Abstract
Comparative genomics were used to assess genetic differences between Staphylococcus aureus strains derived from infected animals versus colonized or infected humans. A total of 77 veterinary isolates were genetically characterized by high-throughput amplified fragment length polymorphism (AFLP). Bacterial genotypes were introduced in a large AFLP database containing similar information for 1,056 human S. aureus strains. All S. aureus strains isolated from animals in close contact with humans (e.g., pet animals) were predominantly classified in one of the five main clusters of the AFLP database (cluster I). In essence, mastitis-associated strains from animals were categorized separately (cluster IVa) and cosegregated with bacteremia-associated strains from humans. Distribution of only 2 out of 10 different virulence genes differed across the clusters. The gene encoding the toxic shock syndrome protein (tst) was more often encountered among veterinary strains (P < 0.0001) and even more in the mastitis-related strains (P<0.0001) compared to human isolate results. The gene encoding the collagen binding protein (cna) was rarely detected among invasive human strains. The virulence potential, as indicated by the number of virulence genes per strain, did not differ significantly between the human- and animal-related strains. Our data show that invasive infections in pets and humans are usually due to S. aureus strains with the same genetic background. Mastitis-associated S. aureus isolated in diverse farm animal species form a distinct genetic cluster, characterized by an overrepresentation of the toxic shock syndrome toxin superantigen-encoding gene.
Staphylococcus aureus can colonize and infect a variety of members of the animal kingdom, including mammals, reptiles, and birds. Infection models for this clinically highly relevant bacterial species in lower organisms, such as insects (Drosophila melanogaster) and worms (Caenorhabditis elegans), have been described as well (28, 40). Various studies address the molecular basis of the apparent host specificity or diversity of S. aureus strains. This is usually approached by determining genome polymorphism with multilocus enzyme electrophoresis (8, 17), pulsed-field gel electrophoresis (PFGE) (46), binary typing (45), or shotgun sequencing (12). In addition, animal studies often involve S. aureus strains with specific mutations in defined virulence factors (6). These studies have demonstrated that strains from humans can cross-infect domestic animals (42) and poultry (35) and vice versa (38). It has been determined that separate host-specific lineages of S. aureus do exist, but the question with regard to cross-species pathogenicity of these lineages still remains unanswered (17).
Genomic studies as mentioned above have revealed extensive genetic variation in natural populations of S. aureus. Pathogenic strains may harbor complete pathogenicity islands (20) or specific “accessory” genes, such as the one encoding the Panton-Valentine leucocidin (22). The evolutionary processes in S. aureus restricting or expanding invasiveness in different hosts are ill defined (9), although, for instance, numbers and combinations of certain virulence genes may be important contributors to pathogenic potential (31). We studied S. aureus host specificity and virulence by comparative genomics (high-throughput amplified fragment length polymorphism [ht-AFLP]). In addition, the distribution and number of virulence genes were compared between clinical and nonclinical strains isolated from different host species.
MATERIALS AND METHODS
Bacterial strains.
Two strain collections were used; the first strain collection comprised 77 S. aureus strains isolated from different infection sites in a variety of animal species, including dogs (n = 8), monkeys and apes (n = 4; different species), pigs (n = 6), birds (n = 5; different species), cats (n = 12), sheep (n = 6), seals (n = 4), goats (n = 8), rabbits (n = 3), cows (n = 7), and horses (n = 10), a rat, a chinchilla, a guinea pig, and an iguana (one isolate each). The second strain collection consisted of 168 human S. aureus strains, which were classified as invasive (n = 56) or colonizing (n = 112). Strains were obtained and selected from a previous study (44). An invasive strain was derived from a patient with a positive nasal culture upon admission to the hospital while suffering from a manifest S. aureus infection by the same strain at a normally sterile site more than 2 days after admission. A colonizing strain was derived from a positive nasal culture from a matched control patient for whom no S. aureus infection was documented during hospitalization. For each invasive isolate, two matched controls were selected. The risk factors for both patient groups were matched as well (same hospital, ward, age, gender, and period of hospitalization) (44). Prior to genomic analysis, bacteria from glycerol stocks, stored at −80°C, were inoculated on Columbia III agar supplemented with 5% sheep blood (Becton Dickinson, Etten-Leur, The Netherlands) and incubated at 37°C for 24 h. All strains were identified as S. aureus by accepted microbiological methods (2).
DNA extraction.
Chromosomal DNA was extracted from bacterial cells with a MagnaPure LC DNA system (DNA isolation kit III; Roche, Almere, The Netherlands) according to the manufacturer's instructions, with the modification that 50 μg/ml lysostaphine (Sigma, Zwijndrecht, The Netherlands) was added during the cell lysis step. The DNA concentration was measured by UV spectroscopy, samples were diluted with distilled water to a final concentration of 10 ng/μl, and DNA was stored at −20°C until use.
ht-AFLP.
The genomes of the animal-associated S. aureus strains (n = 77) were compared by ht-AFLP. Individual AFLP-PCRs were performed essentially as described before in the presence of radioactive nucleotides for the visualization of the fingerprints (25). Ht-AFLP was performed using the restriction enzyme combination MboI and Csp6I (New England Biolabs, Westburg, Leusden, The Netherlands). Each restriction site was filled in by ligation with specific linker oligonucleotide pairs (for MboI, 5′-CTCGTAGACTGCGTACC-3′ and 5′-ATCGGTACGCAGTCTAC-3′; for Csp6I, 5′-ACGATGAGTCCTGAC-3′ and 5′-TAGTCAGGACTCAT-3′). Subsequently, a nonselective preamplification was performed using the MboI primer (5′-GTAGACTGCGTACCGATC-3′) and Csp6I primer (5′-ACGATGAGTCCTGACTAC-3′). In the final amplification, a 33P-labeled MboI primer containing one selective nucleotide (either +C or +G) and a Csp6I primer containing two selective nucleotides (+TA) were used.
AFLP data analysis.
Agglomerative hierarchical cluster analysis was used for two-dimensional data clustering of the AFLP patterns. The unweighted pair group arithmetic mean method was performed for cluster analysis. After inclusion of the AFLP patterns in the AFLP database, the Tanimoto method was used to calculate the similarity matrix. The resulting dendrogram was ordered by average value. Spotfire DecisionSite 7.2 software has been applied to perform statistical analyses.
AFLP database.
This database consists of ht-AFLP fingerprints obtained for 1,056 S. aureus strains from human origin (25) and comprises 829 nonclinical carriage strains from healthy individuals, 74 strains from children with invasive S. aureus disease (bacteremia, arthritis, and abscess), 90 isolates from elderly (>55 years) individuals with S. aureus bacteremia, 40 isolates obtained from lesions of children suffering from impetigo, 21 international epidemic methicillin-resistant S. aureus strains (26), and 2 reference strains (20).
MLST.
A selection of 34 of the animal-associated strains were analyzed by the microarray-mediated multilocus sequence typing (MLST) protocol as described previously (43). Briefly, allele types of seven S. aureus housekeeping genes were defined by specific hybridization of amplified gene fragments to short oligonucleotide probes, synthesized on a Gene Chip array (Affymetrix, Santa Clara, Calif.; bioMérieux, Marcy l'Etoile, France). The resulting allelic sequences were matched with those from the MLST database (www.MLST.net), and an allelic profile with the corresponding sequence type (ST) and clonal complex (CC) could be determined.
PCR of putative virulence factors.
For both strain collections, the genes coding for adhesins fnbA, clfA, clfB, cna, sdrE, and ebpS and for the exoprotein toxins tst, eta, etb, and pvl were amplified using the PCR primers and amplification conditions outlined in Table 1. For the confirmation of the PCR results, a second PCR, targeting a different domain of the same genes, was done. PCR amplifications were performed in a GeneAmp PCR system 9700 (Applied Biosystems, Nieuwerkerk a/d IJssel, The Netherlands). A positive control (Table 1) and a negative control (PCR mixture excluding DNA) were included in each PCR run.
TABLE 1.
PCR primers and conditions for identification of potential virulence genes
| PCR product | Oligonucleotide sequences (5′-3′) | GenBank accession no. | Position in gene | Positive control | PCR conditions | Reference |
|---|---|---|---|---|---|---|
| fnbA | Fw, CAC AAC CAG CAA ATA TAG | AJ629121 | 424-441 | 8325-4 | 1 min at 94°C, 1 min at | 31 |
| Rv, CTG TGT GGT AAT CAA TGT C | 1785-1667 | 50°C, 2 min at 72°C | ||||
| Fw, GGT AAT CAT TCA TTC GAG | 2355-2372 | 8325-4 | 41 | |||
| Rv, TGG CAC ACT GTC GAA GTC | 2561-2544 | |||||
| clfA | Fw, GTA GGT ACG TTA ATC GGT T | Z18852 | 368-386 | Newman | 1 min at 94°C, 1 min at | 31 |
| Rv, CTC ATC AGG TTG TTC AGG | 1951-1934 | 45°C, 2 min at 72°C | ||||
| Fw, GAT TAA GCT TTA CGT TCA AC | 1805-1824 | Newman | 23 | |||
| Rv, GAT TGG TAC CAT TTT TAG GTG | 2948-2928 | |||||
| clfB | Fw, TGC AAG ATC AAA CTG TTC CT | AJ224764 | 425-444 | Newman | 1 min at 94°C, 1 min at | 31 |
| Rv, TCG GTC TGT AAA TAA AGG TA | 1020-1001 | 45°C, 2 min at 72°C | ||||
| Fw, AGG ACA ATC GAA CGA TAC AAC G | 162-183 | Newman | 1 min at 94°C, 1 min at | 32 | ||
| Rv, ACT ACG TAC AGC TCT CGT TCT AAC ACT | 618-592 | 52°C, 1 min at 72°C | ||||
| cna | Fw, AGT GGT TAC TAA TAC TG | M81736 | 1719-1735 | Phillips | 1 min at 94°C, 1 min at | 31 |
| Rv, CAG GAT AGA TTG GTT TA | 3457-3441 | 55°C, 2 min at 72°C | ||||
| Fw, ATG GTA CCA AGA AGA TAC G | 688-705 | Philips | 30 | |||
| Rv, TCT TGA TAC CAA GCT TGT G | 1052-1034 | |||||
| sdrE | Fw, CAG TAA ATG TGT CAA AAG A | AJ005647 | 650-668 | Isolate 476 | 1 min at 94°C, 1 min at | 31 |
| Rv, TTG ACT ACC AGC TAT ATC | 1416-1399 | 45°C, 1 min at 72°C | ||||
| Fw, CTG AAA ACA CTA GTA CAG AAA ATG CA | 158-182 | Isolate 476 | 16 | |||
| Rv, GGT ACT GTT AAA CCT GAA GAA AAG | 1795-1818 | |||||
| ebpS | Fw, CAA TCG ATA GAC ACA AAT TC | U48826 | 40-59 | Isolate 252 | 1 min at 94°C, 1 min at | 31 |
| Rv, CAG TTA CAT CAT CAT GTT TA | 565-546 | 50°C, 1 min at 72°C | ||||
| Fw, CGT CAA TCG ATA GAC ACA AAT | 37-57 | Isolate 252 | 29 | |||
| Rv, CTG TAC CAG CAC CAA TT | 638-621 | |||||
| tst | Fw, AAG CCC TTT GTT GCT TGC G | AY074881 | 36-54 | Mu 50 | 1 min at 94°C, 1 min at | 3 |
| Rv, ATC GAA CTT TGG CCC ATA CTT T | 480-459 | 55°C, 2 min at 72°C | ||||
| Fw, ACC CCT GTT CCC TTA TCA TC | 88-107 | Mu 50 | 24 | |||
| Rv, TTT TCA GTA TTT GTA ACG CC | 394-375 | |||||
| eta | Fw, GCA GGT GTT GAT TTA GCA TT | M17357 | 719-738 | Isolate D72 | 2 min at 94°C, 2 min at | 24 |
| Rv, AGA TGT CCC TAT TTT TGC TG | 811-792 | 57°C, 2 min at 72°C | ||||
| Fw, ACT GTA GGA GCT AGT GCA TTT GT | 308-330 | Isolate D72 | 15 | |||
| Rv, TGG ATA CTT TTG TCT ATC TTT TTC ATC AAC | 496-467 | |||||
| etb | Fw, ACA AGC AAA AGA ATA CAG CG | M17348 | 509-528 | Isolate I 128 | 2 min at 94°C, 2 min at | 24 |
| Rv, GTT TTT GGC TGC TTC TCT TG | 734-719 | 57°C, 2 min at 72°C | ||||
| Fw, CAG ATA AAG AGC TTT ATA CAC ACA TTA C | 574-661 | Isolate I 128 | 15 | |||
| Rv, AGT GAA CTT ATC TTT CTA TTG AAA AAC ACT C | 1183-1153 | |||||
| pvl (lukF) | Fw, ATC ATT AGG TAA AAT GTC TGG ACA TGA TCC A | AB186917 | 579-609 | Isolate D48 | 1 min at 94°C, 1 min at | 33 |
| Rv, GCA TCA AST GTA TTG GAT AGC AAA AGC | 2072-2046 | 55°C, 2 min at 72°C | ||||
| pvl (lukS) | Fw, GCA AGG TTT TAT CAA TTC AAA GAC TAC TT | 234-262 | Isolate D48 | 27 | ||
| Rv, GGG TCA TTT GTT TTG AGA CCA ATA T | 344-320 |
Statistical analysis.
The distribution of putative virulence factors across S. aureus strains originating from different hosts were compared using Fisher's exact test. The odds ratio was defined as the cross-product ratio of the numbers shown in a two-by-two contingency table. Confidence intervals of 95% were used throughout. P values less than 0.05 were considered significant.
RESULTS
Genotyping of S. aureus strains.
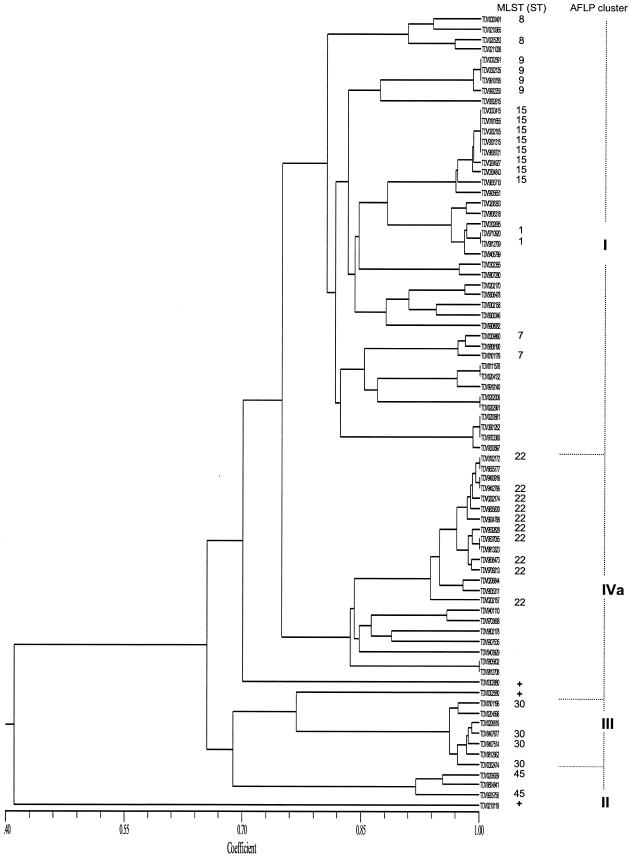
Ht-AFLP was used to determine the genomic variability within the veterinary S. aureus strain collection. AFLP results are summarized in Fig. 1. Four clusters, I, IVa, II, and III (corresponding to the previously defined AFLP clusters for human strains included in the database (25), representing 54.5% (n = 42), 28.6% (n = 22), 9.1% (n = 7), and 3.9% (n = 3) of the veterinary collection, respectively, could be identified. Strains causing infections in pet animals (21 out of 24; cat, dog, guinea pig, and rabbit) were overrepresented in AFLP cluster I (Fisher's exact test; P < 0.0001). AFLP cluster IVa mainly comprised geographically unrelated isolates obtained from farm animals (goats, sheep, and cows) and caused mastitis (14/22 versus 3/55, Fisher's exact test; P < 0.0001), which suggested tissue specificity.
FIG. 1.
The dendrogram shows the level of similarity, expressed by the similarity coefficient, between AFLP patterns from the animal strains (n = 77). AFLP cluster identification is indicated on the right side of the figure. MLST was performed of a selection of 2, 4, 10, and 18 animal strains, representing each AFLP subcluster (defined by an arbitrarily chosen similarity-coefficient level ≥ 0.8) within AFLP clusters III, II, IVa, and I, respectively. Plus signs represent three unique AFLP patterns that were found for 0302880 (horse), 0002580 (ape [gorilla]), and 0210118 (seal).
Integration of the veterinary strains in the AFLP database.
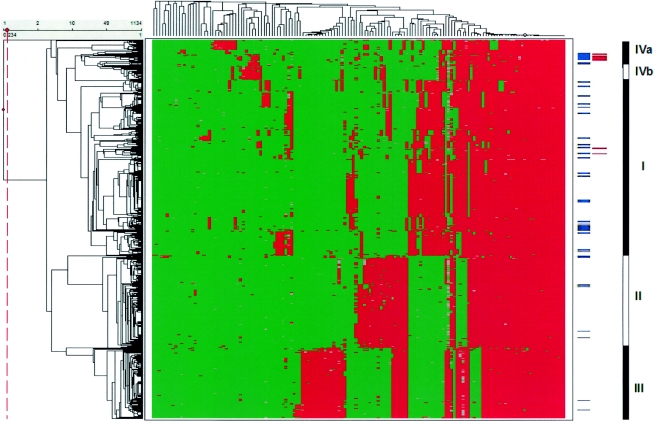
The resulting AFLP patterns obtained for the animal-associated strains were introduced in the AFLP database (25), and the analysis is summarized in Fig. 2. The different AFLP clusters of the animal-related strains matched with the different AFLP clusters of the database. In essence, the strains causing mastitis in the farm animals formed a homogenous subcluster in IVa (see red bars in Fig. 2). In comparison to the human strains (63/1,056), the animal strains (22/77) were significantly overrepresented in AFLP cluster IVa (Fisher's exact test; P < 0.0001) (Table 2). The human isolates in this cluster were also associated with bacteremia (Fisher's exact test; P = 0.0095) (25). None of the animal-related strains were classified in AFLP cluster IVb. S. aureus strains (21 out of 24) isolated from pet animals were predominantly found in major AFLP cluster I (87.5%) but were scattered over its different subclusters (Fig. 2). Strains belonging to AFLP clusters II and III were significantly underrepresented among the animal strains (7/77 and 3/77, respectively) compared to the human strains (263/1,056 and 210/1,056; P = 0.0012 and P = 0.0001, respectively) (Table 2).
FIG. 2.
Agglomerative two-dimensional clustering of strains (n = 1,133) and AFLP markers. AFLP fingerprints of the animal strains (n = 77) are included. The red fields in the figure represent the presence of AFLP markers; green indicates absence. Blue bars at the right side of the figure indicate the position of each animal strain in the analysis; the red bars represent the mastitis-associated strains. The AFLP database is divided in three main strain clusters (indicated as I, II, and III at the far right of the figure) and two minor clusters (indicated as IVa and IVb) (25).
TABLE 2.
Distribution of lineages (AFLP clusters) across the human strains and the veterinary strains
| AFLP cluster | No. (%) of animal strains | No. (%) of human strains | P | Odds ratio | 95% Confidence interval |
|---|---|---|---|---|---|
| I | 42 (54.5) | 480 (45.5) | 0.1256 | 1.44 | 0.9-2.3 |
| II | 7 (9.1) | 263 (24.9) | 0.0012 | 0.3 | 0.14-0.67 |
| III | 3 (3.9) | 210 (19.9) | 0.0001 | 0.16 | 0.05-0.52 |
| IVa | 25 (32.5) | 63 (6.0) | <0.0001 | 7.3 | 4.28-12.6 |
| IVb | 0 | 40 (3.8) | |||
| Total | 77 | 1,056 |
AFLP versus MLST.
MLST analysis was performed for a selection of the animal strains, representing each subcluster in Fig. 1. The animal-related strains classified in AFLP cluster I revealed five different sequence types (ST1, ST7, ST8, ST9, and ST15). In contrast, AFLP clusters II, III, and IVa were more homogenous, showing limited marker variability within each cluster (Fig. 2), and only harbored a single clonal complex (CC30, CC45, and CC22, respectively). Notably, the human-associated strains in cluster IVa are more genetically heterogeneous (25). MLST results agreed with the AFLP classification and were concordant with existing MLST sequence types within each AFLP cluster in the database as determined before (25).
Comparison of the virulence gene distribution between human and veterinary S. aureus isolates.
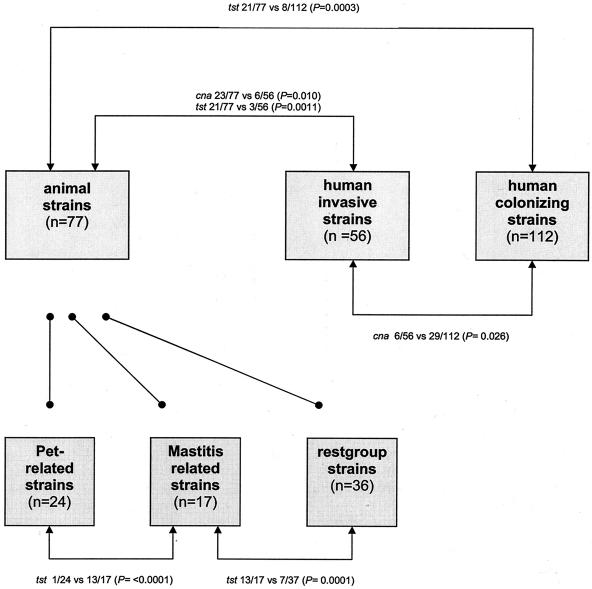
The presence or absence of 10 genes associated with pathogenicity were measured for the animal (n = 77) and human (n = 168) strain collections. Results obtained with both PCR strategies were fully concordant. Virulence gene incidence among the animal-related strains was compared to those of the invasive (n = 56) and colonizing (n = 112) strains. Significant differences in distribution of virulence genes between the groups are outlined in Fig. 3. No significant variation in virulence gene distribution within the different subgroups of the animal strains (mastitis associated, pet animal, and rest group) was measured (data not shown). Most of the genes coding for adhesion proteins were evenly distributed among the human and veterinary strains. The gene encoding collagen binding protein, cna, was underrepresented in the human invasive S. aureus strains (6/56) compared to the human colonizers (29/112, Fisher's exact test; P = 0.026) and the animal strains (23/77, Fisher's exact test; P = 0.01). This gene was not equally distributed among the different genetic lineages of S. aureus, since all animal strains from AFLP cluster II harbored the cna gene (P = 0.0001; data not shown). The exotoxin gene, tst, was found significantly more often in the animal-associated strains (21/77 versus 11/168 in the human strains [P < 0.0001] or versus 3/56 in the human-invasive strains [P = 0.0011]). Strains causing mastitis in animals contribute strongly to tst overrepresentation. The tst gene was present in 13 out of the 17 mastitis-associated strains, which significantly differs from results observed among strains isolated from pets (1/24, P < 0.0001) and among the rest group animals (7/39, P = 0.0001) (data not shown). The bicomponent leucocidin gene, pvl, was only detected in two human-related strains and not at all in the animal isolates. The association between the number of virulence genes and the proportion of animal and human S. aureus strains is stated in Table 3. No difference in the number of virulence genes of the strains causing a variety of infections in the animal host (n = 77) and in the human host (n = 168) was found.
FIG. 3.
A flow diagram of the virulence gene distribution in the four different strain clusters. Only significant differences are noted.
TABLE 3.
Association of the number of virulence genes and the proportion of animal and human S. aureus strains
| No. of virulence genes | No. of animal strains (n = 77) | No. of human strains (n = 168) | P | Odds ratio | 95% Confidence interval |
|---|---|---|---|---|---|
| 2 | 0 | 5 | 0.33 | 0.19 | 0.01-3.5 |
| 3 | 3 | 14 | 0.28 | 0.45 | 0.1-1.6 |
| 4 | 17 | 41 | 0.75 | 0.9 | 0.5-1.7 |
| 5 | 39 | 80 | 0.68 | 1.1 | 0.7-1.9 |
| 6 | 15 | 27 | 0.58 | 1.3 | 0.6-2.5 |
| 7 | 3 | 1 | 0.09 | 6.77 | 0.7-66.2 |
DISCUSSION
Host specificity of S. aureus.
Previous studies have compared host specificity of S. aureus strains isolated from humans and animals. Determinants such as antimicrobial resistance could not discriminate between human and veterinary isolates (5). Other studies identified a certain genetic schism between human and veterinary isolates of S. aureus, based on variability in PFGE (45, 46) or multilocus enzyme electrophoresis patterns (8, 17). In general, these studies compared bovine mastitis-associated isolates with strains of human origin. It was concluded that the overall genetic constitution of these S. aureus strains seemed to indicate that the majority of cases of bovine mastitis are caused by a few “specialized” clones (8, 10, 17, 37). This suggests the existence of bacterial host-specificity factors (12). The aim of the present study was to identify genetic polymorphism of S. aureus associated with host specificity. Moreover, the pathogenic potential, defined by the presence, absence, or number of selected bacterial virulence determinants, was determined.
Genome comparison.
Comparative genomics of the strains was initially done by PFGE. This was not useful for effective determination of the population structure of veterinary S. aureus strains. No host specificity could be determined except for the apparent clonality of goat-specific strains. Ht-AFLP was the prime method used to compare the animal-associated S. aureus strains. The veterinary strains integrated conveniently into the AFLP database, comprising strains isolated from humans. Animals in close contact with humans for reasons of care and treatment (pet animals and animals in contact with humans in a children's farm, riding school, or seal sanctuary or through caretakers in a zoo) were infected with these human-associated strains. Notably, strains isolated from cats and dogs were mainly classified in the heterogeneous AFLP cluster I, scattered over its subclusters that consist of human-related S. aureus strains. The mastitis-related strains, isolated from diverse host species (sheep, goat, and cow) were genetically clustered, signifying tissue specificity.
Cross-infection with S. aureus between humans and domestic animals in the household has been described previously (42). Cats were involved in an outbreak of an epidemic methicillin-resistant S. aureus in a geriatric ward (38). Dogs and children's farm cattle are potential reservoirs for the transmission of S. aureus strains to humans, causing diverse skin infections (34). Transmission of strains from humans to animals has also been observed (1, 35). We here clearly confirm these earlier findings on the basis of an integrated comparison between human and animal isolates and show that many of these veterinary isolates from various animal hosts fall within the same genomic classes.
Host-specific virulence potential.
The presence of certain virulence determinants and subsequent pathogenicity in humans has been observed previously (3, 13, 19, 22, 31). Identical virulence determinants were found in S. aureus strains causing animal infections. Bovine mammary isolates harbor genes encoding superantigens, such as tst (18, 21), exfoliative toxins (7), and enterotoxins (4). These virulence factors were also identified in S. aureus strains causing pneumonia in horses (14, 36, 39) or in poultry (11). However, these studies concern small numbers of strains or single isolation sites. We analyzed 10 different virulence genes and determined their distribution in various S. aureus populations. The tst gene, encoding the exotoxin with superantigen activity, was found significantly more often in the mastitis-associated S. aureus strains. Whether these genes are all actively expressed and whether this signifies tissue specificity is not known but is considered quite likely. The cna gene was not evenly distributed between the different AFLP clusters of the animal-related strains. The prevalence of exotoxin-encoding genes (pvl, eta, and etb) in both strain collections is low. The numbers of virulence genes in the animal- and human-related S. aureus strains were similar on a per strain basis. Apparently, the nature of the virulence genes encountered in an S. aureus strain is primarily an important determinant for host specificity. It has to be noted, however, that the virulence genes selected for the present study were based on those found for human pathogens. Most likely, veterinary pathogens may contain other host-specific virulence genes that are currently unknown.
In conclusion, many S. aureus clones have disseminated widely among humans, colonizing more than 30% of the population and causing a wide variety of severe infections. These same clones have the potency to colonize and infect many different host species. On the other hand, we here identified a tissue-specific clone (udder) responsible for causing disease in diverse host species. The presence of (combinations of) virulence factors plays an important role in host or even tissue specificity in S. aureus infections.
Acknowledgments
The oligoarray-mediated MLST for S. aureus characterization was financially supported by bioMérieux. The AFLP analysis described in this study has been facilitated by a grant provided by the Dutch Ministry of Economic Affairs (BTS 00145).
AFLP is a registered trademark of Keygene N.V., and the AFLP technology is subject to patents and patent applications owned by Keygene N.V.
REFERENCES
- 1.Adekeye, J. D. 1981. Studies on possible cross transmission of mercuric chloride resistant Staphylococcus aureus between dogs and kennel attendants. Int. J. Zoonoses 8:72-76. [PubMed] [Google Scholar]
- 2.Bannerman, T. L. 2003. Staphylococcus, Micrococcus, and other catalase-positive cocci that grow aerobically, p. 384-404. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. P. Yolken (ed.), Manual of clinical microbiology, 8th ed., vol. 1. American Society for Microbiology, Washington, D.C.
- 3.Bohach, G. A., D. J. Fast, R. D. Nelson, and P. M. Schlievert. 1990. Staphylococcal and streptococcal pyrogenic toxins involved in toxic shock syndrome and related illnesses. Crit. Rev. Microbiol. 17:251-272. [DOI] [PubMed] [Google Scholar]
- 4.Cenci-Goga, B. T., M. Karama, P. V. Rossitto, R. A. Morgante, and J. S. Cullor. 2003. Enterotoxin production by Staphylococcus aureus isolated from mastitic cows. J. Food Prot. 66:1693-1696. [DOI] [PubMed] [Google Scholar]
- 5.Cuteri, V., R. Mazzolla, F. Valente, L. Merletti, and C. Valente. 2002. Application of pulsed-field gel electrophoresis (PFGE) to methicillin-resistant strains of Staphylococcus aureus from humans and domestic animals. Infez. Med. 10:25-30. [PubMed] [Google Scholar]
- 6.Dale, S. E., A. Doherty-Kirby, G. Lajoie, and D. E. Heinrichs. 2004. Role of siderophore biosynthesis in virulence of Staphylococcus aureus: identification and characterization of genes involved in production of a siderophore. Infect. Immun. 72:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Endo, Y., T. Yamada, K. Matsunaga, Y. Hayakawa, T. Kaidoh, and S. Takeuchi. 2003. Phage conversion of exfoliative toxin A in Staphylococcus aureus isolated from cows with mastitis. Vet. Microbiol. 96:81-90. [DOI] [PubMed] [Google Scholar]
- 8.Fitzgerald, J. R., W. J. Meaney, P. J. Hartigan, C. J. Smyth, and V. Kapur. 1997. Fine-structure molecular epidemiological analysis of Staphylococcus aureus recovered from cows. Epidemiol. Infect. 119:261-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzgerald, J. R., D. E. Sturdevant, S. M. Mackie, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilot, P., G. Lina, T. Cochard, and B. Poutrel. 2002. Analysis of the genetic variability of genes encoding the RNA III-activating components Agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J. Clin. Microbiol. 40:4060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hazariwala, A., Q. Sanders, C. R. Hudson, C. Hofacre, S. G. Thayer, and J. J. Maurer. 2002. Distribution of staphylococcal enterotoxin genes among Staphylococcus aureus isolates from poultry and humans with invasive staphylococcal disease. Avian Dis. 46:132-136. [DOI] [PubMed] [Google Scholar]
- 12.Herron, L. L., R. Chakravarty, C. Dwan, J. R. Fitzgerald, J. M. Musser, E. Retzel, and V. Kapur. 2002. Genome sequence survey identifies unique sequences and key virulence genes with unusual rates of amino acid substitution in bovine Staphylococcus aureus. Infect. Immun. 70:3978-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hienz, S. A., T. Schennings, A. Heimdahl, and J. I. Flock. 1996. Collagen binding of Staphylococcus aureus is a virulence factor in experimental endocarditis. J. Infect. Dis. 174:83-88. [DOI] [PubMed] [Google Scholar]
- 14.Holbrook, T. C., J. S. Munday, C. A. Brown, B. Glover, P. M. Schlievert, and S. Sanchez. 2003. Toxic shock syndrome in a horse with Staphylococcus aureus pneumonia. J. Am. Vet. Med. Assoc. 222:620-623, 601-602. [DOI] [PubMed] [Google Scholar]
- 15.Jarraud, S., C. Mougel, J. Thioulouse, G. Lina, H. Meugnier, F. Forey, X. Nesme, J. Etienne, and F. Vandenesch. 2002. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 70:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Josefsson, E., K. W. McCrea, D. Ni Eidhin, D. O'Connell, J. Cox, M. Hook, and T. J. Foster. 1998. Three new members of the serine-aspartate repeat protein multigene family of Staphylococcus aureus. Microbiology 144(Pt. 12):3387-3395. [DOI] [PubMed] [Google Scholar]
- 17.Kapur, V., W. M. Sischo, R. S. Greer, T. S. Whittam, and J. M. Musser. 1995. Molecular population genetic analysis of Staphylococcus aureus recovered from cows. J. Clin. Microbiol. 33:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenny, K., R. F. Reiser, F. D. Bastida-Corcuera, and N. L. Norcross. 1993. Production of enterotoxins and toxic shock syndrome toxin by bovine mammary isolates of Staphylococcus aureus. J. Clin. Microbiol. 31:706-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koning, S., A. van Belkum, S. Snijders, W. van Leeuwen, H. Verbrugh, J. Nouwen, M. Op't Veld, L. W. van Suijlekom-Smit, J. C. van der Wouden, and C. Verduin. 2003. Severity of nonbullous Staphylococcus aureus impetigo in children is associated with strains harboring genetic markers for exfoliative toxin B, Panton-Valentine leukocidin, and the multidrug resistance plasmid pSK41. J. Clin. Microbiol. 41:3017-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 21.Kuroishi, T., K. Komine, K. Kai, M. Itagaki, J. Kobayashi, M. Ohta, S. Kamata, and K. Kumagai. 2003. Concentrations and specific antibodies to staphylococcal enterotoxin-C and toxic shock syndrome toxin-1 in bovine mammary gland secretions, and inflammatory response to the intramammary inoculation of these toxins. J. Vet. Med. Sci. 65:899-906. [DOI] [PubMed] [Google Scholar]
- 22.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 23.McDevitt, D., P. Francois, P. Vaudaux, and T. J. Foster. 1994. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol. Microbiol. 11:237-248. [DOI] [PubMed] [Google Scholar]
- 24.Mehrotra, M., G. Wang, and W. M. Johnson. 2000. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J. Clin. Microbiol. 38:1032-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melles, D. C., R. F. Gorkink, H. A. Boelens, S. V. Snijders, J. K. Peeters, M. J. Moorhouse, P. J. van der Spek, W. B. van Leeuwen, G. Simons, H. A. Verbrugh, and A. van Belkum. 2004. Natural population dynamics and expansion of pathogenic clones of Staphylococcus aureus. J. Clin. Investig. 114:1732-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murchan, S., M. E. Kaufman, A. Deplano, M. Struelens, C. S. Elsberg, V. T. Rosdahl, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. B. Van Leeuwen, A. Van Belkum, J. Melo-Cristino, A. Vindel, J. Garaizar, B. M. Hoffman, B. Olsson-Liljequist, U. Ransjo, and B. D. Cookson. 2000. Harmony: establishment of a collection and database of European epidemic MRSA (EMRSA) strains and harmonisation of pulsed-field gel electrophoresis (PFGE), abstr. P084, p. 156. In IMBEM 5, 2000, Noordwijkerhout, The Netherlands.
- 27.Nakagawa, S., I. Taneike, D. Mimura, N. Iwakura, T. Nakayama, T. Emura, M. Kitatsuji, A. Fujimoto, and T. Yamamoto. 2005. Gene sequences and specific detection for Panton-Valentine leukocidin. Biochem. Biophys. Res. Commun. 328:995-1002. [DOI] [PubMed] [Google Scholar]
- 28.Needham, A. J., M. Kibart, H. Crossley, P. W. Ingham, and S. J. Foster. 2004. Drosophila melanogaster as a model host for Staphylococcus aureus infection. Microbiology 150:2347-2355. [DOI] [PubMed] [Google Scholar]
- 29.Park, P. W., J. Rosenbloom, W. R. Abrams, and R. P. Mecham. 1996. Molecular cloning and expression of the gene for elastin-binding protein (ebpS) in Staphylococcus aureus. J. Biol. Chem. 271:15803-15809. [DOI] [PubMed] [Google Scholar]
- 30.Patti, J. M., H. Jonsson, B. Guss, L. M. Switalski, K. Wiberg, M. Lindberg, and M. Hook. 1992. Molecular characterization and expression of a gene encoding a Staphylococcus aureus collagen adhesin. J. Biol. Chem. 267:4766-4772. [PubMed] [Google Scholar]
- 31.Peacock, S. J., C. E. Moore, A. Justice, M. Kantzanou, L. Story, K. Mackie, G. O'Neill, and N. P. Day. 2002. Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect. Immun. 70:4987-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perkins, S., E. J. Walsh, C. C. Deivanayagam, S. V. Narayana, T. J. Foster, and M. Hook. 2001. Structural organization of the fibrinogen-binding region of the clumping factor B MSCRAMM of Staphylococcus aureus. J. Biol. Chem. 276:44721-44728. [DOI] [PubMed] [Google Scholar]
- 33.Prévost, G., B. Cribier, P. Couppié, P. Petiau, G. Supersac, V. Finck-Barbançon, H. Monteil, and Y. Piemont. 1995. Panton-Valentine leucocidin and gamma-hemolysin from Staphylococcus aureus ATCC 49775 are encoded by distinct genetic loci and have different biological activities. Infect. Immun. 63:4121-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rao, P. N., A. S. Naidu, P. R. Rao, and K. Rajyalakshmi. 1987. Prevalence of staphylococcal zoonosis in pyogenic skin infections. Zentbl. Bakteriol. Mikrobiol. Hyg. A 265:218-226. [DOI] [PubMed] [Google Scholar]
- 35.Rodgers, J. D., J. J. McCullagh, P. T. McNamee, J. A. Smyth, and H. J. Ball. 1999. Comparison of Staphylococcus aureus recovered from personnel in a poultry hatchery and in broiler parent farms with those isolated from skeletal disease in broilers. Vet. Microbiol. 69:189-198. [DOI] [PubMed] [Google Scholar]
- 36.Sato, H., Y. Matsumori, T. Tanabe, H. Saito, A. Shimizu, and J. Kawano. 1994. A new type of staphylococcal exfoliative toxin from a Staphylococcus aureus strain isolated from a horse with phlegmon. Infect. Immun. 62:3780-3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlegelova, J., M. Dendis, J. Benedik, V. Babak, and D. Rysanek. 2003. Staphylococcus aureus isolates from dairy cows and humans on a farm differ in coagulase genotype. Vet. Microbiol. 92:327-334. [DOI] [PubMed] [Google Scholar]
- 38.Scott, G. M., R. Thomson, J. Malone-Lee, and G. L. Ridgway. 1988. Cross-infection between animals and man: possible feline transmission of Staphylococcus aureus infection in humans? J. Hosp. Infect. 12:29-34. [DOI] [PubMed] [Google Scholar]
- 39.Shimizu, A., J. Kawano, J. Ozaki, N. Sasaki, S. Kimura, M. Kamada, S. Anzai, H. Saito, and H. Sato. 1991. Characteristics of Staphylococcus aureus isolated from lesions of horses. J. Vet. Med. Sci. 53:601-606. [DOI] [PubMed] [Google Scholar]
- 40.Sifri, C. D., J. Begun, F. M. Ausubel, and S. B. Calderwood. 2003. Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect. Immun. 71:2208-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Signas, C., G. Raucci, K. Jonsson, P. E. Lindgren, G. M. Anantharamaiah, M. Hook, and M. Lindberg. 1989. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc. Natl. Acad. Sci. USA 86:699-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simoons-Smit, A. M., P. H. Savelkoul, J. Stoof, T. M. Starink, and C. M. Vandenbroucke-Grauls. 2000. Transmission of Staphylococcus aureus between humans and domestic animals in a household. Eur. J. Clin. Microbiol. Infect. Dis. 19:150-152. [DOI] [PubMed] [Google Scholar]
- 43.van Leeuwen, W. B., C. Jay, S. Snijders, N. Durin, B. Lacroix, H. A. Verbrugh, M. C. Enright, A. Troesch, and A. van Belkum. 2003. Multilocus sequence typing of Staphylococcus aureus with DNA array technology. J. Clin. Microbiol. 41:3323-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wertheim, H. F., M. C. Vos, A. Ott, A. Voss, J. A. Kluytmans, C. M. Vandenbroucke-Grauls, M. H. Meester, P. H. van Keulen, and H. A. Verbrugh. 2004. Mupirocin prophylaxis against nosocomial Staphylococcus aureus infections in nonsurgical patients: a randomized study. Ann. Intern. Med. 140:419-425. [DOI] [PubMed] [Google Scholar]
- 45.Zadoks, R., W. van Leeuwen, H. Barkema, O. Sampimon, H. Verbrugh, Y. H. Schukken, and A. van Belkum. 2000. Application of pulsed-field gel electrophoresis and binary typing as tools in veterinary clinical microbiology and molecular epidemiologic analysis of bovine and human Staphylococcus aureus isolates. J. Clin. Microbiol. 38:1931-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zadoks, R. N., W. B. van Leeuwen, D. Kreft, L. K. Fox, H. W. Barkema, Y. H. Schukken, and A. van Belkum. 2002. Comparison of Staphylococcus aureus isolates from bovine and human skin, milking equipment, and bovine milk by phage typing, pulsed-field gel electrophoresis, and binary typing. J. Clin. Microbiol. 40:3894-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]