Abstract
The polysaccharide alginate forms a protective capsule for Pseudomonas aeruginosa during chronic pulmonary infections. The structure of alginate, a linear polymer of β1-4-linked O-acetylated d-mannuronate (M) and l-guluronate (G), is important for its activity as a virulence factor. Alginate structure is mediated by AlgG, a periplasmic C-5 mannuronan epimerase. AlgG also plays a role in protecting alginate from degradation by the periplasmic alginate lyase AlgL. Here, we show that the C-terminal region of AlgG contains a right-handed β-helix (RHβH) fold, characteristic of proteins with the carbohydrate-binding and sugar hydrolase (CASH) domain. When modeled based on pectate lyase C of Erwinia chrysanthemi, the RHβH of AlgG has a long shallow groove that may accommodate alginate, similar to protein/polysaccharide interactions of other CASH domain proteins. The shallow groove contains a 324-DPHD motif that is conserved among AlgG and the extracellular mannuronan epimerases of Azotobacter vinelandii. Point mutations in this motif disrupt mannuronan epimerase activity but have no effect on alginate secretion. The D324A mutation has a dominant negative phenotype, suggesting that the shallow groove in AlgG contains the catalytic face for epimerization. Other conserved motifs of the epimerases, 361-NNRSYEN and 381-NLVAYN, are predicted to lie on the opposite side of the RHβH from the catalytic center. Point mutations N362A, N367A, and V383A result in proteins that do not protect alginate from AlgL, suggesting that these mutant proteins are not properly folded or not inserted into the alginate biosynthetic scaffold. These motifs are likely involved in asparagine and hydrophobic stacking, required for structural integrity of RHβH proteins, rather than for mannuronan catalysis. The results suggest that the AlgG RHβH protects alginate from degradation by AlgL by channeling the alginate polymer through the proposed alginate biosynthetic scaffold while epimerizing approximately every second d-mannuronate residue to l-guluronate along the epimerase catalytic face.
Alginate is a viscous polysaccharide produced by brown seaweed and by certain bacteria, including Pseudomonas and Azotobacter species. Alginate is a high-molecular-weight linear copolymer composed of β-d-mannuronic acid (M) and its C5 epimer α-l-guluronic acid (G) linked by β1-4 glycosidic bonds. In bacterial but not in algal alginates, the M residues are modified by the addition of O-acetyl groups at the O-2 and/or O-3 position (70).
In the opportunistic pathogen Pseudomonas aeruginosa, alginate is an important virulence factor, particularly in patients with the genetic disorder cystic fibrosis, where conversion of strains to the alginate-overproducing (mucoid) phenotype often results in chronic pulmonary P. aeruginosa infections (48). Alginate acts as a virulence factor in these infections by contributing to the matrix material of the mucoid P. aeruginosa biofilms (45) and by protecting the bacteria from opsonic phagocytosis (55). Alginate also neutralizes oxygen radicals produced by inflammatory immune cells (38) and stimulates the production of inflammatory cytokines indicative of a Th2-type immune response (47, 49).
The structure of alginate contributes to its activity as a virulence factor. For example, the M/G block structure is important for the chemical and physical properties of the polymer, including its viscosity and its interaction with divalent cations (70, 71). The presence of O-acetyl substitutions on the O-2 and O-3 positions of the d-mannuronate residues (70) is essential for the formation of microcolonies and for the avoidance of antibody-independent and antibody-mediated opsonic phagocytosis (45, 55).
The block structures of alginate vary depending on the organism producing the polymer. Algal and Azotobacter vinelandii alginates contain continuous stretches of G residues (G-blocks), while alginates from pseudomonads contain primarily M residues randomly interspersed with G residues (21, 66, 70). The G-blocks of A. vinelandii alginate chelate calcium and give the polymer its gelling properties (19, 21, 34, 70, 71), important for this organism to build capsules of cysts (60). In P. aeruginosa, G-blocks are not found, most likely because the polymer-level epimerase of pseudomonads, AlgG, is not capable of introducing G residues adjacent to other G residues (66, 70). In A. vinelandii, G-blocks are introduced into the alginate polymer by a series of extracellular mannuronan epimerases, AlgE1 to AlgE7, that are encoded by tandem gene repeats (11, 76). Genes for these extracellular epimerases are not found in P. aeruginosa (75).
The bacterial alginate biosynthetic pathway occurs in four steps: (i) synthesis of the alginate precursor GDP-mannuronic acid, (ii) polymerization of GDP-mannuronic acid into β1-4-linked polymannuronate, (iii) modification of the polymannuronate into its final alginate structure, and (iv) polymer secretion. Precursor synthesis, carried out by AlgA, AlgC, and AlgD, has been well characterized, and the crystal structures of AlgD and AlgC have been solved (56, 72). Less is known about the polymerization step, which may be carried out by Alg8 and Alg44, putative inner membrane proteins (39). The periplasmic proteins AlgX and AlgK, whose functions have not yet been determined, may also play a role in polymerization (26, 43). Enzymes that localize primarily to the periplasm carry out modifications of polymannuronic acid to its structurally mature and functionally active form. These enzymes include AlgI, AlgJ, and AlgF, forming the alginate O-acetylation complex; AlgG, the periplasmic mannuronan epimerase; and AlgL, an alginate lyase (14-16, 43, 64). Polymer secretion is likely mediated by AlgE (6, 57).
Jain et al. (25) proposed a model for alginate biosynthesis in which the alginate biosynthetic proteins form a multicomponent scaffold for polymerization, modification, and export. Evidence supporting this model includes deletion mutation of algG, algK, and algX (25, 26, 59). Strains with these deletions secrete depolymerized alginate, suggesting that removal of any of these components from the biosynthetic scaffold results in access to and degradation of the alginate by the periplasmic alginate lyase AlgL. Further support for the scaffold model is the work of Gimmestad et al. (18), who demonstrated that expression of both a wild-type and epimerase-defective mutant form of AlgG in Pseudomonas fluorescens results in two distinct polymer types, a wild-type M-G alginate and a poly(M) alginate. Those results suggest that one epimerase is required for each alginate molecule synthesized, rather than multiple epimerases acting at random sites on the polymer. Therefore, alginate is likely shuttled across the periplasm through an alginate biosynthetic complex, with the mannuronan epimerase, AlgG, involved in both epimerization and the transfer process.
To further characterize AlgG, its interaction with polymannuronate, and its role in the alginate biosynthetic scaffold, we performed sequence analysis and structural modeling of AlgG as well as phenotypic characterization of AlgG mutant proteins. The results show that AlgG contains a repeating sequence in its C terminus that is predicted to fold into a right-handed β-helix (RHβH), similar to proteins with a carbohydrate-binding and sugar hydrolase (CASH) domain (7). The model of the AlgG RHβH contains a shallow groove that may accommodate the linear polysaccharide, similar to other CASH domain proteins. This groove contains amino acids that are necessary for mannuronan epimerization and transfer of alginate through the periplasm. The structure of AlgG suggests that it may play a protective role as part of the alginate biosynthetic scaffold against alginate degradation by AlgL while performing enzymatic modification of polymannuronic acid.
MATERIALS AND METHODS
Bacterial strains, plasmids, mutagenic oligonucleotides, and media.
The bacterial strains, plasmids, and mutagenic oligonucleotides are listed in Table 1. P. aeruginosa FRD1 and its derivatives were used in this study. P. aeruginosa FRD462 algG4 contains an AlgG S272N mutation (4, 25). This strain has little mannuronan epimerase activity and produces O-acetylated polymannuronic acid. P. aeruginosa FRD1200 ΔalgG::aacCI contains a nonpolar deletion of algG (25). This strain secretes depolymerized alginate, presumably due to disruption of the alginate biosynthetic scaffold and degradation of alginate by AlgL (25). However, it can be complemented in trans by algG, indicating the that the ΔalgG::aacCI insertion is nonpolar on expression of the downstream alg biosynthetic genes (25). P. aeruginosa and Escherichia coli were cultured on L broth (LB) (10 g tryptone, 5 g yeast extract, and 5 g NaCl per liter). Triparental matings were used to transfer plasmids from E. coli to P. aeruginosa using the helper plasmid pRK2013 (13) and Pseudomonas isolation agar (Difco) as the selective medium. Antibiotics, when used, were used at the following concentrations: carbenicillin, 300 μg/ml; ampicillin, 100 μg/ml; tetracycline, 20 μg/ml; and kanamycin, 60 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Mutagenic oligonucleotide | Source or reference |
|---|---|---|---|
| P. aeruginosa | |||
| FRD1 | Cystic fibrosis isolate; Alg+ | This laboratory | |
| FRD462 | algG4 S272N Alg+ | 4 | |
| FRD1200 | ΔalgG::aacCI | 25 | |
| Plasmids | |||
| pRK2013 | ColE1-Tra(RK2)+ Kmr | 13 | |
| pALTER-Ex1 | Phagemid, Tcr | Promega | |
| pMF54 | Ptrc expression vector pKK233-2 with oriVSForiT laclq Apr | 14 | |
| pMF55 | pMF54 with 2.2 kb algG NcoI-NcoI fragment | 14 | |
| pSJ209 | pMF54 with 1.7 kb algG4 S272N in XbaI-Xho sites | 25 | |
| pSAD1 | pALTER-Ex1 with 2.2 kb algG NcoI-NcoI fragment | This study | |
| pSAD47 | algG H326A | TACGGCATCGACCCGGCCGACCGTTCGCAC | |
| pSAD53 | algG D324A | ATCGTCTACGGCATCCGCCCCGCACGACCCGT | |
| pSAD56 | algG P325A | GTCTACGGCAATCGACGCGCACGACCGTTCGCAC | |
| pSAD68 | algG Y321F | CGCGACAACATCGTCGGCGGCATCGACCCG | |
| pSAD70 | algG D327A | TACGGCATCGACCCGCACGCCCGTTCGCACCGCCT | |
| pSAD84 | algG R316A | AAGGGCAACACCTACGCCGACAACATCGTCTAC | |
| pSAD85 | algG N361A | GACAGCTTCATCTTCGCCAACCGCAGCTACGAG | |
| pSAD86 | algG D317A | GGCAACACCTACCGCGCCAACATCGTCTACGGC | |
| pSAD88 | algG D336A | CGCCTGATCATCGCCGCCAACACCGTCCACGGG | |
| pSAD90 | algG N367A | AACCGCAGCTACGAGGCCAAGCTTTCCGGGATC | |
| pSAD92 | algG N362A | AGCTTCATCTTCAACGCCCGCAGCTACGAGAAC | |
| pSAD102 | algG V383A | AGCGAGGGCAACCTGGCCGCCTACAACGAGGTC | |
| pSAD105 | algG Y385F | GGCAACCTGGTGGCCTTCAACGAGGTCTATCGC | |
| pSAD107 | algG Y365F | TTCAACAACCGCAGCTTCGAGAACAAGCTTTCC | |
| pSAD110 | algG S364A | ATCTTCAACAACCGCGCCTACGAGAACAAGCTT | |
| pSAD137 | algG E366A | AACCGCAGCTACGCGAACAAGCTTTCC | |
| pSAD149 | algG with deletion of amino acids 37-137 | ||
| pSAD151 | algG with deletion of amino acids 37-161 |
Abbreviations for phenotypes: Alg+, alginate overproduction; Apr, ampicillin resistance; Kmr, kanamycin resistance; Tra+, transfer by conjugation. Plasmids pSAD47 to pSAD151 contain the gene denoted in the pMF54 vector.
DNA manipulations.
General cloning procedures were carried out as described (2). Site-directed mutations were produced by using the Altered Sites protocol (Promega). For these experiments, the NcoI fragment containing P. aeruginosa algG, described previously (14), was ligated into the pALTER Ex1 phagemid vector (Promega), producing plasmid pSAD1. Single-stranded DNA from pSAD1 was isolated and subjected to mutagenesis as described (Promega) using the oligonucleotides shown in Table 1. Following verification of point mutations by DNA sequencing, the NcoI fragments containing mutant algG genes were ligated into the P. aeruginosa Ptrc expression vector pMF54 (14). Overlap extension PCR was used to produce AlgG with deletions of amino acids 37 to 137 and amino acids 37 to 161 from the N-terminal α-helical region while retaining the native AlgG signal peptide. The primers used for the AlgG 37-137 deletion were 5′-CGACTGCACGGTGCACCAATGCTTC-3′ and 5′-GAAGATCGCCTGGGGCGCCGCCCACGCCTGGCCGTGCAG-3′ to amplify the upstream algG fragment and 5′-CAGGCGTGGGCGGCGCCCCAGGCGATCTTCATCGAAGGC-3′ and 5′-GGGCCATCTAGAGGCCGGCGGCC-3′ to amplify the downstream algG fragment.
The primers used for the AlgG 37 to 161 deletion were 5′-CGACTGCACGGTGCACCAATGCTTC-3′ and 5′-CTCGACCTGTTCCAGCGCCGCCCACGCCTGGCCGTGCAG-3′ for the upstream fragment and 5′-CAGGCGTGGGCGGCGCTGGAACAGGTCGAGCCGGGGGTG-3′ and 5′-TGGGCCATCTAGAGGCCGGCGGCC-3′ for the downstream fragment. The reverse primers introduced XbaI restriction sites into the 3′ end of the PCR products to facilitate ligation of the NcoI-XbaI fragments containing the mutant algG into pMF54. The resulting plasmids were labeled pSAD149 for the AlgG 37 to 137 deletion and pSAD151 for the AlgG 37 to 161 deletion. Plasmids containing algG point mutations and N-terminal deletion mutations were introduced into P. aeruginosa and tested for complementation of the epimerization defect of FRD462 and complementation of the secretion and epimerization defects of FRD1200.
Determination of mannuronan epimerase activity.
P. aeruginosa FRD1, FRD462, and FRD1200 with plasmids containing algG mutations were incubated for 24 h in 10 ml of LB supplemented with 300 μg/ml carbenicillin and 1.0 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Alginates were purified from the culture supernatants as described previously (14), using one precipitation with 2% (wt/vol) cetyl pyridinium chloride and one precipitation with isopropanol.
Alginate pellets were resuspended in 4 ml of saline. Alginate concentrations were assayed by a modification of the Knutson and Jeanes (35) protocols, using Macrocystis pyrifera alginate as a standard. Briefly, partially purified alginates, 20 μl, was mixed with 1.0 ml of borate-sulfuric acid reagent (10 mM H3BO3 in concentrated H2SO4) and 30 μl of carbazole reagent (0.1% in ethanol). The mixtures were heated to 50°C for 30 min, and the concentrations were determined spectrophotometrically at 535 nm.
Alginate epimerization was determined using the alginate lyase assay (4, 14). Alginate lyase of Klebsiella aerogenes cleaves l-guluronate at MG or GG blocks (3). Therefore, the relative number of G residues per gram of alginate can be determining by assaying the abundance of cleaved, unsaturated residues produced by alginate lyase. For this assay, 65 μg of alginate was diluted to 150 μl in deionized water. The alginates were deacetylated with 50 μl of 1 M NaOH for 15 min at 65°C and then neutralized to pH 7.2 with 50 μl of 1 M HCl and 100 μl of lyase buffer (50 mM Tris-HCl, pH 8.0, 10 mM MgCl2 · 6H2O, 5.0 mM CaCl2). Alginate lyase, 10 μl, was added, and the mixture was incubated for 1 h at 25°C. The following were then added to the reaction mix: periodic acid (250 μl of 0.2 M, incubation for 40 min at 25°C), sodium arsenite (100 μl of 2% in 0.5N HCl, incubation for 1 min at 25°C), and thiobarbituric acid (1 ml of 0.6% solution, pH 2.0, incubation for 30 min at 65°C). The samples were cooled for 1 h and centrifuged at 12,000 rpm for 5 min to remove the precipitate. G-residue abundances were measured spectrophotometrically at 535 nm. Results were normalized to one optical density unit at 535 nm/ng uronic acid and compared to FRD1 alginate as a percentage of wild-type activity. Student's t tests were performed for the AlgG mutants and wild-type alginate, with P < 0.05 considered significantly different.
Assays for mannuronan epimerization in vitro.
The in vitro assays for alginate epimerization were similar to those described previously (14). Briefly, 1-liter cultures of E. coli expressing wild-type and mutant forms of AlgG were incubated for 24 h in LB medium supplemented with ampicillin and IPTG. The cultures were centrifuged, and cell pellets were resuspended in 1 ml of lyase buffer. Cells were then subjected to freeze-thawing followed by sonication to lyse the cells. Cell debris was removed by centrifugation, and the supernatants containing AlgG (50 μl) were mixed with 100 μl of polymannuronic acid purified from the supernatants of P. aeruginosa FRD462 algG4. The mixtures were incubated for 2 to 18 h at 25°C. Introduction of l-guluronate residues into the mannuronan was determined by the G-specific alginate lyase assay as described above.
Immunoblot analysis.
Proteins from P. aeruginosa containing plasmids with algG mutations were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (36) using 12% resolving gels. Proteins were electroblotted onto nitrocellulose membrane and then incubated with AlgG antibodies (14). Horseradish peroxidase-conjugated anti-rabbit immunoglobulin G was used as the secondary antibody. Immunoblots were developed using a chemiluminescence protocol as described previously (2).
Preparation of alginate lyases.
The recombinant guluronate-specific alginate lyase of K. aerogenes was expressed in E. coli and prepared from culture supernatants as described previously (3, 14).
Computational analysis.
Initial searches using the SMART database (at http://smart.embl-heidelberg.de/) (65) revealed that AlgG contains the CASH domain (7). The amino acid sequence of AlgG was subsequently submitted to the 3D-PSSM server (at http://www.sbg.bio.ic.ac.uk/∼3dpssm/) (33). This server compares unknown sequences to proteins from the Protein Data Bank (PDB) whose crystal structures have been solved. Unknown proteins are scored on sequence similarity, secondary structural predictions using Psi-Pred (at http://bioinf.cs.ucl.ac.uk/psipred/) (32) and solvent accessibility. The best-fitting proteins are then used as models to thread the unknown protein and create a coordinate files. Support for the predicted right-handed β-helical structure of AlgG was also generated using FFAS (at http://ffas.ljcrf.edu/ffas-cgi/cgi/ffas.pl) and BETAWRAP (at http://betawrap.lcs.mit.edu/) (8).
Comparative modeling was performed by satisfying spatial restraints as implemented in MODELLER (http://salilab.org/modeller/modeller.html) (61). Since the alignment returned by 3D-PSSM had low target-template sequence identity (≈13%), it was necessary to edit the alignments to account for secondary structure predictions (32). Given that the evaluation of three-dimensional models are more reliable than alignment evaluation (40), all models were independently assessed and rebuilt by adjusting the initial alignment. The resulting alignment had ≈15% identity. Models were evaluated by Prosa II empirical mean-force potentials devised to detect problematic areas of structural models (69). The Z-score produced by Prosa II was used to estimate the probability (pG) on a 0 to 1 scale that the model is reliable (http://sanchezlab.org/servers/pg/) (62). High pG values mean that the model is likely to be based on a correct alignment with an appropriate template, although models with pG values very close to 1 may not be accurate in all of the spatial details when the models target-template identities are below 30% (40). The model building and evaluation procedures employed in this work are described in detail at http://www.homepage.montana.edu/∼mdlakic/modeling/methods.html.
The sequences of β-helical repeats were aligned using the secondary structural predictions of AlgG and secondary structures of the known RHβH proteins. Residues that stack in the RHβH fold were aligned. Alignments with the A modules of AlgE1 to AlgE7 C-5 epimerases of Azotobacter vinelandii, coat protein GP1 of Ectocarpus siliculosis virus, and AlgG homologs from P. fluorescens and A. vinelandii were performed using ClustalX multiple sequence alignment (23, 27).
RESULTS
AlgG contains amino acid repeats in its C terminus characteristic of carbohydrate-binding and sugar hydrolase domain proteins.
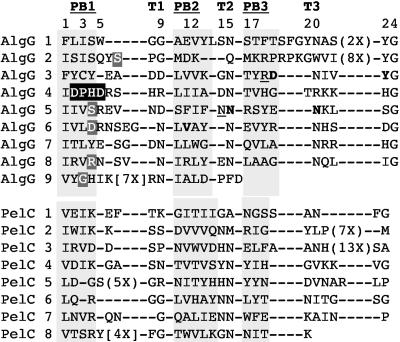
Jain et al. (25) described AlgG as having an α-helical-rich region at its N terminus and a β-rich region in its C terminus. The C terminus of AlgG has a tandem repeating sequence, approximately from amino acids F237 to D462, containing a 24-mer amino acid repeat (Fig. 1). The repeating sequence has the consensus IXZX+XSX+XZZZXNXXXXNXXXG, where Z represents an aliphatic residue, + represents a charged residue, and X represents any amino acid. AlgG contains up to nine of these 24-mer repeats, with six repeats easily identified and three repeats that are more difficult to align due to extended loops in the variable regions (described below). This repeating sequence is characteristic of proteins with carbohydrate-binding and sugar hydrolase (CASH) domains, often found in carbohydrate lyases such as polygalacturonases and pectate lyases (7, 52, 54, 82). This domain is also proposed to exist in surface layer proteins of Archaea and the extracellular mannuronan alginate epimerases of A. vinelandii (7). These proteins are classified in the pectin-lyase superfamily (44). The structures of several of these CASH domain proteins have been solved by X-ray crystallography and shown to form parallel right-handed β-helices (RHβHs).
FIG. 1.
AlgG contains tandem 24-mer peptide repeats in its C-terminal domain. Shown are six of the nine repeats from amino acids F300 to G441. Highlighted in grey are amino acids that are conserved in each position of the repeats. In the consensus sequence, Z represents aliphatic amino acids, + represents positively or negatively charged amino acids, and X represents any amino acid.
A sequence search against the SMART database (65) returned similarity to the CASH domain for the C-terminal region of AlgG (E = 4.1e−11). To determine if AlgG is similar in structure to other CASH domain proteins, we conducted predictive structural modeling by using 3D-PSSM. This program predicted that the C terminus of AlgG folds as a RHβH, with the most probable match to the pectate lyase C of Erwinia chrysanthemi (E = 4.0e−5). The next six proteins that matched AlgG were also RHβH proteins, with E-values ranging from 1.9e−3 to 1.8e−2. The FFAS web-based predictive program also matched AlgG to RHβH proteins, with the most significant matches to polygalacturonase from Erwinia carotovora and pectate lyase A of Aspergillus niger. The scores for these predictions were −9.88 and −9.58, respectively. Scores lower than −9.5 have less than a 3% chance of being false positives. The results from the program BETAWRAP also supported this finding (low probability, P = 2.0e−4, that AlgG similarity to RHβH is by chance).
The most distinctive characteristic of the RHβH fold is that it forms a spring-like structure with a shallow groove on one face that accommodates long-chain linear polysaccharides (54, 82). One rung of the spring is made of one repeating sequence, as in Fig. 1. The length of the helix depends on the number of rungs in the protein and can range from 7 to 11 repeats (7). The components of one rung of a RHβH are three β-sheets (PB1, PB2, and PB3) separated by three turn regions (T1, T2, and T3) (Fig. 2). PB1, PB2, and PB3 of one rung stack with their respective PB1, PB2, and PB3 of the next rung (28, 83). PB1 is associated with a shallow groove. Predictive secondary-structure modeling shows that the AlgG repeats contains these three β-sheets separated by three turn regions and that this secondary structure of the tandem repeats aligns to the structure of several of the crystallized RHβH proteins, including pectate lyase C of Erwinia chrysanthemi, as shown in Fig. 2.
FIG. 2.
Secondary-structure predictions of the tandem repeats of AlgG compared to tandem repeats of pectate lyase C of Erwinia chrysanthemi, whose crystal structure has been solved (81). Highlighted in grey are the β-sheet regions PB1, PB2, and PB3. The turn regions are shown as T1, T2, and T3. Amino acids essential for AlgG activity are highlighted (described below). Shown in white letters highlighted in black are amino acids that are essential for mannuronan epimerase catalytic activity. Shown in bold letters are amino acids that, when changed, result in proteins that are not able to complement P. aeruginosa FRD1200 algG::aacCI to the mucoid phenotype. Underlined are amino acids that, when changed, produce functional epimerases but are recessive to the chromosomally encoded AlgG S272N of P. aeruginosa FRD462 algG4. Shown in white letters highlighted in grey are amino acids at the equivalent positions shown by others to be required for mannuronan epimerization in P. aeruginosa and P. fluorescens (18, 25).
In RHβH proteins, the aliphatic residues of the β-sheets stack in the interior of the helix, creating a hydrophobic core (28). The alignment in Fig. 2 shows that AlgG contains aliphatic residues in the β-sheet regions at positions 1, 3, and 10 to 12 of each repeat. The second and fourth positions in each repeat contain charged and/or aromatic residues, also typical of RHβH PB1 β-sheets. These charged or aromatic residues on the PB1 grooved face often interact with the carbohydrate substrate (1, 37, 63, 68). Asparagine residues dominate the turn regions of AlgG at positions 9, 15, and 20. Asparagine stacking in these turn regions is predicted to be involved in protein structural integrity. Another feature of the RHβH fold is the extended loops observed between PB2 and PB3 or between PB1 and PB2 in some of the repeats. These extended loops are highly diverged among all RHβHs (7, 22). Extended loops likely occur in repeats 1, 2, and 9 of AlgG, making the tertiary structure of these regions difficult to model. In summary, even though the amino acid sequence identity between AlgG and many of the characterized RHβH proteins was generally not better than 15%, the similar features between the repeat sequences of AlgG and these proteins lend support for the C terminus of AlgG folding as a RHβH.
Structural modeling of AlgG.
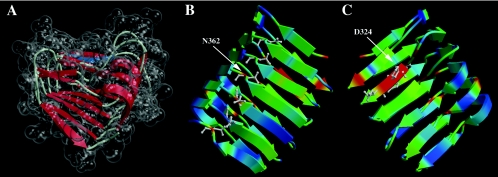
The alignment provided by 3D-PSSM was used as a starting point for protein structural modeling. Several iterations of model building were performed by manually adjusting the alignments and model evaluation (61, 69). The final model has good stereochemistry and can be considered reliable (pG = 0.99). Figure 3A shows the transparent surface of the space-filling model around the schematic secondary-structure elements and indicates the boundaries of the shallow groove of the RHβH (top side of the molecule in Fig. 3A). Two views in Fig. 3B and 3C are related by 180o rotation around the y axis. Shown using the ball and stick representation are the conserved asparagine residues on the opposite side of the shallow groove (Fig. 3B) and the 324-DPHD motif within the groove (Fig. 3C). The models in Fig. 3B and C are colored by sequence conservation (50) between AlgG homologs, with the most conserved amino acids shown in red and the least conserved shown in blue.
FIG. 3.
Three-dimensional model of AlgG from amino acids G286 to A434, showing six rungs of the right-handed β-helix. (A) The model of the RHβH is shown with the transparent space-filling representation encasing schematically drawn secondary-structure elements. This view positions the shallow groove associated with β-sheet 1 (PB1), the putative catalytic face of RHβH proteins, at the top of the molecule. Shown in blue is the 324-DPHD motif that is required for mannuronan epimerase activity. (B) View of AlgG showing turn region T2 and its associated asparagine stacking. Colors correspond to residue conservation among AlgG homologs, with red denoting the most conserved amino acid residues within the AlgG family and blue indicating the least conserved residues. Asparagine residues are shown in the ball-and-stick representation. (C) Rotation (180° around the y axis) view of AlgG showing β-sheet 1 (PB1) that contains the 324-DPHD motif, shown in the ball-and-stick representation. Color coding is based on amino acid conservation as in B.
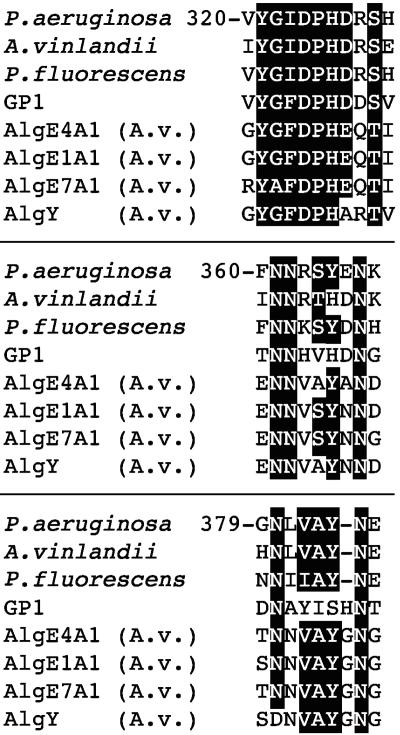
Conserved motifs of C-5 epimerases.
The periplasmic AlgG of P. aeruginosa has little primary sequence identity to the extracellular mannuronan epimerases of A. vinelandii. However, three motifs have been identified that show sequence similarity to these other epimerases (18, 77). These motifs are 324-DPHD, 361-NNRSYEN, and 381-NLVAYN (Fig. 4). Svanem et al. (77) showed that the AlgE7 amino acid corresponding to D324 of P. aeruginosa AlgG is catalytically important for epimerase activity. According to the repeat alignments and structural models, AlgG D324 of the 324-DPHD motif is located on PB1 in the center of the predicted substrate binding groove (shown in blue in Fig. 3A).
FIG. 4.
Conserved amino acid motifs among AlgG proteins, the extracellular epimerases of A. vinelandii (AlgE and AlgY), and the GP1 protein of the Ectocarpus siliculosis virus. White letters highlighted in black show the highly conserved amino acids of these three motifs.
Figure 3B shows the predicted positions of N362 of 361-NNRSYEN motif. This motif occur in T2 of repeat 5, N367 occurs in T3 of the same repeat, and 363-RSY is in PB3 of this repeat. N381 of the 381-NLVAYN motif resides in T1 of repeat 6. The 382-LVAY motif follows in PB2. Based on these structural predictions, the 324-DPHD motif is central to the epimerase active domain (Fig. 3C). The 361-NNRSY and 381-NLVAY motifs would be located on the opposite side of the predicted alginate-binding shallow groove, suggesting that these residues may be involved in asparagine and hydrophobic stacking. The AlgG model threaded to pectate lyase C shows asparagine stacking for the T2 region for amino acids N313, N337, N362, N386, N409, and N432 (Fig. 3B) Asparagine stacking is also predicted for the T3 region with N367, N391, N414, and N437 and for the T1 region for N381, N404, N427, and N455 (Fig. 2). Since the 361-NNRSY and 381-NLVAY motifs are located within these stacking regions, these motifs, although conserved among mannuronan epimerases, are likely important for the structural integrity of AlgG rather than for its catalytic activity.
Mutations that affect mannuronan epimerization and alginate secretion.
To investigate the importance of these conserved motifs in mannuronan epimerization and in alginate secretion, we performed site-directed mutagenesis on algG for key amino acids in domains predicted to be important for AlgG activity. For these experiments, AlgG point mutants were assayed for their ability to complement two algG mutant strains. In strain P. aeruginosa FRD462 algG4, the ability of the mutant proteins to complement the epimerization defect was determined, and in strain P. aeruginosa FRD1200 ΔalgG::aacCI, the ability of the mutant proteins to complement both the epimerization and the alginate secretion defects was determined. The mutations described here fall into several phenotypic classes, including mutations that cause a defect in mannuronan epimerization, mutations that fail to complement the defective alginate biosynthetic scaffold, and mutations that showed a dominant negative phenotype for epimerization.
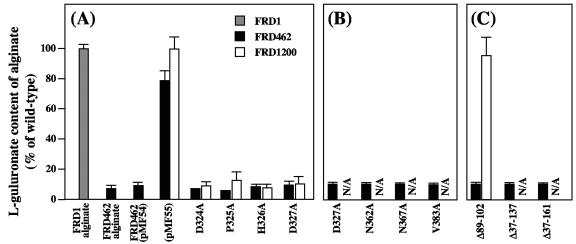
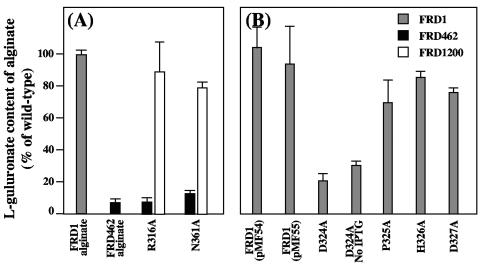
AlgG proteins with mutations in the 324-DPHD motif of the proposed epimerase catalytic domain, D324A, P325A, H326A, and D327A, were all functional in restoring the alginate secretion defect of FRD1200 (Fig. 5A), suggesting that these mutant proteins fold correctly and that they are properly inserted into the alginate biosynthetic scaffold. However, none of these mutant proteins was functional in restoring the epimerization defect of FRD1200. These proteins also failed to complement the epimerase defect of FRD462 (Fig. 5A). Immunoblots demonstrated that full-length AlgG proteins with these mutations were produced in FRD1200 (Fig. 6). Therefore, these mutations likely do not disrupt protein folding but are located in a portion of the protein necessary for mannuronan epimerization and likely the epimerase catalytic center, as predicted by the structural modeling.
FIG. 5.
l-Guluronate content of alginates produced by P. aeruginosa strains FRD1 (grey bars), FRD462 (black bars), and FRD1200 (white bars) expressing mutant forms of AlgG. l-Guluronate content is expressed as a percentage of wild-type FRD1 alginate. pMF54 is the vector control that lacks algG, and pMF55 is the vector control that expresses a wild-type copy of algG. (A) Mutant forms of AlgG where the mutations lie along the proposed epimerase catalytic face. These mutant proteins complement FRD1200 to the mucoid phenotype but do not have epimerase activity in either FRD1200 or FRD462. (B) AlgG mutant proteins that fail to complement epimerase activity in FRD462 and fail to complement FRD1200 to the mucoid phenotype. (C) AlgG mutant proteins containing deletions in the N-terminal region of the protein. Since some of the mutant proteins failed to complement FRD1200 to secrete alginate, the l-guluronate content of the alginate is not applicable (signified by N/A).
FIG. 6.
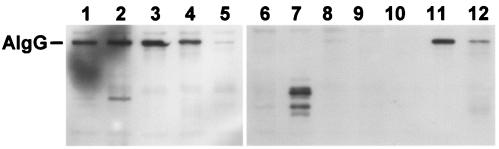
Immunoblot analysis of FRD1200 expressing mutant AlgG proteins. Lanes: (1) wild-type AlgG from plasmid pMF55; (2) P325A proposed catalytic domain mutation; (3) D327A proposed catalytic domain mutation; (4) R316; (5) N362 mutation, recessive to chromosomally encoded S272N of FRD462; (6) pMF54 plasmid lacking algG; (7) D317A; (8) Y321F; (9) N362A; (10) N367A mutation that likely disrupts proper protein folding; (11) V383A; (12) D336A mutation that has no effect on alginate epimerization or secretion.
Mutations that affect protein folding.
The model predicts asparagine stacking in T2 and in T3 as well as hydrophobic stacking in PB2 (Fig. 2 and 3). Mutations N362A in T2 and N367A in T3 of AlgG resulted in proteins that were unable to complement the alginate secretion defect of FRD1200 or the epimerase defect of FRD462 (Fig. 5B). AlgG with mutation V383A in PB2 was also unable to complement FRD1200 to the mucoid phenotype or complement the epimerization defect of FRD462. The results suggest that proteins with these mutations are either not folded correctly or are not properly inserted into the alginate biosynthetic scaffold. Other mutant AlgG proteins that failed to complement the alginate secretion defect of FRD1200 were D317A and Y321F. Figure 6 (lanes 7 to 10) shows the immunoblot of AlgG proteins with mutations predicted to disrupt protein folding. These results show little full-length protein and in some cases AlgG degradation products. Therefore, these proteins are likely produced (since they are expressed from the same vector as the other AlgG proteins) but targeted for degradation due to misfolding.
Mutations in the 361-NNRSYE-365 and 381-NLVAY-385 motifs that do not affect epimerase activity.
Since 361-NNRSYE and 381-NLVAY show sequence conservation in AlgG, the AlgE proteins of A. vinelandii, and the P. aeruginosa alginate lyase AlgL, we tested the ability of AlgG with mutations in other conserved amino acids to restore the mucoid phenotype in FRD1200 and the epimerase defect of FRD462. With the exception of N362A (described above) and V383, which is involved in hydrophobic stacking, none of the other mutants tested here affected mannuronan epimerization. AlgG with mutations S364A, Y365F, and E366A of the 361-NNRSYE motif and Y385F of the 381-NLVAY motif fully complemented FRD1200 to wild-type levels of both alginate production and epimerization and complemented FRD462 to wild-type levels of epimerization (data not shown). Figure 6 (lanes 11 to 12) shows examples of mutant proteins that did not affect mannuronan epimerization, demonstrating that full-length proteins were produced. Therefore, these mutant proteins act as functional epimerases. The results suggest that, although conserved, amino acids in these positions of AlgG do not play a functional role in mannuronan epimerization or in the alginate biosynthetic scaffold.
C terminus of AlgG is not sufficient for complementation of the secretion defect of FRD1200.
The structures obtained from 3D-PSSM did not model the first 120 amino acids of AlgG that follow the signal peptide cleavage site. This region is dominated by α-helices (25). The domain architecture for RHβH proteins has been suggested to exist in the glycosidic hydrolase families 55 and 87 (58) and in the surface layer protein B of Methanosarcina mazei (31). Two crystallized RHβH proteins, the tail spike protein of phage P22 (67) and dextranase from Penicillium minioluteum (37), have been shown to contain multiple domains in addition to the CASH domain.
The α-helical region predicted in the amino terminus of AlgG suggests that it is a separate domain from the RHβH. To test this, we constructed AlgG proteins with deletions in the N-terminal region. An AlgG deletion mutant missing amino acids 89 to 102 fully complemented the epimerase and secretion defects of algG deletion strain FRD1200, suggesting that this region of AlgG is not important for epimerase activity (Fig. 5C). AlgG proteins missing amino acids 37 to 137 and 37 to 161 were unable to complement FRD1200 to the mucoid phenotype (Fig. 5C). By using an in vitro assay with cell extracts of strains with AlgG missing amino acids 37 to 137 and 37 to 161, no epimerase activity was observed compared to the wild-type control. The results indicate that a portion of the N-terminal region is required for epimerase activity and/or for proper folding and insertion of AlgG into the alginate biosynthetic scaffold.
Dominant negative mutations.
Within the 361-NNRSY motif, N362 is predicted to be involved in the asparagine stacking of T2. However, it is possible that the adjacent asparagine, N361, stacks at T2. Therefore, we tested an AlgG N361A mutation for its ability to complement FRD1200 and FRD462. Polymer production and epimerase activity were restored in FRD1200, and full-length AlgG was seen in immunoblots (Fig. 6, lane 5), suggesting that this mutant protein folds correctly. Interestingly, epimerase activity, although restored in FRD1200, was not recovered in FRD462 (Fig. 7A). The results suggest that the chromosomally encoded AlgG S272N protein of FRD462 is dominant negative over the plasmid-encoded AlgG N361A mutant protein and therefore may outcompete the plasmid-encoded AlgG N361A for binding sites on mannuronan or within the proposed alginate scaffold. An AlgG R316A mutation was similar in phenotype and by immunoblot to AlgG N361A (Fig. 7A; Fig. 6, lane 4). The S272N mutation of FRD462 was also dominate negative over AlgG Δ89-102 (Fig. 5C).
FIG. 7.
l-Guluronate content of alginates produced by P. aeruginosa, showing dominant negative phenotype. (A) Point mutants of AlgG that produce a functional epimerase in FRD1200 (white bars) but do not complement epimerase activity in FRD462 (black bars). (B) FRD1 expressing mutant forms of AlgG in trans, demonstrating the dominant negative phenotype of AlgG D324A over wild-type AlgG.
AlgG proteins with mutations in the 324-DPHD motif and AlgG S272N of P. aeruginosa FRD462 algG4 give similar phenotypes in that all of the mutant proteins are able to maintain polymer formation yet cannot epimerize mannuronan. Therefore, these other mutations may also impart a dominant negative phenotype. We tested this hypothesis by expressing AlgG mutant proteins in the wild-type background, P. aeruginosa FRD1. The AlgG D324A mutant reduced alginate epimerization fivefold when expressed in FRD1 (Fig. 7B). Tests without induction by IPTG yielded a threefold reduction in epimerization, suggesting that even with low levels of expression, this mutant protein is dominant over wild-type chromosomally encoded AlgG. AlgG with other mutations in the 324-DPHD motif did not affect epimerization of FRD1 (Fig. 7B).
We also determined if AlgG S272N was dominant negative in the wild-type background. Interestingly, expression of AlgG S272N did not affect epimerization in the wild-type strain. The results suggest that the S272N mutation does not have an enhanced capacity for binding the alginate polymer or the alginate biosynthetic scaffold compared to the wild-type protein. However, AlgG S272N may outcompete AlgG N361A and AlgG R316A for binding sites on alginate or on the alginate biosynthetic scaffold, even though AlgG N361A and AlgG R316A are functional epimerases.
DISCUSSION
The C-terminal region of AlgG is predicted to fold as an RHβH. The RHβH is a common fold among proteins that bind and cleave long-chain linear polysaccharides. To date, the crystal structures of 19 RHβH proteins in the pectin lyase superfamily have been solved (44). This superfamily consists of pectin and pectate lyases (53, 79, 82) and the galacturonases (polygalacturonase and rhamnogalacturonase) (5, 12, 51, 54, 68, 78). Other carbohydrate-binding proteins in this family include the dextranase of Penicillium minioluteum (37), chondroitinase B of Flavobacterium heparinum (24), ι-carrageenase of Alteromonas sp. (42)., methylesterase of Erwinia chrysanthemi (29), the tail spike protein of phage P22 (73), and the P69 pertactin of Bordetella pertussis (10). This structure is found in proteins of all three domains of life, although as yet it has not been found in higher eukaryotes. Most of these proteins cleave polysaccharides that compose the extracellular matrix of plants and animals. The high charge densities of the nonmethylated component of plant pectins, polyglacturonates, and ι-carrageen is similar to the negatively charged alginate. Therefore, this protein fold appears to be a general carbohydrate-binding structure that accommodates high-molecular-weight linear polysaccharides and occurs in several glycosidic hydrolases and also, as shown here, in the P. aeruginosa AlgG epimerase.
Evidence presented here supports the structure of the AlgG C terminus as an RHβH. First, the scores obtained from hidden Markov model searches of the SMART database support this model. 3D-PSSM modeling, BETAWRAP analysis, and FFAS analysis indicate that AlgG has a high probability of folding as an RHβH. Second, the tandem repeats in AlgG are similar to those of other RHβH proteins. Third, each repeat of AlgG is similar in secondary structures to other RHβH proteins, including three β-sheets and three turn regions. Fourth, similar amino acids in each repeat align, including the aliphatic residues in the β-sheets, the asparagines in the turn regions, and the charged residues in PB1. Fifth, AlgG is biologically similar to other members of the RHβH family in that it associate with a linear, negatively charged polymer. Finally, model evaluation confirmed that AlgG is likely to have the RHβH fold. A recent study using hydrophobic cluster analysis showed that an epimerase from the marine brown alga Laminaria digitata has a secondary structure similar to that of P. aeruginosa AlgG (46). Our modeling studies also show a similar fold for the A-module of the extracellular alginate epimerases from A. vinelandii (data not shown). Therefore, this structure appears to be conserved among mannuronan epimerases.
Site-directed mutagenesis of AlgG 324-DPHD indicates that this motif is important for catalytic function and supports the results of Svanem et al. (77) that suggest that the amino acid equivalent of D324 (D152 in A. vinelandii AlgE7) is important for catalytic activity. In the RHβH model for AlgG, this motif lies in the center of the grooved face on PB1 (Fig. 3A). This position corresponds to the location of the catalytic centers for all the known RHβHs, where the reactive residues are either on PB1 or adjacent to it. An alternative explanation of the role of the DPHD motif is that these amino acids (as well as D375 and R422) are important in protein/polymer interactions.
Several RHβH proteins have been cocrystallized with their respective substrates (1, 9, 37, 63, 68). The results of those studies demonstrated that amino acids, often charged or aromatic, are located along the PB1 groove and are in contact with the substrate. It has also been demonstrated in the cellobiohydrolase I of Trichoderma reesei that at least seven sugar residues are in contact with the protein along its binding face, suggesting that many amino acids in the shallow groove are required for polysaccharide/protein interactions (9). Our docking studies indicate that the spacing between helical turns of the AlgG RHβH is such that each repeat could bind one modified sugar residue (data not shown). Therefore, as many as nine uronic acid residues may reside in the AlgG groove at one time. Interestingly, a summary of mutations that affect mannuronan epimerization shown here and those described for equivalent positions of P. fluorescens AlgG (18) all lie along the PB1 groove (shown in Fig. 2). Therefore, this entire groove is required for substrate interaction and/or epimerization.
An interesting observation in these studies is that AlgG with the D324A mutation is dominant negative over wild-type AlgG, suggesting that the mutant protein displaces the wild-type protein from the mannuronan precursor molecule or from the alginate biosynthetic scaffold. Since the mutant gene is plasmid encoded, it may have a higher abundance in the cell periplasm than the chromosomally encoded wild-type protein. If this is the case, the mutant protein may occupy a limited number of sites on the alginate biosynthetic scaffold, thereby displacing the wild-type protein from the complex. When expression of the mutant gene is not induced, we see a return of some epimerase activity, although not a complete restoration of wild-type levels of epimerization (Fig. 7B).
The results suggest that the increased abundance of the mutant AlgG may cause the observed dominant negative phenotype. However, the dominant negative effect is not fully explained by increased abundance, since a dominant negative phenotype was not observed for other mutants of the proposed catalytic domain (AlgG P325A, H326A, and D227A) (Fig. 7B). An alternative explanation is that AlgG D324A may bind to the alginate polymer with greater affinity than the wild-type protein and not allow access of the alginate polymer to the wild-type AlgG. This hypothesis could explain the lack of a dominant negative phenotype of the other catalytic domain mutants if they are defective in polymer binding. Further studies addressing the binding efficiency of these mutant proteins to the substrate will provide insight into the nature of this dominant negative mutation.
It has been proposed that the lyase reaction by β-elimination is mechanistically similar to alginate epimerization, since both reactions form a similar intermediate (17). The pectate and pectin lyases use β-elimination for degrading carbohydrate polymers (24, 41, 63, 84). In those reactions, a proton is abstracted from C-5 by a base and is donated to the glycosidic oxygen at the cleavage site by an acid. This results in the formation of an unsaturated C-4-C-5 bond (20). The base of the reaction is an arginine residue in pectin lyases and a calcium ion in pectate lyases. The acid is often the side chain of an aspartate residue (24, 30, 53, 63). The catalytic residues in AlgG, such as aspartates D324 and D327 and arginines on the catalytic face, would resemble those of the lyases. In the study by Svanem et al. (77), the aspartate corresponding to D324 of AlgG from the A. vinelandii AlgE7 was shown to be important for both epimerase and lyase activity, providing evidence for the similar mechanisms of these two reactions. Those results combined with the studies here indicate why the AlgG epimerase may be structurally similar to a class of proteins involved in polysaccharide cleavage.
The NNRSY motif and the NLVAY motif have been identified previously (18) and appear to be conserved among all of the C-5 epimerases, including the extracellular epimerases of A. vinelandii. Interestingly, a similar motif exists in AlgL and in the alginate lyase A1-III of Sphingomonas sp. (80). Structural analysis of AI-III demonstrated protein-carbohydrate interactions with N191 of this motif and H192 as the catalytic center (84). The results initially suggested that the NNRSY motif of AlgG may play a similar catalytic role, since the lyases and epimerases have similar reaction chemistries. However, the structure of alginate lyase from Sphingomonas sp. forms an α6/α5 barrel consisting of many α-helices and therefore is structurally distinct from AlgG.
Since N362 and N367 of AlgG lie on the opposite side of the RHβH of the proposed catalytic face, these amino acids are likely involved in asparagine stacking and not in epimerization. As such, mutant proteins AlgG N362A and AlgG N367A were not functional in complementing the secretion defect of FRD1200, suggesting that these mutations caused protein misfolding. In addition, the AlgG S364A and AlgG Y365F mutations of this conserved domain had no effect on AlgG activity in vivo, supporting the hypothesis that this motif is not the epimerase catalytic center.
Three different types of stacking side chains occur in the RHβH: hydrogen bond stacking by asparagine residues, hydrophobic stacking by aliphatic residues, and planar stacking by aromatic residues. Many RHβH structures have asparagine stacks (30). AlgG is unique in that asparagine stacking would occur at all three turn regions. Site-directed mutations of N362 and N367 disrupt alginate secretion and epimerization. Since these amino acids occur in T2 and T3, the mutations likely disrupt the structural stability of the protein. Aromatic stacking may have been disrupted with the Y321F mutation, which would stack with Y254, and Y298 (Fig. 2). The mutation V383A likely disrupts aliphatic stacking of PB2.
The predicted α-helices of the N terminus of AlgG suggest that this protein contains multiple domains, similar to several other glycosidic hydrolases. The dextranase of P. minioluteum (37), the tail spike protein of phage P22 (67, 74), and surface layer protein B of M. mazei (31) contain domains in addition to the RHβH. In these examples, the other protein domain is involved in protein-protein interactions. Our results indicate that the N-terminal portion of AlgG may be required for alginate polymerization and/or epimerization. Therefore, the N terminus of AlgG may play a role in protein/protein interactions necessary for the alginate biosynthetic scaffold. Future studies addressing the role of AlgG in the alginate biosynthetic scaffold are necessary to determine the functional role of the N-terminal domain of AlgG.
Acknowledgments
This work was supported by Public Health Service grant AI-46588 (M.J.F.) from the National Institute of Allergy and Infectious Diseases and in part by grant 1P20/RR-16455 from the National Center for Research Resources.
REFERENCES
- 1.Armand, S., M. J. Wagemaker, P. Sanchez-Torres, H. C. Kester, Y. van Santen, B. W. Dijkstra, J. Visser, and J. A. Benen. 2000. The active site topology of Aspergillus niger endopolygalacturonase II as studied by site-directed mutagenesis. J. Biol. Chem. 275:691-696. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1993. Current protocols in molecular biology, vol. 2. Greene Publishing Associates, Inc., New York, NY.
- 3.Caswell, R. C., P. Gacesa, K. E. Lutrell, and A. J. Weightman. 1989. Molecular cloning and heterologous expression of a Klebsiella pneumoniae gene encoding alginate lyase. Gene 75:127-134. [DOI] [PubMed] [Google Scholar]
- 4.Chitnis, C. E., and D. E. Ohman. 1990. Cloning of Pseudomonas aeruginosa algG, which controls alginate structure. J. Bacteriol. 172:2894-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho, S. W., S. Lee, and W. Shin. 2001. The X-ray structure of Aspergillus aculeatus polygalacturonase and a modeled structure of the polygalacturonase-octagalacturonate complex. J. Mol. Biol. 311:863-878. [DOI] [PubMed] [Google Scholar]
- 6.Chu, L., T. B. May, A. M. Chakrabarty, and T. K. Misra. 1991. Nucleotide sequence and expression of the algE gene involved in alginate biosynthesis by Pseudomonas aeruginosa. Gene 107:1-10. [DOI] [PubMed] [Google Scholar]
- 7.Ciccarelli, F. D., R. R. Copley, T. Doerks, R. B. Russell, and P. Bork. 2002. CASH-a beta-helix domain widespread among carbohydrate-binding proteins. Trends Biochem. Sci. 27:59-62. [DOI] [PubMed] [Google Scholar]
- 8.Cowen, L., P. Bradley, M. Menke, J. King, and B. Berger. 2002. Predicting the beta-helix fold from protein sequence data. J. Comput. Biol. 9:261-276. [DOI] [PubMed] [Google Scholar]
- 9.Divne, C., J. Stahlberg, T. T. Teeri, and T. A. Jones. 1998. High-resolution crystal structures reveal how a cellulose chain is bound in the 50 Å long tunnel of cellobiohydrolase I from Trichoderma reesei. J. Mol. Biol. 275:309-325. [DOI] [PubMed] [Google Scholar]
- 10.Emsley, P., I. G. Charles, N. F. Fairweather, and N. W. Isaacs. 1996. Structure of Bordetella pertussis virulence factor P.69 pertactin. Nature 381:90-92. [DOI] [PubMed] [Google Scholar]
- 11.Ertesvag, H., H. K. Hoidal, I. K. Hals, A. Rian, B. Doseth, and S. Valla. 1995. A family of modular type mannuronan C-5-epimerase genes controls alginate structure in Azotobacter vinelandii. Mol. Microbiol. 16:719-731. [DOI] [PubMed] [Google Scholar]
- 12.Federici, L., C. Caprari, B. Mattei, C. Savino, A. Di Matteo, G. De Lorenzo, F. Cervone, and D. Tsernoglou. 2001. Structural requirements of endopolygalacturonase for the interaction with PGIP (polygalacturonase-inhibiting protein). Proc. Natl. Acad. Sci. USA 98:13425-13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Figurski, D., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franklin, M. J., C. E. Chitnis, P. Gacesa, A. Sonesson, D. C. White, and D. E. Ohman. 1994. Pseudomonas aeruginosa AlgG is a polymer level alginate C-5-mannuronan epimerase. J. Bacteriol. 176:1821-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franklin, M. J., and D. E. Ohman. 1993. Identification of algF in the alginate biosynthetic gene cluster of Pseudomonas aeruginosa which is required for alginate acetylation. J. Bacteriol. 175:5057-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franklin, M. J., and D. E. Ohman. 2002. Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J. Bacteriol. 184:3000-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gacesa, P., R. Caswell, and P. Kille. 1989. Bacterial alginases. Antibiot. Chemother. 42:67-71. [DOI] [PubMed] [Google Scholar]
- 18.Gimmestad, M., H. Sletta, H. Ertesvag, K. Bakkevig, S. Jain, S. J. Suh, G. Skjak-Braek, T. E. Ellingsen, D. E. Ohman, and S. Valla. 2003. The Pseudomonas fluorescens AlgG protein but not its mannuronan C-5 epimerase activity is needed for alginate polymer formation. J. Bacteriol. 185:3515-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grant, G. T., E. R. Morris, D. A. Rees, P. J. C. Smith, and D. Thom. 1973. Biological interactions between polysaccharides and divalent cations: the egg-box model. FEBS Lett. 32:195-198. [Google Scholar]
- 20.Greiling, H., H. W. Stuhlsatz, T. Eberhard, and A. Eberhard. 1975. Studies on the mechanism of hyaluronate lyase action. Connect. Tissue Res. 3:135-139. [DOI] [PubMed] [Google Scholar]
- 21.Haug, A., B. Larsen, and O. Smidsrod. 1974. Uronic acid sequence in alginate from different sources. Carbohydr. Res. 32:217-223. [Google Scholar]
- 22.Henrissat, B., S. E. Heffron, M. D. Yoder, S. E. Lietzke, and F. Jurnak. 1995. Functional implications of structure-based sequence alignment of proteins in the extracellular pectate lyase superfamily. Plant Physiol. 107:963-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgins, D. G., and P. M. Sharp. 1988. CLUSTAL: a package fore performing multiple sequence alignment on a microcomputer. Gene 73:237-244. [DOI] [PubMed] [Google Scholar]
- 24.Huang, W., A. Matte, Y. Li, Y. S. Kim, R. J. Linhardt, H. Su, and M. Cygler. 1999. Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccharide product at 1.7 Å resolution. J. Mol. Biol. 294:1257-1269. [DOI] [PubMed] [Google Scholar]
- 25.Jain, S., M. J. Franklin, H. Ertesvag, S. Valla, and D. E. Ohman. 2003. The dual roles of AlgG in C-5-epimerization and secretion of alginate polymers in Pseudomonas aeruginosa. Mol. Microbiol. 47:1123-1133. [DOI] [PubMed] [Google Scholar]
- 26.Jain, S., and D. E. Ohman. 1998. Deletion of algK in mucoid Pseudomonas aeruginosa blocks alginate polymer formation and results in uronic acid secretion. J. Bacteriol. 180:634-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 28.Jenkins, J., O. Mayans, and R. Pickersgill. 1998. Structure and evolution of parallel beta-helix proteins. J. Struct. Biol. 122:236-246. [DOI] [PubMed] [Google Scholar]
- 29.Jenkins, J., O. Mayans, D. Smith, K. Worboys, and R. W. Pickersgill. 2001. Three-dimensional structure of Erwinia chrysanthemi pectin methylesterase reveals a novel esterase active site. J. Mol. Biol. 305:951-960. [DOI] [PubMed] [Google Scholar]
- 30.Jenkins, J., and R. Pickersgill. 2001. The architecture of parallel beta-helices and related folds. Prog. Biophys. Mol. Biol. 77:111-175. [DOI] [PubMed] [Google Scholar]
- 31.Jing, H., J. Takagi, J. H. Liu, S. Lindgren, R. G. Zhang, A. Joachimiak, J. H. Wang, and T. A. Springer. 2002. Archaeal surface layer proteins contain beta propeller, PKD, and beta helix domains and are related to metazoan cell surface proteins. Structure (Cambridge) 10:1453-1464. [DOI] [PubMed] [Google Scholar]
- 32.Jones, D. T. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195-202. [DOI] [PubMed] [Google Scholar]
- 33.Kelley, L. A., R. M. MacCallum, and M. J. Sternberg. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299:499-520. [DOI] [PubMed] [Google Scholar]
- 34.Kloareg, B., and R. S. Quatrano. 1988. Structure of the cell walls of marine algae and ecophysiological functions of the matrix polysaccharides. Oceanogr. Mar. Biol. Annu. Rev. 26:259-315. [Google Scholar]
- 35.Knutson, C. A., and A. Jeanes. 1968. A new modification of the carbazole analysis: application to heteropolysaccharides. Anal. Biochem. 24:470-481. [DOI] [PubMed] [Google Scholar]
- 36.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 37.Larsson, A. M., R. Andersson, J. Stahlberg, L. Kenne, and T. A. Jones. 2003. Dextranase from Penicillium minioluteum: reaction course, crystal structure, and product complex. Structure (Cambridge) 11:1111-1121. [DOI] [PubMed] [Google Scholar]
- 38.Learn, D. B., E. P. Brestel, and S. Seetharama. 1987. Hypochlorite scavenging by Pseudomonas aeruginosa alginate. Infect. Immun. 55:1813-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maharaj, R., T. B. May, S. K. Wang, and A. M. Chakrabarty. 1993. Sequence of the alg8 and alg44 genes involved in the synthesis of alginate by Pseudomonas aeruginosa. Gene 136:267-269. [DOI] [PubMed] [Google Scholar]
- 40.Marti-Renom, M. A., A. C. Stuart, A. Fiser, R. Sanchez, F. Melo, and A. Sali. 2000. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29:291-325. [DOI] [PubMed] [Google Scholar]
- 41.Mayans, O., M. Scott, I. Connerton, T. Gravesen, J. Benen, J. Visser, R. Pickersgill, and J. Jenkins. 1997. Two crystal structures of pectin lyase A from Aspergillus reveal a pH driven conformational change and striking divergence in the substrate-binding clefts of pectin and pectate lyases. Structure 5:677-689. [DOI] [PubMed] [Google Scholar]
- 42.Michel, G., L. Chantalat, E. Fanchon, B. Henrissat, B. Kloareg, and O. Dideberg. 2001. The iota-carrageenase of Alteromonas fortis. A beta-helix fold-containing enzyme for the degradation of a highly polyanionic polysaccharide. J. Biol. Chem. 276:40202-40209. [DOI] [PubMed] [Google Scholar]
- 43.Monday, S. R., and N. L. Schiller. 1996. Alginate synthesis in Pseudomonas aeruginosa: the role of AlgL (alginate lyase) and AlgX. J. Bacteriol. 178:625-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murzin, A. G., S. E. Brenner, T. Hubbard, and C. Chothia. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247:536-540. [DOI] [PubMed] [Google Scholar]
- 45.Nivens, D. E., D. E. Ohman, J. Williams, and M. J. Franklin. 2001. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J. Bacteriol. 183:1047-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nyvall, P., E. Corre, C. Boisset, T. Barbeyron, S. Rousvoal, D. Scornet, B. Kloareg, and C. Boyen. 2003. Characterization of mannuronan C-5-epimerase genes from the brown alga Laminaria digitata. Plant Physiol. 133:726-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pedersen, S. S. 1992. Lung infection with alginate-producing, mucoid Pseudomonas aeruginosa in cystic fibrosis. APMIS Suppl. 28:1-79. [PubMed] [Google Scholar]
- 48.Pedersen, S. S., N. Hoiby, F. Espersen, and C. Koch. 1992. Role of alginate in infection with mucoid Pseudomonas aeruginosa in cystic fibrosis. Thorax 47:6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pedersen, S. S., A. Kharazmi, F. Espersen, and N. Hoiby. 1990. Pseudomonas aeruginosa alginate in cystic fibrosis sputum and the inflammatory response. Infect. Immun. 58:3363-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pei, J., and N. V. Grishin. 2001. AL2CO: calculation of positional conservation in a protein sequence alignment. Bioinformatics 17:700-712. [DOI] [PubMed] [Google Scholar]
- 51.Petersen, T. N., S. Kauppinen, and S. Larsen. 1997. The crystal structure of rhamnogalacturonase A from Aspergillus aculeatus: a right-handed parallel beta helix. Structure 5:533-544. [DOI] [PubMed] [Google Scholar]
- 52.Pickersgill, R., G. Harris, L. Lo Leggio, O. Mayans, and J. Jenkins. 1998. Superfamilies: the 4/7 superfamily of beta alpha-barrel glycosidases and the right-handed parallel beta-helix superfamily. Biochem. Soc. Trans. 26:190-198. [DOI] [PubMed] [Google Scholar]
- 53.Pickersgill, R., J. Jenkins, G. Harris, W. Nasser, and J. Robert-Baudouy. 1994. The structure of Bacillus subtilis pectate lyase in complex with calcium. Nat. Struct. Biol. 1:717-723. [DOI] [PubMed] [Google Scholar]
- 54.Pickersgill, R., D. Smith, K. Worboys, and J. Jenkins. 1998. Crystal structure of polygalacturonase from Erwinia carotovora ssp. carotovora. J. Biol. Chem. 273:24660-24664. [DOI] [PubMed] [Google Scholar]
- 55.Pier, G. B., F. Coleman, M. Grout, M. Franklin, and D. E. Ohman. 2001. Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect. Immun. 69:1895-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Regni, C. A., P. A. Tipton, and L. J. Beamer. 2000. Crystallization and initial crystallographic analysis of phosphomannomutase/phosphoglucomutase from Pseudomonas aeruginosa. Acta Crystallogr. D Biol. Crystallogr. 56:761-762. [DOI] [PubMed] [Google Scholar]
- 57.Rehm, B. H., G. Boheim, J. Tommassen, and U. K. Winkler. 1994. Overexpression of algE in Escherichia coli: subcellular localization, purification, and ion channel properties. J. Bacteriol. 176:5639-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rigden, D. J., and O. L. Franco. 2002. Beta-helical catalytic domains in glycoside hydrolase families 49, 55 and 87: domain architecture, modelling and assignment of catalytic residues. FEBS Lett. 530:225-232. [DOI] [PubMed] [Google Scholar]
- 59.Robles-Price, A., T. Y. Wong, H. Sletta, S. Valla, and N. L. Schiller. 2004. AlgX is a periplasmic protein required for alginate biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 186:7369-7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sadoff, H. L. 1975. Encystment and germination in Azotobacter vinelandii. Bacteriol. Rev. 39:516-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 62.Sanchez, R., and A. Sali. 1998. Large-scale protein structure modeling of the Saccharomyces cerevisiae genome. Proc. Natl. Acad. Sci. USA 95:13597-13602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scavetta, R. D., S. R. Herron, A. T. Hotchkiss, N. Kita, N. T. Keen, J. A. Benen, H. C. Kester, J. Visser, and F. Jurnak. 1999. Structure of a plant cell wall fragment complexed to pectate lyase C. Plant Cell. 11:1081-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schiller, N. L., S. R. Monday, C. M. Boyd, N. T. Keen, and D. E. Ohman. 1993. Characterization of the Pseudomonas aeruginosa alginate lyase gene (algL): cloning, sequencing, and expression in Escherichia coli. J. Bacteriol. 175:4780-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schurks, N., J. Wingender, H. C. Flemming, and C. Mayer. 2002. Monomer composition and sequence of alginates from Pseudomonas aeruginosa. Int. J. Biol. Macromol. 30:105-111. [DOI] [PubMed] [Google Scholar]
- 67.Seckler, R. 1998. Folding and function of repetitive structure in the homotrimeric phage P22 tailspike protein. J. Struct. Biol. 122:216-222. [DOI] [PubMed] [Google Scholar]
- 68.Shimizu, T., T. Nakatsu, K. Miyairi, T. Okuno, and H. Kato. 2002. Active-site architecture of endopolygalacturonase I from Stereum purpureum revealed by crystal structures in native and ligand-bound forms at atomic resolution. Biochemistry 41:6651-6659. [DOI] [PubMed] [Google Scholar]
- 69.Sippl, M. J. 1993. Recognition of errors in three-dimensional structures of proteins. Proteins 17:355-362. [DOI] [PubMed] [Google Scholar]
- 70.Skjåk-Bræk, G., H. Grasdalen, and B. Larsen. 1986. Monomer sequence and acetylation pattern in some bacterial alginates. Carbohydr. Res. 154:239-250. [DOI] [PubMed] [Google Scholar]
- 71.Smidsrod, O., and K. I. Draget. 1996. Chemistry and physical properties of alginates. Carbohydr. Eur. 14:6-13. [Google Scholar]
- 72.Snook, C. F., P. A. Tipton, and L. J. Beamer. 2003. Crystal structure of GDP-mannose dehydrogenase: a key enzyme of alginate biosynthesis in P. aeruginosa. Biochemistry 42:4658-4668. [DOI] [PubMed] [Google Scholar]
- 73.Steinbacher, S., U. Baxa, S. Miller, A. Weintraub, R. Seckler, and R. Huber. 1996. Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors. Proc. Natl. Acad. Sci. USA 93:10584-10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Steinbacher, S., S. Miller, U. Baxa, N. Budisa, A. Weintraub, R. Seckler, and R. Huber. 1997. Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 Å, fully refined structure of the endorhamnosidase at 1.56 Å resolution, and the molecular basis of O-antigen recognition and cleavage. J. Mol. Biol. 267:865-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 76.Svanem, B. I., G. Skjak-Braek, H. Ertesvag, and S. Valla. 1999. Cloning and expression of three new Azotobacter vinelandii genes closely related to a previously described gene family encoding mannuronan C-5 epimerases. J. Bacteriol. 181:68-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Svanem, B. I., W. I. Strand, H. Ertesvag, G. Skjak-Braek, M. Hartmann, T. Barbeyron, and S. Valla. 2001. The catalytic activities of the bifunctional Azotobacter vinelandii mannuronan C-5-epimerase and alginate lyase AlgE7 probably originate from the same active site in the enzyme. J. Biol. Chem. 276:31542-31550. [DOI] [PubMed] [Google Scholar]
- 78.van Santen, Y., J. A. Benen, K. H. Schroter, K. H. Kalk, S. Armand, J. Visser, and B. W. Dijkstra. 1999. 1.68-Å crystal structure of endopolygalacturonase II from Aspergillus niger and identification of active site residues by site-directed mutagenesis. J. Biol. Chem. 274:30474-30480. [DOI] [PubMed] [Google Scholar]
- 79.Vitali, J., B. Schick, H. C. Kester, J. Visser, and F. Jurnak. 1998. The tree-dimensional structure of Aspergillus niger pectin lyase B at 1.7-Å resolution. Plant Physiol. 116:69-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wong, T. Y., L. A. Preston, and N. L. Schiller. 2000. Alginate lyase: review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu. Rev. Microbiol. 54:289-340. [DOI] [PubMed] [Google Scholar]
- 81.Yoder, M. D., and F. Jurnak. 1995. The refined three-dimensional structure of pectate lyase C from Erwinia chrysanthemi at 2.2 angstrom resolution (implications for an enzymatic mechanism). Plant Physiol. 107:349-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yoder, M. D., N. T. Keen, and F. Jurnak. 1993. New domain motif: the structure of pectate lyase C, a secreted plant virulence factor. Science 260:1503-1507. [DOI] [PubMed] [Google Scholar]
- 83.Yoder, M. D., S. E. Lietzke, and F. Jurnak. 1993. Unusual structural features in the parallel beta-helix in pectate lyases. Structure 1:241-251. [DOI] [PubMed] [Google Scholar]
- 84.Yoon, H. J., W. Hashimoto, O. Miyake, K. Murata, and B. Mikami. 2001. Crystal structure of alginate lyase A1-III complexed with trisaccharide product at 2.0 Å resolution. J. Mol. Biol. 307:9-16. [DOI] [PubMed] [Google Scholar]