 3 synthase can reinitiate synthesis following polysaccharide ejection.
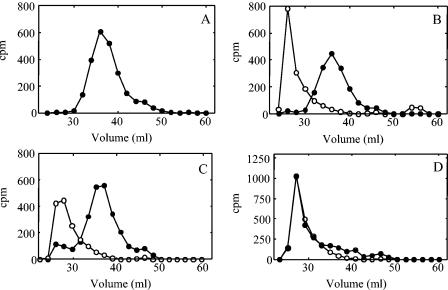
To determine if the type 3 synthase could reinitiate polymer synthesis following ejection of its nascent polysaccharide chain, we used membranes from JD614 to first synthesize low-molecular-weight polysaccharide-enzyme complexes labeled with [14C]GlcUA (Fig. 8A). These complexes were then incubated with either 100 μM UDP-Glc, 100 μM UDP-GlcUA, or no UDP-sugars prior to the addition of 20 μM of UDP-[3H]Glc and UDP-GlcUA in a complete reaction mixture. The membranes were then washed extensively prior to size exclusion chromatography. Preincubation of the complexes with either nucleotide sugar alone resulted in formation of a new 3H-labeled high-molecular-weight product, while the size of the majority of the 14C-labeled products remained unchanged (Fig. 8B and C). These data indicated that the majority of the 14C-labeled polymer had been ejected from the enzyme during the incubation with the single nucleotide sugars but was retained on the membrane and that the synthase had initiated synthesis of a new 3H-labeled polysaccharide chain in the final polymerization reaction. The extension of a small amount of the 14C-labeled product incubated with UDP-GlcUA (Fig. 8C) reflects the lower rate of polysaccharide ejection from the enzyme with UDP-GlcUA (15). Preincubation of the polysaccharide-enzyme complex with no UDP-sugars resulted in all of the 3H being incorporated into polymer that coeluted with the 14C as high-molecular-weight polysaccharide (Fig. 8D). These data were consistent with the addition of [3H]Glc to the 14C-prelabeled polymer. Together, these data indicate that the type 3 synthase in S. pneumoniae membranes is capable of initiating synthesis of a new polysaccharide chain following ejection of the nascent chain.
FIG. 8.
Reinitiation of polysaccharide synthesis following ejection of the nascent polymer. [14C]GlcUA-labeled polysaccharide-enyzme complexes were prepared in a 500-μl reaction mixture consisting of 100 mM HEPES (pH 7.5), 10 mM MnCl2, 100 μM UDP-Glc, 100 μM UDP-[14C]GlcUA (27 mCi/mmol), and JD614 membranes (1.5 mg of protein). The reaction was incubated for 5 min at 35°C and stopped by placing it on ice. The volume of the reaction was brought to 2 ml with wash buffer (100 mM HEPES [pH 7.5] containing 10% glycerol), and the membranes were washed four times. (A) An aliquot of the 14C-labeled product was chromatographed on a Sephacryl S-500 column. Aliquots of the labeled complex were incubated for 10 min at 35°C in 100-μl-volume reactions containing 100 mM HEPES (pH 7.5), 10 mM MnCl2, and either (B) 100 μM UDP-Glc, (C) 100 μM UDP-GlcUA, or (D) no UDP-sugars. Membranes were sedimented by centrifugation and washed once as described above. The volume was then brought to 100 μl so that the reaction contained 100 mM HEPES (pH 7.5), 10 mM MnCl2, 20 μM UDP-[3H]Glc (20 μCi/mmol), and 20 μM UDP-GlcUA. The reactions were incubated for an additional 5 min at 35°C and terminated by placing them on ice and adding 200 μl of ice-cold wash buffer. The membranes were sedimented by centrifugation, washed, and analyzed by chromatography on a Sephacryl S-500 column. 14C (•) and 3H (○) levels were measured in the even-numbered fractions. The values compensate for a 17% overlap of 14C and a 0.2% overlap of 3H in the corresponding channels.
Following release of nascent polysaccharide chains, release of newly synthesized polymer by WU2 membranes mimics that of JD614 and E. coli membranes.
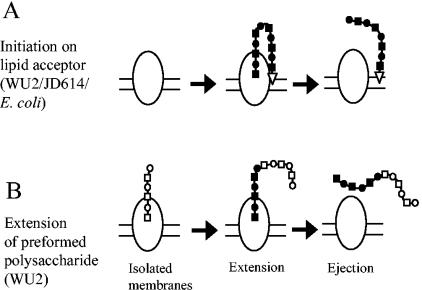
Approximately 20 to 40% of the polysaccharide synthesized using WU2 membranes is released from the membrane following incubation with a single nucleotide sugar (Fig. 6) (15). These data indicate that the releasable polysaccharide, although actively engaged with the synthase, is not attached to the membrane. Following ejection and release of this polymer, the type 3 synthase in WU2 membranes was capable of synthesizing new polymer in the same manner as JD614, above (data not shown). This newly synthesized polymer from WU2 membranes, like polymer synthesized using JD614 and E. coli membranes, was not released from the membrane following incubation with a single nucleotide sugar but was ejected from the enzyme. These observations indicated that the releasable portion of the WU2 polymer was detached from the membrane, and all the newly synthesized polymer was initiated on a lipid acceptor that serves as a membrane anchor. Models for the synthesis and release of type 3 polysaccharide, and the differences observed using WU2, JD614, and E. coli membranes, are shown in Fig. 9 and discussed further below.
FIG. 9.
Models of type 3 polysaccharide synthesis and ejection. (A) Polysaccharide synthesis initiates on a lipid acceptor (open triangle) with membranes from WU2, JD614, or recombinant E. coli. Following ejection, the polysaccharide remains tethered to the membrane via the lipid acceptor. (B) Preformed polysaccharide (open symbols) of S. pneumoniae strain WU2 that is no longer associated with the membrane is extended (closed symbols) in the presence of both nucleotide sugars. Following ejection, the polysaccharide is released into the medium. Some of the preformed polysaccharide in WU2 may still contain its lipid anchor; thus, synthesis and ejection would resemble the scheme shown in panel A. Following ejection, all new polysaccharide synthesis occurs on the lipid acceptor as described in the legend to panel A, regardless of the initial acceptor.
DISCUSSION
The type 3 synthase from S. pneumoniae is a member of a group of processive β-glycosyltransferases that form multiple glycosidic linkages (22). Many of the enzymes in this class, including the type 3 synthase (10), the hyaluronan synthases (for a review, see reference 38), and the cellulose synthases (24, 29-31) have been extensively studied; however, initiation of polymer synthesis has only recently been investigated. One model suggests that synthesis initiates de novo from the nucleotide sugars (28). Recent studies on the cellulose synthase CesA from cotton (Gossypium hirsutum), however, demonstrated that this enzyme utilizes sitosterol-β-glucoside to initiate cellulose formation (27). In addition, we demonstrated that the recombinant type 3 synthase in E. coli membranes catalyzes synthesis of the type 3 polysaccharide on an endogenous glycerophosphate lipid (9). Here, we demonstrate that synthesis initiates on phosphatidylglycerol in both the S. pneumoniae and recombinant E. coli systems.
When incubated in the presence of a single nucleotide sugar, the type 3 synthase catalyzes the addition of a single Glc or GlcUA onto an endogenous lipid acceptor (reference 9 and this study). Characterization of the GlcUA- and Glc-labeled products using acid and alkaline hydrolysis and sensitivity to PLD digestion indicated that the lipid acceptor for these single-sugar additions was a glycerophosphate lipid. Following digestion of the glycolipids with PLD, the resulting products did not migrate on paper as their free monosaccharides, suggesting that the glycolipids contained an additional residue. Consistent with this possibility, the PLD-treated Glc-labeled product eluted prior to a Glc standard, whereas the PLD-treated GlcUA-labeled product eluted prior to cellobiose when chromatographed on Sephadex G-10. The larger size of the GlcUA-labeled product suggested that the order of sugar addition to the lipid acceptor is Glc followed by GlcUA. The capacity of the type 3 synthase in membranes from S. pneumoniae and E. coli to incorporate relatively large amounts of GlcUA into a lipid product (9) suggested that the membranes contain large amounts of glucosylated lipid acceptor. Mass spectrometry analysis of an anionic lipid band isolated from S. pneumoniae and E. coli strains expressing the type 3 synthase indicated the presence of glycosylated phosphatidylglycerol. This lipid is not present in native E. coli membranes (21; this study) and was not observed in an S. pneumoniae strain in which the synthase had been insertionally inactivated, indicating that its synthesis is dependent on the type 3 synthase. Since phosphatidylglycerol is the most abundant glycerophosphate lipid in S. pneumoniae membranes (36), it is somewhat unexpected that the levels of Glc incorporation into the lipid product were relatively low compared to those of GlcUA. One explanation for this observation is that phosphatidylglycerol is a less-effective substrate for Glc addition than a terminal GlcUA on an oligosaccharide or polysaccharide, whereas GlcUA is readily added to the glucosylated phosphatidylglycerol. Perhaps the negatively charged phosphate attached to glycerol mimics the carboxyl group of GlcUA, thus allowing phosphatidylglycerol to function as a weak acceptor for Glc addition.
Two lines of evidence demonstrate that type 3 polysaccharide is initiated on a glycerophosphate lipid. First, a portion of the type 3 polysaccharide synthesized using JD614 membranes in a pulse reaction was reduced in size by treatment with PLD. Second, 95% of the polymer synthesized using JD614 membranes remained tethered to the membrane following incubation with a single nucleotide sugar, even though it had been ejected from the enzyme. We previously reported that 40 to 60% of the polymer synthesized in vitro using WU2 membranes was ejected from the synthase and released from the membrane upon incubation with a single nucleotide sugar (15). We have demonstrated here that despite retention on the membrane, nearly all of the polymer is actually ejected from the enzyme under these conditions. The remaining polymer thus appears to be tethered to the membrane. As shown in Fig. 9, the difference in the amount of polysaccharide released from the membrane by WU2 and its derivatives that contain mutations that affect substrate concentration and capsule levels may reflect the initial acceptor during polymer synthesis. For WU2, approximately half of the polysaccharide synthesized in vitro initiates on preformed polysaccharide that is engaged with the enzyme but is no longer associated with the membrane, possibly due to loss of the lipid moiety (9, 15). Strains like JD614, however, contain mutations that result in a decrease in the UDP-GlcUA concentration below a critical level. These skewed substrate concentrations likely result in not only a decrease in capsule production but also an increased rate of polysaccharide ejection from the enzyme. Thus, in membranes isolated from these strains, the majority of the enzyme is either not engaged with preformed polysaccharide and initiates synthesis de novo from the lipid acceptor or is engaged with short preformed chain’s that still contain the lipid moiety. The rate of synthesis starting from the lipid acceptor may be lower than that of synthesis from an oligosaccharide acceptor, which may explain why the synthase from JD614 membranes exhibits only 60% of the polymer synthesis activity of the parent strain. Similar to JD614, none of the type 3 polysaccharide synthesized in vitro with E. coli membranes containing the type 3 synthase was released from the membrane upon incubation with a single nucleotide sugar; however, all the polysaccharide was ejected from the enzyme (9). The synthase isolated from E. coli, which produces little or no UDP-GlcUA, also would not be actively engaged with polymer upon addition to the in vitro synthesis assay, and thus, all the polymer is initiated de novo on the lipid acceptor (9).
Type 3 polysaccharide in S. pneumoniae cultures can be found both cell associated and soluble in the medium (18). Unlike most capsule types in S. pneumoniae, type 3 capsule is not covalently linked to the cell wall (33), suggesting an alternative mechanism for attachment to the cell surface. We previously proposed that the cell-associated polysaccharide was due to its interaction with the synthase and that the soluble form was generated following an ejection reaction (15). With the evidence presented here, it appears that the cell association of the polymer may occur via the lipid anchor on the reducing end of the polymer. The soluble form of the polysaccharide could then be generated by breakage of the polysaccharide chains and release from the enzyme or breakage of the chains following release. Alternatively, a dedicated mechanism for cleaving the polysaccharide or the lipid primer may exist. If the latter occurs, finding a way to inhibit formation of the soluble form of polysaccharide could provide insights into its role in type 3 S. pneumoniae pathogenesis.
Acknowledgments
This work was supported by Public Health Service grants GM53017 and T32 HL07553 from the National Institutes of Health.
Mass spectrometry analyses were performed by the Comprehensive Cancer Mass Spectrometry Shared Facility of the University of Alabama at Birmingham.
REFERENCES
- 1.Abeyta, M., G. G. Hardy, and J. Yother. 2003. Genetic alteration of capsule type but not PspA type affects accessibility of surface-bound complement and surface antigens of Streptococcus pneumoniae. Infect. Immun. 71:218-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arrecubieta, C., R. Lopez, and E. Garcia. 1996. Type 3-specific synthase of Streptococcus pneumoniae (Cap3B) directs type 3 polysaccharide biosynthesis in Escherichia coli and in pneumococcal strains of different serotypes. J. Exp. Med. 184:449-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashwell, G. 1966. New colorimetric methods of sugar analysis. Methods Enzymol. 8:85-95. [Google Scholar]
- 4.Avery, O. T., and R. Dubos. 1931. The protective action of a specific enzyme against type III pneumococcus infections in mice. J. Exp. Med. 54:73-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender, M. H., R. T. Cartee, and J. Yother. 2003. Positive correlation between tyrosine phosphorylation of CpsD and capsular polysaccharide production in Streptococcus pneumoniae. J. Bacteriol. 185:6057-6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briles, D. E., J. L. Claflin, K. Schroer, J. Davie, P. Baker, J. Kearney, and R. Barletta. 1981. Antiphosphocholine antibodies found in normal mouse serum are protective against intravenous infection with type 3 Streptococcus pneumoniae. J. Exp. Med. 153:694-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, E. J. 1985. Interaction of gram-positive microorganisms with complement. Curr. Top. Microbiol. Immunol. 121:159-187. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, J. A., G. J. Davies, V. Bulone, and B. Henrissat. 1997. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem. J. 326:929-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cartee, R. T., W. T. Forsee, J. W. Jensen, and J. Yother. 2001. Expression of the Streptococcus pneumoniae type 3 synthase in Escherichia coli: assembly of type 3 polysaccharide on a lipid primer. J. Biol. Chem. 276:48831-48839. [DOI] [PubMed] [Google Scholar]
- 10.Cartee, R. T., W. T. Forsee, J. S. Schtuzbach, and J. Yother. 2000. Mechanism of type 3 capsular polysaccharide synthesis in Streptococcus pneumoniae. J. Biol. Chem. 275:3907-3914. [DOI] [PubMed] [Google Scholar]
- 11.Dennis, E. A. 1983. The enzymes, p. 307-353. In P. D. Boyer (ed.), vol. 16. Academic Press, Inc., Orlando, Fla.
- 12.Dillard, J., M. Vandersea, and J. Yother. 1995. Characterization of the cassette containing genes for type 3 capsular polysaccharide biosynthesis in Streptococcus pneumoniae. J. Exp. Med. 181:973-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dillard, J., and J. Yother. 1994. Genetic and molecular characterization of capsular polysaccharide biosynthesis in Streptococcus pneumoniae type 3. Mol. Microbiol. 12:959-972. [DOI] [PubMed] [Google Scholar]
- 14.Duck-Chong, C. G. 1979. A rapid sensitive method for determining phospholipid phosphorus involving digestion with magnesium nitrate. Lipids 14:492-497. [Google Scholar]
- 15.Forsee, W. T., R. T. Cartee, and J. Yother. 2000. Biosynthesis of type 3 capsular polysaccharide in Streptococcus pneumoniae: enzymatic chain release by an abortive translocation process. J. Biol. Chem. 275:25972-25978. [DOI] [PubMed] [Google Scholar]
- 16.Griffith, F. 1928. The significance of pneumococcal types. J. Hyg. 27:113-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guidolin, A., J. K. Morona, R. Morona, D. Hansman, and J. C. Paton. 1994. Nucleotide sequence analysis of genes essential for capsular polysaccharide biosynthesis in Streptococcus pneumoniae type 19F. Infect. Immun. 62:5384-5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy, G. G., M. J. Caimano, and J. Yother. 2000. Capsule biosynthesis and basic metabolism in Streptococcus pneumoniae are linked through the cellular phosphoglucomutase. J. Bacteriol. 182:1854-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Hardy, G. G., A. D. Mager, C. L. Ventura, M. J. Caimano, and J. Yother. 2001. Essential role for cellular phosphoglucomutase in virulence of type 3 Streptococcus pneumoniae. Infect. Immun. 69:2309-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henrichsen, J. 1995. Six newly recognized types of Streptococcus pneumoniae. J. Clin. Microbiol. 33:2759-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imamura, S., and Y. Horiuti. 1979. Purification of Streptomyces chromofuscus phospholipase D by hydrophobic affinity chromatography on palmitoyl cellulose. J. Biochem. (Tokyo) 85:79-95. [DOI] [PubMed] [Google Scholar]
- 21.Jorasch, P., D. C. Warnecke, B. Lindner, U. Zahringer, and E. Heinz. 2000. Novel processive and nonprocessive glycosyltransferases from Staphylococcus aureus and Arabidopsis thaliana synthesize glycoglycerolipids, glycophospholipids, glycosphingolipids, and glycosterols. Eur. J. Biochem. 267:3770-3783. [DOI] [PubMed] [Google Scholar]
- 22.Keenleyside, W., and C. Whitfield. 1997. A novel pathway for O-polysaccharide biosynthesis in Salmonella enterica serovar Borreze. J. Biol. Chem. 271:28581-28592. [DOI] [PubMed] [Google Scholar]
- 23.Kolkman, M. A. B., W. Wakarchuk, P. J. M. Nuijten, and B. A. M. van der Zeijst. 1997. Capsular polysaccharide synthesis in Streptococcus pneumoniae serotype 14: molecular analysis of the complete cps locus and identification of genes encoding glycosyltransferases required for the biosynthesis of the tetrasaccharide subunit. Mol. Microbiol. 26:197-208. [DOI] [PubMed] [Google Scholar]
- 24.Koyama, M., W. Helbert, T. Imai, J. Sugiyama, and B. Henrissat. 1997. Parallel-up structure evidences the molecular directionality during biosynthesis of bacterial cellulose. Proc. Natl. Acad. Sci. USA 94:9091-9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Llull, D., E. Gracia, and R. Lopez. 2001. Tts, a processive β-glucosyltransferase of Streptococcus pneumoniae, directs the synthesis of the branched type 37 capsular polysaccharide in pneumococcus and other gram-positive species. J. Biol. Chem. 276:21053-21061. [DOI] [PubMed] [Google Scholar]
- 26.Magee, A. D., and J. Yother. 2001. Requirement for capsule in colonization by Streptococcus pneumoniae. Infect. Immun. 69:3755-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng, L., Y. Kawagoe, P. Hogan, and D. Delmer. 2002. Sitosterol-β-glucoside as primer for cellulose synthesis in plants. Science 295:147-150. [DOI] [PubMed] [Google Scholar]
- 28.Saxena, I. M., R. M. Brown, Jr., M. Fevre, R. A. Geremia, and B. Henrissat. 1995. Multidomain architecture of β-glycosyl transferases: implications for mechanism of action. J. Bacteriol. 177:1419-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saxena, I. M., K. Kudlicka, K. Okuda, and R. M. Brown, Jr. 1994. Characterization of genes in the cellulose-synthesizing operon (acs operon) of Acetobacter xylinum: implications for cellulose crystallization. J. Bacteriol. 176:5735-5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saxena, I. M., F. C. Lin, and R. M. Brown, Jr. 1990. Cloning and sequencing of the cellulose synthase catalytic subunit gene of Acetobacter xylinum. Plant Mol. Biol. 15:673-683. [DOI] [PubMed] [Google Scholar]
- 31.Saxena, I. M., F. C. Lin, and R. M. Brown, Jr. 1991. Identification of a new gene in an operon for cellulose biosynthesis in Acetobacter xylinum. Plant Mol. Biol. 16:947-954. [DOI] [PubMed] [Google Scholar]
- 32.Smith, E. B., G. T. Mills, H. P. Bernheimer, and R. Austrian. 1960. The synthesis of type III pneumococcal capsular polysaccharide from uridine nucleotides by a cell-free extract of Diplococcus pneumoniae type III. J. Biol. Chem. 235:1876-1880. [PubMed] [Google Scholar]
- 33.Sorensen, U. B. S., J. Henrichsen, H. C. Chen, and S. C. Szu. 1990. Covalent linkage between the capsular polysaccharide and the cell wall peptidoglycan of Streptococcus pneumoniae revealed by immunochemical methods. Microb. Pathog. 8:325-334. [DOI] [PubMed] [Google Scholar]
- 34.Tlapak-Simmons, V. L., C. A. Baron, R. Gotschall, D. Haque, W. M. Canfield, and P. H. Weigel. 2005. Hyaluronan biosynthesis by class I streptococcal hyaluronan synthases occurs at the reducing end. J. Biol. Chem. 280:13012-13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tolmasky, M. E., R. J. Staneloni, and L. F. Leloir. 1982. Lipid bound saccharides in Rhizobium meliloti. J. Biol. Chem. 257:6751-6757. [PubMed] [Google Scholar]
- 36.Trombe, M., M. Laneelle, and G. Laneelle. 1979. Lipid composition of aminopterin-resistant and sensitive strains of Streptococcus pneumoniae. Biochim. Biophys. Acta 574:290-300. [DOI] [PubMed] [Google Scholar]
- 37.van Dam, J. E., A. Fleer, and H. Snippe. 1990. Immunogenicity and immunochemistry of Streptococcus pneumoniae capsular polysaccharides. Antonie Leeuwenhoek 58:1-47. [DOI] [PubMed] [Google Scholar]
- 38.Weigel, P., V. Hascall, and M. Tammi. 1997. Hyaluronan synthases. J. Biol. Chem. 272:13997-14000. [DOI] [PubMed] [Google Scholar]
- 39.Whitfield, C., and I. S. Roberts. 1999. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol. Microbiol. 31:1307-1319. [DOI] [PubMed] [Google Scholar]
- 40.Winkelstein, J. A. 1981. The role of complement in the host's defense against Streptococcus pneumoniae. Rev. Infect. Dis. 3:289-298. [DOI] [PubMed] [Google Scholar]
- 41.Wood, W. B., and M. R. Smith. 1949. Inhibition of surface phagocytosis by capsular “slime layer” of pneumococcus type III. J. Exp. Med. 90:85-99. [DOI] [PMC free article] [PubMed] [Google Scholar]