Abstract
δ-Aminolevulinic acid, the biosynthetic precursor of tetrapyrroles, is synthesized from glutamate via the tRNA-dependent five-carbon pathway in the green sulfur bacterium Chlorobium vibrioforme. The enzyme glutamyl-tRNA reductase (GTR), encoded by the hemA gene, catalyzes the first committed step in this pathway, which is the reduction of tRNA-bound glutamate to produce glutamate 1-semialdehyde. To characterize the GTR protein, the hemA gene from C. vibrioforme was cloned into expression plasmids that added an N-terminal His6 tag to the expressed protein. The His-tagged GTR protein was purified using Ni affinity column chromatography. GTR was observable as a 49-kDa band on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. The native molecular mass, as determined by gel filtration chromatography, appeared to be approximately 40 kDa, indicating that native GTR is a monomer. However, when the protein was mixed with 5% (vol/vol) glycerol, the product had an apparent molecular mass of 95 kDa, indicating that the protein is a dimer under these conditions. Purified His6-GTR was catalytically active in vitro when it was incubated with Escherichia coli glutamyl-tRNAGlu and purified recombinant Chlamydomonas reinhardtii glutamate-1-semialdehyde aminotransferase. The expressed GTR contained 1 mol of tightly bound heme per mol of pep tide subunit. The heme remained bound to the protein throughout purification and was not removed by anion- or cation-exchange column chromatography. However, the bound heme was released during SDS-PAGE if the protein was denatured in the presence of β-mercaptoethanol. Added heme did not inhibit the activity of purified expressed GTR in vitro. However, when the GTR was expressed in the presence of 3-amino-2,3- dihydrobenzoic acid (gabaculine), an inhibitor of heme synthesis, the purified GTR had 60 to 70% less bound heme than control GTR, and it was inhibited by hemin in vitro.
δ-Aminolevulinic acid (ALA), the first universal precursor in the biosynthesis of chlorophylls, hemes, and other tetrapyrroles, can be formed by two biosynthetic pathways. In animals, yeasts and fungi, and α-proteobacteria, ALA is synthesized by condensation of glycine and succinyl coenzyme A, which is mediated by the enzyme ALA synthase (5, 10). In plants, algae, and most bacteria, ALA is synthesized from glutamate in a three-step reaction (reviewed in reference 3). In the first step, glutamyl-tRNA synthetase activates glutamate by forming glutamyl-tRNA. The activated glutamate is then reduced by glutamyl-tRNA reductase (GTR) in the presence of NADPH to form glutamate 1-semialdehyde (GSA). Finally, GSA is transaminated to ALA by GSA aminotransferase (GSAT). Biosynthesis of ALA from glutamate is very widespread in nature and is probably the pathway that arose earlier.
Although genes encoding GTR have been detected in many plants, algae, and prokaryotes, including both bacteria and archaea, the enzyme itself has not been extensively characterized. GTR from most sources is unstable in vitro, and the assay for GTR activity is difficult because the substrate, glutamyl-tRNA, is unavailable commercially and the product, GSA, is very unstable and difficult to quantitate. Most assays employ a coupled enzyme system in which glutamyl-tRNA is generated in situ either during or immediately before the start of the GTR assay and the product, GSA, is enzymatically converted to ALA as it is formed by including GSAT in the assay mixture. Moreover, GTR recognizes and discriminates among tRNAGlu molecules from various sources (1, 7, 17, 18, 32), and the cognate tRNA for the GTR being studied is often not available.
To investigate the structure, mechanism, and regulation of expression of GTR, we cloned the hemA gene, which encodes GTR, from the green sulfur bacterium Chlorobium vibrioforme into expression vectors and expressed the protein with and without an N-terminal His6 tag. His tagging facilitated purification of protein by Ni affinity column chromatography. In this way we obtained highly purified protein in quantities sufficient for characterizing the structure and enzymatic properties of GTR. Here we report on some properties of the purified protein.
MATERIALS AND METHODS
Expression of GTR in Escherichia coli cells.
The C. vibrioforme hemA gene was cloned initially in the pYA1 plasmid and was subsequently subcloned into the pYA3 plasmid (2, 12). In these constructs the enzyme was transcribed from its native promoter. The hemA gene was recloned into the pET30 (Novagen, Madison, WI) and pQE30 (QIAGEN, Valencia, CA) expression vectors to enhance its expression by transcription from the lac promoter. The pET30 and pQE30 vectors have T7 and T5 lac promoters, respectively. These vectors also allowed addition of an N-terminal His6 tag to the protein, which facilitated fast and efficient purification by Ni affinity column chromatography. Because expression was under control of the lac promoter, it was inducible by isopropyl β-d-thiogalactopyranoside (IPTG).
The cloning was performed in two steps. The open reading frame of pYA3 was amplified by PCR with primers containing additional nucleotides with two different restriction sites at each end, using Pfu DNA polymerase. Oligonucleotides 5′-CATATGAACATCATTTCAGTTGGTGTC-3′ and 5′-AAGCTTTTATTGGAGTGACTGGTTTGG-3′ were used to add NdeI and HindIII restriction sites for insertion into the pET30 vector. Oligonucleotides 5′-GAGCTCATGAACATCATTTCAGTTGG-3′ and 5′-AAGCTTTTATTGGAGTGACTGGTTTGG-3′ were used to add SacI and HindIII restriction sites for insertion into the pQE30 vector. The resulting PCR products were inserted into the pPCR-Script Amp SK+ plasmid (Stratagene, La Jolla, CA) by using a PCR Script Amp cloning kit (Stratagene).
These plasmids were amplified and then digested with the appropriate restriction enzymes, and the inserts were ligated into the matching sites of the pET30 and pQE30 vectors, resulting in inserts with the correct orientation and position of the His tag.
Both recombinant plasmids were transformed into E. coli strain BL21-Gold (Stratagene). BL21-Gold was also used for expression of pET30-hemA. However, E. coli strain SG13009 (QIAGEN) was used to express pQE30-hemA. This strain contains pREP4, a low-copy-number plasmid which confers kanamycin resistance and expresses the lac repressor protein encoded by the lac1 gene.
The transformed BL21-Gold cells (containing pET30-hemA) were grown in LB medium containing 100 μg/ml ampicillin at 37°C until the A600 of the culture was 0.6. SG13009 cells containing pQE30-hemA were grown in the presence of 100 μg/ml ampicillin and 50 μg/ml kanamycin. In both cases, protein expression was induced by addition of various concentrations of ITPG for different times. After induction, the cells were harvested by centrifugation and washed twice with extraction buffer (50 mM NaPO4, pH 8.0, 300 mM NaCl, 10 mM imidazole).
Purification of the expressed proteins.
Cell pellets from 2-liter cultures were resuspended in 4 ml of extraction buffer with 0.2 volume of glass powder (diameter, 5 mm), and the suspension was sonicated with a Sonifier cell disruptor (Heat Systems-Ultrasonics, Plainville, NY) at 0°C using 10 20-s periods with 30-s cooling intervals. The resulting extracts were clarified by centrifugation at 36,000 × g for 30 min. The supernatants were mixed with 1.5 ml of Ni-nitrilotriacetic acid (NTA) resin and kept on ice for 1 h with gentle shaking. Each slurry was transferred into a 1-ml polypropylene column, and the flowthrough was collected. The column was washed five times with at least 5 ml of wash buffer (50 mM NaPO4, pH 8.0, 300 mM NaCl, 20 mM imidazole). The bound protein was eluted in approximately 2 ml of elution buffer (50 mM NaPO4, pH 8.0, 300 mM NaCl, 250 mM imidazole) as described in the QIA Expressionist manual (QIAGEN). The purity of the protein was analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (21).
GTR activity determination.
Because both the substrate (glutamyl-tRNA) and the product (GSA) of GTR are unstable and difficult to measure sensitively, a coupled assay was used, in which glutamyl-tRNA was generated in the reaction tube, using an E. coli tRNAGlu and E. coli aminoacyl-tRNA synthetase mixture (Sigma, St. Louis, MO), and GSA was converted to ALA by GSAT as it was being generated. The GSAT used was His6-tagged Chlamydomonas reinhardtii GSAT that was expressed and purified in our lab (L. A. Nogaj and S. I. Beale, unpublished data).
The reaction mixture (0.5 ml) consisted of assay buffer (1.0 M glycerol, 50 mM Tricine, pH 7.9, 15 mM MgCl2, 5 mM ATP, 5 mM levulinic acid, 1 mM l-glutamate, 1 mM dithiothreitol [DTT], 1 mM NADPH, 20 μM pyridoxal phosphate), 100 U of E. coli aminoacyl-tRNA synthetases, 100 μg of His6-tagged C. reinhardtii GSAT, and 10 to 400 μg of expressed GTR protein. The reaction was started by adding 0.15 to 0.20 U of E. coli tRNAGlu. The reaction mixture was incubated at 30°C, and the reaction was terminated by adding 25 μl of 100% (wt/vol) trichloroacetic acid and mixing. After the mixture was cooled for 10 min on ice, the precipitate was removed by centrifugation for 10 min at 13,000 × g at 4°C. The supernatant was neutralized (by adding 150 μl of 500 mM Na3PO4) to pH 6.8, ethylacetoacetate (25 μl) was added, and the solution was mixed and heated at 95°C for 15 min to obtain ALA pyrrole (13) After reaction with an equal volume of Ehrlich Hg reagent (26), the product was quantified spectrophotometrically at 553 nm using a ɛ553 value of 8.0 × 104 M−1.
Esterase activity determination.
The hydrolysis of p-nitrophenyl acetate by GTR was measured directly with a spectrophotometer (Cary 219; Varian, Palo Alto, CA) equipped with a magnetic stirrer. For a typical kinetic experiment, 3 ml of assay buffer (50 mM Tricine, pH 7.9, 1.0 M glycerol, 15 mM MgCl2) was mixed with 15 μl of substrate solution (7. 4 mM p-nitrophenyl acetate in 100% acetonitrile) and equilibrated in a quartz cuvette for 1 min with constant stirring. Then 10 to 100 μl of protein (from a concentrated solution containing about 5 to 10 mg/ml) was added. The change in the absorption at 320 nm was monitored for at least 3 to 4 min.
Heme detection in polyacrylamide gels.
Normal SDS-PAGE was performed with the purified GTR protein, which was denatured by boiling for 10 min in loading buffer (50 mM Tris-HCl, pH 6.8, 2% [wt/vol] SDS, 0.1% [wt/vol] bromophenol blue, 10% [wt/vol] glycerol, 100 mM DTT) either in the presence or in the absence of 2.5% (vol/vol) β-mercaptoethanol. One part of the gel was stained with Coomassie brilliant blue to visualize the protein pattern on the gel, and another part of the gel was stained for heme-associated peroxidase activity by the method of Thomas et al. (25). A 6.3 mM solution of 3,3′,5,5′-tetramethylbenzidine (TMBZ) was freshly prepared in methanol. Immediately before use, 3 parts of the TMBZ solution was mixed with 7 parts of 0.25 M sodium acetate, pH 5.0. The SDS-PAGE gel was immersed in this mixture and kept at room temperature in the dark with gentle shaking. After 1 h, H2O2 was added to a final concentration of 30 mM. The staining was visible within 3 min.
Fast protein liquid column chromatography.
The native molecular weight of the expressed GTR protein was determined by Superdex G-300 HR 10/30 gel filtration column chromatography using a model GP250 fast protein liquid chromatography instrument (Pharmacia). The equilibration buffer was 50 mM Tris-HCl, pH 7.5, 100 mM KCl, and the protein standards used were obtained from Sigma. Purified protein was loaded either in Ni column elution buffer (without glycerol) or after it was mixed with 10% (vol/vol) glycerol, as described in the Results. For Mono-S and Mono-Q column chromatography, linear gradients of buffer A (10 mM Tris-HCl, pH 8, 125 mM NaCl, 10% [vol/vol] glycerol) and buffer B (10 mM Tris-HCl, pH 8, 500 mM NaCl, 10% [vol/vol] glycerol) were used. C. vibrioforme GTR precipitated if the NaCl concentration was below 125 mM. Therefore, 125 mM NaCl was used in buffer A, even though this NaCl concentration lowered the binding capacity of the protein on both Mono-S and Mono-Q columns. Elution of the protein was monitored by light absorption at 280 nm, and elution of heme was monitored by light absorption at 400 nm. The protein concentrations in fractions were determined by a dye-binding assay (4), using bovine serum albumin as the standard.
Absorption spectra.
The absorption spectra of the purified proteins were determined in alkaline pyridine (25% [vol/vol] pyridine in 0.1 M NaOH). The heme concentration was determined by measuring the absorption at 418.5 nm in alkaline pyridine and using a ɛ418.5 value of 1.915 × 105 M−1 (24). For reducing the pure hemin and the heme bound to protein, a saturating concentration of sodium dithionite was added, with mixing, to the alkaline pyridine solution, and the spectra were measured immediately.
Other procedures.
The DNA was visualized on a 1% (wt/vol) agarose gel after ethidium bromide staining, and DNA sequencing was done by Davis Sequencing (Davis, CA).
Nucleotide sequence accession number.
The new sequence of the C. vibrioforme hemA gene determined in this study has been deposited in the GenBank database under accession number AF080069.
RESULTS
Corrected C. vibrioforme hemA sequence.
The hemA gene of C. vibrioforme was one of the first GTR-encoding genes to be sequenced (12) (GenBank accession number M59194). A recent comparison of the predicted translation product of this sequence with other published GTR sequences revealed significant differences at the C terminus (S. P. Gough, personal communication). It was possible that these differences could have been caused by a sequencing error in the original published sequence. Therefore, the original pYA3 clone was resequenced. Compared with the new sequence, the original sequence had two missing bases near the 3′ end of the open reading frame, which caused a frameshift, incorrect amino acids in the C-terminal one-fifth of the predicted polypeptide, and premature termination. The new sequence predicts a translation product that is much more consistent with other GTR proteins at the C terminus. The predicted translation product contains 424 amino acids and has a molecular weight of 47,971 and an isoelectric point of 8.75.
Expression of GTR.
Expression vectors pET30 and pQE30 containing the hemA gene from C. vibrioforme were designed to produce GTR without and with a His6-containing N-terminal extension, respectively. The His tag was added to facilitate purification of the protein by Ni affinity column chromatography.
When E. coli cells transformed with the pET30 or pQE30 vector containing the C. vibrioforme hemA gene were grown and the GTR protein was induced with IPTG, the cells developed a brownish color. Extracts of these cells contained a protein with a molecular mass of approximately 49 kDa, as determined by SDS-PAGE (data not shown). This molecular mass is similar to the predicted molecular mass of GTR (47,971 Da) plus an additional 2,179 Da for the His6-containing N-terminal extension.
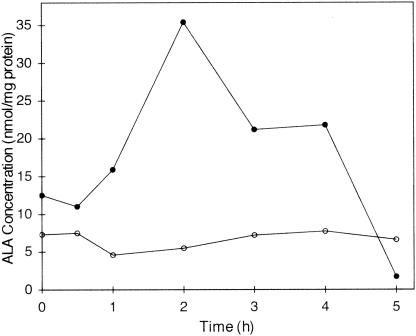
GTR could be expressed in both the pET30 and pQE30 vectors, but the quantity of expressed GTR was severalfold higher in the pQE30 vector than in the pET30 vector. For all other experiments described here, the pQE30 construct was used. Several concentrations of IPTG, ranging from 0.05 mM to 1 mM, were tested to optimize GTR expression. The best results were obtained when the GTR was induced in the presence of 0.5 mM IPTG. The optimum induction time for protein expression was found to be 4 h after IPTG addition. At later times, degradation of protein was observed. During the expression of GTR, ALA synthesis was apparently accelerated, resulting in a severalfold increase in the concentration of ALA in the E coli cells (Fig. 1). Several hours after the addition of IPTG, both the quantity of expressed GTR and the accumulated ALA concentration decreased.
FIG. 1.
Time course of accumulation of ALA in E. coli cells during GTR expression from the pQE30 vector. The cells were not induced (○) or were induced (•) with 0.5 mM IPTG.
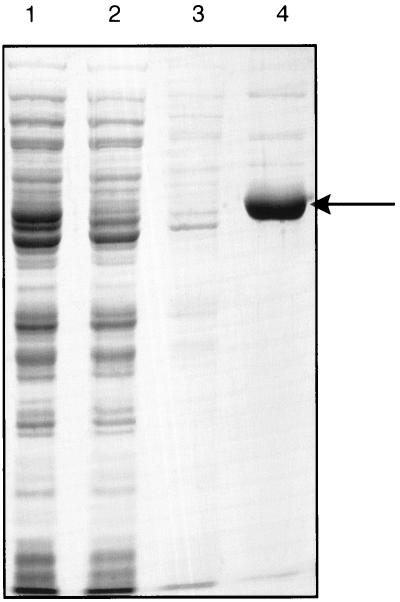
The protein having the predicted polypeptide molecular weight, as determined by SDS-PAGE, was retained by the Ni chelating column (Fig. 2), indicating that the expressed protein was soluble and that it contained a His6 tag.
FIG. 2.
Purification of C. vibrioforme GTR. GTR was expressed from the pQE30 vector and was purified by Ni-NTA agarose column chromatography under native conditions. Proteins were separated by SDS-PAGE and visualized by Coomassie blue staining. Lane 1, cell lysate; lane 2, flowthrough from the Ni-NTA agarose column; lane 3, washthrough; lane 4, eluate containing GTR. The arrow indicates the position of GTR at approximately 49 kDa.
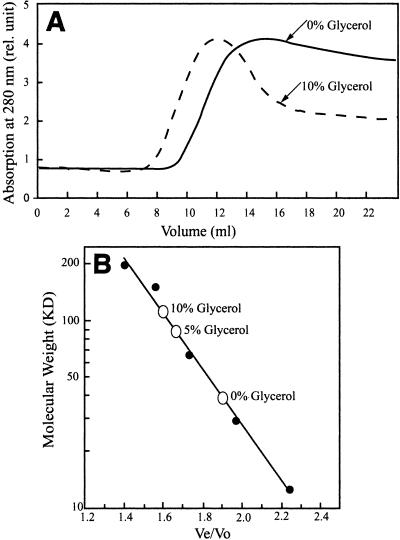
Glycerol-dependent dimerization.
The reported native molecular masses of GTRs from various sources vary over a wide range and are not consistent with peptide molecular masses predicted from gene sequences. The purified C. vibrioforme GTR, in the native form, had an apparent molecular mass of approximately 40 kDa (Fig. 3), indicating that it is a monomer. However, when the protein was mixed with 5 to 10% (vol/vol) glycerol, the molecular mass of the product was approximately double the predicted polypeptide molecular mass, indicating that the protein is a dimer under these conditions.
FIG. 3.
(A) Gel filtration of expressed recombinant C. vibrioforme GTR through Superdex 300 HR 10/30 in the absence and presence of 10% (vol/vol) glycerol. (B) Standard calibration curve for the column using proteins having known molecular weights. The open circles indicate the elution positions of purified C. vibrioforme GTR at different concentrations of glycerol.
Enzyme activity.
The purified C. vibrioforme GTR was enzymatically active. The activity had a nearly linear relationship with protein concentration over the range from 20 to 350 μg protein in a 0.5-ml reaction mixture (data not shown).
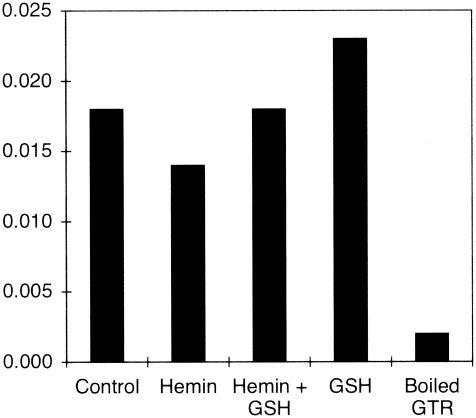
GTRs from some sources are allosterically inhibited by heme (6, 19, 20, 29, 30). Purified C. vibrioforme GTR was not inhibited by heme at concentrations up to 4 μM, the highest concentration tested (Fig. 4). The sensitivity to inhibition by heme of GTR activity in extracts of the unicellular green alga Chlorella vulgaris was reported to be enhanced by the presence of reduced glutathione (GSH). However, the presence of 5 mM GSH did not cause the activity of the purified C. vibrioforme GTR to become sensitive to heme inhibition.
FIG. 4.
GTR activity in the presence and absence of 4 μM heme and 5 mM GSH.
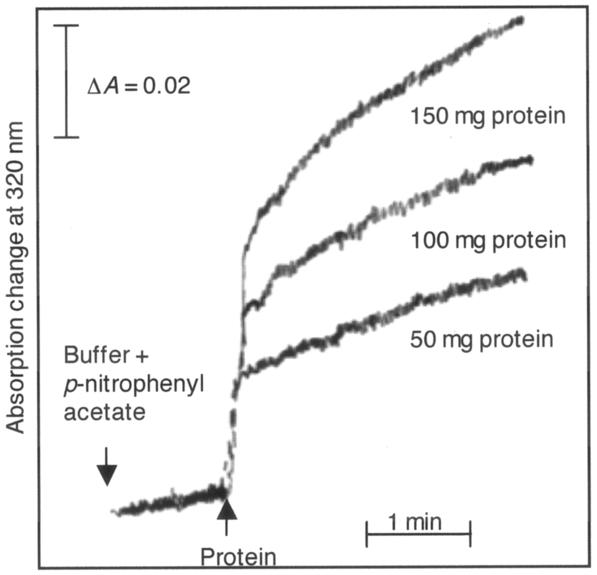
Esterase activity is an activity that is typical of enzymes that form a covalent acyl-enzyme intermediate involving an active site cysteinyl residue (for example, glyceraldehyde dehydrogenase, thiol proteinases, and aldehyde dehydrogenase). It was previously reported that GTR also has esterase activity (15, 23). His6-tagged C. vibrioforme GTR clearly exhibited esterase activity with the artificial substrate p-nitrophenyl acetate (Fig. 5). An initial high rate of hydrolysis followed by a lower steady-state rate is common for esterase reactions and indicates that the rate-limiting step of the reaction is the release of the second product (acetate) from the enzyme.
FIG. 5.
Esterase activity of expressed recombinant C. vibrioforme GTR. The time course for the formation of p-nitrophenol from p-nitrophenyl acetate was determined at 320 nm in the presence of different concentrations of the GTR protein.
Bound heme.
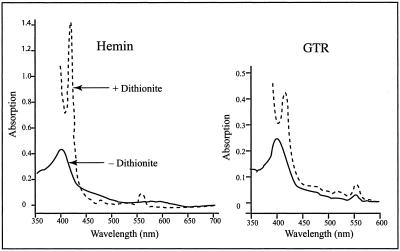
Purified C. vibrioforme GTR was light brown. One possible source of the color was bound heme. The absorption spectrum of hemin dissolved in alkaline pyridine had a peak at 400 nm and a broad shoulder at about 550 nm (Fig. 6). When the heme was reduced with sodium dithionite, the 400-nm peak became very sharp and shifted toward a longer wavelength (418.5 nm) and another sharp peak appeared at 550 nm. The absorption spectrum of the purified GTR protein also had a peak at around 400 nm in alkaline pyridine, indicating the presence of bound heme. When sodium dithionite was added, the absorption spectrum closely resembled that of reduced heme. From the heme absorption at 418.5 nm of the reduced heme in alkaline pyridine, it was calculated that the protein contains 1 mol heme per mol of polypeptide. In four separate preparations, the molar ratio of heme to protein in purified GTR ranged from 0.80 to 1.10.
FIG. 6.
Absorption spectra of reference hemin and purified GTR dissolved in alkaline pyridine before (solid line) and after (dashed line) dithionite addition.
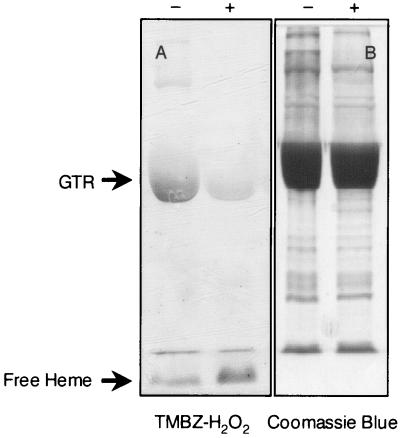
Heme remained bound to the GTR protein even after passage through gel filtration and anion-exchange (Mono-Q) or cation-exchange (Mono-S) columns (data not shown). Heme remained bound to GTR even during SDS-PAGE, as determined by heme-catalyzed peroxidase activity (Fig. 7). However, most of the bound heme was released upon denaturation of the protein in the presence of β-mercaptoethanol, indicating that the heme was not covalently bound (other than possibly by thiol-based bonds) to the GTR protein.
FIG. 7.
Polyacrylamide gels stained for heme-associated peroxidase activity with TMBZ-H2O2 (A) and for protein with Coomassie blue (B). Before protein samples were loaded onto the gel, they were denatured by boiling either in the presence of 100 mM DTT (lane −) or in the presence of 100 mM DTT plus 2.5% (vol/vol) β-mercaptoethanol (lane +).
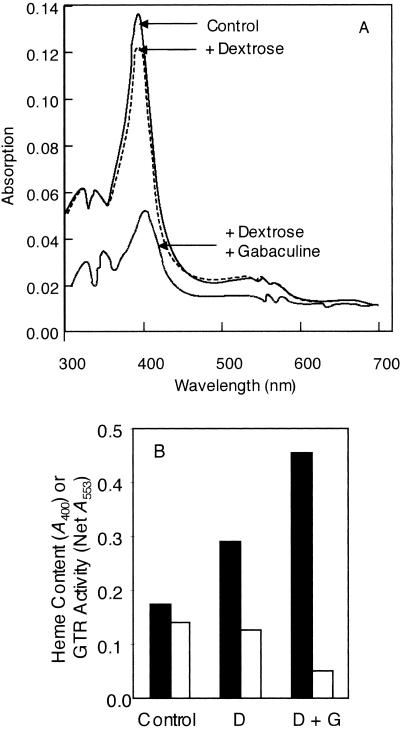
The amount of heme bound to GTR could be lowered by expressing the protein in the presence of an inhibitor of heme synthesis. GTR was expressed in the presence of 3-amino-2,3-dihydrobenzoic acid (gabaculine), an inhibitor of GSAT (1, 9). Because gabaculine inhibits the heme biosynthetic pathway, E. coli cells grown in the presence of gabaculine would be predicted to be heme deficient, and therefore there would be less heme available to bind to the expressed GTR. However, the heme deficiency also would lead to slower growth because heme is also required for respiration. Therefore, for expression in the presence of gabaculine, dextrose was also added to the LB medium to allow fermentative metabolism during the growth and GTR expression phases. When the cells were grown and induced in the presence of added 1% (wt/vol) dextrose alone or dextrose plus 100 μM gabaculine, the GTR expression level was much less than that in control cells without these additions to the medium. Nevertheless, we were able to purify His6-tagged GTR expressed in all three different situations. The A400 of purified GTR protein expressed in the presence of gabaculine and dextrose indicated that it had 70% less bound heme than GTR expressed in unsupplemented medium (Fig. 8). Dextrose alone also slightly lowered the heme content of expressed GTR.
FIG. 8.
(A) Absorption spectra of purified GTR in elution buffer (300 mM NaCl, 250 mM imidazole, 50 mM NaPO4, pH 8.0). Protein was expressed in normal LB medium or in medium supplemented with 1% (wt/vol) dextrose or dextrose plus 100 μM gabaculine. (B) Comparison of the relationship of the amount of heme bound to GTR (solid bars) to GTR activity (open bars). The expression of GTR was induced in cells in normal LB medium (Control) or in medium supplemented with 1% (wt/vol) dextrose (D) or dextrose plus 100 μM gabaculine (D + G).
The in vitro GTR activity and bound heme content were inversely related. The in vitro specific activity of the GTR protein expressed in the presence of dextrose plus gabaculine was almost threefold higher than the in vitro specific activity of GTR expressed in control cells (Fig. 8).
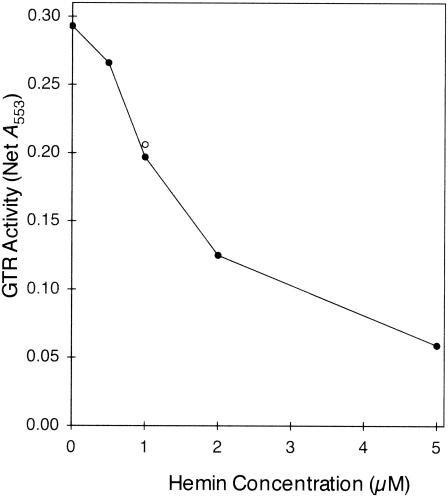
In contrast to the GTR activity expressed in control cells, the GTR activity expressed in the presence of dextrose and gabaculine was inhibited by heme (Fig. 9). The activity was inhibited more than 50% by 2 μM heme. These results indicate that the reason that GTR purified from cells expressing the protein in normal LB medium was insensitive to inhibition by added heme is that heme was already occupying all the inhibitory sites of the enzyme. The sensitivity to inhibition by subsaturating concentrations of heme (1 μM) was not increased by the presence of 5 mM GSH.
FIG. 9.
Inhibition by heme of GTR purified from cells that were induced in medium containing 1% (wt/vol) dextrose and 100 μM gabaculine. The open circle indicates the activity in the presence of 2 μM heme plus 5 mM GSH.
The specific activity of GTR purified from cells expressing the protein in the presence of dextrose plus gabaculine (60 nmol/mg protein in a 30-min assay; 2 nmol min−1 mg protein−1) was 4-fold higher than the activity previously reported for the purified E. coli GTR (22) and 160-fold higher than the activity reported for the purified Methanopyrus kandleri GTR (15).
DISCUSSION
GTR catalyzes the first committed step of tetrapyrrole biosynthesis, and it is therefore likely to be a key point of regulation of tetrapyrrole formation. Among the known regulatory mechanisms is allosteric feedback inhibition by heme for GTRs from plants, algae, and some bacteria (6, 19, 20, 29, 30).
To date, the most extensively characterized GTR at the structural level is the enzyme from the hyperthermophilic archaeon M. kandleri (15, 16). Notably, this is the only GTR whose structure has been determined by X-ray crystallography (16). Although much valuable information about GTR has been derived from studies of this enzyme, it must be noted that it may be an atypical GTR. First, because M. kandleri is a hyperthermophile, kinetic characterization of its GTR derived from experiments performed at mesothermic temperatures may not reflect the in vivo properties of the enzyme, but characterization at higher temperatures, at which the substrates and products are very unstable, is very difficult. Second, because M. kandleri does not synthesize or contain heme, in vitro effects of heme on its GTR are difficult to interpret. Therefore, to gain a better understanding of GTR, it is necessary to characterize the enzyme from additional sources.
GTR from C. vibrioforme is particularly well suited for such studies. First, this enzyme functions in vitro with commercially available E. coli tRNAGlu as well as or better than with cognate tRNAGlu from C. vibrioforme (20). Second, C. vibrioforme GTR is active when it is expressed with an N-terminal His tag, and the His-tagged enzyme remains stable through purification to a nearly homogeneous state. Finally, purified C. vibrioforme GTR has a higher specific activity than GTR from any other source thus far reported.
In view of the report that M. kandleri GTR is homodimeric and cannot be dissociated into monomers without denaturation of the protein (16), it was unexpected that C. vibrioforme GTR is monomeric but becomes dimeric in solutions containing glycerol. It is unlikely that this difference, compared to the M. kandleri GTR, is due to the presence of the His tag, because structural studies of the M. kandleri GTR indicated that the dimerization domain is at the C terminus (16), whereas the His tag on the C. vibrioforme GTR is located at the N terminus. Further characterization of GTRs from additional sources is required to determine whether the C. vibrioforme GTR is unusual in its glycerol-dependent dimerization or if the very tight dimeric state of the M. kandleri GTR is associated with its derivation from a hyperthermophilic species.
Although initial observations of C. vibrioforme GTR suggested that the enzyme is not sensitive to allosteric inhibition by heme, it was later determined that the expressed enzyme, as purified by Ni affinity column chromatography, contains tightly bound heme at a 1:1 ratio with polypeptides. It was not possible to remove this heme without denaturing the protein. However, when the GTR was expressed under conditions under which the host cells were heme deficient, there was much less bound heme and the GTR activity was sensitive to heme inhibition. Based on a comparison of the activities of the enzyme expressed in heme-deficient and heme-replete cells, it is clear that the heme-containing GTR is only about 30% as active as the mostly heme-free GTR. However, even saturating concentrations of heme do not inhibit the activity further. It is possible that the apparent insensitivity or low sensitivity to heme inhibition reported for some other GTR enzymes (8, 14) might also be due to the presence of saturating amounts of heme already bound to the enzyme when it is isolated. Previously, purified recombinant barley GTR was reported to contain a tightly bound heme molecule that could be reduced by NADPH and oxidized by air (27). The role of this heme and its effect on enzyme activity have not been reported.
The presence of GSH, which increases the sensitivity of the C. vulgaris GTR to heme inhibition severalfold (31), does not affect the inhibition of C. vibrioforme GTR by subsaturating heme concentrations. Further study of GTRs from additional sources is necessary to determine whether the GSH effect on heme inhibition is unique to the C. vulgaris GTR or is characteristic of eukaryotic (chloroplastic) GTRs. It should be noted that the concentration of heme required for 50% inhibition of purified C. vibrioforme GTR in this study (approximately 2 μM) is significantly greater than the concentration (0.2 μM) required for 50% inhibition of ALA formation from glutamate in an unfractionated C. vibrioforme extract (20). Therefore, it is possible that unidentified substances in C. vibrioforme extracts may influence heme inhibition of GTR.
Although GTR has been shown to be the rate-limiting enzyme for ALA synthesis from glutamate in vitro, using unfractionated cell extracts (19) it has been difficult to establish whether this is also true in vivo, because overexpressed GTR is inhibited by low concentrations of heme and in some cases it is rapidly degraded (28), so that effects of its overexpression in otherwise wild-type cells are not observed. Antisense repression of GTR expression in Arabidopsis thaliana was shown to decrease heme and chlorophyll synthesis (11), but whether GTR is rate limiting at its normal expression level is not known. Because C. vibrioforme GTR can be highly expressed in E. coli cells and its activity is not completely inhibited by heme, its overexpression in E. coli cells caused ALA to accumulate, thereby directly demonstrating that GTR is the rate-limiting enzyme for ALA synthesis.
As was previously reported for GTRs from M. kandlerii and E. coli, C. vibrioforme GTR has intrinsic esterase activity. This suggests a common catalytic mechanism for the GTRs, in which an active site cysteine residue acts as an acceptor for the activated glutamyl α carbonyl group of glutamyl-tRNA. As proposed for the M. kandlerii and E. coli enzymes, the bound thioester would then be the immediate substrate for reduction to an aldehyde by NADPH.
In conclusion, C. vibrioforme GTR was expressed in a highly active form with an N-terminal His tag that allowed rapid purification almost to homogeneity. Some of its properties are similar to those of other GTRs, and some are significantly different. The protein undergoes glycerol-dependent dimerization and contains 1 mol of tightly bound heme per mol of polypeptide. The heme-bound form is catalytically active and is not further inhibited by heme. Higher activity is observed, smaller amounts of heme are present, and inhibition by heme occurs with GTR that is expressed in heme-deficient cells. The intrinsic esterase activity suggests that the activity of C. vibrioforme GTR, like that reported for the M. kandleri and E. coli enzymes, involves formation of a thioester bond between the activated glutamate carboxyl group and a protein cysteine residue. Accumulation of ALA in E. coli cells expressing C. vibrioforme GTR indicated that the GTR step is the rate-limiting step of ALA synthesis.
Acknowledgments
This work was supported by National Science Foundation grant MCB-9808578 to S.I.B.
We thank S. P. Gough for bringing to our attention the possibility of an error in the previously published C. vibrioforme hemA sequence, R. D. Willows for help with and advice concerning DNA sequencing, and L. A. Nogaj for critical comments on the manuscript.
REFERENCES
- 1.Avissar, Y. J., and S. I. Beale. 1989. Biosynthesis of tetrapyrrole pigment precursors: pyridoxal requirement of the aminotransferase step in the formation of δ-aminolevulinate from glutamate in extracts of Chlorella vulgaris. Plant Physiol. 89:852-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avissar, Y. J., and S. I. Beale. 1990. Cloning and expression of a structural gene from Chlorobium vibrioforme that complements the hemA mutation in Escherichia coli. J. Bacteriol. 172:1656-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beale, S. I. 1999. Enzymes of chlorophyll biosynthesis. Photosynth. Res. 60:43-73. [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Gibson, K. D. 1958. Biosynthesis of δ-aminolaevulic acid by extracts of Rhodopseudomonas spheroides. Biochim. Biophys. Acta 28:451. [DOI] [PubMed] [Google Scholar]
- 6.Gough, S. P., and C. G. Kannangara. 1979. Biosynthesis of δ-aminolevulinate in greening barley leaves. III. The formation of δ-aminolevulinate in tigrina mutants of barley. Carlsberg Res. Commun. 44:403-416. [DOI] [PubMed] [Google Scholar]
- 7.Jahn, D., G. P. O'Neill, E. Verkamp, and D. Söll. 1992. Glutamate tRNA: involvement in protein-synthesis and aminolevulinate formation in Chlamydomonas reinhardtii. Plant Physiol. Biochem. 30:245-253. [Google Scholar]
- 8.Jahn, D., U. Michelsen, and D. Söll. 1991. Two glutamyl-tRNA reductase activities in Escherichia coli. J. Biol. Chem. 266:2542-2548. [PubMed] [Google Scholar]
- 9.Kannangara, C. G., and A. Schouboe. 1985. Biosynthesis of δ-aminolevulinate in greening barley leaves. VII. Glutamate 1-semialdehyde accumulation in gabaculine treated leaves. Carlsberg Res. Commun. 50:179-191. [DOI] [PubMed] [Google Scholar]
- 10.Kikuchi, G., A. Kumar, P. Talmage, and D. Shemin. 1958. The enzymatic synthesis of δ-aminolevulinic acid. J. Biol. Chem. 233:1214-1219. [PubMed] [Google Scholar]
- 11.Kumar, A. M., and D. Söll. 2000. Antisense HEMA1 RNA expression inhibits heme and chlorophyll biosynthesis in Arabidopsis. Plant Physiol. 122:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majumdar, D., Y. J. Avissar, J. H. Wyche, and S. I. Beale. 1991. Structure and expression of the Chlorobium vibrioforme hemA gene. Arch. Microbiol. 156:281-289. [DOI] [PubMed] [Google Scholar]
- 13.Mauzerall, D., and S. Granick. 1956. The occurrence and determination of δ-aminolevulinic acid and porphobilinogen in urine. J. Biol. Chem. 219:435-446. [PubMed] [Google Scholar]
- 14.Mayer, S. M., J. D. Weinstein, and S. I. Beale. 1987. Enzymatic conversion of glutamate to δ-aminolevulinate in soluble extracts of Euglena gracilis. J. Biol. Chem. 262:12541-12549. [PubMed] [Google Scholar]
- 15.Moser, J., S. Lorenz, C. Hubschwerlen, A. Rompf, and D. Jahn. 1999. Methanopyrus kandleri glutamyl-tRNA reductase. J. Biol. Chem. 274:30679-30685. [DOI] [PubMed] [Google Scholar]
- 16.Moser, J., W.-D. Schubert, V. Beier, I. Bringemeier, D. Jahn, and D. W. Heinz. 2001. V-shaped structure of glutamyl-tRNA reductase, the first enzyme of tRNA-dependent tetrapyrrole biosynthesis. EMBO J. 20:6583-6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Neill, G. P., D. M. Peterson, A. Schön, M.-W. Chen, and D. Söll. 1988. Formation of the chlorophyll precursor δ-aminolevulinic acid in cyanobacteria requires aminoacylation of a tRNAGlu species. J. Bacteriol. 170:3810-38166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Randau, L., S. Schauer, A. Ambrogelly, J. C. Salazari, J. Moser, S. Sekine, S. Yokoyama, D. Söll, and D. Jahn. 2004. tRNA recognition by glutamyl-tRNA reductase. J. Biol. Chem. 279:34931-34937. [DOI] [PubMed] [Google Scholar]
- 19.Rieble, S., and S. I. Beale. 1988. Enzymatic transformation of glutamate to δ-aminolevulinic acid by soluble extracts of Synechocystis sp. 6803 and other oxygenic prokaryotes. J. Biol. Chem. 263:8864-8871. [PubMed] [Google Scholar]
- 20.Rieble, S., J. G. Ormerod, and S. I. Beale. 1989. Transformation of glutamate to δ-aminolevulinic acid by soluble extracts of Chlorobium vibrioforme. J. Bacteriol. 171:3782-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook, J., and Russell, D. W. 2001. Molecular cloning, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Schauer, S., C. Lüer, and J. Moser. 2003. Large scale production of biologically active Escherichia coli glutamyl-tRNA reductase from inclusion bodies. Protein Expr. Purif. 31:271-275. [DOI] [PubMed] [Google Scholar]
- 23.Schauer, S., S. Chaturvedi, L. Randau, J. Moser, M. Kitabatake, S. Lorenz, E. Verkamp, W.-D. Schubert, T. Nakayashiki, M. Murai, K. Wall, H.-U. Thomann, D. W. Hieinz, H. Inokuchi, D. Söll, and D. Jahn. 2002. Escherichia coli glutamyl-tRNA reductase. Trapping the thioester intermediate. J. Biol. Chem. 277:48658-48663. [DOI] [PubMed] [Google Scholar]
- 24.Smith, K. M. 1975. Porphyrins and metalloporphyrins. Elsevier, Amsterdam, The Netherlands.
- 25.Thomas, P. E., D. Ryan, and W. Levin. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Ann. Biochem. 75:168-176. [DOI] [PubMed] [Google Scholar]
- 26.Urata, G., and S. Granick. 1963. Biosynthesis of α-aminoketones and the metabolism of aminoacetone. J. Biol. Chem. 238:811-820. [PubMed] [Google Scholar]
- 27.Vothknecht, U. C., C. G. Kannangara, and D. von Wettstein. 1996. Expression of catalytically active barley glutamyl tRNAGlu reductase in Escherichia coli as a fusion protein with glutathione S-transferase. Proc. Natl. Acad. Sci. USA 93:9287-9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, L. Y., M. Elliott, and T. Elliott, T. 1999. Conditional stability of the HemA protein (glutamyl-tRNA reductase) regulates heme biosynthesis in Salmonella typhimurium. J. Bacteriol. 181:1211-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, W.-Y., D.-D. Huang, D. Stachon, S. P. Gough, and C. G. Kannangara. 1984. Purification, characterization, and fractionation of the δ-aminolevulinic acid synthesizing enzymes from light-grown Chlamydomonas reinhardtii cells. Plant Physiol. 74:569-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinstein, J. D., and S. I. Beale. 1985. Enzymatic conversion of glutamate to δ-aminolevulinate in soluble extracts of the unicellular green alga, Chlorella vulgaris. Arch. Biochem. Biophys. 237:454-464. [DOI] [PubMed] [Google Scholar]
- 31.Weinstein, J. D., R. W. Howell, R. D. Leverette, S. Y. Grooms, P. S. Brignola, S. M. Mayer, and S. I. Beale. 1993. Heme inhibition of δ-aminolevulinic acid synthesis is enhanced by glutathione in cell-free extracts of Chlorella. Plant Physiol. 101:657-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinstein, J. D., S. M. Mayer, and S. I. Beale. 1986. Stimulation of δ-aminolevulinic acid formation in algal extracts by heterologous RNA. Plant Physiol. 82:1096-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]