Abstract
In Escherichia coli the sulfur-containing amino acid homocysteine (Hcy) is the last intermediate on the methionine biosynthetic pathway. Supplementation of a glucose-based minimal medium with Hcy at concentrations greater than 0.2 mM causes the growth of E. coli Frag1 to be inhibited. Supplementation of Hcy-treated cultures with combinations of branched-chain amino acids containing isoleucine or with isoleucine alone reversed the inhibitory effects of Hcy on growth. The last intermediate of the isoleucine biosynthetic pathway, α-keto-β-methylvalerate, could also alleviate the growth inhibition caused by Hcy. Analysis of amino acid pools in Hcy-treated cells revealed that alanine, valine, and glutamate levels are depleted. Isoleucine could reverse the effects of Hcy on the cytoplasmic pools of valine and alanine. Supplementation of the culture medium with alanine gave partial relief from the inhibitory effects of Hcy. Enzyme assays revealed that the first step of the isoleucine biosynthetic pathway, catalyzed by threonine deaminase, was sensitive to inhibition by Hcy. The gene encoding threonine deaminase, ilvA, was found to be transcribed at higher levels in the presence of Hcy. Overexpression of the ilvA gene from a plasmid could overcome Hcy-mediated growth inhibition. Together, these data indicate that in E. coli Hcy toxicity is caused by a perturbation of branched-chain amino acid biosynthesis that is caused, at least in part, by the inhibition of threonine deaminase.
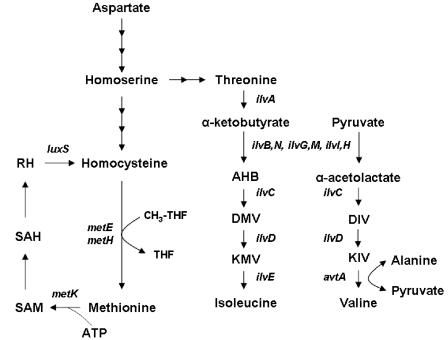
Homocysteine (Hcy) is a sulfur-containing amino acid that is found as a normal metabolite in all three domains of life. In Escherichia coli it is the last intermediate on the methionine biosynthetic pathway. The final step of this pathway involves the methylation of Hcy to produce methionine, the methyl group being derived from methyl tetrahydrofolate (Fig. 1). Methionine is essential to the process of protein translation and is also an essential methyl donor in the cell, after it has been converted to S-adenosylmethionine (SAM) (6). Thus, Hcy lies at an important metabolic junction in E. coli; it enables single carbon units to be shuttled from the reduced folate pool to the principal methyl donor in the cell, SAM (Fig. 1). In humans Hcy plays essentially the same biochemical role (reviewed by van der Put et al. [32]), although the methionine produced in the methyl transfer reaction is not sufficient to supply the body with enough methionine for protein synthesis. Hcy can also act as a precursor for cysteine biosynthesis in humans but not in E. coli.
FIG. 1.
Role of Hcy in methionine biosynthesis in E. coli. The final step in the biosynthesis of methionine from homoserine involves the transfer of a methyl group from the methyl tetrahydrofolate (CH3-THF) to Hcy. The single carbon unit is derived from serine. The reaction is catalyzed by either of two enzymes, the vitamin B12-dependent enzyme MetH, which functions anaerobically when B12 is present in the growth medium, or the B12-independent enzyme MetE (6). MetK catalyzes the synthesis of SAM from methionine and ATP. The biosynthesis of the branched-chain amino acids isoleucine and valine is also depicted. Gene names are indicated in italics. AHB, α-aceto-α-hydroxybutyrate; DIV, α,β-dihydroxy-isovalerate; DMV, α-β-dihydroxy-β-methylvalerate; KIV, α-ketoisovalerate; KMV, α-keto-β-methylvalerate; RH, ribosylhomocysteine; SAH, S-adenosylhomocysteine. Note that there are three isozymes of acetohydroxy acid synthase (AHAS) in non-K-12 strains of E. coli, encoded by ilvB ilvN (AHAS I), ilvG ilvM (AHAS II), and ilvI ilvH (AHAS III), respectively. In Frag1, which is a K-12 strain of E. coli, AHAS II is nonfunctional because of a frameshift mutation in ilvG (18).
In recent years Hcy has been increasingly well studied in the context of human biology. Elevated serum levels of Hcy are associated with several serious diseases, including cardiovascular disease (1, 34), birth defects, and pregnancy complications (32, 33), as well as neurodegenerative diseases such as Alzheimer's disease (26). In addition, Hcy is known to induce apoptotic death in human endothelial cells grown in tissue culture (39). The molecular basis for the link between disease and high serum Hcy levels remains uncertain, although a number of explanations have been postulated (9). Hcy can trigger oxidative stress in human cells through the production of hydrogen peroxide (29), although in some cell types oxidative stress is not detected (20). Tissue damage through homocysteinylation of lysine residues on serum proteins has also been suggested (8, 11). S-Nitrosylated Hcy, which arises when Hcy reacts with nitric oxide (NO), can become translationally incorporated into protein through mischarged methionyl tRNA, leading to protein damage (12). Vascular disease linked to elevated levels of Hcy has also been suggested to result from the oxidative inactivation of NO, which acts as a potent vasodilator (35).
In E. coli Hcy is produced either by the action of cystathionine-β-lyase on cystathionine or by the action of LuxS on S-ribosylhomocysteine (25, 38), which is produced when S-adenosylmethionine donates its methyl group (6) (Fig. 1). Low concentrations of Hcy (≥0.5 mM) supplemented into minimal growth medium are inhibitory to the growth of E. coli (23). The basis for this toxicity is not known, although Hcy is known to compete with methionine and isoleucine for binding to methionyl- and isoleucyl-tRNA synthetases, respectively (13). Misactivated Hcy is rejected by methionyl- and isoleucyl-tRNA synthetases prior to transfer to the tRNA molecule, and this occurs by a cyclization reaction that leads to the synthesis of homocysteine thiolactone (13).
Hcy differs from cysteine only in the length of its side chain; Hcy has two carbon atoms in its side chain, whereas cysteine has only one. A number of early reports in the literature indicate that cysteine can cause growth inhibition of E. coli (14, 24). These findings have never been satisfactorily explained, although it is known that cysteine inhibits the activity of threonine deaminase (TD) (7), an enzyme required for the first step of isoleucine biosynthesis in E. coli (Fig. 1). Furthermore, cysteine induces amino acid starvation in E. coli when it is growing in a minimal medium (27).
In the present study we investigated the molecular basis for the toxicity of Hcy in E. coli K-12. We looked at the effects of inhibitory levels of Hcy on growth and on the size of amino acid pools. We found that the inhibitory effects of Hcy can be reversed by supplementation of the growth medium with the branched-chain amino acid isoleucine. This observation can be explained by the finding that TD is inhibited by Hcy. Overexpression of this enzyme partially relieves the inhibitory effects of Hcy. Together, these findings demonstrate that in E. coli the principal inhibitory effect of Hcy is on the biosynthesis of isoleucine.
MATERIALS AND METHODS
Bacterial strains, growth media, and chemicals.
E. coli Frag1 (F− thi rha lac gal) was used throughout. Salmonella enterica serovar Typhimurium LT2, nontoxigenic E. coli O157 strain P1432, and E. coli MG1655 were used in growth experiments with Hcy and were from our freezer stocks. E. coli J1 (a wild-type commensal isolate) was a gift from Ian Booth (University of Aberdeen). E. coli strains YG191 (MG1655 ΔbrnQ) and YG192 (MG1655 ΔlivHMGF) are described by Koyanagi et al. (15) and were a gift from Hidehiko Kumagai (Kyoto University, Kyoto, Japan). Growth experiments were carried out in McIlvaine's minimal medium (MM), a citrate-phosphate-buffered medium with a pH of 6.0. The medium contains 20 mM citric acid, 56 mM Na2HPO4, 5 mM K2HPO4, 0.4 mM MgSO4 · 7H2O, 7.6 mM (NH4)2SO4, 3 μM thiamine, and 6 μM (NH4)2SO4 · FeSO4 · 6H2O. The carbon source was glucose at 0.04% (wt/vol) or 0.4% (wt/vol). The medium was supplemented with Hcy (0.2 to 20 mM), isoleucine (0.15, 0.5, or 8 mM), leucine (0.15 mM), or valine (0.17 mM), where indicated. Kanamycin (50 μg ml−1) was added to MM where indicated. Overnight cultures were grown with limiting glucose (0.04% [wt/vol]) and then supplemented with 0.4% (wt/vol) glucose. After one doubling, this culture was used to inoculate 20 to 25 ml of fresh medium (0.4% [wt/vol] glucose) to an optical density at 600 nm (OD600) of 0.05. This growth protocol gives very reproducible growth rates between experiments and eliminates the lag phase from the growth curve. These cultures were then grown at 37°C with shaking in 250-ml conical flasks, and growth was monitored by measuring the OD600 of 1-ml samples. Specific growth rates (μ) were calculated from the doubling times (g) by using the relationship μ = ln2/g. Because of the biphasic nature of the growth curve, g was calculated from the slope of the initial part of the growth curve (typically 40 to 250 min). All chemicals used in the present study were obtained from either Sigma Aldrich or BDH unless otherwise stated.
Extract preparation and TD assay.
Crude cell extracts were prepared from cells grown either to an OD600 of ∼0.3 in 200 ml of MM or to stationary phase in LB. Cultures were harvested by centrifugation at 12,000 × g for 10 min at 4°C and washed once with MM or LB (depending on the growth medium that was used). Pellets were then resuspended in 4 ml of buffer containing 50 mM Tris-hydrochloride (pH 7.6), 50 mM MgCl2 · 6H2O, 25 mM KCl, 1 mM 2-mercaptoethanol, and 5% (vol/vol) glycerol. The cell suspension was disrupted by sonication using an MSE Soniprep 150 set at an amplitude of 16 μm for six 30-s pulses with a 30-s rest between pulses. The tubes used were kept on ice during the sonication procedure. The extracts were clarified by centrifugation at 14,000 × g for 20 min, and the supernatant was stored at −80°C. Protein concentrations were determined by the Bio-Rad RC DC kit, using bovine serum albumin as a standard.
TD assays were carried out by the method of Harris (7) with some modifications. Briefly, 800 μl of assay buffer containing 0.1 M Tris-HCl (pH 8.0), 0.1 M NH4Cl, 0.1 mM pyridoxal phosphate, and 40 mM threonine was placed into each tube. After addition of 0.1 ml of cell extract (final protein concentration, 0.01 to 0.5 mg ml−1), the tubes were incubated in a 37°C water bath for 20 min after which 0.1 ml of 50% (vol/vol) trichloroacetic acid was added, followed by 3 ml of 0.025% (wt/vol) 2,4-dinitrophenylhydrazine in 0.5 M HCl. After 15 min in a 25°C water bath, 1 ml of 40% (wt/vol) NaOH was added, and the resulting color was measured by recording the absorbance at 540 nm. The amount of product formed was determined by comparison with a standard curve prepared using known concentrations of α-ketobutyrate. Control experiments revealed that this assay is influenced slightly by the presence of Hcy. To control for this effect, blanks were prepared by adding appropriate concentrations of Hcy to cell extracts with no added substrate (threonine) and proceeding through the assay as described above. These blanks were then used to zero the spectrophotometer at 540 nm prior to reading the test assays (i.e., a separate blank was used for each concentration of Hcy tested).
Cloning and overexpression of ilvA.
The ilvA gene was amplified by PCR from E. coli Frag1 using primers ilvAFOR1, and ilvAREV1 (Table 1). This PCR product was 1,588 bp in length and consisted of the entire coding sequence of ilvA combined with a 3′ sequence encoding six histidine residues, which was included to facilitate the future purification of TD. This product was ligated into the multicloning site of vector pBAD202/D-TOPO (Invitrogen Life Technologies), yielding plasmid pNLT01. In this plasmid the ilvA gene is transcribed from a constitutively active vector-associated promoter but not from the arabinose-inducible PBAD promoter that is also present on the vector. The insert in pNLT01 was sequenced on both strands, and the sequence was compared to the ilvA sequence of E. coli K-12 MG1655, GenBank entry U00096. The sequences were identical except that the leucine residue at position 358 in the MG1655 protein sequence was found to be a glutamine residue in the Frag1 sequence. Frag1 was transformed with pNLT01, selecting for kanamycin-resistant transformants, and the resulting strain was used to prepare crude cell extracts containing high levels of TD activity for the Hcy inhibition studies.
TABLE 1.
PCR primers used in this study
| Primer | Sequence (5′-3′)a |
|---|---|
| ilvAFOR1 | CACCTAGAGGAGATATACATACCCATGGCTGACTCGCAACCCCTGb |
| ilvAREV1 | CTAGTGATGGTGATGGTGATGACCCGCCAAAAAGAACCTGAACGc |
| ilvAFOR2 | CCGCAAATCAAAGTGATC |
| ilvAREV2 | GAATTCGAAGCTGTAGAG |
| polAFOR | GAAGAGTTGTTGGGGCTG |
| polAREV | CAGACGAAGGGTGAGCTC |
Primers were designed by using published sequences from the Colibri website (http://genolist.pasteur.fr/Colibri/).
Underlined sequence is the start site in ilvAFOR1.
Underlined sequence is the 3′ His tag in ilvAREV1.
SDS-PAGE.
Crude cell extracts were prepared from overnight LB cultures of Frag1 and Frag1/pNLT01 as described above. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed by the method of Laemmli (16) in a discontinuous system with a 13% acrylamide gel. The gel was stained with Coomassie blue (GelCode Blue Stain Reagent; Pierce) and destained according to the manufacturer's protocol. After destaining, the gel was imaged by using a HP Scanjet 5300C scanner.
Amino acid pool measurements.
Cells were grown to an OD600 of 0.2 in MM before being treated with 0.5 mM Hcy, with 0.5 mM Hcy and 0.15 mM isoleucine, or with no addition (control). Growth was continued for 1 h (approximate OD600 of 0.4) before cells were harvested for amino acid pool analysis by filtration of 20 ml through a cellulose nitrate filter (Whatman; 0.45-μm pore size). The filters were immediately washed with an equal volume of an ice-cold 0.5 M NaCl solution. The filters were then placed in a sterile test tube containing 1 ml of ice-cold 0.1% (vol/vol) trifluoroacetic acid containing 25 μM norleucine as an internal standard. The test tubes were vortexed thoroughly and then left on ice for 30 min to allow for extraction of the amino acids. The filters were then removed, and the samples were stored at −20°C. Aliquots (100 μl) were dried under vacuum (SAVANT Speedvac concentrator) and then reconstituted in 200 μl of 0.1 M HCl before being passed through a 10-kDa molecular cutoff filter (Millipore). An aliquot of 20 μl was then loaded onto the 420A Applied Biosystems amino acid analyzer. For this analysis, cultures were grown in duplicate, and two samples were prepared from each flask.
Reverse transcriptase PCR.
Cells were grown in MM to an OD600 of 0.2 before the addition of Hcy (0.5 mM) to half of the samples. The flasks were returned to the incubator for 1 h, before 20 ml of cells was removed and added to a sterile centrifuge tube containing the RNA stabilization reagent RSR-3 (95% ethanol, 5% phenol [vol/vol]) (2). Cells were then harvested by centrifugation (15 min, 4°C, 8,200 × g), the supernatant was decanted, and the pellets were stored at −20°C until required. RNA was prepared by using the GenElute Mammalian Total RNA Miniprep kit (Sigma) according to the manufacturer's instructions. The RNA was then treated to remove DNA contamination by using the DNA-free kit (Ambion) according to the manufacturer's protocol. cDNA was synthesized from 20 μl of diluted RNA by using Expand reverse transcriptase with random primer p(dN)6 (both supplied by Roche). Aliquots (2 μl) of the resulting cDNA were subjected to 18, 24, or 30 cycles of PCR and run on agarose gels. Primers polAFOR and polAREV for the gene encoding DNA polymerase A (polA) were used as controls to detect contaminating DNA and to allow normalization of the cDNA template concentration. Non-reverse-transcribed RNA was used as a template for PCRs to ensure complete removal of genomic DNA. Specific primers for ilvA, ilvAFOR2 and ilvAREV2 (Table 1), were used in conjunction with cDNA generated from E. coli Frag1 grown in the presence or absence of 0.5 mM Hcy. RNA and cDNA were prepared twice, from independent cultures, and the reverse transcription-PCRs were performed twice on each cDNA preparation.
RESULTS
Hcy is toxic for E. coli.
In an earlier study one of us showed that the growth inhibition of E. coli in the presence of acetic acid is due, at least partly, to the accumulation of toxic levels of Hcy within the cell (23). We sought to investigate the basis for the toxicity of Hcy further. We examined the effect on growth of supplementing a range of Hcy concentrations into cultures of E. coli Frag1, growing in MM, with glucose (0.4% [wt/vol]) as the sole carbon source. Growth was inhibited slightly by the addition of 0.2 mM Hcy, whereas complete growth inhibition was observed in cultures treated with 20 mM Hcy (Fig. 2, top panel). Homocystine, the oxidized form of Hcy, had no inhibitory effect on the growth of Frag1 when added at a concentration of 0.5 mM (data not shown). We tested whether sensitivity to Hcy was unique to E. coli Frag1, which is a K-12 strain. Several other strains, including E. coli MG1655, E. coli J1 (a human commensal isolate), a nontoxigenic E. coli O157 isolate, and Salmonella enterica serovar Typhimurium strain LT2, all showed significant growth inhibition by 6 mM Hcy, although the extent of the inhibition varied from strain to strain (60, 56, 30 and 30% inhibition, respectively [data not shown]).
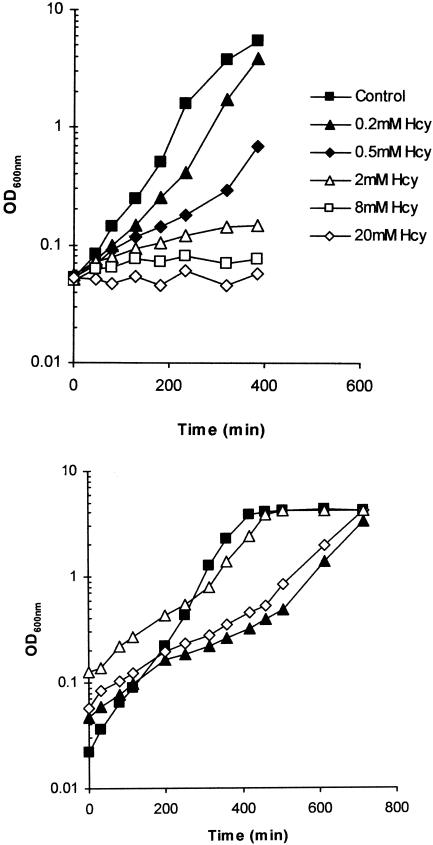
FIG. 2.
Growth inhibition of E. coli Frag1 by Hcy. (Top) Cultures of Frag1 were grown in MM (0.4% [wt/vol] glucose) and supplemented with 0 (control), 0.2, 0.5, 2.0, 8.0, or 20.0 mM Hcy. Growth was monitored by recording the OD600 at suitable time intervals after inoculation. (Bottom) Frag1 was inoculated into MM (0.4% [wt/vol] glucose) supplemented with 1 mM Hcy at a starting optical density (OD600) of 0.128 (▵), 0.047 (▴), or 0.057 (◊). The control culture (▪) had no added Hcy and had a starting OD600 of 0.022. Vertical dashed lines indicate the time points that Hcy-treated cultures showed a transition from slow inhibited growth to faster growth rates.
With concentrations of Hcy that inhibited the growth of Frag1 significantly (e.g., 1 mM), we repeatedly observed a biphasic pattern of growth inhibition. In these cultures the greatest inhibition was observed early during growth, while cultures achieved growth rates similar to the control later in exponential phase (Fig. 2, bottom panel). This effect was probably due to cultures metabolizing the Hcy, since the length of time taken to achieve the faster growth rate was indirectly related to the inoculum size; cultures that began with a higher level of starting inoculum achieved the second faster phase of growth sooner than cultures inoculated with fewer cells (Fig. 2, bottom panel). If cells were simply adapting to the presence of Hcy in the growth medium, the length of time to adapt and achieve a faster growth rate would not be expected to depend on the number of cells present in the inoculum.
In E. coli Hcy can be metabolized to homocysteine thiolactone through the editing activity of methionyl tRNA synthetase. This reaction is inhibited strongly in the presence of methionine, since methionine competes efficiently with Hcy for binding to the enzyme (10). To determine whether the inhibitory effects of Hcy required the formation of homocysteine thiolactone, we performed growth experiments with Hcy in the presence or absence of either 0.2 or 2 mM methionine. Cultures of Frag1 growing in MM and supplemented with either methionine concentration were inhibited by Hcy to the same extent as control cultures (data not shown). Homocysteine thiolactone (0.5 mM) was also added directly to the growth medium, and no inhibitory effect on growth was observed (data not shown). Together, these results suggested that Hcy toxicity did not require the formation of homocysteine thiolactone.
Hcy toxicity is relieved by branched-chain amino acids.
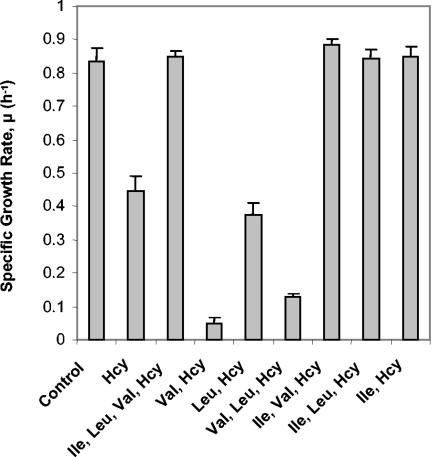
A number of reports have shown that cysteine, which is an amino acid closely related to homocysteine, is also toxic for E. coli (7, 24, 27). Supplementation of cultures with branched-chain amino acids can overcome this inhibition (7). We investigated whether the toxicity of Hcy could also be relieved by branched-chain amino acids. Frag1 was grown in MM with or without 0.5 mM Hcy, and cultures were supplemented with each of the branched-chain amino acids, isoleucine (0.15 mM), leucine (0.15 mM), and valine (0.17 mM), either together, in pairs, or individually. Any combination of supplemented amino acids containing isoleucine, including the addition of isoleucine alone, gave full relief from the growth inhibition observed in the presence of 0.5 mM Hcy (Fig. 3). Indeed, the growth curves of cultures containing Hcy plus isoleucine were indistinguishable from the untreated controls (data not shown). The addition of leucine alone gave no relief against Hcy toxicity (Fig. 3). The addition of valine alone (or in combination with leucine) increased the extent of growth inhibition (Fig. 3), and this inhibitory effect was also observed in the absence of Hcy (data not shown). The toxicity of valine for E. coli K-12 is well documented and is caused by the feedback inhibition of acetohydroxy acid synthase, an enzyme that catalyzes the first reaction of the valine biosynthetic pathway and the second reaction of the isoleucine biosynthetic pathway (Fig. 1), and thus starvation for isoleucine (4). Control cultures without Hcy showed no change in growth rate when supplemented with branched-chain amino acids (except for the inhibitory effects of valine), indicating that these amino acids were not limiting for growth in MM (data not shown).
FIG. 3.
Isoleucine relieves the inhibitory effect of Hcy on growth. Cultures of Frag1 were grown in MM (0.4% [wt/vol] glucose) and supplemented with either 0 (control) or 0.5 mM Hcy. The indicated amino acids were added at a final concentration of 0.15 mM (Ile and Leu) or 0.17 mM (Val). The growth rates were calculated from the growth curves as described in Materials and Methods. Growth curves were performed in triplicate, and error bars indicate the standard deviation from the mean.
Supplementation with isoleucine could not fully relieve the inhibition caused by higher concentrations of Hcy (i.e., >2 mM). For example, the specific growth rate (μ) was reduced from 0.78 h−1 (±0.02, n = 3) in the untreated control to 0.07 h−1 (±0.02, n = 3) in the presence of 8 mM Hcy, and supplementation with 0.15 mM isoleucine partially relieved this inhibition, giving a specific growth rate of 0.38 h−1 (±0.01, n = 3). Increasing the isoleucine concentration further, to 0.5 mM or even 8 mM, did not increase the extent of the growth relief (data not shown). At 20 mM Hcy we observed no relief of growth inhibition in the presence of 0.15 mM isoleucine. These results suggest that at high concentrations of Hcy the inhibitory mode of action is likely to be more complex than at lower concentrations, perhaps involving the inhibition of multiple targets within the cell.
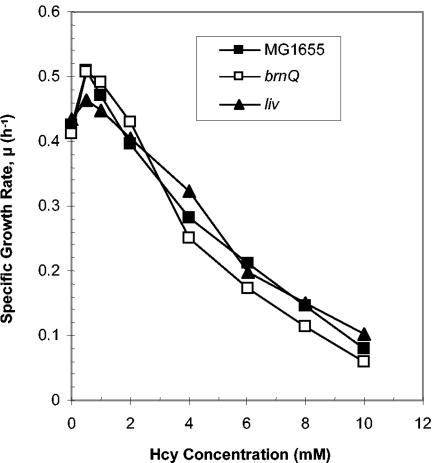
We considered the possibility that the relieving effect of isoleucine on Hcy-inhibited growth could be produced if isoleucine prevented the transport of Hcy into the cell. This situation might arise if these two amino acids were transported into the cell by a shared uptake system. If this was the case, then the presence of isoleucine in the growth medium might competitively inhibit the uptake of Hcy, thereby alleviating its toxicity. Isoleucine is transported into the cell by two distinct uptake systems, an ATP-dependent transport system that consists of six distinct protein subunits (encoded by livJ and livKHMGF), known as the LIV-I/LS system, and a single component system that does not require ATP, known as BrnQ (or LIV-II) (30). The uptake system for Hcy remains unknown at present. The finding that leucine, which is a substrate for both the LIV-I/LS and the BrnQ uptake systems, does not alleviate Hcy-mediated growth inhibition (Fig. 3) suggests that Hcy is not transported via either of these systems. To further test this conclusion, we compared the Hcy sensitivity of a wild-type E. coli strain (MG1655) to two isogenic mutants that were deficient for either of the two isoleucine uptake systems (ΔlivHMGF and ΔbrnQ [15]). All three strains were grown in MM with a range of Hcy concentrations (0 to 10 mM), and their growth rates were measured. Neither mutant showed any significant difference in sensitivity to Hcy compared to the parent (Fig. 4). These data suggest that neither transport system is involved in the uptake of Hcy, a finding that makes it very unlikely that isoleucine relieves Hcy-mediated growth inhibition by preventing Hcy transport into the cell.
FIG. 4.
Hcy sensitivity does not depend on the isoleucine transport systems. Cultures of E. coli MG1655 (▪) or YG191 (MG1655 ΔbrnQ; □) or YG192 (MG1655, ΔlivHMGF; ▴) were grown in MM (0.4% [wt/vol] glucose) with a range of Hcy concentrations, and the growth rates were determined.
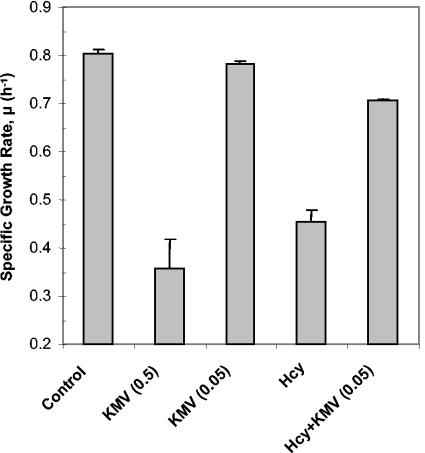
We then investigated whether other intermediates on the branched-chain amino acid biosynthetic pathway were able to confer protection against the inhibitory effects of Hcy. Only two compounds were commercially available: α-ketobutyrate and α-keto-β-methylvalerate (KMV) (Fig. 1). In the absence of added Hcy, α-ketobutyrate inhibited the growth of Frag1 cultures significantly, even at concentrations as low as 15 μM (data not shown). The toxicity of α-ketobutyrate for E. coli is well known, although there is still some uncertainty about the basis for its toxic effect (3, 17, 30). The toxicity of α-ketobutyrate made it impossible to assess the impact of this compound on the inhibitory effect of Hcy. KMV was also found to inhibit the growth of Frag1 when added to cultures at a concentration of 0.5 mM. However, supplementation of cultures with 50 μM KMV was not inhibitory to growth (Fig. 5). When 50 μM KMV was added to cultures of Frag1 containing 0.5 mM Hcy, there was a significant increase in the growth rates compared to cultures containing Hcy alone. Indeed, the growth rates were almost restored to the rates observed in the untreated controls (Fig. 5). Taken together with the relieving effect of isoleucine, this result suggests strongly that Hcy causes a perturbation of isoleucine biosynthesis in E. coli.
FIG. 5.
KMV relieves the inhibitory effect of Hcy on growth. Cultures of Frag1 were grown in MM (0.4% [wt/vol] glucose) and supplemented with either 0.5 mM KMV [KMV (0.5)], 50 μM KMV [KMV (0.05)], 0.5 mM Hcy (Hcy), or 0.5 mM Hcy plus 50 μM KMV [Hcy+KMV (0.05)]. The growth rates were determined from the growth curves as described in Materials and Methods. Error bars represent the standard deviation from the mean (n = 4).
Hcy perturbs intracellular amino acid pools.
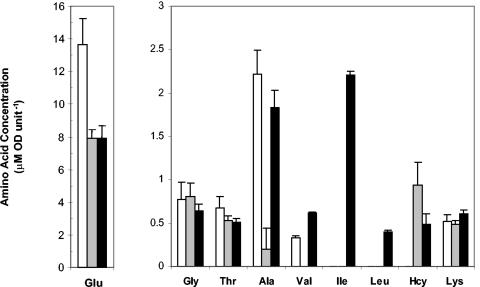
The relieving effect of isoleucine and KMV on Hcy toxicity suggested that Hcy might inhibit some step on the isoleucine biosynthetic pathway, leading to isoleucine-limited growth. We examined the effect of 0.5 mM Hcy on the intracellular pools of amino acids of Frag1 cells growing in MM. Several of the low-abundance amino acids were present at levels below the detection limit of our analytical method. However, the data gave a number of important insights into the effects of Hcy on the intracellular amino acid pools (Fig. 6). In the presence of 0.5 mM Hcy, (i) Hcy accumulated intracellularly; (ii) the levels of glycine, threonine, and lysine remained unchanged; and (iii) the glutamate, alanine, and valine pools were all depleted in the presence of Hcy compared to the untreated control (Fig. 6). The isoleucine and leucine levels for both the control and the Hcy-treated cultures were below the detection limit, making it impossible to determine whether Hcy led to their depletion.
FIG. 6.
Amino acid pools are perturbed in the presence of Hcy. Cultures of Frag1 were grown in MM with 0.4% glucose and grown to an OD600 of 0.2. Amino acid pools were analyzed 60 min after either no addition (□), the addition of 0.5 mM Hcy (░⃞), or the addition of 0.5 mM Hcy plus 0.15 mM Ile (▪) as described in Materials and Methods. Error bars indicate the standard deviation from the mean (n = 4).
The amino acid pools were also measured in cultures treated with both Hcy and isoleucine. The isoleucine and leucine pools were both higher in the presence of isoleucine, whereas there was no effect on the pools of glutamate, glycine, threonine, or lysine (Fig. 6). A decrease in the intracellular levels of Hcy was observed when isoleucine was present. Strikingly, the pools of both valine and alanine were restored in cultures supplemented with isoleucine compared to the Hcy-treated cultures (Fig. 6). It is likely that the alanine depletion arises as a result of the reduced pool of valine since alanine is synthesized directly from valine in a transamination reaction catalyzed by valine-pyruvate aminotransferase (36).
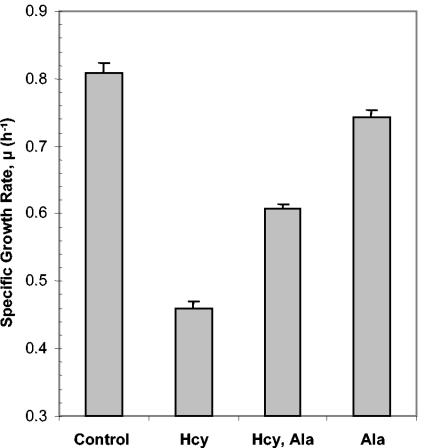
These data suggested that, in addition to isoleucine starvation, Hcy-treated cells may also be limited for valine and alanine. The toxicity of valine for E. coli K-12 (Fig. 3) made it difficult to test whether valine was limiting for growth. However, the effect of alanine on Hcy-mediated growth inhibition was investigated. Supplementation of cultures containing 0.5 mM Hcy with 0.5 mM alanine resulted in a significant increase in growth rate compared to cultures containing Hcy alone, although growth was not restored to the same rate as the untreated control (Fig. 7). This result indicated that Hcy-treated cells are limited for alanine, as well as for isoleucine. Unlike isoleucine, however, alanine could not reverse fully the inhibitory effects of Hcy. Taken together with the amino acid pool data, these data demonstrate that Hcy causes a perturbation of branched-chain amino acid biosynthesis in E. coli.
FIG. 7.
Alanine provides limited relief from the inhibitory effect of Hcy on growth. Cultures of Frag1 were grown in MM (0.4% [wt/vol] glucose) either with no additions (control), 0.5 mM Hcy (Hcy), or 0.5 mM alanine (Ala) or with a combination of the two (Hcy, Ala). The growth rates were determined from the growth curves as described in Materials and Methods. Error bars represent the standard deviation from the mean (n = 4).
Hcy inhibits the activity of TD.
In E. coli five enzymes are required for the biosynthesis of isoleucine from threonine, and three of these enzymes are shared by the pathway leading to valine biosynthesis (Fig. 1). Two enzymes are used primarily by the isoleucine biosynthetic pathway: TD (the product of the ilvA gene), which catalyzes the conversion of threonine to α-ketobutyrate, and transaminase B (the product of the ilvE gene), which catalyzes the conversion of KMV to isoleucine (Fig. 1) (30). Transaminase B can also catalyze the final step on the valine biosynthetic pathway, but its principal role is in isoleucine biosynthesis, as evidenced by the finding that isoleucine supplementation alone can restore the growth defect of an ilvE mutant (36). The finding that KMV can protect against the inhibitory effects of Hcy (Fig. 5) suggested that transaminase B was unlikely to be the target of Hcy inhibition, since only intermediates below the inhibited step would be expected to confer relief. The fact that cysteine is known to inhibit the activity of TD (7), prompted us to investigate whether Hcy affects the activity of TD in E. coli Frag1.
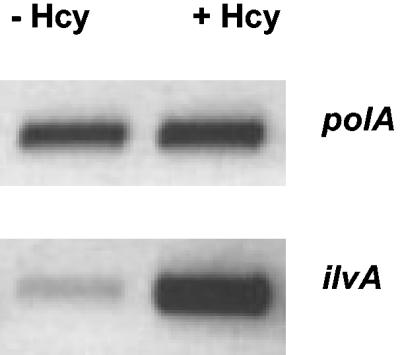
First, we investigated the levels of TD activity in cells growing exponentially (OD600 of 0.3) in MM with or without added Hcy (0.5 mM). Crude whole-cell extracts were prepared from these cultures, and the TD activity was measured from each by the method of Harris (7). Extracts from cultures grown with Hcy had a TD activity ∼3-fold higher than the untreated control (29 ± 4 versus 10 ± 2 nmol mg−1 min−1; n = 4). This result suggested that cells treated with Hcy might express the gene encoding TD, ilvA, at a higher level than untreated cells. Reverse transcriptase PCR was used to assess the relative abundance of ilvA mRNA in cells grown with or without added Hcy. The ilvA transcript was found to be significantly more abundant (∼15-fold) in cells treated with Hcy than in the control (Fig. 8). These data suggest that Hcy results in higher levels of ilvA transcription, and this is responsible for the higher levels of TD activity observed in Hcy-treated cells.
FIG. 8.
Reverse transcriptase PCR (RT-PCR) analysis of the transcription of ilvA. cDNA concentrations were normalized by using primers against polA. RNA was prepared from cells grown in the presence (+Hcy) or absence (−Hcy) of 0.5 mM Hcy. Hcy was added to cultures at an OD600 of ∼0.2 and incubated for 1 h before RNA extraction. The DNA products obtained after 30 cycles of RT-PCR are shown.
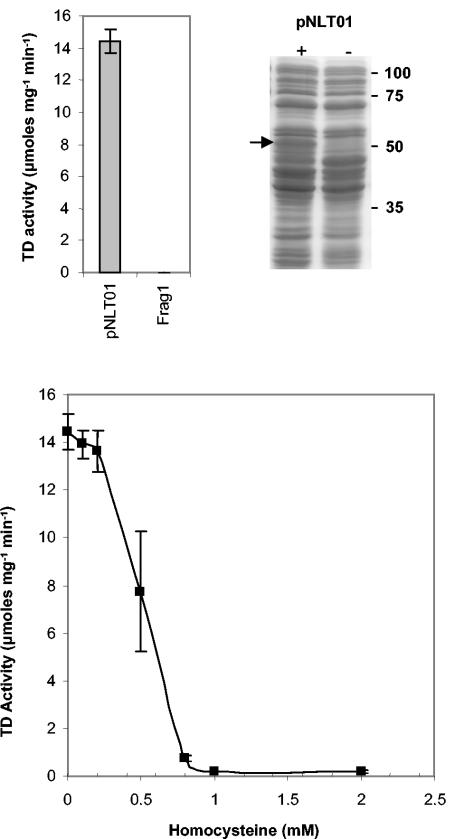
Next, we investigated the effect of increasing Hcy concentrations on the activity of TD in vitro. First, the ilvA gene was cloned into pBAD202/D-TOPO, generating plasmid pNLT01, from which the ilvA gene is constitutively expressed (as described in Materials and Methods). Frag1 was transformed with pNLT01, and this strain was used to generate extracts containing high levels of TD activity. Extracts from Frag1/pNLT01 had ∼450-fold higher TD activity than Frag1 grown under the same conditions (Fig. 9, top left panel). On SDS-PAGE gels a protein band migrating at approximately 55 kDa was apparent in extracts prepared from Frag1/pNLT01, which was not detectable in extracts from Frag1. This band is likely to correspond to TD, which has a predicted molecular mass of 56 kDa (Fig. 9, top right panel). These protein extracts were used to determine the effect of Hcy on TD activity in vitro. Hcy had no significant effect on TD activity at concentrations up to 0.2 mM. Above this concentration TD was inhibited significantly, with ca. 50% inhibition at 0.5 mM Hcy and 100% inhibition at ≥1 mM Hcy (Fig. 9, bottom panel). These data indicate that Hcy can inhibit the activity of TD, and they suggest a possible explanation for the ability of isoleucine to relieve the inhibitory effects of Hcy on growth.
FIG. 9.
TD activity is inhibited by Hcy. (Top left) Cells carrying pNLT01 have increased TD activity. Crude cell extracts were prepared from either Frag1 or Frag1/pNLT01 grown to stationary phase in LB. TD activity was measured from each extract in triplicate, and errors bars indicate the standard deviation from the mean. Assays contained protein concentrations of 0.04 mg ml−1 for Frag1 and 0.01 mg ml−1 for Frag1/pNLT01 extracts. (Top right) Cells carrying pNLT01 express a protein of the size predicted for TD. Proteins extracted from Frag1 and Frag1/pNLT01 were separated by SDS-PAGE. Numbers indicate the sizes of molecular mass markers in kilodaltons. A band with increased intensity migrating at ca. 55 kDa is apparent in extracts from Frag1/pNLT01 and is indicated by an arrow. (Bottom) TD activity was measured as a function of increasing Hcy concentration. Assays were performed in triplicate, and errors bars indicate the standard deviation from the mean. All assays contained 0.01 mg of protein ml−1.
Overexpression of TD protects against Hcy.
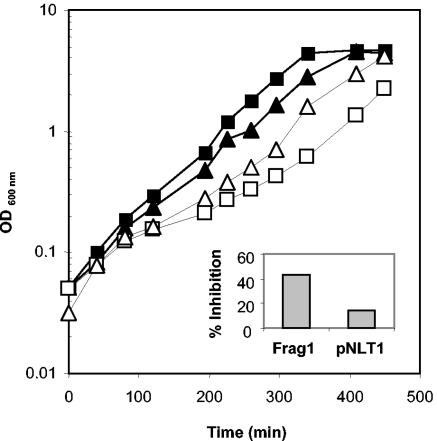
If Hcy was restricting the flux through the isoleucine biosynthetic pathway by inhibiting TD activity, we reasoned that a strain overexpressing this enzyme might be less susceptible to the inhibitory effects of Hcy. To test this, we measured the extent of growth inhibition caused by Hcy (0.5 mM) in Frag1 compared to Frag1/pNLT01, which has a TD activity ∼64-fold higher (10 ± 1 versus 641 ± 2 nmol mg−1 min−1) in MM. The presence of pNLT01 was associated with a slightly reduced growth (ca. 10%) compared to Frag1 without the plasmid (Fig. 10). The growth rates of Frag1 cultures were reduced by ca. 42% in the presence of Hcy, whereas the growth rate of Frag1/pNLT01 was reduced by only 14% under the same conditions (Fig. 10). Frag1 cells that were transformed with the vector only (i.e., with no insert) were also inhibited to the same extent as Frag1 in the presence of 0.5 mM Hcy (data not shown). These data suggest that high levels of TD activity can protect E. coli cells, at least partially, from the inhibitory effects of Hcy. Furthermore, it supports the idea that Hcy toxicity in E. coli results from a perturbation of branched-chain amino acid biosynthesis caused at least partly by the inhibition of TD.
FIG. 10.
Overexpression of TD protects against the inhibitory effects of Hcy. Cultures of Frag1 (squares) and Frag1/pNLT01 (triangles) were grown in MM with 0.4% glucose either in the presence (open symbols) or absence (closed symbols) of 0.5 mM Hcy. Cultures of Frag1/pNLT01 were supplemented with kanamycin (50 μg ml−1). The growth rates were calculated from these curves and were used to calculate the percentage inhibition caused by Hcy. These values are plotted on the inset graph.
DISCUSSION
We confirm here the toxicity of Hcy for E. coli and establish that this toxicity is due to a perturbation of branched-chain amino acid biosynthesis. The ability of isoleucine (Fig. 3) and KMV (Fig. 5) to relieve Hcy-mediated growth inhibition suggests that Hcy blocks a step on the isoleucine biosynthetic pathway, leading to isoleucine starvation. The finding that Hcy inhibits the activity of TD in vitro (Fig. 9, bottom panel) and that increased TD activity protects against the toxic effects of Hcy (Fig. 10) suggests that this block occurs at the first committed step of the isoleucine biosynthetic pathway, the conversion of threonine to α-ketobutyrate. The finding that alanine confers a limited protection against the inhibitory effects of Hcy (Fig. 7) suggests that the presence of Hcy perturbs both the isoleucine and the valine arms of the branched-chain amino acid biosynthesis pathway (Fig. 1). This conclusion is supported by amino acid pool data, which show that both valine and alanine pools are depleted in Hcy-treated cells (Fig. 6).
The ability of isoleucine alone to restore the growth of cells exposed to 0.5 mM Hcy suggests that the principal effect of Hcy on cell physiology is the perturbation of isoleucine biosynthesis. However, higher concentrations of Hcy (e.g., 8 mM) cause growth inhibition that cannot be fully reversed by the inclusion of isoleucine in the growth medium. Increasing the levels of isoleucine further does not give greater levels of protection against high Hcy levels. These results suggest that Hcy may have more complex effects on cell physiology at higher concentrations. Hcy is known to compete with methionine and isoleucine for binding to their respective tRNA synthetases (10, 13), and it could be that at higher Hcy concentrations this competition depletes the cell of charged tRNA species, resulting in reduced rates of protein synthesis. Under these conditions homocysteine thiolactone is likely to accumulate in the cytoplasm due to the editing function of the methionyl and isoleucyl tRNA synthetases (13). Although homocysteine thiolactone is known to trigger increased expression of RpoS (5), it does not appear to be inhibitory to growth, at least not under the growth conditions used in the present study.
It is not clear whether the inhibitory effect of Hcy on TD serves any physiological purpose within the cell. The only obvious connection between the methionine and isoleucine biosynthetic pathways is that they both require homoserine as a precursor (Fig. 1). The intracellular concentration of Hcy reaches a high level when 0.5 mM Hcy is present extracellularly, being roughly equivalent to the threonine pool. This concentration of Hcy is probably not often encountered within the cell since the cell would rarely encounter extracellular concentrations in the millimolar range. However, it is known that high levels of Hcy can accumulate within the cytoplasm when E. coli is exposed to inhibitory levels of acetic acid (23). In this case Hcy accumulation is thought to result from the inhibition of the final step of methionine biosynthesis, the methylation of Hcy (Fig. 1). It is possible that the cell uses Hcy accumulation as a cue that methionine biosynthesis is not proceeding efficiently and therefore protein synthesis cannot proceed (since the initiation of protein synthesis requires formylmethionine tRNAfmet). If protein synthesis is perturbed, then perhaps it makes physiological sense to downregulate amino acid biosynthesis in parallel. Although this idea remains speculative at present, there is evidence that an imbalance in methionine biosynthesis can lead to major changes in cell physiology. For example, methionine biosynthesis is blocked in E. coli cells that overexpress a putative efflux pump called HsrA, and this block leads to elevated expression of RpoS, the stress-inducible sigma factor (5).
The way in which Hcy enters the cell is not known at present. The amino acid pool data show that it does accumulate intracellularly (Fig. 6), and this is consistent with a previous report (23). It seems likely that it is transported into the cell by an amino acid uptake system. Growth experiments with mutants lacking either of the two isoleucine uptake systems indicate that these transport systems are probably not involved in transporting Hcy (Fig. 4). The absence of a relieving effect with leucine, which is also a substrate for both isoleucine transport systems, supports this conclusion. The uptake system for the related amino acid methionine is also unlikely to be involved since the presence of methionine in the growth medium has no effect on the toxicity of Hcy. The close similarity of Hcy to the sulfur-containing amino acid cysteine might suggest that the cysteine uptake system is the most likely route of entry for Hcy. Surprisingly, the uptake systems for l-cysteine and l-cystine in E. coli remain unknown, although d-cysteine is believed to be transported by a member of the ATP-binding cassette superfamily, encoded by the fliY, yecC, and yecS genes (28). The transport of cystine by E. coli requires the presence of reduced glutathione, and the transported cystine is reduced to cysteine within the cytoplasm, but the transporter remains to be identified (21). In the present study we found that homocystine, the oxidized form of Hcy, is not inhibitory to the growth of E. coli Frag1, whereas the same concentration of cystine is (N. L. Tuite and C. P. O'Byrne, unpublished data). This result suggests that homocystine is not accumulated by E. coli Frag1 and indicates that homocystine is not a substrate for the cystine uptake system.
The level of TD activity was elevated substantially (threefold) in cells treated with Hcy compared to the untreated controls. Since Hcy inhibits the activity of TD in vitro, it seems likely that this increase is due to elevated levels of the TD protein in Hcy-treated cells. This idea is consistent with the finding that the ilvA transcript is present at significantly higher levels in cells treated with 0.5 mM Hcy (Fig. 8). The ilvA gene is located in a five-gene operon (ilvGMEDA) that is primarily regulated by transcriptional attenuation, although the leucine-responsive regulatory protein, Lrp, also plays a role (22). Transcriptional deattenuation occurs when any of the three branched-chain amino acids are limiting (19). Therefore, the increase in ilvA transcription that occurs in the presence of Hcy is probably explained by transcriptional deattenuation resulting from reduced intracellular pools of isoleucine and valine. This effect may also be exacerbated by the fact that Hcy competes, albeit poorly, with isoleucine for binding to isoleucyl tRNA synthetase (13). This might further reduce the intracellular levels of charged isoleucyl tRNA and thereby promote transcriptional deattenuation.
If the activity of TD is elevated in Hcy-treated cells, why is this not sufficient to overcome Hcy toxicity? The amino acid pool analysis shows that Hcy accumulates intracellularly in Hcy-treated cells, and it is likely that the internal concentration is simply too high to allow TD to act optimally. This fact is not reflected in the enzyme assays since any Hcy present in the crude-cell extracts will be diluted out to subinhibitory levels (at least 20-fold) by the assay buffer. Furthermore, the amino acid pool data indicate that the pool of isoleucine is low (below the detection limit of the assay), even in untreated cells (Fig. 6). It seems possible therefore that even a small inhibitory effect of Hcy on TD activity would be sufficient to make isoleucine limiting for growth.
Branched-chain amino acid biosynthesis in E. coli is unique in that the enzymes catalyzing three of the biochemical steps are shared between the isoleucine branch and the valine/leucine branch of the pathway (Fig. 1). The amino acid pool analysis performed on cells treated with 0.5 mM Hcy revealed that valine and alanine pools are depleted compared to the untreated control (Fig. 6). It is not clear why this reduction occurs, but it seems likely that both pools are reduced for the same reason, since alanine is synthesized primarily from valine in E. coli (36). One possibility is that, in addition to inhibiting TD activity, Hcy might also inhibit an enzyme that is shared by both the isoleucine branch and the valine/leucine branch of the pathway. An alternative and arguably simpler possibility is that some enzyme, shared between the isoleucine and valine/leucine branches of the pathway, is not expressed efficiently when TD is inhibited. It is known that efficient expression of ilvC, which encodes acetohydroxy acid isomeroreductase, requires the presence of α-aceto-α-hydroxybutyrate, which is a substrate for this enzyme (37) (Fig. 1). When TD is inhibited by Hcy the concentration of this intermediate is likely to be lowered. Evidence from gene macroarray experiments indicates that ilvC is transcribed at a reduced level (<50%) in the presence of Hcy (K. R. Fraser, A. Bhagwat, and C. P. O'Byrne, unpublished data). It is possible that the reduced level of IlvC (acetohydroxy acid isomeroreductase) leads to a decrease in the rate of valine biosynthesis, which causes a drop in the size of the valine pool and a concomitant drop in the pool of alanine. Although this model might account for the decrease in valine and alanine levels in cells treated with 0.5 mM Hcy, it does not account for the increase in these pools that is observed when isoleucine is added to the growth medium (Fig. 6). One possibility is that the presence of isoleucine has the effect of reducing the flux of intermediates through the isoleucine branch of the pathway (by the feedback inhibition of TD [31]), and this increases the flux of intermediates through the valine branch of the pathway because the shared enzymes are no longer competing for substrate.
The question of how Hcy might inhibit the activity of TD remains to be determined. Within the cell Hcy is metabolized either to methionine (Fig. 1) or to homocysteine thiolactone through the editing activity of methionyl tRNA synthase (10). In humans, one of the ways in which Hcy is believed to cause tissue damage is through the homocysteinylation of proteins. This is believed to occur through a reaction between Hcy thiolactone and the ɛ amino group of lysine residues (11). Thus far, there is no evidence that this reaction occurs in E. coli. The finding that methionine fails to prevent Hcy toxicity suggests that homocysteine thiolactone is unlikely to be responsible for the inhibitory effects observed. Hcy could directly inhibit TD activity by interacting with the enzyme, perhaps by competing with the substrate, threonine, for binding to the active site. Cysteine is also known to inhibit TD activity but, despite attempts to examine the kinetic properties of TD in the presence of cysteine, the underlying basis for this inhibition also remains unclear (7). Detailed kinetic studies with the purified enzyme will be required in order to establish the nature of the inhibition.
Acknowledgments
We are grateful to Ian Davidson for his assistance with the amino acid pool analysis. We also thank Emily Starr and Emily Rabbitte for help with some preliminary growth experiments. We thank Hidehiko Kumagai and Ian Booth for providing strains used in the present study.
This research was supported by a grant from the Irish Research Council for Science, Engineering, and Technology and by the NUI Galway Millennium Fund.
REFERENCES
- 1.Anderson, J. L., J. B. Muhlestein, B. D. Horne, J. F. Carlquist, T. L. Bair, T. E. Madsen, and R. R. Pearson. 2000. Plasma homocysteine predicts mortality independently of traditional risk factors and C-reactive protein in patients with angiographically defined coronary artery disease. Circulation 102:1227-1232. [DOI] [PubMed] [Google Scholar]
- 2.Bhagwat, A. A., R. P. Phadke, D. Wheeler, S. Kalantre, M. Gudipati, and M. Bhagwat. 2003. Computational methods and evaluation of RNA stabilization reagents for genome-wide expression studies. J. Microbiol. Methods 55:399-409. [DOI] [PubMed] [Google Scholar]
- 3.Danchin, A., L. Dondon, and J. Daniel. 1984. Metabolic alterations mediated by 2-ketobutyrate in Escherichia coli K-12. Mol. Gen. Genet. 193:473-478. [DOI] [PubMed] [Google Scholar]
- 4.De Felice, M., M. Levinthal, M. Iaccarino, and J. Guardiola. 1979. Growth inhibition as a consequence of antagonism between related amino acids: effect of valine in Escherichia coli K-12. Microbiol. Rev. 43:42-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodrich-Blair, H., and R. Kolter. 2000. Homocysteine thiolactone is a positive effector of sigma(S) levels in Escherichia coli. FEMS Microbiol. Lett. 185:117-121. [DOI] [PubMed] [Google Scholar]
- 6.Greene, R. C. 1996. Biosynthesis of methionine, p. 542-560. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 7.Harris, C. L. 1981. Cysteine and growth inhibition of Escherichia coli: threonine deaminase as the target enzyme. J. Bacteriol. 145:1031-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jakubowski, H. 2002. Homocysteine is a protein amino acid in humans. Implications for homocysteine-linked disease. J. Biol. Chem. 277:30425-30428. [DOI] [PubMed] [Google Scholar]
- 9.Jakubowski, H. 2004. Molecular basis of homocysteine toxicity in humans. Cell Mol. Life Sci. 61:470-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakubowski, H. 1990. Proofreading in vivo: editing of homocysteine by methionyl-tRNA synthetase in Escherichia coli. Proc. Natl. Acad. Sci. USA 87:4504-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jakubowski, H. 1999. Protein homocysteinylation: possible mechanism underlying pathological consequences of elevated homocysteine levels. FASEB J. 13:2277-2283. [PubMed] [Google Scholar]
- 12.Jakubowski, H. 2000. Translational incorporation of S-nitrosohomocysteine into protein. J. Biol. Chem. 275:21813-21816. [DOI] [PubMed] [Google Scholar]
- 13.Jakubowski, H., and A. R. Fersht. 1981. Alternative pathways for editing non-cognate amino acids by aminoacyl-tRNA synthetases. Nucleic Acids Res. 9:3105-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanzaki, S., and Y. Anraku. 1971. Transport of sugars and amino acids in bacteria. IV. Regulation of valine transport activity by valine and cysteine. J. Biochem. 70:215-224. [DOI] [PubMed] [Google Scholar]
- 15.Koyanagi, T., T. Katayama, H. Suzuki, and H. Kumagai. 2004. Identification of the LIV-I/LS system as the third phenylalanine transporter in Escherichia coli K-12. J. Bacteriol. 186:343-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 17.LaRossa, R. A., T. K. Van Dyk, and D. R. Smulski. 1987. Toxic accumulation of alpha-ketobutyrate caused by inhibition of the branched-chain amino acid biosynthetic enzyme acetolactate synthase in Salmonella typhimurium. J. Bacteriol. 169:1372-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawther, R. P., D. H. Calhoun, C. W. Adams, C. A. Hauser, J. Gray, and G. W. Hatfield. 1981. Molecular basis of valine resistance in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 78:922-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawther, R. P., and G. W. Hatfield. 1980. Multivalent translational control of transcription termination at attenuator of ilvGEDA operon of Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 77:1862-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Outinen, P. A., S. K. Sood, P. C. Liaw, K. D. Sarge, N. Maeda, J. Hirsh, J. Ribau, T. J. Podor, J. I. Weitz, and R. C. Austin. 1998. Characterization of the stress-inducing effects of homocysteine. Biochem. J. 332(Pt. 1):213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park, S., and J. A. Imlay. 2003. High levels of intracellular cysteine promote oxidative DNA damage by driving the Fenton reaction. J. Bacteriol. 185:1942-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhee, K. Y., B. S. Parekh, and G. W. Hatfield. 1996. Leucine-responsive regulatory protein-DNA interactions in the leader region of the ilvGMEDA operon of Escherichia coli. J. Biol. Chem. 271:26499-26507. [DOI] [PubMed] [Google Scholar]
- 23.Roe, A. J., C. O'Byrne, D. McLaggan, and I. R. Booth. 2002. Inhibition of Escherichia coli growth by acetic acid: a problem with methionine biosynthesis and homocysteine toxicity. Microbiology 148:2215-2222. [DOI] [PubMed] [Google Scholar]
- 24.Rowley, D. 1953. Inhibition of E. coli strains by amino-acids. Nature 171:80-81. [DOI] [PubMed] [Google Scholar]
- 25.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 26.Seshadri, S., A. Beiser, J. Selhub, P. F. Jacques, I. H. Rosenberg, R. B. D'Agostino, P. W. Wilson, and P. A. Wolf. 2002. Plasma homocysteine as a risk factor for dementia and Alzheimer's disease. N. Engl. J. Med. 346:476-483. [DOI] [PubMed] [Google Scholar]
- 27.Sorensen, M. A., and S. Pedersen. 1991. Cysteine, even in low concentrations, induces transient amino acid starvation in Escherichia coli. J. Bacteriol. 173:5244-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soutourina, J., S. Blanquet, and P. Plateau. 2001. Role of d-cysteine desulfhydrase in the adaptation of Escherichia coli to d-cysteine. J. Biol. Chem. 276:40864-40872. [DOI] [PubMed] [Google Scholar]
- 29.Starkebaum, G., and J. M. Harlan. 1986. Endothelial cell injury due to copper-catalyzed hydrogen peroxide generation from homocysteine. J. Clin. Investig. 77:1370-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Umbarger, H. E. 1996. Biosynthesis of the branched-chain amino acids, p. 442-457. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 31.Umbarger, H. E. 1956. Evidence for a negative-feedback mechanism in the biosynthesis of isoleucine. Science 123:848.. [DOI] [PubMed] [Google Scholar]
- 32.van der Put, N. M., H. W. van Straaten, F. J. Trijbels, and H. J. Blom. 2001. Folate, homocysteine, and neural tube defects: an overview. Exp. Biol. Med. 226:243-270. [DOI] [PubMed] [Google Scholar]
- 33.Vollset, S. E., H. Refsum, L. M. Irgens, B. M. Emblem, A. Tverdal, H. K. Gjessing, A. L. Monsen, and P. M. Ueland. 2000. Plasma total homocysteine, pregnancy complications, and adverse pregnancy outcomes: the Hordaland Homocysteine Study. Am. J. Clin. Nutr. 71:962-968. [DOI] [PubMed] [Google Scholar]
- 34.Vollset, S. E., H. Refsum, A. Tverdal, O. Nygard, J. E. Nordrehaug, G. S. Tell, and P. M. Ueland. 2001. Plasma total homocysteine and cardiovascular and noncardiovascular mortality: the Hordaland Homocysteine Study. Am. J. Clin. Nutr. 74:130-136. [DOI] [PubMed] [Google Scholar]
- 35.Weiss, N., Y. Y. Zhang, S. Heydrick, C. Bierl, and J. Loscalzo. 2001. Overexpression of cellular glutathione peroxidase rescues homocyst(e)ine-induced endothelial dysfunction. Proc. Natl. Acad. Sci. USA 98:12503-12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whalen, W. A., and C. M. Berg. 1982. Analysis of an avtA::Mu d1(Ap lac) mutant: metabolic role of transaminase C. J. Bacteriol. 150:739-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wild, J., J. M. Smith, and H. E. Umbarger. 1977. In vitro synthesis of beta-galactosidase with ilv-lac fusion deoxyribonucleic acid as template. J. Bacteriol. 132:876-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winzer, K., K. R. Hardie, N. Burgess, N. Doherty, D. Kirke, M. T. Holden, R. Linforth, K. A. Cornell, A. J. Taylor, P. J. Hill, and P. Williams. 2002. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148:909-922. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, C., Y. Cai, M. T. Adachi, S. Oshiro, T. Aso, R. J. Kaufman, and S. Kitajima. 2001. Homocysteine induces programmed cell death in human vascular endothelial cells through activation of the unfolded protein response. J. Biol. Chem. 276:35867-35874. [DOI] [PubMed] [Google Scholar]