Abstract
Streptococcus mutans, a normal inhabitant of dental plaque, is considered a primary etiological agent of dental caries. Two virulence determinants of S. mutans are its acidogenicity and aciduricity (the ability to produce acid and the ability to survive and grow at low pH, respectively). Citric acid is ubiquitous in nature; it is a component of fruit juices, bones, and teeth. In lactic acid bacteria citrate transport has been linked to increased survival in acidic conditions. We identified putative citrate transport and metabolism genes in S. mutans, which led us to investigate citrate transport and metabolism. Our goals in this study were to determine the mechanisms of citrate transport and metabolism in S. mutans and to examine whether citrate modulates S. mutans aciduricity. Radiolabeled citrate was used during citrate transport to identify citrate metal ion cofactors, and thin-layer chromatography was used to identify metabolic end products of citrate metabolism. S. mutans was grown in medium MM4 with different citrate concentrations and pH values, and the effects on the growth rate and cell survival were monitored. Intracellular citrate inhibited the growth of the bacteria, especially at low pH. The most effective cofactor for citrate uptake by S. mutans was Fe3+. The metabolic end product of citrate metabolism was aspartate, and a citrate transporter mutant was more citrate tolerant than the parent.
Streptococcus mutans is a normal inhabitant of dental plaque and is considered the major etiological agent of dental caries, one of the most prevalent dental diseases. Two virulence determinants of S. mutans linked to its cariogenicity are its acidogenicity and aciduricity (the ability to produce acid and the ability to survive and grow at low pH, respectively). Its acidogenicity and aciduricity allow S. mutans to outcompete other organisms within dental plaque, which can lead to the formation of carious lesions.
Citric acid is ubiquitous in nature; for example, fruit juices contain between 5 and 8% citric acid, and teeth are composed of 0.3% citric acid by weight (3, 16). Citric acid is also a cause of tooth enamel erosion (3). Citrate is widely used as a food preservative and also has bactericidal activity against coagulase-negative Staphylococcus and Staphylococcus aureus (11).
Many bacteria have transport systems that allow the uptake of citrate. In the genera Lactococcus and Leuconostoc citrate transport is mediated by homologous citrate/lactate antiporters designated CitP (5, 6). This transport leads to the generation of a proton motive force and to increased cell survival in acidic conditions (5, 6). In these bacteria cometabolism of citrate with glucose results in heterofermentation and the production of the end products CO2, diacetyl, acetoin, and butanediol. Some bacteria, including Klebsiella pneumoniae, are even able to grow in an anaerobic environment utilizing citrate as the sole carbon source (reviewed in reference 4). K. pneumoniae possesses two distinct citrate transporters; under aerobic conditions a proton-dependent CitH citrate tranporter is expressed, whereas under anaerobic conditions a sodium ion-dependent CitS is expressed (12).
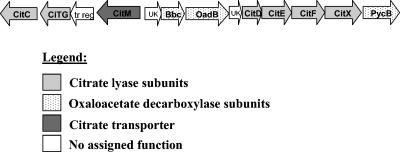
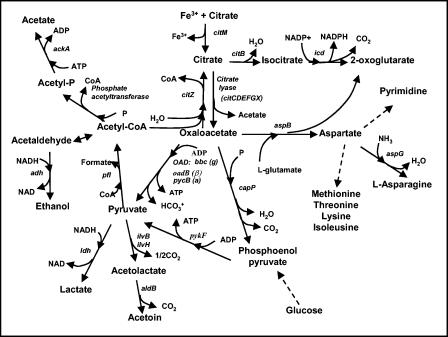
Unlike K. pneumoniae, S. mutans is unable to survive when citrate is the sole source of carbon, as determined by negative growth on Simmons citrate agar. When citrate is available, S. mutans also does not produce CO2 gas, which is a common by-product of citrate metabolism in many other lactic acid bacteria. Despite these observations the relationship between S. mutans and citrate has been largely unstudied. Interestingly, the S. mutans genome contains orthologs of many citrate metabolic genes that are present in bacterial species known to utilize citrate (3). These genes encode the subunits of citrate lyase (citCDEFGX) and oxaloacetate decarboxylase [bbc (α), oadB (β), and pycB (γ)] (2). The S. mutans genome also harbors a putative Mg2+-dependent citrate transporter which we designated citM. This citrate metabolism locus is illustrated in Fig. 1, and the possible metabolic outcomes for citrate are shown in Fig. 2. In this study, the first of its kind with S. mutans, we focused on the mechanisms of citrate uptake and metabolism by S. mutans and the effect of citrate on cell survival at low pH.
FIG. 1.
Graphic representation of the putative citrate metabolism locus in S. mutans. Genes encoding the putative citrate transporter, citrate lyase subunits, and oxaloacetate decarboxylase subunits are shown.
FIG. 2.
Putative citrate utilization pathway of S. mutans. CoA, coenzyme A.
MATERIALS AND METHODS
Bacterial strains and growth conditions. The S. mutans strains used in this study are listed in Table 1. For transport assays S. mutans UA159 cells were grown in Berman's broth (2% Trypticase peptone, 0.1% yeast extract, 25 mM potassium phosphate, 34 mM NaCl, 0.5% thioglycolic acid, 1 mM MgSO4, 0.1 mM MnSO4) (14). For growth curve analysis cells were grown in defined minimal medium MM4 (7). When needed, erythromycin was added at a final concentration of 5 μg/ml.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant characteristics | Reference |
|---|---|---|
| UA159 | Wild type | 2 |
| SMCitM | UA159::CitM−, Emr (putative Mg2+ citrate transporter) | This study |
| SMOAD | UA159::Oad−, Emr (oxaloacetate decarbox- ylase B) | This study |
| SMCLY | UA159::Cly−, Emr (citrate lyase a) | This study |
| SMASPB | UA159::AspB−, Emr (aspartate transaminase) | This study |
Construction of isogenic mutants.
Knockouts in the S. mutans citM, citF, oadB, and aspB genes were generated using PCR ligation mutagenesis (10). Briefly, on the basis of the nucleotide sequence of each gene two sets of primers were designed. The first set of primers was directed toward the 5′ flanking region of the gene of interest, and the second set was directed toward the 3′ flanking region of the gene. Using UA159 genomic DNA as the template, the two fragments were amplified by PCR. The two amplicons were subsequently ligated to an erythromycin resistance cassette. The entire fragment was then transformed into genetically competent S. mutans UA159 cells previously exposed to synthetic S. mutans competence-stimulating peptide (10). The fragment was integrated into the S. mutans chromosome via a double-crossover event. The resulting erythromycin-resistant transformants were evaluated by PCR to confirm integration at the desired loci. The primers used for mutagenesis are listed in Table 2.
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence (5′ to 3′) | Description |
|---|---|---|
| Erm 1 | GGCGCGCCCGGGCCCAAAATTTGTTTGAT | ermAM cassette |
| Erm2 | GGCCGGCCAGTCGGCAGCGACTCATAGAAT | ermAM cassette |
| CitM1 | TCTGTTGTAGGAGTGATGGGGG | citM mutagenesis |
| CitM2a | GGCGCGCCCTCAGTGCCGTAAAAGGCGAC | citM mutagenesis |
| CitM3b | GGCCGGCCCTTAACGGGCTTAGACATGGGC | citM mutagenesis |
| CitM4 | TCGGTAACGATCTCTCGTAGGC | citM mutagenesis |
| OAD1 | CAATCTGAATGGGAGCCTATTTCG | oad mutagenesis |
| OAD2a | GGCGCGCCCCGCCGTTAATAACCTGAGTTAGGAC | oad mutagenesis |
| OAD3b | GGCCGGCCCGCAGTAGGTGCTAATGTTTCAGGAC | oad mutagenesis |
| OAD4 | TGACCATCAGGATAACGGCG | oad mutagenesis |
| CLY1 | TTCTGTTATCGCAGGTGGCTTAC | cly mutagenesis |
| CLY2a | GGCGCGCCCAAGGAGCATCTCGCAGCATAGC | cly mutagenesis |
| CLY3b | GGCCGGCCCCGCTATTGTTGGTAATCCTCAACC | cly mutagenesis |
| CLY4 | TGTCCGCCAGCTTCCTTAGTAG | cly mutagenesis |
| ASPB1 | TGCTGGATGAAGCGAAACTTACTC | aspB mutagenesis |
| ASPB2a | GGCGCGCCAGGTGTTACAAAATCAGGCTGTCC | aspB mutagenesis |
| ASPB3b | GGCCGGCCCAGGTGTCGCTTTGGTAACAGG | aspB mutagenesis |
| ASPB4 | GCATTGCCCGATAAGATTGGAC | aspB mutagenesis |
| CitMRT1 | AGTTGTTATCCTTGTCTTCGCTGC | citM real-time PCR |
| CitMRT2 | CGATGACTAAGCCCCAGAAACC | citM real-time PCR |
Underlined bases indicate the AscI restriction sequence.
Underlined bases indicate the FseI restriction sequence.
Growth curves.
Single colonies of a strain were inoculated into THYE and incubated overnight at 37°C in 5% CO2. Overnight cultures were subsequently inoculated 1:10 into anaerobic medium MM4 at pH 7 and incubated overnight at 37°C under an atmosphere containing 10% CO2, 10% H2, and 80% N2 (COHN atmosphere). The overnight subcultures were then inoculated 1:10 in quadruplicate into microtiter plates containing anaerobic medium MM4 (pH 7) containing the appropriate concentrations of citrate (0 mM, 2 mM, 4 mM, 6 mM, or 8 mM). The plates were then sealed and placed into a Bioscreen C automated growth monitor (Lab Systems). The bacteria were incubated for 3 days at 37°C. Data were obtained every 20 min after 20 s of shaking prior to measurement of the optical density at 600 nm. Growth curves and doubling times were calculated from these data.
Citrate killing curves.
Single colonies of a strain were inoculated into THYE and incubated overnight at 37°C in a 5% CO2 atmosphere. Overnight cultures were subsequently inoculated 1:100 into anaerobic medium MM4 at pH 7 with the COHN atmosphere. Two aliquots of the overnight subcultures were then centrifuged at 4,000 × g for 10 min. The pellets were then suspended in 10 times the original volume of medium MM4 at pH 5 containing either no citrate or 8 mM citrate and incubated at 37°C in the COHN atmosphere. An aliquot of each culture was removed and serially diluted in 10 mM potassium phosphate buffer, pH 7.2. Twenty microliters of each dilution was spotted in triplicate onto THYE-1% agar plates that were incubated at 37°C in the COHN atmosphere for 2 days. The first plate was considered time zero. Subsequent aliquots were removed at 5 h, 1 day, 2 days, and 3 days, serially diluted, and plated.
Citrate adaptation.
Single colonies of a strain were inoculated into THYE and incubated overnight at 37°C in a 5% CO2 atmosphere. Overnight cultures were subsequently inoculated 1:10 into anaerobic medium MM4 at pH 7 and incubated overnight at 37°C under the COHN atmosphere. Two aliquots of the overnight subcultures were then centrifuged at 4,000 × g for 10 min. The pellets were then suspended in 10 times the original volume of medium MM4 at pH 7 containing either no citrate or 8 mM citrate and incubated at 37°C in the COHN atmosphere for 18 h. Following incubation, two aliquots from each subculture were centrifuged at 16,000 × g for 5 min. The pellets were then suspended in 10 times the original volume of medium MM4 at pH 5 containing either no citrate or 8 mM citrate and incubated at 37°C in the COHN atmosphere. An aliquot was removed and serially diluted in 10 mM potassium phosphate buffer, pH 7.2. Twenty microliters of each dilution was spotted in triplicate onto THYE-1% agar plates that were incubated at 37°C in the COHN atmosphere for 2 days. The first plate was considered time zero. Subsequent aliquots were removed, serially diluted, and plated at 24 h and 48 h.
Citrate transport assay.
Citrate transport assays were performed as described by Krom et al. (9), with minor modifications. Single colonies of S. mutans UA159 were inoculated into 10 ml of modified Berman's broth at pH 6.5 supplemented with 0.1% raffinose and 10 mM citrate and incubated overnight at 37°C in a 5% CO2 atmosphere. Overnight cultures were subsequently diluted fivefold in fresh Berman's broth at pH 6.5 supplemented with 0.35% raffinose and 10 mM citrate and incubated at 37°C until the cells reached the mid-logarithmic phase (optical density at 600 nm, ∼0.4 to 0.6). The cells were then harvested by centrifugation and washed twice with cold 50 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.5. The PIPES buffer had previously been treated with15 g/liter Chelex 100 (Sigma Chemicals) for 18 h to remove residual metal ions. Cells were then resuspended in 10 ml of the same buffer supplemented with the appropriate amount of metal ions. Aliquots (100 μl) were incubated at 37°C and allowed to equilibrate for 15 min. At time zero [1,5-14C]citrate (final concentration, 4.5 μM; 112 mCi/mmol) was added. Uptake was stopped by addition of 2 ml of ice-cold 0.1 M LiCl, and cells were immediately filtered through a 0.2-μm-pore-size nitrocellulose filter. The filters were washed once with 2 ml of ice-cold 0.1 M LiCl and submerged in scintillation fluid, and the internalized radioactivity was counted with a liquid scintillation counter. To obtain dry weights for standardization, quadruplicate 1-ml aliquots of resuspended cells were filtered through dried preweighed filters, dried for 24 h, and reweighed.
Quantitative real-time PCR analysis of citM expression.
To determine the effects of the presence of citrate or pH on S. mutans citM transcription levels, quantitative real-time PCR was performed. S. mutans UA159 cells were grown in Berman's broth (pH 5.0, pH 6.0, and pH 7.0) to the mid-log phase. Total RNA was harvested as described by Hanna et al. (8). A First Strand cDNA synthesis kit (MBI Fermentas) was used according to the manufacturer's specifications to generate cDNA from 1 μg of DNase-treated RNA. The resulting single-stranded cDNA was diluted to a concentration of 50 ng/μl for real-time PCR analysis. To ensure that there was no contaminating DNA, two negative control reaction mixtures were set up, one without template RNA and the other lacking reverse transcriptase.
For amplification and detection of real-time PCR products, we used a Quantitect SYBR Green PCR kit (QIAGEN). The real-time PCR mixtures contained 100 ng of template DNA, 250 nM of each primer, and 2× SYBR Green mixture (which contained SYBR Green, deoxynucleoside triphosphates, MgCl2, and Hotstar Taq polymerase). Real-time PCR was carried out with a Cepheid Smart cycler (Cepheid, Sunnyvale, CA) using citM-specific primers (Table 2). For each reaction the cycle threshold (Ct) value was determined; this value reflected the starting amount of cDNA in each sample. The initial DNA concentrations were determined by comparing experimental Ct values to a standard curve for citM which was generated by plotting these Ct values against the absorbance of serially diluted known quantities of amplicons. The data were normalized using gyrA expression results, which did not vary for the conditions examined (data not shown).
Citrate metabolism assay.
S. mutans cells were grown in Berman's broth containing 10 mM citrate to the mid-log phase (optical density at 600 nm, ∼ 0.4). Cells were subsequently washed and resuspended in potassium phosphate, 10 mM MgCl2 buffer at the appropriate pH. The mixture was then incubated at 37°C for 10 min to allow equilibration, after which 2.5 mM [1,5-14C]citrate (1 Ci mol−1) was added. The cells were allowed to metabolize the citrate for 1 h, and this was followed by centrifugation to separate the cells from the extracellular metabolites. Cells were then lysed using glass beads and a Fast Prep machine to release the intracellular metabolic end products. The intracellular extract was treated with trichloroacetic acid to precipitate proteins and nucleic acids, and this was followed by an EDTA treatment to chelate metal ions. The extract was then neutralized with potassium hydroxide (KOH). The extracellular metabolites were also treated in the same fashion as the intracellular extract, with additions of trichloroacetic acid, EDTA, and KOH. Both extracts were spotted onto 60-Å silica gel thin-layer chromatography (TLC) plates. Radiolabeled citrate, aspartate, glutamate, lysine, and histidine standards (Amersham Biosciences) were also spotted onto the TLC plates. The plates were then placed into a developing chamber that had previously been charged with an n-butanol-acetic acid-water (4:1:1) eluent and allowed to resolve. Following migration for 4 to 6 h, the plates were air dried, and the spots were visualized by autoradiography. This procedure was repeated for the SMCitM, SMCLY, SMOAD, and SMASPB strains. Nonradiolabeled citrate, oxaloacetate, and aspartate standards were also spotted onto the TLC plates, and after resolution the standards were detected colorimetrically using a 10% solution of bromocresol green in ethanol.
RESULTS
Effect of citrate on growth.
In lactic acid bacteria citrate provides increased growth yields and acid tolerance (5, 6). To determine if similar benefits could be realized in S. mutans, strain UA159 and the putative citrate metabolic knockout strains SMCitM, SMCLY, SMOAD, and SMASPB were grown in defined minimal medium MM4 in the presence of citrate, as described previously (Table 3). The knockout genes were selected on the basis of their putative roles in citrate catabolism, as shown in Fig. 2. Citrate at a concentration of 4 mM resulted in no statistically significant change in the growth rate of any of the strains except SMASPB. Increasing the citrate concentration to 8 mM resulted in a drastic increase in the generation time and completely inhibited the growth of SMAPSB. Interestingly, of the strains that could grow in the presence of 8 mM citrate, wild-type strain UA159 was most affected by the citrate, as shown by the 5.5-fold increase in the doubling time compared with the 3-fold increase in the doubling time for the isogenic mutants SMCitM, SMOAD, and SMCLY.
TABLE 3.
Growth rates of citrate metabolic mutant strains
| Strain | Doubling time (min)
|
||
|---|---|---|---|
| No citrate | 4 mM citrate | 8 mM citrate | |
| UA159 | 135.4 ± 4.7 | 130.1 ± 2.7 | 748.8 ± 130.7a,b |
| SMCitM | 141.6 ± 5.4 | 131.3 ± 2.6 | 402.8 ± 69.7a |
| SMOAD | 138.4 ± 5.9 | 127.5 ± 2.2 | 458.1 ± 107.5a |
| SMCLY | 131 ± 2.9 | 139.1 ± 3.6 | 411.9 ± 40.0a |
| SMASPB | 132.5 ± 1.5 | 155.3 ± 6.5a,c | NGd |
Significantly different from growth without citrate (P < 0.01).
Significantly different from growth of the other strains (P < 0.01).
Significantly different from growth of the other strains (P < 0.05).
NG, no growth.
Citrate enhances killing of S. mutans at low pH.
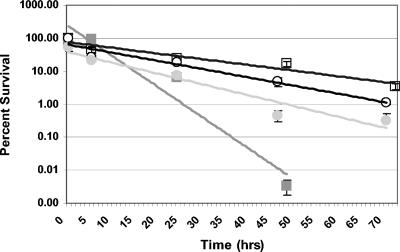
Citrate cometabolism with glucose provides a growth advantage to Lactococcus lactis at a low pH (6). Therefore, the role of citrate in survival under acidic conditions was investigated for S. mutans. No growth was observed at pH 5.0 in medium MM4 broth, with or without citrate, using the protocol described above. The rate of cell death was then investigated in medium MM4 with and without citrate to determine what effect citrate had at low pH. It was observed that citrate markedly increased the rate at which S. mutans was killed at pH 5.0 for both strain UA159 and CitM− cells (Fig. 3). The CitM− cells were more resistant to citrate killing than UA159 cells when 8 mM citrate was added to the medium (Fig. 3).
FIG. 3.
Relative survival of S. mutans at pH 5 in medium MM4 with 8 mM citrate (solid symbols) and without citrate (open symbols). Samples were serially diluted and plated on THYE agar plates. The level of survival of each strain compared to the survival during growth without citrate was determined by dividing the number of colonies present at each time by the number of colonies present at time zero without citrate. Symbols: ▪ and □, UA159; • and ○, SMCitM.
Adaptation to citrate enhances survival at low pH.
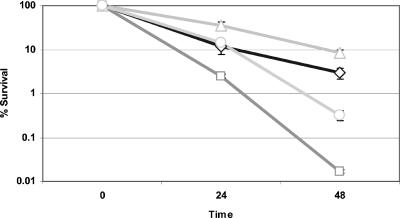
As shown in Fig. 3, citrate exacerbated killing of S. mutans at low pH. Investigations into whether S. mutans could adapt to citrate were subsequently undertaken. S. mutans cells were subcultured at neutral pH in the presence or absence of citrate before they were subjected to killing at pH 5 with or without citrate. Cells that were previously exposed to citrate (citrate adapted) exhibited greater survival at pH 5 than cells that were not exposed to citrate (unadapted) (Fig. 4). Citrate enhanced killing of both the adapted and unadapted S. mutans cells, and more adapted cells than unadapted cells survived (Fig. 4).
FIG. 4.
Effect of incubating S. mutans UA159 cells in the presence of citrate prior to exposure to pH 5 with and without citrate. Cells were grown in medium MM4 at pH 7 in the absence or presence of 8 mM citrate and subsequently exposed to medium MM4 at pH 5 with 8 mM citrate. After this the cells were serially diluted and plated on THYE agar plates. The level of survival for each strain was determined by dividing the number of colonies present at each time by the number of colonies present at time zero. Symbols: ◊, cells grown at pH 7 without citrate and then incubated at pH 5 without citrate; □, cells grown at pH 7 without citrate and then incubated at pH 5 with 8 mM citrate; ▵, cells grown at pH 7 with 8 mM citrate and then incubated at pH 5 without citrate; ○, cells grown at pH 7 with 8 mM citrate and then incubated at pH 5 with 8 mM citrate.
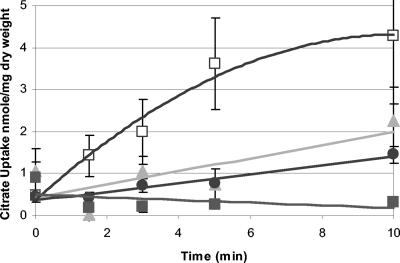
S. mutans transports citrate with divalent metal ion cofactors.
The citM gene product was designated a putative Mg2+ citrate transporter, but it exhibited only limited homology (36% identity at the amino acid level) to the Bacillus subtilis Mg2+ citrate transporter CitM, which is known to catalyze the uptake of citrate in a complex with Mg2+ (2, 9). When all divalent metal ions were depleted from the reaction mixture, very little citrate was transported by S. mutans (Fig. 5). When 1 μM FeCl3 or 5 mM MnCl2 was added to the reaction mixture, citrate uptake was increased (Fig. 5). When 5 mM CaCl2, MgCl2, or 5 mM NiCl2 was added to the reaction mixture, the rate of uptake was not greater than the rate of uptake without metal cations (data not shown). Negligible amounts of citrate entered SMCitM cells, in which the citrate transport gene was mutated (Fig. 5). These observations indicated that Fe3+ was the best cofactor tested for facilitating citrate transport via CitM.
FIG. 5.
Uptake of [1,5-14C]citrate by S. mutans in the presence of different metal cations. The uptake of [14C]citrate was measured in 50 mM PIPES, pH 6.5, in the presence of 1 μM Fe3+ (□) or 5 mM Mn2+ (▴) or without added metal ions (•). The uptake of [1,5-14C]citrate by SMCitM was measured in the presence of 1 μM Fe3+ (▪). [1,5-14C]citrate uptake was not detected with 5 mM Mg2+, Ni2+, or Ca2+ as a cofactor (data not shown).
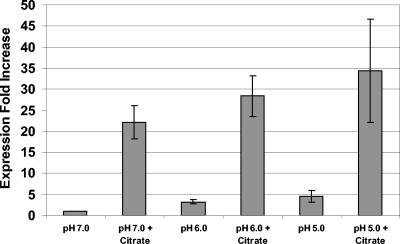
citM expression is increased by citrate.
Our transport data indicated that citrate was transported by S. mutans in a complex with ferric ions and that this process was mediated by the CitM transporter. To determine if citM expression was modulated by exposure to citrate or by pH, a quantitative real-time PCR expression analysis was performed. The expression of citM was up-regulated 25-fold at pH 7.0 upon exposure to citrate, as shown in Fig. 6. To examine the hypothesis that citrate uptake and metabolism may be involved in acid tolerance, citM expression levels were measured at lower pH values (pH 6.0 and pH 5.0). Our data indicated that the mean expression increased as the pH was reduced, as shown in Fig. 6. However, the citM expression levels at pH 6.0 and pH 5.0 were not significantly different from the expression levels at pH 7.0.
FIG. 6.
Expression of citM in response to pH and citrate. The data indicate the increased expression compared to the expression at pH 7 without citrate, normalized to gyrA expression levels. The values are means of five replicates.
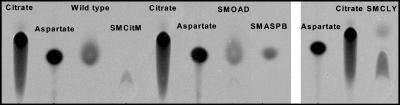
S. mutans metabolizes citrate.
Although many lactic acid bacteria are capable of citrate metabolism, to our knowledge the ability of S. mutans to metabolize citrate has not been examined previously. To determine if S. mutans was capable of citrate metabolism, wild-type UA159 cells were incubated in the presence of [1,5-14C]citrate, and metabolites were then separated on silica TLC plates. Our results indicated that S. mutans converts internalized citrate into the amino acid aspartate (as shown for the wild type in Fig. 7). However, when nonradiolabeled standards and colorimetric detection were used, aspartate resolved the same as oxaloacetate. This suggested that the spot identified in Fig. 7 as aspartate could also be oxaloacetate or, more likely, a combination of the two metabolites (colorimetric detection not shown). Lowering the external pH or oxygen concentration had no effect on the metabolic end product composition (data not shown).
FIG. 7.
Autoradiograph of intracellular metabolites separated by TLC from S. mutans strains. The cells were resuspended in 50 mM PIPES, pH 6.5, and allowed to take up 2.5 mM [1,5-14C]citrate for 1 h. Radiolabeled [1,5-14C]citrate and [14C]aspartate standards were also spotted on the plate. The strains examined were S. mutans UA159 (Wild type), SMCitM, SMCLY, SMOAD, and SMASPB.
Knockout strains SMCitM, SMOAD, SMCLY, and SMASPB were used to determine if the inactivated genes encoded enzymes involved in citrate metabolism. Like UA159, these strains were incubated in the presence of [1,5-14C] citrate, and the metabolites were separated via TLC. The metabolites of the oxaloacetate decarboxylase mutant, SMOAD, separated in the same way that the metabolites of the wild type separated, as shown in Fig. 7, indicating that this enzyme is not involved in citrate metabolism. The metabolites of the aspartate mutant, SMASP, also separated in the same way that the metabolites of the wild type separated, but since oxaloacetate could not be resolved from aspartate using the techniques employed, a firm conclusion could not be drawn. Citrate was found to accumulate inside the citrate lyase mutant, SMCLY, as shown in Fig. 7, and therefore we concluded that citrate lyase is responsible for the conversion of citrate to oxaloacetate. The SMCitM mutant was unable to transport citrate as no spot corresponding to citrate was visualized by TLC, as shown in Fig. 7.
DISCUSSION
This is the first known description of citrate transport and metabolism by S. mutans. Several bacterial species, including B. subtilis, K. pneumoniae, and L. lactis, have been shown to both transport and metabolize citrate (9, 12, 6). This study demonstrated that S. mutans does, in fact, transport and metabolize citrate. In contrast to the advantages that citrate provides to many other lactic acid bacteria, in S. mutans citrate appears to affect growth and survival negatively at concentrations that are physiologically plausible. The addition of citrate increased the killing of the SMCitM transport knockout mutant at pH 5 much less than the addition of citrate increased the killing of UA159. The results reflected by these killing curves (Fig. 3) indicated that citrate killing occurs via both intracellular and extracellular processes. Citrate could therefore potentially be used as an S. mutans growth inhibitor. In fact, this may explain the observed effectiveness of citrate as an additive in toothpaste (1).
Ferric citrate transport has been well documented in gram-negative bacteria. We believe that this is the first known report of a gram-positive citrate transporter that preferentially utilizes ferric ions as a cofactor. Citrate chelates ferric ions and readily precipitates them out of solution; therefore, transport analyses were performed using 1 μM FeCl3 instead of the 5 mM used for other metal ions. S. mutans is known to have a nutritional requirement for trace metals, including magnesium, manganese, and iron (3). S. mutans requires iron at a concentration of 3.6 μM for optimal growth (3), and this concentration is far greater that the concentration present in human saliva (0.01 to 1.0 μM) (18). An S. mutans ABC transporter, encoded by the sloABCR operon, has previously been identified as an Fe2+ transporter (17). Experiments have shown that iron increases the dental plaque pH in vivo; however, the mechanisms are not known (13, 15). Iron has an important role in the cariogenesis of S. mutans, and CitM may play a part in iron acquisition.
This study is also the first study to describe citrate metabolism by S. mutans. This metabolism results in the conversion of citrate into aspartate. Aspartate production in the SMOAD mutant can be explained since the Oad enzyme is not part of the citrate-to-aspartate pathway. The precise metabolic end product of SMASPB was not determined; however, it can be inferred that AspB is involved in citrate metabolism as an AspB mutant was more sensitive to growth in the presence of citrate than the parent strain. Citrate lyase is responsible for the conversion of citrate to oxaloacetate as citrate was seen to accumulate in the SMCLY strain. Citrate does appear to enter central metabolism and does not seem to provide any growth or survival benefit under acidic conditions. Interestingly, however, the presence of citrate during growth at neutral pH affords S. mutans enhanced acid tolerance, as well as tolerance to citrate at low pH. S. mutans may, in fact, exploit citrate's ability to chelate essential ferric iron to transport it into the cell.
Acknowledgments
We thank Jason Tanzer for helpful advice with the experimental design.
This work was supported by operating grant MT-15431 from the Canadian Institute for Health Research. D.G.C. is the recipient of a Canada Research Chair. B.K. is a CIHR strategic training fellow in Cell Signaling in Mucosal Inflammation & Pain (STP-53877).
REFERENCES
- 1.Adams, S. E., A. J. Theobald, N. M. Jones, M. G. Brading, T. F. Cox, A. Mendez, D. M. Chesters, D. G. Gillam, C. Hall, and J. Holt. 2003. The effect of a toothpaste containing 2% zinc citrate and 0.3% Triclosan on bacterial viability and plaque growth in vivo compared to a toothpaste containing 0.3% Triclosan and 2% copolymer. Int. Dent. J. 53:398-403. [DOI] [PubMed] [Google Scholar]
- 2.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aranha, H., R. C. Strachan, J. E. L. Arceneaux, and B. R. Byers. 1982. Effect of trace metals on growth of Streptococcus mutans in a Teflon chemostat. Infect. Immun. 35:456-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bott, M. 1997. Anaerobic citrate metabolism and its regulation in enterobacteria. Arch. Microbiol. 167:78-88. [PubMed] [Google Scholar]
- 5.Cogan, T. M. 1987. Co-metabolism of citrate and glucose by Leuconostoc spp.: effects on growth, substrates and products. J. Appl. Bacteriol. 63:551-558. [Google Scholar]
- 6.Garcia-Quintas, N., C. Magni, D. de Mendoza, and P. Lopez. 1998. The citrate transport system of Lactococcus lactis subsp. lactis biovar diacetylactis is induced by acid stress. Appl. Environ. Microbiol. 64:850-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamilton, I. R., and G. Svensäter. 1998. Acid-regulated proteins induced by Streptococcus mutans and other oral bacteria during acid shock. Oral Microbiol. Immunol. 13:292-300. [DOI] [PubMed] [Google Scholar]
- 8.Hanna, M. M., R. J. Ferguson, Y.-H. Li, and D. G. Cvitkovitch. 2001. uvrA is an acid-inducible gene involved in the adaptive response to low pH in Streptococcus mutans. J. Bacteriol. 183:5964-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krom, B. P., J. B. Warner, W. N. Konings, and J. S. Lolkema. 2000. Complementary metal ion specificity of the metal-citrate transporters CitM and CitH of Bacillus subtilis. J. Bacteriol. 182:6374-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau, P. C., C. K. Sung, J. H. Lee, D. A. Morrison, and D. G. Cvitkovitch. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193-205. [DOI] [PubMed] [Google Scholar]
- 11.Lee, Y.-L., L. Thrupp, J. Owens, T. Cesario, and E. Shanbrom. 2001. Bactericidal activity of citrate against Gram-positive cocci. Lett. Appl. Microbiol. 33:349-351. [DOI] [PubMed] [Google Scholar]
- 12.Lolkema, J. S., H. Enequist, and M. E. Van Der Rest. 1994. Transport of citrate catalyzed by the sodium-dependent citrate carrier of Klebsiella pneumoniae is obligatory coupled to the transport of two sodium ions Eur. J. Biochem. 220:469-475. [DOI] [PubMed] [Google Scholar]
- 13.Miguel, J. C., W. H. Bowen, and S. K. Pearson. 1997. Effects of iron salts in sucrose on dental caries in rats. Arch. Oral Biol. 42:377-383. [DOI] [PubMed] [Google Scholar]
- 14.Noji, S., Y. Sato, R. Suzuki, and S. Taniguchi. 1988. Effect of intercellular pH and potassium ions on primary transport system for glutamate/aspartate in Streptococcus mutans. Eur. J. Biochem. 175:491-495. [DOI] [PubMed] [Google Scholar]
- 15.Oppermann, R. V., and G. Rolla. 1980. Effect of some polyvalent cations on the acidogenicity of dental plaque in vivo. Caries Res. 14:422-427. [DOI] [PubMed] [Google Scholar]
- 16.Pierre, J. L., and I. Gautier-Luneau. 2000. Iron and citric acid: a fuzzy chemistry of ubiquitous biological relevance. Biometals 13:91-96. [DOI] [PubMed] [Google Scholar]
- 17.Spatafora, G., M. Moore, S. Landgren, E. Stonehouse, and S. Michalek. 2001. Expression of Streptococcus mutans fimA is iron-responsive and regulated by a DtxR homologue. Microbiology 147:1599-1610. [DOI] [PubMed] [Google Scholar]
- 18.Spatafora, G. A., and M. W. Moore. 1998. Growth of Streptococcus mutans in an iron-limiting medium. Methods Cell Sci. 20:217-221. [Google Scholar]