Abstract
In gram-negative bacteria, a pathway for aerobic degradation of phenylacetic acid (PAA) that proceeds via phenylacetyl-coenzyme A (CoA) and hydrolytic ring fission plays a central role in the degradation of a range of aromatic compounds. In contrast, the PAA pathway and its role are not well characterized in gram-positive bacteria. A cluster including 13 paa genes encoding enzymes orthologous to those of gram-negative bacteria was identified on the chromosome of Rhodococcus sp. strain RHA1. These genes were transcribed during growth on PAA, with 11 of the genes apparently in an operon yielding a single transcript. Quantitative proteomic analyses revealed that at least 146 proteins were more than twice as abundant in PAA-grown cells of RHA1 than in pyruvate-grown cells. Of these proteins, 29 were identified, including 8 encoded by the paa genes. Knockout mutagenesis indicated that paaN, encoding a putative ring-opening enzyme, was essential for growth on PAA. However, paaF, encoding phenylacetyl-CoA ligase, and paaR, encoding a putative regulator, were not essential. paaN was also essential for growth of RHA1 on phenylacetaldehyde, phenylpyruvate, 4-phenylbutyrate, 2-phenylethanol, 2-phenylethylamine, and l-phenylalanine. In contrast, growth on 3-hydroxyphenylacetate, ethylbenzene, and styrene was unaffected. These results suggest that the range of substrates degraded via the PAA pathway in RHA1 is somewhat limited relative to the range in previously characterized gram-negative bacteria.
Rhodococci are aerobic, gram-positive actinomycetes of high G+C content and are capable of morphological differentiation in response to their environment (e.g., cocci or filaments). These widely occurring organisms are of considerable environmental and biotechnological importance due to their broad metabolic diversity and array of unique enzymatic capabilities. These characteristics are of interest for potential pharmaceutical, environmental, chemical, and energy applications. Rhodococci are well suited for bioremediation due to their capacity for long-term survival in soil, their exceptional ability to degrade hydrophobic pollutants even in the presence of more readily degraded substrates, and their ability to accumulate high levels of heavy metals (11, 20).
Rhodococcus sp. strain RHA1 was originally isolated from gamma-hexachlorocyclohexane-contaminated soil and is one of the best-characterized polychlorinated biphenyl degraders (13). RHA1 uses multiple enzyme systems, including at least three ring-hydroxylating dioxygenases, to degrade biphenyl (9) and has a duplicate set of genes encoding degradation of phthalate (16). A draft assembly of the 9.7-Mb genome (for the current assembly, see http://www.rhodococcus.ca/) revealed a large number of genes putatively involved in aromatic-compound degradation, including a cluster of genes described in this report that encodes a phenylacetic acid (PAA) pathway.
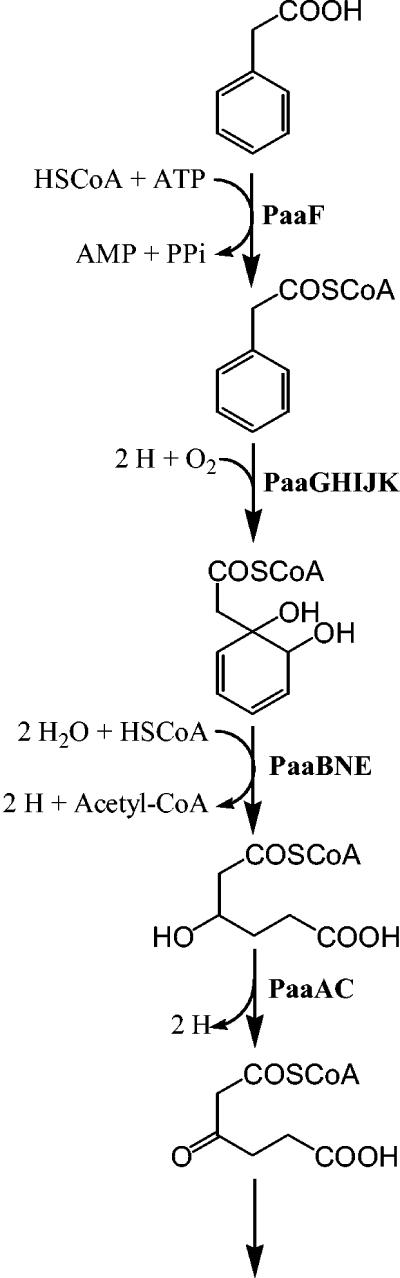
PAA arises from the deamination of the aromatic amino acid phenylalanine and is widely distributed in the environment. An aerobic PAA degradation pathway (Fig. 1) has been described for some gram-negative bacteria, such as Pseudomonas putida U and Escherichia coli (12). In this pathway, PAA is first transformed to phenylacetyl-coenzyme A (CoA), which subsequently undergoes ring hydroxylation, hydrolytic-ring opening, and further degradation. Until recently, aryl-CoA derivatives were considered to be specific to anaerobic catabolic pathways. As such, the PAA pathway has been proposed to be a hybrid pathway. A similar hybrid pathway responsible for the aerobic degradation of benzoate via benzoyl-CoA was recently described for Burkholderia xenovorans LB400 (4, 23). The roles of many catabolic proteins of these pathways remain poorly defined.
FIG. 1.
PAA catabolism pathway (based on that described in reference 8), showing the proposed functions of most paa gene products.
In P. putida U and E. coli, the PAA pathway acts as the key terminal part of the phenylacetyl-CoA catabolon, a complex functional unit that integrates peripheral catabolic pathways and catalyzes the transformation of related compounds via a common metabolite. The phenylacetyl-CoA catabolons of these organisms include a range of compounds, some of them important pollutants, such as styrene and ethylbenzene. This catabolon is also significant for biotechnological applications, such as the enzymatic synthesis of penicillins, the production of intermediates (such as 2-hydroxyphenylacetate or 2-hydroxybenzoate) necessary for the synthesis of many chemical compounds, and the production of bioplastics (12). There is evidence for the existence of the PAA pathway in gram-positive bacteria, including the cloning of putative phenylacetyl-CoA ligase genes from some gram-positive strains (1) and the growth of Rhodococcus opacus PD630 on PAA, and evidence that PD630 degrades phenyldecane via PAA (2). However, in contrast to the PAA pathway in gram-negative bacteria, the pathway in gram-positive bacteria is not characterized, nor is its role well understood.
In this study, we used proteomic, transcriptional, and gene disruption approaches for the first time to demonstrate a functional PAA pathway and the genes responsible in a gram-positive bacterium. We also determined the pathway's role in the degradation of a range of additional aromatic substrates.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Rhodococcus sp. strain RHA1 was kindly provided by M. Fukuda. RHA1 and its mutants were grown with shaking at 200 rpm at 30°C in W minimal salts medium (19) with various organic substrates. Pyruvate and 2-phenylethanol were added directly to the culture medium, and the remaining compounds were added from stock solutions dissolved in NaOH (PAA, 2-hydroxyphenylacetate, 3-hydroxyphenylacetate, and phenylpyruvate) or in water (benzoate, phthalate, mandelate, l-phenylalanine, 4-phenylbutyrate, and tropic acid). Volatile substrates (styrene, ethylbenzene, benzene, phenylacetaldehyde, and biphenyl) were supplied as vapors saturating the culture headspace. For reverse transcriptase PCR (RT-PCR) and proteomic analyses, 500-ml cultures were incubated in 2-liter Erlenmeyer flasks with 20 mM PAA or 20 mM pyruvate. Cells were harvested in mid-exponential growth phase (optical density at 600 nm, 2.0) by centrifugation for 10 min at 4°C and 8,000 × g. For proteomic analyses, the cell pellets were flash frozen in liquid nitrogen and stored at −80°C, and subsequent cell manipulation steps were performed at 4°C.
E. coli strains were grown with shaking at 200 rpm at 37°C in Luria-Bertani broth (LB). When appropriate, antibiotics were added at the following concentrations: 50 μg/ml apramycin (Apr), ampicillin (Amp), kanamycin (Km), or nalidixic acid or 25 μg/ml chloramphenicol (Cm). E. coli WM276/fosmid RF0011cE07 (Cmr) was provided by the Genome Sciences Centre (Vancouver) and contains a fosmid with a 36.6-kb insert containing the paa gene cluster. E. coli BW25113/pKD20 and E. coli DH10b/pUZ8002 (Kmr) (7) were provided by B. Gust of the John Innes Institute. The plasmid pIJ773 (4.3 kb) (7) was used as the source for a disruption cassette containing the apramycin resistance gene used for PCR-targeted gene replacement. The plasmid pKD20 contains the λ-Red (gam, bet, exo) recombination system (3). The sequences of the PCR primers used in this work are available upon request.
Molecular biology techniques.
Plasmid DNA was obtained by rapid alkaline lysis, and DNA manipulations and other molecular biology techniques were done essentially as described previously (18). Genomic DNA extraction was based on a procedure developed for Listeria monocytogenes (6). Cell pellets from 3-ml overnight cultures in LB were washed in 1.5 ml of 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and suspended in 300 μl of 0.01 N sodium phosphate buffer in 20% (wt/vol) sucrose (pH 7.0) containing 2.5 mg/ml of lysozyme for an overnight incubation at 37°C. Then, 100 μl of 10% sodium dodecyl sulfate (wt/vol) and 200 μl of Tris-EDTA plus proteinase K at 5 mg/ml were added to the sample and incubated at 37°C for 30 min. Phenol-chloroform extraction, ethanol precipitation, and spooling were performed as described previously (18). RNA was obtained from mid- exponential growth phase cultures by use of RNAwiz (Ambion) and by treating the cultures two or three times with DNase (Ambion) until no sign of DNA was detected by PCR. Synthesis of cDNA was done with Superscript III reverse transcriptase (Invitrogen) at 55°C, and the resulting cDNA was treated for 1 h at 37°C with RNase A plus RNase H before being used for RT-PCR. RT-PCRs were carried out in the presence of 4% (vol/vol) dimethyl sulfoxide for 30 cycles (45 s at 94°C, 3 min at 55°C, and 45 s at 72°C) using 0.2 μg cDNA in a 50-μl reaction mixture. Nucleotide and protein sequence similarity searches were done by using the BLASTP, BLASTN, and BLASTX programs from the EMBL server. DNA secondary structure was analyzed using GeneBee (http://www.genebee.msu.su/).
Isolation of mutants.
The paaF, paaN, and paaR genes were replaced using the λ-Red-based methodology (3) as modified for Streptomyces spp. (7) and modified for Rhodococcus strain RHA1 (16). Fosmid clones containing the target genes were selected from an RHA1 library (22) and used to transform E. coli BW25113 containing the λ-Red recombination plasmid pKD46 (Ampr). Gene replacement cassettes consisting of Aprr oriT flanked by sequences from both sides of the target gene were amplified by PCR and introduced into E. coli BW25113(pKD46) containing the appropriate fosmid. Successful transformants (containing the Cmr Aprr mutagenized fosmid) were verified by PCR. Recombinant fosmids were isolated, introduced into DH10B/pUZ8002, and then transferred to Rhodococcus sp. strain RHA1 by intergeneric conjugation. Selection for double recombination was performed with LB plates containing nalidixic acid plus Apr incubated at 30°C for 5 days. The resulting mutant strains had only the target gene deleted with no transcription termination sequence added. These strains were verified by PCR as described above and by Southern blot analysis. Southern blot analysis was performed with 10 μg EcoRI-digested genomic DNA, which was transferred to a nylon membrane and hybridized, in the presence of 50% formamide at 30°C, with digoxigenin-labeled PCR probes amplified from RHA1 (FiF-FiR primers for paaF, RiR-RiR for paaR, and NiR-NiR for paaN).
Proteomic analysis.
Cytoplasmic proteins were analyzed by two-dimensional (2D) gel electrophoresis followed by 2D gel quantitative analysis and peptide mass fingerprint analysis essentially as previously described (16). Briefly, cells were thoroughly washed, suspended in a lysis buffer, and disrupted using a bead beater. The cell-free protein extract obtained was either stored at −80°C or used immediately. The first dimension was run using nonlinear IPG strips (Immobiline DryStrips; 24 cm, pH 3 to 7) and 90 μg of protein extract. The IPG strips were then equilibrated and run into 24- by 20-cm gels with 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis using the ETTAN DALTtwelve System (Amersham Biosciences). Protein spots were detected using Sypro Ruby stain, and gels were imaged using a Typhoon 9400 (excitation wavelength, 488 nm; emission wavelength, 610 nm) (Amersham Biosciences). Images were differentially analyzed using Progenesis Workstation software (Nonlinear Dynamics, Durham, NC). The signal intensity of each spot was averaged over gels obtained from three different cultures. Only spots with a minimum normalized volume of 0.002 or greater were further analyzed. For proteins appearing on the gel as a horizontal series of spots, likely due to carbamylation, the pI and molecular weight of only the major spot in the series were recorded, and the differences in expression were calculated based on the summed signal intensities of all the spots in the series. Protein spots whose intensities increased at least twofold above that of the control (pyruvate-grown cells) were recorded as more abundant. Proteins were identified based on peptide mass fingerprint analyses by use of a Voyager-DE STR matrix-assisted laser desorption ionization-time-of-flight mass spectrometer (Applied Biosystems). Proteins were identified using the Mascot search engine (www.matrixscience.com) and a database generated by in silico digestion of the total RHA1 proteome predicted from the genome assembly as described previously (16).
Analysis of aromatic metabolites.
Culture supernatant (40 ml) was passed through a high-capacity C18 Maxi-Clean 300-mg solid-phase extraction cartridge (Alltech, Deerfield, IL) previously conditioned with methanol and equilibrated with water. The cartridge was rinsed with 2 ml water and then eluted with 2.5 ml methanol. The methanol eluate was evaporated under a stream of nitrogen, and the residue was redissolved in methanol and evaporated again. The residue was taken up in 1 ml of dry methanol. Half of the residue was derivatized with diazomethane, using ethyl acetate (20%) as a cosolvent. Both underivatized and derivatized samples were analyzed by gas chromatography-mass spectroscopy (electron ionization mode) using an Agilent Technologies 6890N network gas chromatograph system equipped with an Agilent 5973 mass selective detector. Compounds were identified by inspection of fragmentation patterns and comparison to the NIST 98 mass spectral library.
RESULTS
paa gene cluster in RHA1.
Analysis of the RHA1 draft genome sequence (http://www.rhodococcus.ca/) revealed a chromosomal gene cluster encoding a putative PAA biodegradation pathway (Fig. 2). RHA1 was subsequently found to grow on 15 mM PAA as the sole organic substrate. The deduced proteins encoded by the RHA1 paa gene cluster (Table 1) as well as the pathway itself (Fig. 1) are similar to those described for P. putida U and E. coli. Several systems for nomenclature of the paa genes have been used, and we have adopted the consensus nomenclature proposed by Luengo et al. (12).
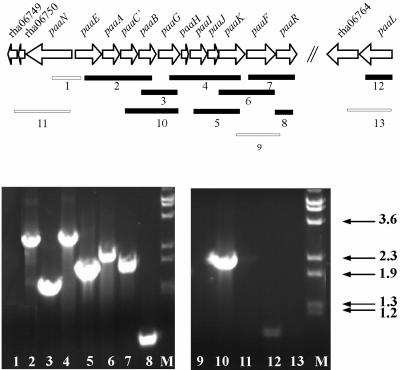
FIG. 2.
paa gene cluster in Rhodococcus sp. strain RHA1 and transcription of the paa genes during growth on PAA. Filled bars indicate RT-PCR amplicons detected, and open bars indicate negative RT-PCR assays. Numbers relate gel lanes (bottom) to amplicons (top). RT-PCRs were performed at least three times with different cDNA preparations with consistent results, and a representative experiment is shown. Positive controls with genomic DNA as the template verified all PCRs and the amplicon sizes (not shown). Negative controls without RT verified that DNA did not contaminate any of the RNA templates (not shown). M, size marker of λ DNA, labeled in kilobases, digested with BstEII.
TABLE 1.
ORFs in a chromosomal fragment of RHA1 containing the genes encoding the PAA pathway
| Gene (ORF no.) | No. of aa | % GC | EMBL-EBI accession no. (% aa identity); organism | Predicted function |
|---|---|---|---|---|
| paaL (rha06765) | 516 | 66.2 | Q8ENN9 (46.7); Oceanobacillus iheyensis | Na+-dependent symporter |
| (rha06764) | 425 | 72.1 | Q7WFH4 (45); Bordetella bronchiseptica RB50 | Amidase |
| paaR (rha06762) | 342 | 69.3 | P72312 (43.3); Rhodococcus rhodochrous | Nitrilase regulator |
| paaF (rha06761) | 432 | 66.0 | Q82NP4 (70); Streptomyces avermitilis | Aerobic phenylacetate-CoA ligase (EC 6.2.1.30) |
| paaK (rha06760) | 365 | 67.9 | Q93JC1 (71.7); Streptomyces coelicolor A3 | Hydroxylating complex V |
| paaJ (rha06759) | 169 | 67.9 | Q93JC2 (69.8); Streptomyces coelicolor A3 | Hydroxylating complex IV |
| paaI (rha06758) | 305 | 68.9 | Q93JC3 (54.4); Streptomyces coelicolor A3 | Hydroxylating complex III |
| paaH (rha06757) | 101 | 66.3 | Q93JC4 (69.3); Streptomyces coelicolor A3 | Hydroxylating complex II |
| paaG (rha06756) | 317 | 65.5 | Q93JC5 (84.5); Streptomyces coelicolor A3 | Hydroxylating complex I |
| paaB (rha06755) | 263 | 73.6 | Q9L4S8 (48); Streptomyces collinus | 2-Cyclohexenylcarbonyl-CoA isomerase |
| paaC (rha06754) | 284 | 69.7 | Q8FRT3 (61.6); Corynebacterium efficiens YS-314 | Putative 3-hydroxybutyryl-CoA dehydrogenase |
| paaA (rha06753) | 257 | 67.8 | Q8FRT4 (64.6); Corynebacterium efficiens YS-314 | Putative enoyl-CoA hydratase I |
| paaE (rha06752) | 406 | 73.4 | Q8FRT5 (68.1); Corynebacterium efficiens YS-314 | Putative beta-ketoadipyl CoA thiolase |
| paaN (rha06751) | 634 | 69.1 | Q92TG5 (68); Rhizobium meliloti | Putative aldehyde dehydrogenase protein (EC 1.2.1.-) ring-opening enzyme |
| (rha06750) | 105 | 64.5 | Q93EX2 (52); Rhodococcus ruber | EthD, from the ethyl tert-butyl ether degradation cluster |
| (rha06749) | 137 | 69.8 | Q93JC6 (65.7); Streptomyces coelicolor A3 | Putative PAA degradation protein (thioesterase) |
The RHA1 genome also contained paralogs of three paa genes, including 1 paralog of paaC, 18 of paaE, and 9 of paaF. The product of open reading frame (ORF) rha07533 shares 42% sequence identity with the 3-hydroxybutyryl-CoA dehydrogenase encoded by paaC. ORF rha07533 belongs to a chromosomal cluster of unknown function whose genes were predicted to encode a formamidase, an isocitrate lyase, a methionine synthase, an alcohol dehydrogenase, and a MerR-type transcriptional regulator. The 18 paralogs of the thiolase-encoding paaE include pcaF, which encodes β-ketoadipate:succinyl-CoA thiolase (16). Finally, the paralogs of paaF share 15 to 28% sequence identity with the phenylacetate-CoA ligase. The ORF with the highest identity, rha10245, is annotated as a potential phenylacetate-CoA ligase-encoding gene. A more detailed annotation of these genes is available at http://www.rhodococcus.ca/.
Gene expression analysis by RT-PCR.
Most of the paa genes appear to lie within a single large operon (Fig. 2). Using GeneBee, we predicted that the secondary structure of the putative polycistronic mRNA from paaE to paaR would include a weak hairpin (20 bp, −18 kcal/mol) between paaB and paaG and a strong hairpin downstream of paaR (300 bp, −94 kcal/mol). RT-PCR analysis of RHA1 indicates that, in cells grown on PAA, most or all of the paa genes are cotranscribed with at least one other paa gene (Fig. 2). Two transcripts, including paaBG (Fig. 2), indicate that the hairpin between these genes does not terminate all transcripts. We failed to obtain a product spanning paaKFR (lane 9), which we cannot explain. However, the product including paaFR (lane 7) suggests that paaR is cotranscribed with some or all of the other paa genes. These results suggest a polycistronic, 9.9-kb transcript including paaEACBGHIJKFR. However, smaller transcripts may occur in addition to or instead of the 9.9-kb transcript. The paaN and paaL genes appear to be individually transcribed, as there was no evidence of cDNA from a paaL-rha06764 transcript and only a very faint band for the paaN-rha06749 transcript.
PAA proteome.
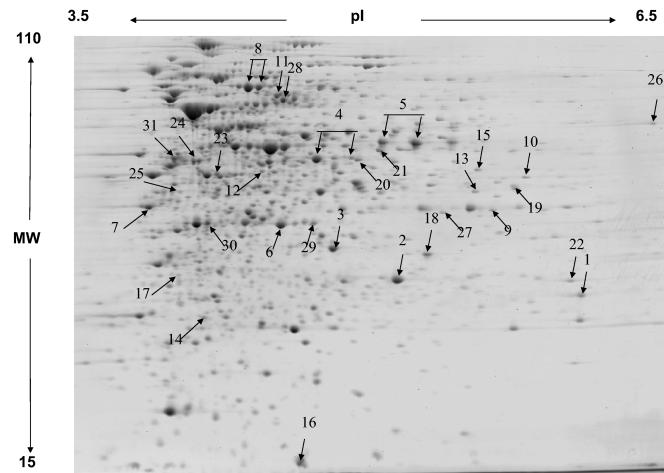
A total of approximately 1,430 protein spots were resolved in the proteome of RHA1 grown on PAA (Fig. 3). A quantitative comparison of the proteomes revealed that 75 proteins were at least twofold more abundant during growth on PAA than on pyruvate and that 71 proteins were apparently unique to the PAA proteome. A total of 29 of the upregulated proteins were identified by mass spectometric analysis. The calculated and observed pIs and molecular weights of all identified proteins together with the averaged normalized signal intensities observed under each condition are presented in Table S1 (supplemental material available at http://www.rhodococcus.ca/publications/supplementary/JBact05A.pdf). Eight of the 13 predicted paa gene products (PaaABCEFGIN) were identified (Fig. 3; Table 2). These proteins were detected only in PAA-grown cells, not in pyruvate-grown controls. Of the five predicted Paa proteins that were not found, three have properties that are incompatible with detection by 2D gel electrophoresis, being either too small (PaaH) or too basic (PaaR and PaaL) (Table S1).
FIG. 3.
2D gel analysis of the proteome of RHA1 grown on PAA. The major proteins associated with phenylacetate catabolism are indicated with arrows. The numbers correspond to spot numbers in Table 2, where the protein names and corresponding ORFs are listed. MW, molecular weight (in thousands).
TABLE 2.
Proteins more abundant in the proteome of RHA1 grown on phenylacetate than in that of RHA1 grown on pyruvate
| Protein | ORF no. | Spot no.a | No. of peptides matched | Sequence coverage (%) | Normalized signal intensityb
|
Difference (fold)c | t-test P value | |
|---|---|---|---|---|---|---|---|---|
| Pyruvate | Phenylacetate | |||||||
| PaaA | rha06753 | 1 | 8 | 45 | NDd | 0.14 | 70 | NAe |
| PaaB | rha06755 | 2 | 7 | 36 | ND | 0.79 | 395 | NA |
| PaaC | rha06754 | 3 | 8 | 39 | ND | 0.61 | 305 | NA |
| PaaE | rha06752 | 4 | 13 | 51 | ND | 1.02 | 510 | NA |
| PaaF | rha06761 | 5 | 9 | 25 | ND | 0.87 | 435 | NA |
| PaaG | rha06756 | 6 | 5 | 22 | ND | 0.43 | 215 | NA |
| PaaI | rha06758 | 7 | 7 | 35 | ND | 0.61 | 305 | NA |
| PaaN | rha06751 | 8 | 22 | 45 | ND | 1.6 | 800 | NA |
| 3-Hydroxybutyryl-CoA dehydrogenasef | rha07533 | 9 | 10 | 36 | ND | 0.059 | 30 | NA |
| Acyl-CoA dehydrogenase | rha06610 | 12 | 8 | 30 | 0.027 | 0.094 | 4 | 0.003553 |
| Malate dehydrogenase | rha08393 | 24 | 8 | 32 | 0.055 | 0.33 | 6 | 0.000109 |
| Succinate dehydrogenase flavoprotein subunit | rha02921 | 28 | 11 | 23 | 0.17 | 0.47 | 3 | 0.001381 |
| Succinate-CoA ligase alpha subunit | rha08148 | 29 | 9 | 38 | 0.017 | 0.16 | 9 | 0.000713 |
Spot numbers correspond to those in Fig. 3.
Spot signal intensities were normalized and averaged over three replicate gels (each from a different culture).
Difference was calculated as a ratio of averaged normalized signal intensities. For spots that were not detected under one of the conditions, a value of 0.002 was used as the normalized signal intensity (see Materials and Methods). Standard errors are 15%.
ND, not detected.
NA, not applicable, because the protein spot was not detectable in the pyruvate control proteome.
Shares 42% sequence identity with PaaC.
In addition to the proteins encoded by the paa gene cluster, others encoded by genes elsewhere on the chromosome were associated with growth on PAA. These included the PaaC paralog encoded by rha07533, which was not detected in pyruvate-grown cells (Table 2). The paaC-encoded protein was more abundant than its paralog. Other proteins that were more abundant in PAA-grown cells included three enzymes involved in the tricarboxylic acid (TCA) cycle: malate dehydrogenase, succinate dehydrogenase, and succinate-CoA ligase. This observation is consistent with the proposed pathway for phenylacetate degradation, the last step of which generates two intermediates of the TCA cycle: acetyl-CoA and succinyl-CoA. We have previously observed (16) that in RHA1 the glyoxylate shunt is upregulated on pyruvate, which supplies only acetyl-CoA to the TCA cycle. This observation is consistent with the relatively higher expression of TCA cycle enzymes on PAA than on pyruvate.
Analysis of knockout mutants.
Three genes were selected as targets for knockout mutagenesis in order to confirm their predicted roles in PAA degradation (Fig. 1). Disruption of paaN (RHA1_001) completely prevented growth on PAA in liquid medium but had no significant effect on growth on pyruvate or in rich medium (LB). In contrast, the paaF mutant (RHA1_002) and the paaR mutant (RHA1_003) were able to grow on PAA. However, RHA1_002 grew with a longer lag phase than did the wild type (WT) or RHA1_003 when inoculated from cultures grown in a different substrate (e.g., LB medium). The doubling times on PAA of both RHA1_002 (5.7 ± 0.1 h) and RHA1_003 (5.9 ± 0.1 h) were slightly greater than that of the WT (5.3 ± 0.2 h).
The RHA1 WT and mutants were tested for growth on various additional aromatic substrates to determine which are degraded via the PAA pathway. The RHA1 WT grew on 14 aromatic substrates tested (Table 3) but failed to grow on 2-hydroxyphenylacetate (0.1 to 100 mM) or tropic acid (6 to 30 mM) on either agar plates or liquid medium. The paaN mutant was unable to grow on six compounds in addition to PAA. Of those six compounds, the paaF mutant was unable to grow on one and impaired in growth on two. The paaR mutant was able to grow on the same 14 compounds as the WT at similar rates and to similar final optical densities.
TABLE 3.
Growth of Rhodococcus sp. strain RHA1 and three mutant strains on different substratesa
| Growth substrate (concn)b | Growth ofc:
|
|||
|---|---|---|---|---|
| WT RHA1 | RHA1_001 (paaN) | RHA1_002 (paaF) | RHA1_003 (paaR) | |
| PAA (15 mM) | + | − | + | + |
| Styrene (v) | + | + | + | + |
| Ethylbenzene (v) | + | + | + | + |
| Benzene (v) | + | + | + | + |
| Phenylacetaldehyde (v) | + | − | (+) | + |
| Biphenyl (v) | + | + | + | + |
| Mandelate (130 mM) | + | + | + | + |
| Benzoate (20 mM) | + | + | + | + |
| Phthalate (20 mM) | + | + | + | + |
| 3-Hydroxyphenylacetate (20 mM) | + | + | + | + |
| 4-Phenylbutyrate (30 mM) | + | − | + | + |
| Phenylpyruvate (20 mM) | + | − | + | + |
| l-Phenylalanine (12 mM) | + | − | + | + |
| 2-Phenylethylamine (3 mM) | + | − | − | + |
| 2-Phenylethanol (6.2 mM) | + | − | (+) | + |
All tests were repeated three times.
v, volatile compounds added as vapors, according to Materials and Methods;
+, growth; −, no growth; (+), poor growth (only isolated colonies grew from streak).
Analysis of metabolites.
When the paaN mutant (RHA1_001), which is unable to grow on PAA as a sole organic substrate, was grown on 20 mM pyruvate plus 0.15 mM PAA, a yellow color was observed in the medium after 2 days. The yellow color was not observed if RHA1_001 was grown without PAA or if the WT was grown on pyruvate plus PAA. The medium of RHA1_001 grown on pyruvate plus PAA was found to contain tropone (m/z = 106, 78, 51), 2-coumaranone (m/z = 134, 106, 78, 51), and, after derivatization, the methyl ester of 2-methoxyphenylacetate (m/z = 180, 121, 91), in addition to a large amount of undegraded PAA. The medium of the WT had only traces of 2-coumaranone and tropone and had no remaining PAA. Both tropone and 2-coumaranone are yellow and likely contribute to the color observed. After the paaF mutant (RHA1_002) was grown on pyruvate with PAA for 2 days, the medium contained undegraded PAA but no other metabolites, confirming a partial defect of this strain in PAA degradation.
DISCUSSION
This study provided conclusive evidence that the paa genes of RHA1 encode a functional PAA degradation pathway. First, most of the paa genes were transcribed in PAA-grown cells (Fig. 2). Second, eight of the predicted Paa proteins were expressed in PAA-grown cells while being nondetectable in pyruvate-grown controls (Fig. 3; Table 2). Third, the paaN gene of RHA1 was essential for growth on PAA, as was previously reported for two gram-negative bacteria (8, 17). Fourth, the paaN mutant produced metabolites consistent with the predicted role of PaaN in the PAA pathway (Fig. 1). In agreement with our results, a paaN mutant of Azoarcus evansii was previously reported to accumulate tropone (17), and paaN mutants of E. coli were previously reported to accumulate 2-hydroxyphenylacetate (5, 8). While we did not detect the latter metabolite, we hypothesize that the 2-coumaranone we detected was formed from 2-hydroxyphenylacetate and that the methyl ester of 2-methoxyphenylacetate we detected was formed by derivatization of 2-coumaranone. Thus, the metabolites found in this study are all potential derivatives of the expected substrate for PaaN. The phenotype of the paaN mutant indicates that RHA1 does not have an alternative pathway for PAA catabolism or an alternative mechanism for transformation of phenylacetyl-CoA.
Although the paaF mutant of RHA1 was able to grow on PAA, its phenotype is consistent with involvement of PaaF in the PAA pathway. Unlike WT RHA1, the paaF mutant failed to completely remove PAA from the medium while growing on pyruvate. It appears that RHA1 possesses one or more CoA ligases that can partially complement PaaF. This possibility is supported by the existence of at least nine paralogs of paaF in the RHA1 genome, as well as over 50 other genes whose putative products show some sequence identity to known CoA ligases. The phenotype of the paaF mutant (Table 3) suggests that CoA ligases other than PaaF cannot transform 2-phenylethylamine and are limited in the transformation of phenylacetaldehyde and 2-phenylethanol. In contrast to RHA1, P. putida U (15) and E. coli (8) both require PaaF for growth on phenylacetate.
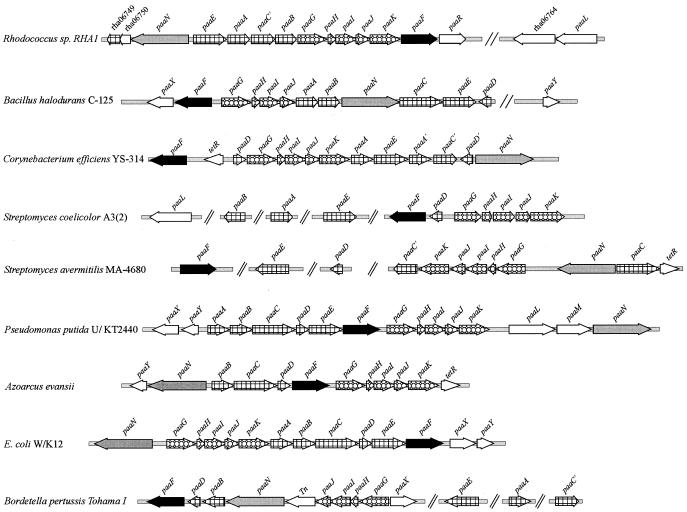
The organization of the paa genes differs among different organisms, but some features are common to all described paa clusters (Fig. 4). Genes encoding two core functional units of the pathway that are consistently clustered include paaGHIJK (encoding a ring-hydroxylating system) and, usually, paaABCE (encoding a β-oxidation system). Other genes commonly occurring in paa gene clusters include paaN (encoding a putative ring-opening enzyme) and paaF (encoding an acetyl-CoA ligase). Apart from these genes, some paa gene clusters also contain genes encoding a transport system (e.g., paaLM) and genes encoding a regulatory system (e.g., paaXY) (5, 12, 15). Most of the components described above were found in a single gene cluster in Rhodococcus sp. strain RHA1. This clustering is in contrast to the gene organization of at least two other actinomycetes, Streptomyces avermitilis and Streptomyces coelicolor, in which there are homologs of several paa genes but in which only those encoding the ring-hydroxylating system, paaGHIJK, are clustered.
FIG. 4.
Comparison of paa gene organizations in Rhodococcus sp. strain RHA1 and other bacteria. The gene names are in accordance with those listed in reference 12. Tn, transposase; TetR, TetR family regulator; paaR, an AraC-type regulator. Variants of paa genes are indicated with a prime (e.g., C′, A′, and D′). All sequences, except that of RHA1, were obtained from The Institute for Genomic Research (http://www.tigr.org/tdb) or NCBI (http://www.ncbi.nlm.nih.gov).
The paaC gene of RHA1 putatively encodes a 284-amino acid (aa) dehydrogenase similar to those in S. avermitilis, Corynebacterium efficiens, and Bordetella pertussis (Fig. 4). Homologs of these PaaC proteins are found in other gram-positive and gram-negative bacteria but are distinct in that they encode larger proteins (ca. 500 aa). A paralog of PaaC, encoded by rha07533, was detected only during growth on PAA (spot 9 in Fig. 3 and Table 2).
Associated with the paa gene cluster of RHA1 but independently transcribed is paaL (Fig. 2), which encodes a putative permease having ca. 40% aa identity with PAA permeases in Pseudomonas putida (AAC24338), Pseudomonas sp. strain Y2 (CAE45115, CAE45115, and CAD76939), and Chromobacterium violaceum (AAQ58580). For comparative purposes, it is difficult to identify phenylacetate transport systems in relatives of RHA1, such as S. avermitilis and S. coelicolor, because they lack clustered paa genes. In RHA1, there is no paaM gene adjacent to paaL or elsewhere, but this is not surprising since a porin encoded by paaM is presumably unnecessary in a gram-positive organism, which lacks an outer cell membrane.
In some genomes (E. coli, Klebsiella pneumoniae, and Bacillus halodurans), a paa regulator gene is downstream of paaF (12). Downstream of paaF in RHA1 is paaR (Fig. 4), whose product has the highest similarity to an AraC-type transcriptional regulator (Table 1) (10). The product of paaR may serve the function of the regulatory system encoded by paaXY in some gram-negative bacteria. Other actinomycetes, such as S. coelicolor, S. avermitilis, and Corynebacterium efficiens, also lack homologs of paaXY associated with paa genes, but the last two have genes encoding TetR-type regulators, which are negative regulators associated with paa genes. Our results (Table 3; Fig. 2 and 3) neither confirm nor contradict the possibility of a role for PaaR in regulating the paa genes. If PaaR does regulate paa gene expression, the phenotype of the paaR mutant suggests that it is likely a negative regulator.
It was recently suggested that the PAA degradation pathway is widely distributed and that paa genes may have horizontally transferred across bacterial phyla (1). The average GC content of the RHA1 paa gene cluster is the same as that of the RHA1 genome, and there are no transposase genes or other evidence of horizontal gene transfer in regions flanking the paa gene cluster of RHA1. Thus, despite the similarity of the Paa proteins to those in organisms with lower GC content, there is no evidence that the paa genes were recently obtained by RHA1 from another organism.
Seven substrates, including PAA, were determined to be part of the phenylacetyl-CoA catabolon of RHA1. These seven substrates did not support growth of the paaN mutant (Table 3). Predictably, six substrates that were not expected to be degraded via the PAA pathway—pyruvate, benzene, biphenyl, mandelate, benzoate, and phthalate—supported very similar growth levels of RHA1 and the mutant strains. However, the paaN mutant also grew on styrene, ethylbenzene, and 3-hydroxyphenylacetate, despite the fact that these compounds are degraded via the PAA pathway in certain gram-negative bacteria (12, 17). It is likely that RHA1 degrades ethylbenzene via a pathway also used for biphenyl degradation (unpublished data). RHA1 might degrade styrene via 3-vinylcatechol, as has been proposed for Rhodococcus rhodochrous NCIMB 13259 (14, 21). Our results suggest that the phenylacetyl-CoA catabolon in RHA1 may be more limited than those of other organisms. However, the evidence does not exclude the possibility that the PAA pathway is partially responsible for the degradation of styrene, ethylbenzene, and 3-hydroxyphenylacetate (i.e., that different pathways might act simultaneously).
Acknowledgments
We thank Manisha Dosanjh for developing the mutagenesis method and Christine Florizone for assistance with proteomic analyses.
This work was supported by a Genome Canada/Genome BC grant and by the Universidad Complutense Madrid (UCM) project PR1/03-11648. J.M.N.-L. was a recipient of a UCM-Flores Valles scholarship.
REFERENCES
- 1.Abe-Yoshizumi, R., U. Kamei, A. Yamada, M. Kimura, and S. Ichihara. 2004. The evolution of the phenylacetic acid degradation pathway in bacteria. Biosci. Biotechnol. Biochem. 68:746-748. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez, H. M., H. Luftmann, R. A. Silva, A. C. Cesari, A. Viale, M. Wältermann, and A. Steinbüchel. 2002. Identification of phenyldecanoic acid as a constituent of triacylglycerols and wax ester produced by Rhodococcus opacus PD630. Microbiology 148:1407-1412. [DOI] [PubMed] [Google Scholar]
- 3.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denef, V. J., J. Park, T. V. Tsoi, J.-M. Rouillard, H. Zhang, J. A. Wibbenmeyer, W. Verstraete, E. Gulari, S. A. Hashsham, and J. M. Tiedje. 2004. Biphenyl and benzoate metabolism in a genomic context: outlining genome-wide metabolic networks in Burkholderia xenovorans LB400. Appl. Environ. Microbiol. 70:4961-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrández, A., B. Miñambres, B. García, E. R. Olivera, J. M. Luengo, J. L. García, and E. Díaz. 1998. Catabolism of phenylacetic acid in Escherichia coli: characterization of a new aerobic hybrid pathway. J. Biol. Chem. 273:25974-25986. [DOI] [PubMed] [Google Scholar]
- 6.Flamm, R. K., D. J. Hinrichs, and M. F. Thomashow. 1984. Introduction of pAMβ1 into Listeria monocytogenes by conjugation and homology between native L. monocytogenes plasmids. Infect. Immun. 44:157-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ismail, W., M. El-Said Mohamed, B. L. Wanner, K. A. Datsenko, W. Eisenreich, F. Rohdich, A. Bacher, and G. Fuchs. 2003. Functional genomics by NMR spectroscopy: phenylacetate catabolism in Escherichia coli. Eur. J. Biochem. 270:3047-3054. [DOI] [PubMed] [Google Scholar]
- 9.Kitagawa, W., A. Suzuki, T. Hoaki, E. Masai, and M. Fukuda. 2001. Multiplicity of aromatic ring hydroxylation dioxygenase genes in a strong PCB degrader, Rhodococcus sp. strain RHA1 demonstrated by denaturing gradient gel electrophoresis. Biosci. Biotechnol. Biochem. 65:1907-1911. [DOI] [PubMed] [Google Scholar]
- 10.Komeda, H., Y. Hori, M. Kobayashi, and S. Shimizu. 1996. Transcriptional regulation of the Rhodococcus rhodochrous J1 nitA gene encoding a nitrilase. Proc. Natl. Acad. Sci. USA 93:10572-10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulakov, L. A., and M. J. Larkin. 2002. Genetic organization of Rhodococcus, p. 15-46. In A. Danchin (ed.), Genomics of GC-rich gram-positive bacteria. Caister Academic Press, Wymondham, United Kingdom.
- 12.Luengo, J. M., J. L. Garcia, and E. R. Olivera. 2001. The phenylacetyl-CoA catabolon: a complex catabolic unit with broad biotechnological applications. Mol. Microbiol. 39:1434-1442. [DOI] [PubMed] [Google Scholar]
- 13.Masai, E., A. Yamada, J. M. Healy, T. Hatta, K. Kimbara, M. Fukuda, and K. Yano. 1995. Characterization of biphenyl catabolic genes of gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Leary, N. D., K. E. O'Connor, and A. D. Dobson. 2002. Biochemistry, genetics and physiology of microbial styrene degradation. FEMS Microbiol. Rev. 26:403-417. [DOI] [PubMed] [Google Scholar]
- 15.Olivera, E. R., B. Miñambres, B. Garcia, C. Muñiz, M. A. Moreno, A. Ferrández, E. Díaz, J. L. García, and J. M. Luengo. 1998. Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: the phenylacetyl-CoA catabolon. Proc. Natl. Acad. Sci. USA 95:6419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patrauchan, M. A., C. Florizone, M. Dosanjh, W. W. Mohn, J. Davies, and L. D. Eltis. 2005. Catabolism of benzoate and phthalate in Rhodococcus strain RHA1: redundancies and convergence. J. Bacteriol. 187:4050-4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rost, R., S. Haas, E. Hammer, H. Herrmann, and G. Burchhardt. 2002. Molecular analysis of aerobic phenylacetate degradation in Azoarcus evansii. Mol. Genet. Genomics 267:656-663. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Seto, M., E. Masai, M. Ida, T. Hatta, K. Kimbara, M. Fukuda, and K. Yano. 1995. Multiple polychlorinated biphenyl transformation systems in the gram-positive bacterium Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:4510-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Geize, R., and L. Dijkhuizen. 2004. Harnessing the catabolic diversity of rhodococci for environmental and biotechnological applications. Curr. Opin. Microbiol. 7:255-261. [DOI] [PubMed] [Google Scholar]
- 21.Warhurst, A. M., K. F. Clarke, R. A. Hill, R. A. Holt, and C. A. Fewson. 1994. Metabolism of styrene by Rhodococcus rhodochrous NCIMB 13259. Appl. Environ. Microbiol. 60:1137-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warren, R., W. W. L. Hsiao, H. Kudo, M. Myhre, M. Dosanjh, A. Petrescu, H. Kobayashi, S. Shimizu, K. Miyauchi, E. Masai, G. Yang, J. M. Stott, J. E. Schein, H. Shin, J. Khattra, D. Smailus, Y. S. Butterfield, A. Siddiqui, R. Holt, M. A. Marra, S. J. M. Jones, W. W. Mohn, F. S. L. Brinkman, M. Fukuda, J. Davies, and L. D. Eltis. 2004. Functional characterization of a catabolic plasmid from polychlorinated-biphenyl-degrading Rhodococcus sp. strain RHA1. J. Bacteriol. 186:7783-7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zaar, A., J. Gescher, W. Eisenreich, A. Bacher, and G. Fuchs. 2004. New enzymes involved in aerobic benzoate metabolism in Azoarcus evansii. Mol. Microbiol. 54:223-238. [DOI] [PubMed] [Google Scholar]