Abstract
Outer membrane cytochromes are often proposed as likely agents for electron transfer to extracellular electron acceptors, such as Fe(III). The omcF gene in the dissimilatory Fe(III)-reducing microorganism Geobacter sulfurreducens is predicted to code for a small outer membrane monoheme c-type cytochrome. An OmcF-deficient strain was constructed, and its ability to reduce and grow on Fe(III) citrate was found to be impaired. Following a prolonged lag phase (150 h), the OmcF-deficient strain developed the ability to grow in Fe(III) citrate medium with doubling times and yields that were ca. 145% and 70% of those of the wild type, respectively. Comparison of the c-type cytochrome contents of outer membrane-enriched fractions prepared from wild-type and OmcF-deficient cultures confirmed the outer membrane association of OmcF and revealed multiple changes in the cytochrome content of the OmcF-deficient strain. These changes included loss of expression of two previously characterized outer membrane cytochromes, OmcB and OmcC, and overexpression of a third previously characterized outer membrane cytochrome, OmcS, during growth on Fe(III) citrate. The omcB and omcC transcripts could not be detected in the OmcF-deficient mutant by either reverse transcriptase PCR or Northern blot analyses. Expression of the omcF gene in trans restored both the capacity of the OmcF-deficient mutant to reduce Fe(III) and wild-type levels of omcB and omcC mRNA and protein. Thus, elimination of OmcF may impair Fe(III) reduction by influencing expression of OmcB, which has previously been demonstrated to play a critical role in Fe(III) reduction.
Dissimilatory Fe(III)-reducing bacteria can obtain energy by coupling the oxidation of organic compounds to the reduction of Fe(III) (31). Molecular analysis of the composition of microbial communities has shown that the Geobacteraceae, a family of dissimilatory Fe(III)-reducing bacteria in the delta subdivision of the class Proteobacteria, are the predominant Fe(III)-reducing microorganisms in a wide variety of sedimentary environments in which Fe(III) reduction is the principal terminal electron-accepting process (1, 42, 48, 49, 54, 55). Some of these bacteria can use alternate electron acceptors, such as the radionuclide U(VI) (32), vanadium (43), humic substances (30), and insoluble graphite electrodes (5). The Geobacteraceae can play an important role both in the bioremediation of organic contaminants, radionuclides, and toxic metals and in biological electricity production (1, 5, 6, 29, 43, 49).
In well-studied bacteria, such as Escherichia coli and Bacillus subtilis, most c-type cytochromes either are located in the periplasm or are associated with the cytoplasmic membrane and are involved in the generation of proton motive force (21, 57). However, two genera of dissimilatory Fe(III)-reducing bacteria, Geobacter and Shewanella, have multiple outer membrane c-type cytochromes (4, 16, 27, 33, 38, 39). Outer membrane c-type cytochromes and other outer membrane-associated proteins are postulated to play an important role in the reduction of insoluble electron acceptors, such as Fe(III) oxide, Mn(IV) oxide, and graphite electrodes, because direct contact is required for the reduction of these acceptors (8, 40). Furthermore, most of the Fe(III) reductase activity of Geobacter sulfurreducens (16) and Shewanella oneidensis (38, 39) has been reported to be associated with the outer membrane.
The genome of G. sulfurreducens contains over 100 putative c-type cytochrome genes, and at least 30 of the cytochromes are predicted to be localized in the outer membrane (36). In order to elucidate their physiological roles, the genes for these cytochromes were systematically disrupted and their phenotypes were analyzed. One of these putative outer membrane cytochromes, OmcF, was found to play a critical role in Fe(III) reduction.
Here we report the results of the phenotypic analysis of an OmcF-deficient mutant, as well as evidence that deletion of the omcF gene affects the expression of other outer membrane cytochromes, including OmcB, which was previously determined to play a critical role in Fe(III) reduction.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli strain DH5α [supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 recA1] (15, 38) and E. coli strain TOP10 [F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara-leu)7697 galU galK rpsL (Str) endA1 nupG] (Invitrogen Co., Carlsbad, CA) were used for DNA manipulations and PCR product subcloning, respectively. G. sulfurreducens strains DLBK01 (omcF::kan) and DLBK01/pRG5-omcF (omcF::kan/pRG5-omcF) were produced from G. sulfurreducens strain DL1 (7) as described below. G. sulfurreducens strains were routinely cultured anaerobically in either acetate-fumarate or acetate-Fe(III) citrate medium as previously described (11).
DNA manipulations.
Genomic DNA was purified using the MasterPure Complete DNA & RNA purification kit (Epicentre Technologies, Madison, WI.). Plasmid DNA purification, PCR product purification, and gel extraction were carried out using Mini Plasmid purification kits, PCR purification kits, and the Qiaquick gel extraction kit (QIAGEN Inc., Valencia, CA). DNA cloning and other DNA manipulations were carried out as described by Sambrook et al. (50). Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs, Inc. (Beverly, MA). All primers were purchased from Sigma-Genosys (The Woodlands, TX). Taq DNA polymerase (QIAGEN Inc., Valencia, CA) was used for all PCR amplifications.
Deletion of the omcF gene via single-step gene replacement.
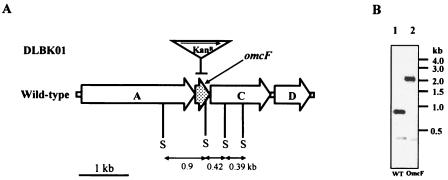
Single-step gene replacement was performed essentially as previously described (28). To disrupt the omcF gene, a linear 2.2-kb DNA fragment, containing the kanamycin resistance marker (Kanr) flanked by ca. 0.5 kb of sequence upstream and downstream of omcF (Table 1 and Fig. 1), was generated by recombinant PCR (28, 37). The sequence upstream of omcF was amplified with primers 4017-1 and 4017-2 (Table 1). The sequence downstream of omcF was amplified with primers 4017-5 and 4017-6 (Table 1). The kanamycin resistance cassette was amplified from plasmid pBBR1MCS-2 (24) with primers 4017-3 and 4017-4 (Table 1). Following recombinant PCR with the three primary PCR products, the final 2.2-kb fragment was amplified with distal primers 4017-1 and 4017-6. The PCR conditions were similar to those described previously (11, 28), except that the annealing temperature was 60°C. Electroporation, mutant isolation, and genotype confirmation were performed as previously described (11, 28). One mutant strain, designated DLBK01 (omcF::kan), was chosen as the representative strain (Fig. 1). The genotype of this strain was confirmed by Southern blot analysis (Fig. 1B), performed as previously described (11, 27). The 2.2-kb probe used for Southern blot analysis was amplified from the OmcF-deficient mutant (strain DLBK01) with primers 4017-1 and 4017-6 (Table 1) and labeled with digoxigenin (Roche Diagnostics Corp., Indianapolis, IN). Hybridization was detected with a digoxigenin nucleic acid detection kit (Roche Diagnostics Co., Indianapolis, IN). To ensure that the omcB and omcC genes had not been accidentally disrupted during deletion of the omcF gene, omcB- and omcC-specific fragments were amplified from the OmcF-deficient mutant with primers 8916 and 8908-2 and primers 8914 and 8915, respectively (Table 1).
TABLE 1.
Primers used in this study
| Primer | Use | Sequence (5′ to 3′) | Positiona | Reference |
|---|---|---|---|---|
| 4017-1 | Recombinant PCR and Southern blot analysis | ACGAAGGGGTCGAGGGAGAC | −514 to −497 of omcF | This study |
| 4017-2 | Recombinant PCR | GACGATTGCCCCCCCTCG | −18 to −1 of omcF | This study |
| 4017-3 | Recombinant PCR | CGAGGGGGGGCAATCGTCAGTGCCACCTGGGATGAATGb | −18 to −1 of omcF, −274 to −255 of Kanr in pBBR1MCS-2 | This study |
| 4017-4 | Recombinant PCR | CTCATGGGAAGCTCGCCACGATGGCAGGTTGGGCGTCGCc | −1 to 18 from omcF stop codon, −51 to −33 from Kanr stop codon in pBBR1MCS-2 | This study |
| 4017-5 | Recombinant PCR | CGTGGCGAGCTTCCCATGAG | −1 to 18 from omcF stop codon | This study |
| 4017-6 | Recombinant PCR and Southern blot analysis | CCTTTTCAGCGGGGAGCGAC | −567 to −548 from omcF stop codon | This study |
| ComF2 | Expression of the omcF gene in trans | CGAGAAAGACGAGAAGAAGG | −63 to −44 of omcF | This study |
| ComR3 | Expression of the omcF gene in trans | AAGCTTCGGAACGAGGGCTCTCATGGGAd | −14 to 8 from omcF stop codon | This study |
| 8916 | RT-PCR and Northern blot analysis of omcB | GGACTGCGCACCATCAAGG | 580 to 598 of omcB | 26 |
| 8908-2 | RT-PCR and Northern blot analysis of omcB | GGTCAGCAGGCCACCGG | 998 to 1004 of omcB | 27 |
| 8914 | RT-PCR and Northern blot analysis of omcC | CCTGCCATGAGACCGTTGCC | 285 to 302 of omcC | 26 |
| 8915 | RT-PCR and Northern blot analysis of omcC | GGGTGTTGTGGTAGAAGGG | 879 to 897 of omC | 9 |
| RT4017F | RT-PCR and Northern blot analysis of omcF | ATGAGAGGGCTTGCCCCGGTG | 1 to 21 of omcF | This study |
| RT4017R | RT-PCR and Northern blot analysis of omcF | TGGGAAGCTCGCCACGACG | 294 to 312 of omcF | This study |
| RT4018F | RT-PCR analysis of orfA | GCATGGCCACCACCAACTTC | 1037 to 1056 of orfA | This study |
| RT4018R | RT-PCR analysis of orfA | CTCCTCGATCCGGTTACTCC | 1631 to 1650 of orfA | This study |
| RT4016F | RT-PCR analysis of orfC | CCATCGGCGAGAAGCAGGAC | 421 to 420 of orfC | This study |
| RT4016R | RT-PCR analysis of orfC | TCGGCAATGAAAAGGATGAG | 901 to 920 of orfC | This study |
Unless indicated otherwise, the A of the ATG start codon is considered position 1.
The omcF bases are underlined, and pBBR1MCS-2 bases are indicated by boldface type.
The underlining indicates positions −1 to 18 from the omcF stop codon, and the boldface type indicates positions −51 to −33 from the Kanr stop codon in pBBR1MCS-2.
HindIII site is underlined.
FIG. 1.
(A) Structure of gene cluster containing omcF (DL1) and mutation scheme for OmcF-deficient strain DLBK01. The omcF gene was replaced with a kanamycin cassette in mutant DLBK01 (omcF::kan). The gene replacement is indicated by a horizontal bar. The transcriptional orientation of the kanamycin resistance cassette (Kanr) is the same as that of omcF and is indicated by a horizontal arrow. “S” indicates SmaI sites. The current annotation (36) of the genes surrounding omcF is as follows: orfA, ATP-dependent protease; orfC, putative membrane protein; and orfD, SPFH/band 7 domain protein. (B) Confirmation of omcF::kan genotype by Southern blot analysis. Wild-type and OmcF-deficient mutant genomic DNA were cleaved with SmaI, blotted, and probed with a 2.2-kb DNA fragment amplified from the omcF::kan strain with primers 4017-1 and 4017-6 (Table 1). The expected labeled fragments are 0.9 kb and 0.42 kb long for the wild type (WT) and 2.1 kb and 0.39 kb long for the mutant.
Expression of the omcF gene in trans.
An expression vector, pRG5, was constructed for complementation of G. sulfurreducens knockout mutants. The aadA spectinomycin resistance cassette was amplified from pSJS985 (51) with primers RGspcRV1 (CGATGATATCGCACAGGATGACGCCTAAC) and RGspcRV2 (GCGATGATATCGAAGCGGCGTCGGCTTG) (EcoRV sites underlined) and digested with EcoRV. The cassette was ligated into the AfeI site of pCM66 (34), creating the vector pRGCM66spec. The promoter, multiple cloning site, and kanamycin resistance cassette were then excised from pRGCM66spec with PciI and XhoI and replaced with a PciI/XhoI-digested DNA fragment, containing the taclac promoters and the multiple cloning site of pCD342 (12), which was amplified with primers RGG1 (GCGATCTCGAGCCAGTGAGACGGGCAACAGCTG; XhoI site underlined) and RGG2 (GCGATACATGTCGCCAAAAGTTGGCCCAGGGCTTC; PciI site underlined). The resulting plasmid was designated pRG5. The complete omcF coding sequence was amplified with primers ComF2 and ComR3 (Table 1) using the following conditions: 96°C for 4 min, followed by 30 cycles of 96°C for 30 s, 60°C for 30 s, and 72°C for 30 s and a final extension at 72°C for 10 min. The omcF coding sequence was cloned into pCR 2.1-TOPO (Invitrogen, Carlsbad, CA), excised with EcoRI and HindIII, and inserted into the EcoRI and HindIII sites of pRG5 to generate the omcF expression vector, pRG5-omcF. The omcF gene was sequenced to screen for PCR artifacts. The simultaneous presence of both plasmid pRG5-omcF and the omcF::kan mutation in this strain was confirmed by PCR screening with primers ComF2 and ComR3 (for pRG5-omcF) and primers 4017-1 and 4017-6 (for the omcF::kan mutation) (Table 1).
Detection and identification of proteins.
The membrane fraction of G. sulfurreducens was isolated as previously described (22, 41). Outer membrane-enriched fractions were prepared by treating crude membranes with a Sarkosyl (sodium N-lauroylsarcosinate) solution (1%, wt/vol) to extract inner membrane proteins (22, 41). Outer membrane proteins were analyzed by Tris-Tricine denaturing polyacrylamide gel electrophoresis (3), and c-type cytochromes were detected by staining with N,N,N′,N′-tetramethylbenzidine as previously described (14, 56). SeeBlue Plus prestained protein standards were purchased from Invitrogen Co. (Carlsbad, CA). The Tris-Tricine gel image was digitized using a Scanjet 7400C series scanner (Hewlett-Packard, Palo Alto, CA).
Protein bands of interest were manually excised, and in-gel tryptic digestion was performed in a previously described digestion buffer (10). Digests were concentrated using ZipTip C18 pipette tips (Millipore, MA, Boston) according to the recommended protocol, except that 1% formic acid was used instead of trifluoroacetic acid. Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) was carried out (58) with a Bruker Reflex III TOF mass spectrometer equipped with a 26-sample SCOUT source and video system, a nitrogen UV laser (maximum wavelength, 337 nm), and a dual-channel plate detector (Bruker Daltonik, Bremen, Germany). A sample solution (1 μl) was placed on the target, and 1 μl of a freshly prepared saturated solution of α-cyano-4-hydroxycinnamic acid in acetonitrile-H2O (2:1) with 0.1% trifluoroacetic acid was added. For recording of the spectra, an acceleration voltage of 20 kV was used and the detector voltage was adjusted to 1.7 kV. External calibration was carried out using a mixture of three synthetic peptides with molecular masses between 1,046 and 3,500 Da. The MALDI-TOF MS was carried out at the Center of Mass Spectrometry, University of Massachusetts, Amherst.
RT-PCR and Northern analyses.
Total RNA was purified from mid-log cultures using RNeasy Mini kits (QIAGEN Inc., Valencia, CA) with on-column DNase digestion. cDNA for reverse transcriptase PCR (RT-PCR) was synthesized using random hexamers (Life Technologies, Rockville, MD) or gene-specific primers (Table 1). RT-PCR was performed using the Thermoscript RT-PCR system (Life Technologies, Rockville, MD) according to the manufacturer's instructions. As a control for potential DNA contamination, amplification was performed with RNA that had not been treated with reverse transcriptase. For Northern analysis, total RNA was blotted and probed with the Northern Max-Gly kit (Ambion Inc., Austin, TX) according to the manufacturer's instructions. The probes used for identifying omcB and omcC transcripts (9, 26) were amplified with primers listed in Table 1 and were labeled with [α-32P]dATP using the NEBlot kit (New England Biolabs Inc., Beverly, MA).
Analytical techniques.
Growth of fumarate cultures was assessed by measuring turbidity at 600 nm with a Genesys 2 spectrophotometer (Spectronic Instruments, Rochester, NY). Cell densities of Fe(III)-grown cultures were determined by epifluorescence microscopy using acridine orange staining (19, 47). G. sulfurreducens cells were quantified using the SimplePCI software, version 5.3 (C-Imaging Systems, Compix Inc., Mars, PA). Cell suspension experiments were carried out as previously described (27). Protein concentrations were determined by the bicinchoninic acid method with bovine serum albumin as a standard (53).
Protein sequence accession numbers.
The GenBank accession numbers for the G. sulfurreducens proteins described in this report are as follows: OmcF, GI-39997527; OmcB, GI-39997831; OmcC, GI-39997825; OrfA, GI-39997528; OrfC, GI-39997526; and OrfD, GI-39997525.
RESULTS
Analysis of the omcF gene.
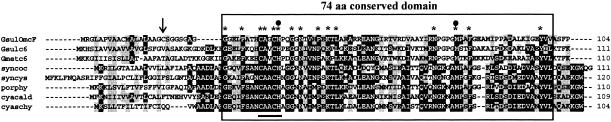
The genome of G. sulfurreducens contains more than 100 genes predicted to encode c-type cytochromes based on the presence of the CXXCH heme c binding motif (36). Thirty-one of these cytochromes are predicted to be localized in the outer membrane (http://psort.nibb.ac.jp./form.html). The smallest of these putative outer membrane c-type cytochromes is the product of the omcF gene, which encodes a 104-amino-acid protein with a single heme-binding motif and a prokaryotic membrane lipoprotein lipid attachment site and signal sequence (Fig. 2). Following heme incorporation and cleavage of the signal sequence, the molecular mass of OmcF is predicted to be approximately 9.4 kDa.
FIG. 2.
Alignment of the amino acid sequence of OmcF with those of other related c-type cytochromes identified by BLAST analysis. Identical residues are indicated by asterisks, and the heme-binding domain (CXXCH) is underlined. The lipid attachment site of OmcF is indicated by a vertical arrow. Solid circles indicate the amino acid residues that serve as axial ligands for iron (60). A 74-amino-acid (aa) domain that is shared by OmcF (G27 to V100) and the various cyanobacterial and algal c6 cytochromes is enclosed in a box. Similar amino acid residues are highlighted. G. sulfurreducens OmcF and other related cytochromes were aligned using the Bioedit program (version 7.0.1) (18). Heme-binding sites and signal sequences were identified with Prosite scan (http://us.expasy.org/prosite) and the motifs program of GCG (Wisconsin Package, version 10.2; Genetics Computer Group, Madison, WI). The cleavage site for the putative OmcF signal peptide was predicted using the LipoP 1.0 server (www.cbs.dtu.dk/services/lipoP). Abbreviations (National Center for Biotechnology Information accession numbers are indicated in parentheses): GsulOmcF, G. sulfurreducens OmcF (GI-39997527); Gsulc6, G. sulfurreducens cytochrome c family protein (GI-39997837); Gmetc6, G. metallireducens cytochrome c mono- and diheme variants (GI-48846289); syncoc, Synechococcus elongatus PCC 7942 cytochrome c6 precursor (GI-117934); syncys, Synechocystis sp. strain PCC 6803 cytochrome c6 precursor (GI-538865); porphy, Porphyra yezoensis cytochrome c6 precursor (GI-25008338); cyacald, Cyanidium caldarium cytochrome c6 precursor (GI-14285419); cyaschy, Cyanidioschyzon merolae strain 10D cytochrome c553 (GI-30409324).
A BLAST search of the National Center for Biotechnology Information database revealed that the closest relatives of OmcF are found in the Geobacteraceae. The genomes of G. sulfurreducens and Geobacter metallireducens encode related monoheme c-type cytochromes which are 57% and 50% similar to OmcF, respectively (Fig. 2). Outside the Geobacteraceae, OmcF is most similar to the soluble monoheme c6 cytochromes of cyanobacteria and planktonic algae (Fig. 2). The c6 cytochromes serve as electron donors to cytochrome c oxidase in cyanobacteria (2, 13) and transfer electrons from cytochrome b6f to photosystem I in both cyanobacteria and unicellular algae (2, 13, 23). OmcF and the c6 family of c-type cytochromes share a 74-amino-acid domain (Fig. 2, G27 to V100 of OmcF) which includes 15 strictly conserved residues, including the heme-binding motif and a methionine residue (M78) that serves as an axial ligand for heme iron (60). The 74-amino-acid domain of OmcF is 64 to 65% similar to those found in the related soluble cytochromes of G. metallireducens and G. sulfurreducens and 53 to 59% similar to those of various algal and cyanobacterial c6 cytochromes (Fig. 2).
Expression of omcF.
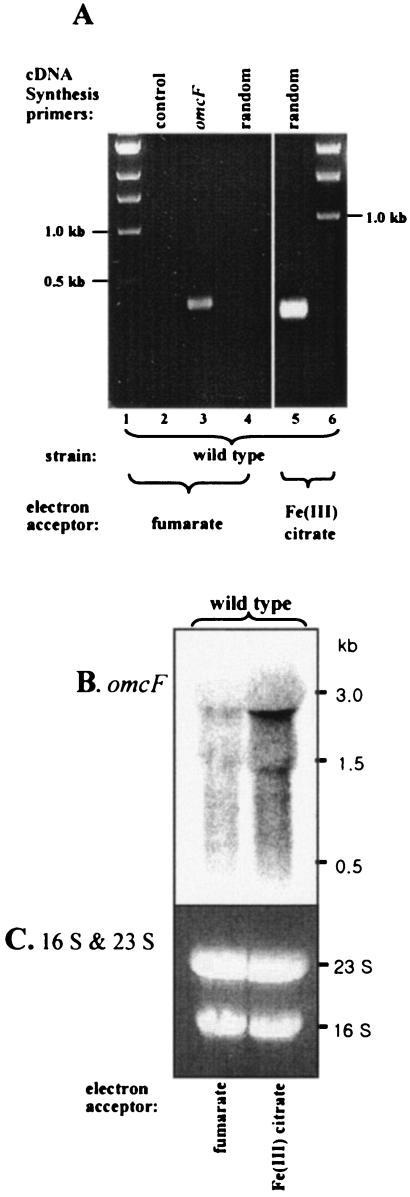
Expression of omcF was detected in mRNA prepared from Fe(III) citrate-grown cells by RT-PCR when cDNA was synthesized with either omcF-specific or random primers. In contrast, omcF expression could be detected in mRNA prepared from fumarate-grown cells only when cDNA was synthesized with omcF-specific primers (Fig. 3A). Because the levels of omcF cDNA synthesized with specific primers are likely to be much higher than those generated with random primers, this result suggested that omcF expression is significantly higher during growth on Fe(III) citrate than during growth on fumarate. The results of the Northern blot analysis (Fig. 3B) were consistent with this conclusion, and a smeared omcF transcript at roughly 3 kb was significantly more abundant during growth on Fe(III) citrate than during growth fumarate. This result is consistent with proteomic studies which detected OmcF only in Fe(III) citrate-grown cells, not in fumarate-grown cells (Ding, unpublished data).
FIG. 3.
RT-PCR (A) and Northern blot (B) analyses of omcF expression. Total RNA was extracted from acetate-fumarate or acetate-Fe(III) citrate-grown wild-type cultures. (A) The following primers were used for cDNA synthesis: lane 2, none; lane 3, omcF-specific primers RT4017F and RT4017R (Table 1); lane 4, random hexamers; lane 5, random hexamers. (B) The Northern blot was hybridized with 32P-labeled probes specific for omcF as indicated. Equal amounts of mRNA (20 μg/lane) were loaded. (C) Ethidium bromide-stained gel showing the positions of 16S and 23S rRNA, as a loading control.
Characterization of OmcF-deficient (omcF::kan) mutant.
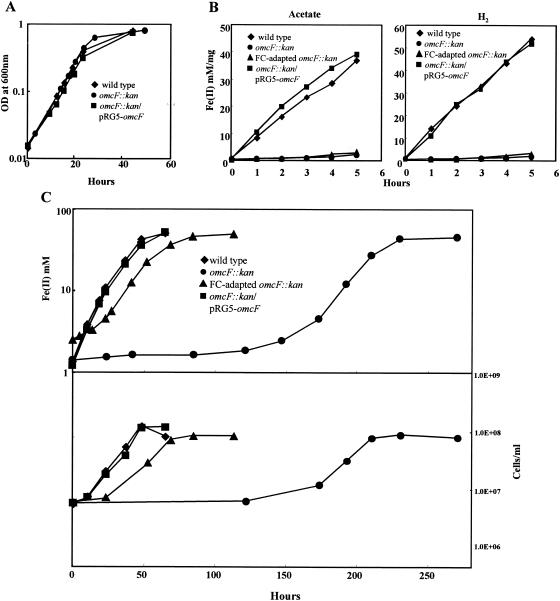
To investigate the physiological function of OmcF, a deletion mutant (omcF::kan, strain DLBK01) was constructed by homologous recombination (Fig. 1). The OmcF-deficient mutant grew as well as the wild type when fumarate was provided as the electron acceptor (Fig. 4A). However, when washed suspensions of the OmcF-deficient mutant were suspended in bicarbonate buffer and supplied with Fe(III) citrate as the electron acceptor, the rates of Fe(III) reduction with acetate and hydrogen as the electron donors were only 4.4% and 2.7% of the rates observed for the wild type, respectively (Fig. 4B). The ability of the OmcF-deficient mutant to reduce Fe(III) was also impaired under growth conditions (Fig. 4C). When acetate-fumarate-grown wild-type cells were inoculated into acetate-Fe(III) citrate medium, they completely reduced the Fe(III) in the medium within 3 days. In contrast, Fe(III) reduction by the OmcF-deficient mutant was not detectable until 6 days (150 h) after inoculation (Fig. 4C). Expression of the omcF gene in trans fully restored the ability of acetate-fumarate-grown cultures of the mutant to reduce Fe(III) in cell suspension assays (Fig. 4B) and also enabled the mutant to grow in Fe(III) citrate medium with a lag phase, doubling time, and maximum cell density that were comparable to those of the wild-type strain (Fig. 4C).
FIG. 4.
Characterization of the OmcF-deficient mutant. (A) Growth of wild-type, OmcF-deficient (omcF::kan), and complemented (omcF::kan/pRG5-omcF) strains in acetate-fumarate medium. Log-phase (A600, ∼0.5) acetate-fumarate-grown cultures were inoculated (3% inoculum) into fresh medium at time zero. OD at 600nm, optical density at 600 nm. (B) Reduction of Fe(III) citrate by wild-type, OmcF-deficient, FC-adapted OmcF-deficient, and complemented OmcF-deficient cell suspensions. Acetate or hydrogen was supplied as the electron donor. Log-phase (A600, ∼0.5) fumarate-grown cultures were harvested and used for this assay. (C) Growth of wild-type, OmcF-deficient (omcF::kan), and complemented (omcF::kan/pRG5-omcF) strains in acetate-Fe(III) citrate medium. Log-phase (A600, ∼0.5) acetate-fumarate-grown cultures were inoculated (3% inoculum) into acetate-Fe(III) citrate medium. The FC-adapted omcF::kan strain is an OmcF-deficient strain that was transferred four times in acetate-Fe(III) citrate medium and then four times in acetate-fumarate medium. The data are the means for triplicate cultures or incubations.
After approximately 150 h of incubation in Fe(III) citrate medium, the OmcF-deficient mutant developed the ability to grow using Fe(III) citrate as an electron acceptor. However, the doubling time and the final cell yield for the OmcF-deficient mutant were ca. 145% and 70%, respectively, of those for the wild type (15.3 h and 1.11 × 108 cells/ml for the mutant and 10.3 h and 1.55 × 108 cells/ml for the wild-type strain). During four subsequent transfers in Fe(III) citrate medium, the mutant grew with a much shorter lag phase but consistently grew more slowly than the wild type (data not shown). In order to determine if acquisition of the ability to reduce Fe(III) citrate was reversible, the adapted mutant was transferred four times (3% inoculum) in acetate-fumarate medium. Despite extensive culturing in acetate-fumarate medium, the adapted mutant retained the ability to grow on acetate-Fe(III) citrate medium with approximately the same lag phase (20 h), doubling time (15.2 h), and cell yield (1.06 × 108 cells/ml) observed for the adapted mutant prior to culturing on fumarate. However, it was clear that the adaptation had not restored the original pathway for Fe(III) reduction found in the wild-type strain, as washed cell suspensions of the fumarate-grown adapted mutant reduced Fe(III) citrate only at rates comparable to those of the unadapted mutant (Fig. 4B).
The long lag phase required for the initial growth of the OmcF-deficient mutant in Fe(III) citrate medium and the stability of the adapted phenotype suggested that growth of the OmcF-deficient mutant on Fe(III) citrate might have resulted from selective growth of a subpopulation of phase variants or cells with a secondary mutation. Due to the possibility of genetic differences between the OmcF-deficient strain cultured exclusively on acetate-fumarate medium and the strain that was subjected to selective pressure in acetate-Fe(III) citrate medium, the OmcF-deficient strain that had been adapted for growth on Fe(III) citrate was designated the Fe(III) citrate-adapted (FC-adapted) strain.
Cytochrome content of the OmcF-deficient mutants.
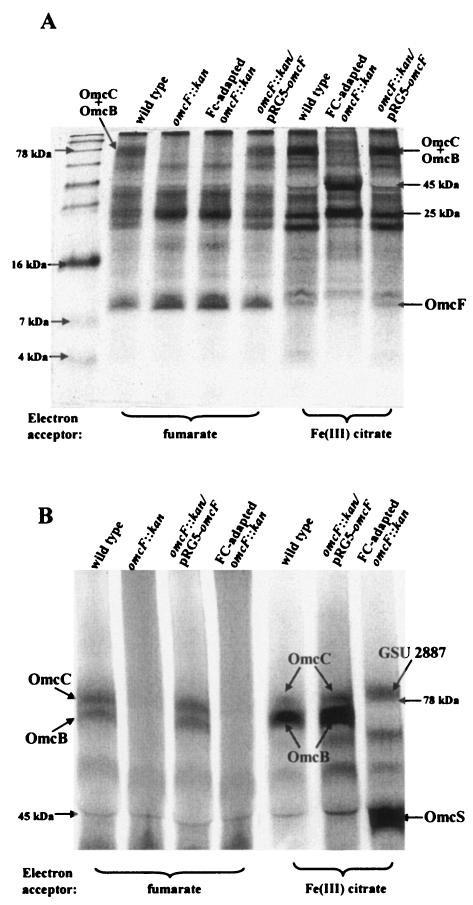
In order to gain insight into the subcellular localization of OmcF, the cytochrome protein compositions of outer membraneenriched fractions prepared from the wild-type and the OmcF-deficient cultures were compared. No difference in the abundance of low-molecular-mass (<10-kDa) cytochromes was vis-ible in outer membrane-enriched fractions prepared from the fumarate-grown cultures of the wild type and either the original or the FC-adapted OmcF-deficient mutant. However, outer membrane-enriched fractions prepared from the FC-adapted OmcF-deficient mutant grown with Fe(III) citrate as the electron acceptor (Fig. 4C) lacked a low-molecular-weight c-type cytochrome that was present in wild-type Fe(III)-grown cells (Fig. 5A). This cytochrome was also present in the complemented mutant cultured on Fe(III) citrate. When this band was excised, digested with trypsin, and analyzed by MALDI-TOF MS, three peptides corresponding to OmcF (DVAAYIR, TLARARR, and TLARARREANGIRTVR) were identified. Thus, consistent with predictions based on sequence analysis, OmcF appears to be an outer membrane-associated c-type cytochrome. As expected from the expression studies discussed above and other proteomic studies (Ding, unpublished data), OmcF was substantially more abundant during growth on Fe(III) than during growth on fumarate.
FIG. 5.
Heme staining and Tricine-polyacrylamide gel electrophoresis of outer membrane-enriched fractions prepared from wild-type, OmcF-deficient (omcF::kan), FC-adapted OmcF-deficient (FC-adapted omcF::kan), and complemented (omcF::kan/pRG5-omcF) strains. Outer membrane-enriched fractions were prepared from cultures grown in acetate-Fe(III) citrate or acetate-fumarate medium by Sarkosyl extraction (41). Outer membrane proteins (10 μg for panel A, 30 μg for panel B) were resolved on a 15% (A) or 7.5% (B) Tris-Tricine polyacrylamide gel and stained for heme. In order to resolve OmcB and OmcC (27), the gel shown in panel B was run until all proteins having molecular masses less than 30 kDa were off the gel.
Differences in the higher-molecular-weight cytochromes present in the wild-type and OmcF-deficient outer membrane-enriched fractions were apparent (Fig. 5). Of particular interest was a heme-staining band migrating at ca. 78 kDa that was missing from the outer membrane-enriched fraction of the OmcF-deficient mutant (Fig. 5A) during growth on both fumarate and Fe(III) citrate. This band was resolved into two discrete bands following electrophoresis on a lower-percentage Tricine gel (Fig. 5B). The electrophoretic mobilities and expression patterns of these two bands resembled those of two previously described outer membrane cytochromes, OmcB and OmcC (27). The band with the lower electrophoretic mobility yielded tryptic peptides corresponding to OmcB (AYLGTTRP, GILFFNAHPYFYR, CADCHDGTTAVATNSDTAFAESR, and DVMGAAFNANLLIHDPGGYAHNR), and the other band yielded peptide sequences corresponding to OmcC (AITDADGILGFVNSHYLTAGGQLFGTTGYEYATR, LAGADRPNTIFLDWGQSAHGGK, TTASGTTIEGYVIR, and GILFFNAHPYFYR). Another high-molecular-weight heme-staining band with an electrophoretic mobility similar to that of OmcC was present in outer membrane-enriched fractions prepared from the F(III) citrate-grown OmcF-deficient strain (Fig. 5B). However, when this band was analyzed by MALDI-TOF MS, no peptide corresponding to OmcC was detected. Instead, three peptides corresponding to the putative outer membrane c-type cytochrome GSU2887 (GenBank accession number GI-39997978) were identified (LYVSSISSSR, LYVSSISSSRTLR, and VVLRLYVSSISSSSRTLR). Restoration of omcF expression in trans restored expression of OmcB and OmcC to nearly wild-type levels. Thus, OmcF expression appeared to be required for expression of OmcB and OmcC during growth on both fumarate and Fe(III) citrate.
In addition to GSU2887, deletion of OmcF resulted in overexpression of a 45-kDa cytochrome during growth on Fe(III) citrate (Fig. 5A and B). MALDI-TOF MS analysis of a tryptic digest of this protein yielded the amino acid sequences HPAGNGAKFGATIAGLYNSYK, SLSGSYAFANQVPAAVAPSTYNR, and FNLAYEFTTIADASGNSIYGTDPNTSSLQGR. These sequences identified the cytochrome as OmcS, a previously studied outer membrane c-type cytochrome (35).
Changes in the expression patterns of two additional cytochromes, which had molecular masses of ca. 25 kDa and 21 kDa, were visible during growth of the OmcF-deficient mutant on both fumarate and Fe(III) citrate (Fig. 5A). The 25-kDa cytochrome was overexpressed in the OmcF-deficient mutant, whereas expression of the 21-kDa cytochrome was lower in the OmcF-deficient mutant than in the wild type. In both cases, alterations in expression were more pronounced during growth on Fe(III) citrate. Restoration of omcF expression in trans restored wild-type expression patterns for all four cytochromes (Fig. 5A and B).
Expression of omcB and omcC.
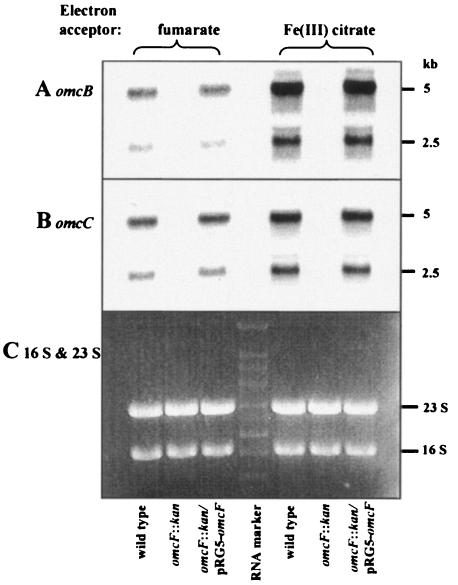
In order to gain further insight into the mechanism by which deletion of omcF reduced the abundance of OmcB and OmcC, Northern analysis of omcB and omcC expression was performed. The omcB and omcC genes are located in a tandem chromosomal duplication consisting of two three-gene clusters: orf1-orf2-omcB and orf1-orf2-omcC (27). Both omcB and omcC are transcribed from two independent promoters in their individual clusters, which results in the production of two transcripts for each gene, a 5-kb transcript that includes all three genes and a 2.5-kb transcript consisting of omcB or omcC alone (26). The two omcB and omcC transcripts were visible in mRNA prepared from wild-type cells growing on either fumarate or Fe(III) citrate but were absent in the OmcF-deficient strains growing in either medium (Fig. 6A and B). Restoration of omcF expression in trans restored the levels of the omcB and omcC transcripts to the levels present in wild-type cells (Fig. 6A and B). Similar results were obtained when omcB expression and omcC expression were evaluated by RT-PCR (data not shown). The failure to detect omcB and omcC transcripts in the OmcF-deficient mutant did not appear to be due to the quality of the mRNA prepared from the OmcF-deficient mutant, as the levels of 16S and 23S rRNA present in the various preparations were comparable (Fig. 6C) and expression of the open reading frames immediately upstream and downstream of omcF was detected by RT-PCR (data not shown).
FIG. 6.
Northern blot analysis of omcB and omcC expression. Total RNA was extracted from acetate-fumarate- or acetate-Fe(III) citrate-grown cultures of wild-type, OmcF-deficient (omcF::kan), and complemented (omcF::kan/pRG5-omcF) strains and hybridized with 32Plabeled probes for omcB (A) and omcC (B). Equal amounts of mRNA (5 μg/lane) were loaded for each strain, and RNA quantification was confirmed by ethidium bromide staining (C). The probes were specific for bp 508 to 1004 of omcB and bp 285 to 897 of omcC.
DISCUSSION
Our results indicate that OmcF, a small, outer membrane, c-type cytochrome, plays an important role in Fe(III) reduction in G. sulfurreducens. Elimination of OmcF alters the abundance of as many as six outer membrane c-type cytochromes. This includes the loss of OmcB, an outer membrane cytochrome that is required for optimal Fe(III) reduction (27). This is the first example of a requirement for one outer membrane cytochrome for the production of another outer membrane cytochrome that is necessary for Fe(III) reduction in G. sulfurreducens.
Potential roles of OmcF.
The most readily apparent phenotype of the OmcF-deficient mutant was its inability to reduce Fe(III) citrate. Deletion of another outer membrane c-type cytochrome, OmcB, also results in impairment of Fe(III) citrate reduction (27). Like the OmcF-deficient mutant, the OmcB-deficient mutant adapted to grow with Fe(III) citrate as the electron acceptor and eventually reduced Fe(III) nearly as fast as the wild type, but the cell yields were about one-half those of the wild type (25). Adaptation of the OmcB-deficient mutant to reduce Fe(III) was associated with increased production of other outer membrane c-type cytochromes, most notably OmcS (25). It is interesting that the reduced capacity of the OmcF-deficient mutant to grow with Fe(III) citrate as an electron acceptor was also associated with the loss of OmcB and that adaptation of the OmcF-deficient mutant was associated with increased production of OmcS. The lack of OmcS during growth of the FC-adapted OmcF-deficient mutant on fumarate and the inability of the fumarate-grown FC-adapted OmcF-deficient mutant to reduce Fe(III) in cell suspension further suggest that increased production of OmcS was an important part of the adaptation of the OmcF-deficient mutant to grow on Fe(III).
These results suggest that the inability of the OmcF-deficient mutant to reduce Fe(III) may be due to the lack of OmcB. However, this cannot be concluded with certainty because the presence of another outer membrane c-type cytochrome (molecular mass, ca. 21 kDa) which has an unknown function was also diminished in the mutant. Production of OmcC, a homolog of OmcB, was also lower in the OmcF-deficient mutant, but genetic studies indicate that OmcC is unlikely to play a critical role in Fe(III) reduction (27).
The mechanism by which the loss of OmcF results in loss of OmcB and other cytochromes is not known. The similarity of OmcF to cytochrome c6 of cyanobacteria and planktonic algae suggests that it may serve as an important electron carrier in G. sulfurreducens (2, 13, 23). If OmcF is part of a complex with one or more of the cytochromes that were not present in the OmcF-deficient mutant, then the absence of OmcF might result in accumulation of partially folded components of the complex, which, as seen in E. coli (44-46, 59), could activate an envelope stress response influencing gene expression.
Alternatively, OmcF may be part of a signal transduction pathway that ultimately influences either the transcriptional activation or the stability and/or processing of the transcripts of a number of genes, including omcB and omcC. Heme-containing proteins, such as OmcF, can serve as sensors of redox potential (15, 17, 20, 52), which can be influenced by the relative abundance of Fe(III) and Fe(II). Another possibility is that OmcF is part of a signaling pathway involving proteolysis. Northern analysis suggested that the omcF transcript is polycistronic (Fig. 6B). The gene immediately upstream of omcF (orfA) is homologous (57% similar) to the gene encoding the regulatory Lon protease of E. coli.
Considerable additional investigation is required to determine the physiological role of OmcF. However, this study clearly demonstrated that OmcF is one of several cytochromes that play a critical role in Fe(III) reduction in G. sulfurreducens. Thus, Fe(III) reduction is likely to be a complex process involving multiple steps and complexes with many potential levels of regulation.
Acknowledgments
This research was supported by the Office of Science (BER), U.S. Department of Energy (cooperative agreement DE-FC02-02ER63446 and grant DE-FG02-02ER63423). This work was also supported by the Postdoctoral Fellowship Program of the Korea Science & Engineering Foundation (KOSEF).
We are grateful for the excellent technical support provided by Betsy Blunt.
REFERENCES
- 1.Anderson, R. T., H. V. Vrionis, I. Ortiz-Bernad, C. T. Resch, P. E. Long, R. Dayvault, K. Karp, S. Marutzky, D. R. Metzler, A. Peacock, D. C. White, M. Lowe, and D. R. Lovley. 2003. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69:5884-5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ardelean, I., H. C. P. Matthijs, M. Havaux, F. Joset, and R. Jeanjean. 2002. Unexpected changes in photosystem I function in a cytochrome c6-deficient mutant of the cyanobacterium Synechocystis PCC 6803. FEMS Microbiol. Lett. 213:113-119. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1999. Current protocols in molecular biology, vol. 2. John Wiley & Sons, Inc., New York, N.Y.
- 4.Beliaev, A. S., D. A. Saffarini, J. L. Mclaughlin, and D. Hunnicutt. 2001. MtrC, an outer membrane decahaem c cytochrome required for metal reduction in Shewanella putrefaciens MR-1. Mol. Microbiol. 39:722-730. [DOI] [PubMed] [Google Scholar]
- 5.Bond, D. R., D. E. Holmes, L. M. Tender, and D. R. Lovley. 2002. Electrode-reducing microorganisms that harvest energy from marine sediments. Science 295:483-485. [DOI] [PubMed] [Google Scholar]
- 6.Bond, D. R., and D. R. Lovley. 2003. Electricity production by Geobacter sulfurreducens attached to electrodes. Appl. Environ. Microbiol. 69:1548-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caccavo, F., Jr., D. J. Lonergan, D. R. Lovley, M. Davis, J. F. Stolz, and M. J. McInerney. 1994. Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganisms. Appl. Environ. Microbiol. 60:3752-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Childers, S. E., S. Ciufo, and D. R. Lovley. 2002. Geobacter metallireducens accesses insoluble Fe(III) oxide by chemotaxis. Nature 416:767-769. [DOI] [PubMed] [Google Scholar]
- 9.Chin, K., A. Esteve-Nunez, C. Leang, and D. R. Lovley. 2004. Direct correlation between rates of anaerobic respiration and levels of mRNA for key respiratory genes in Geobacter sulfurreducens. Appl. Environ. Microbiol. 70:5183-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen, A. M., K. Rumpel, G. H. Coombs, and J. M. Wastling. 2002. Characterization of global protein expression by two-dimensional electrophoresis and mass spectrometry: proteomics of Toxoplasma gondii. Int. J. Parasitol. 32:39-51. [DOI] [PubMed] [Google Scholar]
- 11.Coppi, M. V., C. Leang, S. J. Sandler, and D. R. Lovley. 2001. Development of a genetic system for Geobacter sulfurreducens. Appl. Environ. Microbiol. 67:3180-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dehio, M., A. Knorre, C. Lanz, and C. Dehio. 1998. Construction of versatile high-level expression vectors from Bartonella henselae and the use of green fluorescent protein as a new expression marker. Gene 215:223-229. [DOI] [PubMed] [Google Scholar]
- 13.Duran, R. V., M. Hervas, M. A. De la Rosa, and J. A. Navarro. 2004. The efficient functioning of photosynthesis and respiration in Synechocystis sp. PCC 6803 strictly requires the presence of either cytochrome c6 or plastocyanin. J. Biol. Chem. 279:7229-7233. [DOI] [PubMed] [Google Scholar]
- 14.Francis, R. T., Jr., and R. R. Becker. 1984. Specific indication of hemoproteins in polyacrylamide gels using a double-staining process. Anal. Biochem. 136:509-514. [DOI] [PubMed] [Google Scholar]
- 15.Fu, R., J. D. Wall, and G. Voordouw. 1994. DcrA, a c-type heme-containing methyl-accepting protein from Desulfovibrio vulgaris Hildenborough, senses the oxygen concentration or redox potential of the environment. J. Bacteriol. 176:344-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaspard, S., F. Vazquez, and C. Holliger. 1998. Localization and solubilization of the iron(III) reductase of Geobacter sulfurreducens. Appl. Environ. Microbiol. 64:3188-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilles-Gonzalez, M., and G. Gonzalez. 2004. Signal transduction by heme-containing PAS-domain proteins. J. Appl. Physiol. 96:774-783. [DOI] [PubMed] [Google Scholar]
- 18.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 19.Hobbie, J. E., R. J. Daley, and S. Jasper. 1977. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hou, S., T. Freitas, R. W. Larsen, M. Piatibratov, V. Sivozhelezov, A. Yamamoto, E. A. Meleshkevitch, M. Zimmer, G. W. Ordal, and M. Alam. 2001. Globin-coupled sensors: a class of heme-containing sensors in Archaea and Bacteria. Proc. Natl. Acad. Sci. USA 98:9353-9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, C. W. 1988. Membrane-associated energy conservation in bacteria: a general introduction, p. 1-82. In C. Anthony (ed.), Bacterial energy transduction. Academic Press, Ltd., London, United Kingdom.
- 22.Kaufmann, F., and D. R. Lovley. 2001. Isolation and characterization of a soluble NADPH-dependent Fe(III) reductase from Geobacter sulfurreducens. J. Bacteriol. 183:4468-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerfeld, C. A., and D. Krogman. 1998. Photosynthetic cytochromes c in cyanobacteria, algae, and plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49:397-425. [DOI] [PubMed] [Google Scholar]
- 24.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, I. I., and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 25.Leang, C., L. A. Adams, K.-J. Chin, B. A. Methe, J. Webster, K. P. Nevin, M. L. Sharma, and D. R. Lovley. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 26.Leang, C., and D. R. Lovley. 2005. Regulation of two highly similar genes, omcB and omcC, in a 10-kb chromosomal duplication in Geobacter sulfurreducens. Microbiology 151:1761-1767. [DOI] [PubMed]
- 27.Leang, C., M. V. Coppi, and D. R. Lovley. 2003. OmcB, a c-type polyheme cytochrome, involved in Fe(III) reduction in Geobacter sulfurreducens. J. Bacteriol. 185:2096-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lloyd, J. R., C. Leang, A. L. Hodges-Myerson, M. V. Coppi, S. Ciufo, B. Methe, S. J. Sandler, and D. R. Lovley. 2003. Biochemical and genetic characterization of PpcA, a periplasmic c-type cytochrome in Geobacter sulfurreducens. Biochem. J. 369:153-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lloyd, J. R., and D. R. Lovley. 2001. Microbial detoxification of metals and radionuclides. Curr. Opin. Biotechnol. 12:248-253. [DOI] [PubMed] [Google Scholar]
- 30.Lonergan, D. J., H. L. Jenter, J. D. Coates, E. J. Philips, T. M. Schmidt, and D. R. Lovley. 1996. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J. Bacteriol. 178:2402-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lovley, D. R. 1993. Dissimilatory metal reduction. Annu. Rev. Microbiol. 47:263-291. [DOI] [PubMed] [Google Scholar]
- 32.Lovley, D. R., E. J. P. Phillips, Y. A. Gorby, and E. R. Landa. 1991. Microbial reduction of uranium. Nature 350:413-416. [Google Scholar]
- 33.Magnuson, T. S., N. Isoyama, A. L. Hodges-Myerson, G. Davidson, M. J. Maroney, G. G. Geesey, and D. R. Lovley. 2001. Isolation, characterization and gene sequence analysis of a membrane-associated 89 kDa Fe(III) reducing cytochrome c from Geobacter sulfurreducens. Biochem. J. 359:147-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marx, C. J., and M. E. Lidstrom. 2001. Development of improved versatile broad-host-range vectors for use in methylotrophs and other Gram-negative bacteria. Microbiology 147:2065-2075. [DOI] [PubMed] [Google Scholar]
- 35.Mehta, T., M. V. Coppi, S. E. Childers, and D. R. Lovley. Unpublished data.
- 36.Methe, B. A., K. E. Nelson, J. A. Eisen, I. T. Paulsen, W. Nelson, J. F. Heidelberg, D. Wu, M. Wu, N. Ward, M. J. Beanan, R. J. Dodson, R. Madupu, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, M. Gwinn, J. F. Kolonay, S. A. Sullivan, D. H. Haft, J. Selengut, T. M. Davidsen, N. Zafar, O. White, B. Tran, C. Romero, H. A. Forberger, J. Weidman, H. Khouri, T. V. Feldblyum, T. R. Utterback, S. E. V. Aken, D. R. Lovley, and C. M. Fraser. 2003. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science 302:1967-1969. [DOI] [PubMed] [Google Scholar]
- 37.Murphy, K. C., K. G. Campellone, and A. R. Poteete. 2000. PCR-mediated gene replacement in Escherichia coli. Gene 246:321-330. [DOI] [PubMed] [Google Scholar]
- 38.Myers, C., and J. Myers. 1993. Ferric reductase is associated with the membranes of anaerobically grown Shewanella putrefaciens MR-1. FEMS Microbiol. Lett. 108:15-22. [Google Scholar]
- 39.Myers, C. R., and J. M. Myers. 2003. Cell surface exposure of the outer membrane cytochromes of Shewanella oneidensis MR-1. Lett. Appl. Microbiol. 37:254-258. [DOI] [PubMed] [Google Scholar]
- 40.Nevin, K. P., and D. R. Lovley. 2002. Mechanisms for Fe(III) oxide reduction in sedimentary environments. Geomicrobiol. J. 19:141-159. [Google Scholar]
- 41.Nikaido, H. 1994. Isolation of outer membranes. Methods Enzymol. 235:225-234. [DOI] [PubMed] [Google Scholar]
- 42.North, N. N., S. L. Dollhopf, L. Petrie, J. D. Istok, D. L. Balkwill, and J. E. Kostka. 2004. Change in bacterial community structure during in situ biostimulation of subsurface sediment cocontaminated with uranium and nitrate. Appl. Environ. Microbiol. 70:4911-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ortiz-Bernad, I., R. T. Anderson, H. V. Vrionis, and D. R. Lovley. 2004. Vanadium respiration by Geobacter metallireducens: novel strategy for in situ removal of vanadium from groundwater. Appl. Environ. Microbiol. 70:3091-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raffa, R. G., and T. L. Ravio. 2002. A third envelope stress signal transduction pathway in E. coli. Mol. Microbiol. 45:1599-1611. [DOI] [PubMed] [Google Scholar]
- 45.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55:591-624. [DOI] [PubMed] [Google Scholar]
- 46.Raivio, T. L., and T. J. Silhavy. 1997. Transduction of envelope stress in E. coli by the Cpx two-component system. J. Bacteriol. 179:7724-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rapposch, S., P. Zangerl, and W. Ginzinger. 2000. Influence of fluorescence of bacteria stained with acridine orange on the enumeration of microorganisms in raw milk. J. Dairy Sci. 83:2753-2758. [DOI] [PubMed] [Google Scholar]
- 48.Roling, W., F. M., B. M. van Breukelen, B. L. Braster, and H. van Verseveld, W. 2001. Relationships between microbial community structure and hydrochemistry in a landfill leachate-polluted aquifer. Appl Environ Microbiol 67:4619-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rooney-Varga, J. N., R. T. Anderson, J. L. Fraga, D. Ringelberg, and D. R. Lovley. 1999. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 65:3056-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press., Cold Spring Harbor, N.Y.
- 51.Sandler, S. J., and A. J. Clark. 1994. RecOR suppression of recF mutant phenotypes in Escherichia coli K-12. J. Bacteriol. 176:3661-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sasakura, Y., S. Hirata, S. Sugiyama, S. Suzuki, S. Taguchi, M. Watanabe, T. Matsui, I. Sagami, and T. Shimizu. 2002. Characterization of a direct oxygen sensor heme protein from Escherichia coli. Effects of the heme redox states and mutations at the heme-binding site on catalysis and structure. J. Biol. Chem. 277:23821-23827. [DOI] [PubMed] [Google Scholar]
- 53.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 54.Snoeyenbos-West, O., K. Nevin, R. Anderson, and D. Lovley. 2000. Enrichment of Geobacter species in response to stimulation of Fe(III) reduction in sandy aquifer sediments. Microb. Ecol. 39:153-167. [DOI] [PubMed] [Google Scholar]
- 55.Stein, L., M. La Duc, T. Grundl, and K. Nealson. 2001. Bacterial and archaeal populations associated with freshwater ferromanganous micronodules and sediments. Environ. Microbiol. 3:10-18. [DOI] [PubMed] [Google Scholar]
- 56.Thomas, P. E., D. Ryan, and W. Levin. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75:168-176. [DOI] [PubMed] [Google Scholar]
- 57.Thony-Meyer, L. 1997. Biogenesis of respiratory cytochromes in bacteria. Microbiol. Mol. Biol. Rev. 61:337-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vanrobaeys, F., B. Devreese, E. Lecocq, L. Rychlewski, L. De Smet, and J. Van Beeumen. 2003. Proteomics of the dissimilatory iron-reducing bacterium Shewanella oneidensis MR-1, using a matrix-assisted laser desorption/ionization-tandem-time of flight mass spectrometer. Proteomics 3:2249-2257. [DOI] [PubMed] [Google Scholar]
- 59.Walsh, N. P., B. M. Alba, B. Bose, C. A. Gross, and R. T. Sauer. 2003. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113:61-71. [DOI] [PubMed] [Google Scholar]
- 60.Yamada, S., S. Y. Park, H. Shimizu, Y. Koshizuka, K. Kadokura, T. Satoh, K. Suruga, M. Ogawa, Y. Isogai, T. Nishio, Y. Shiro, and T. Oku. 2000. Structure of cytochrome c6 from the red alga Porphyra yezoensis at 1.57 A resolution. Acta Crystallogr. Sect. D 56:1577-1582. [DOI] [PubMed] [Google Scholar]