Abstract
The global dissemination of the multiply-antibiotic-resistant Salmonella enterica serovar Typhimurium DT104 clone with the resistance genes located in a class 1 integron, here designated In104, within genomic island SGI1 is a significant public health issue. Here, we have shown that SGI1 and variants of it carrying different combinations of resistance genes are found in several Salmonella enterica serovars. These are serovars Cerro, Derby, Dusseldorf, Infantis, Kiambu, and Paratyphi B dT+ isolated from human infections and serovar Emek from sewage effluent. Two new variants, SGI1-I and SGI1-J, both of which include the dfrA1-orfC cassette array, were identified.
Most multiply-antibiotic-resistant (MR) Salmonella enterica serovar Typhimurium DT104 isolates exhibit resistance to ampicillin, chloramphenicol, florfenicol, streptomycin, spectinomycin, sulfonamides, and tetracycline but, because screening for spectinomycin and florfenicol resistance is not routine, are often incorrectly described as “pentaresistant.” Multidrug resistance in the majority of DT104 strains is due to the presence of five genes aadA2, sul1, floR, tetA(G), and blaP1 (variant form blaPSE-1 or blaCARB-2) that confer resistance to streptomycin and spectinomycin, sulfonamides, chloramphenicol and florfenicol, tetracyclines, and β-lactam antibiotics, respectively (4, 5). These isolates represent a single clone that has emerged as a dominant serovar Typhimurium clone globally and has been isolated from humans, most meat-producing animals, exotic birds, and various foods, including meat, dairy products, and salad ingredients (43, 45).
Molecular typing studies have shown that this MR DT104 clone appeared simultaneously in the United Kingdom, Europe, and the United States during the mid-1980s (9). MR DT104 with the same resistance profile (ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline) had been isolated from humans in Asia prior to 1982 (22), but it is not currently known if these are identical to the globally disseminated clone. MR DT104 is now the second most prevalent Salmonella serotype isolated from humans in the United Kingdom, and the incidence of this clone has increased significantly in the United States, South Africa, the United Arab Emirates, Trinidad, the Philippines, the Irish Republic, Canada, Israel, Japan, and most European countries (36, 43, 45). However, MR DT104 has not taken hold in Australia, where it has been detected only infrequently in imported foods and travelers returning from overseas (D. Lightfoot, unpublished observations).
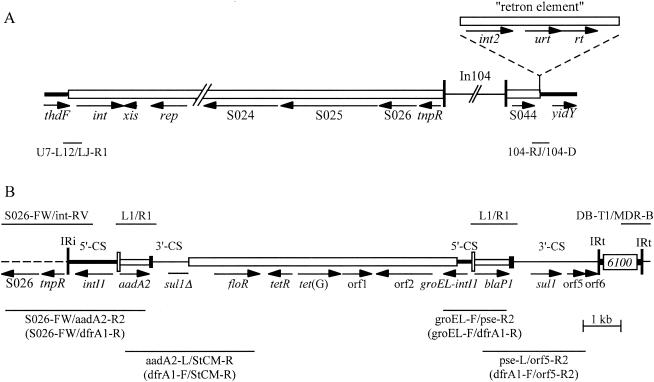
The resistance genes in the major MR DT104 clone all lie within the boundaries of a large (13 kb) class 1 integron with a complex structure, here named In104 (Fig. 1). The sequence of this region (4, 5) has revealed that the integron has a backbone similar to that of In4 (28, 29), but it includes duplications of parts of the integron conserved segments (CS), specifically, part of the intI1 gene from the 5′-CS and part of the 3′-CS (qacEΔ1 and partial sul1 genes) that normally flank integrated gene cassettes (16, 17). As a consequence, In104 includes two attI1 sites, into which gene cassettes can be incorporated (8, 16, 17, 35), with the aadA2 gene cassette in one and the blaP1 cassette in the other (37, 39). This arrangement produces a characteristic pair of PCR products of 1.0 kb and 1.2 kb using standard primers that amplify the cassette-containing regions, and these regions have often been called InC and InD (37, 39), assuming there are two integrons rather than one. The floR and tetRA(G) genes lie between the two integron-derived regions (5) together with an open reading frame, orf2, that is potentially part of an unusual type of insertion sequence recently named CR3 (32).
FIG. 1.
Structure of SGI1 regions. (A) The SGI1 region of serovar Typhimurium DT104 (from GenBank accession no. AF261825) is drawn to scale except for In104, which is to scale in B. The SGI1 backbone and the retron element (shown above) are represented by open boxes, and the chromosomal and In104 regions as lines of different thicknesses. Vertical bars indicate the IR bounding In104. Fragments amplified by PCR are shown as thin lines below, with the primer pairs used (Table 1) indicated underneath. (B) In104 region. Dashed lines represent the surrounding SGI1 backbone and narrow vertical bars represent the IRs (IRi and IRt). The 5′-CS, 3′-CS, and tni regions of class 1 integrons are indicated by lines of different thicknesses, with attI1 as a tall open box and gene cassettes as open boxes with a black bar at one end, indicating the 59-be. IS6100 and the central, non-integron-derived region are open boxes. Arrows indicate the position and orientation of genes and ORFs. Primer pairs used to detect the boundaries of In104 or to determine the size of the cassette array and the location of cassettes are indicated above or below.
In104 lies within a genomic island known as SGI1 (4) (GenBank accession no. AF261825). It appears to have moved to this location by transposition because there is a 5-bp direct duplication flanking the 25-bp inverted repeats (IRs) of the integron and it is located in a presumptive res site adjacent to a resolvase gene (tnpR) in the SGI1 backbone, which is where class 1 integrons are most commonly found (18, 29). SGI1 has been completely sequenced and is a 43-kb genomic island located in the chromosome between the thdF and yidY genes (4). Based on the fact that an int gene encoding a site-specific recombinase belonging to the tyrosine recombinase family was found within SGI1, at the left-hand end adjacent to the thdF gene, SGI1 is likely to have been integrated into the chromosome by site-specific recombination (4).
A second integrated element that also includes an int gene and that has been called a retron-phage was found between SGI1 and the yidY gene in DT104 strains (4) but is not usually present in other S. enterica serovars (3, 10, 12, 13, 24). SGI1 was subsequently found in serovar Typhimurium strains typed as DT1, DT12, DT120, and U302 (3, 6, 19), but it has been shown that the DT12 and DT120 strains probably arose by changes in the phage susceptibility of the SGI1-containing DT104 strain (19). However, SGI1 has also been found in other serovars with diverse sources; Agona isolated from poultry in Belgium (3, 10), Paratyphi B dT+ (formerly biovar Java) from tropical fish in Singapore (24), Albany from food fish in Thailand (12), Meleagridis from an unspecified animal in the United States (14), and Newport, isolated from a human (13), confirming that it is not confined to the serovar Typhimurium or to DT104 strains and suggesting that it may be able to spread by horizontal transfer.
Variations in the structure of the integron, all of which could have arisen by homologous recombination, have also been identified (3, 6, 10, 12, 13). One type of variation involves loss of some of the resistance genes, which can occur via homologous recombination between one or other pair of duplicated regions, namely the 5′-CS or 3′-CS, causing loss of either aadA2, floR, and tet(G) or floR, tet(G), and blaP1 (3, 6). Other variations involve the exchange of one or both of the cassette arrays to introduce different gene cassettes (10, 12, 13), which can also occur via homologous recombination (30, 31). A further source of variation (3) is the acquisition of the dfrA10 trimethoprim resistance gene (27) together with the sequences that flank it in a different type of complex class 1 integron that has a duplicated 3′-CS and a potential insertion sequence, CR1, that is related to CR3 (3, 32). Acquisition of this segment introduces an additional copy of the sul1 gene and has been shown experimentally to occur via homologous recombination (30).
Here we have screened a set of resistant and MR S. enterica isolates belonging to 20 different serovars and derived from various sources for the presence of SGI1. All strains were isolated in Australia and originated from human infections, acquired either locally and overseas, or from sick animals or environmental sources in Australia. The structure of the class 1 integron in isolates containing SGI1 was analyzed and complete maps were derived using a variety of methods.
MATERIALS AND METHODS
Strain collection.
A total of 76 S. enterica strains isolated in Australia in 1999 or 2001 and submitted to the Microbiological Diagnostic Unit for typing were examined. Of these, 39 isolates were from humans with infections, with 16 isolates recorded as acquired overseas (recently returned travelers), and the remainder presumed to be acquired locally. The rest of the collection consisted of 36 isolates from Australian animals, pigs (10 isolates), cattle (9 isolates), chickens (9 isolates), ducks (5 isolates), horses (2 isolates), and cats (1 isolate), all presumed to be associated with disease, and one isolate from sewage effluent. A total of 20 different serovars, determined using standard procedures (33), were included in the collection. The collection contained 14 isolates of serovars Typhimurium (six from humans; four bovine; two porcine; two equine), and Hadar (nine human; five duck), seven serovar Sofia (two human; five chicken), six each of serovars Infantis (two human; three chicken; one cat) and Bovismorbificans (one human; five bovine), four Derby (two human; two porcine) and Agona (two human; two porcine), three Kiambu (two human; one chicken), two each of Blockley (human), Havana (one human; one porcine), Ohio (porcine), Paratyphi B dT+ (biovar Java) (human), Singapore (human), Stanley (human), single isolates of Cerro, Dusseldorf, Enteritidis, Montevideo (all human), and Seftenberg (porcine), and a single serovar Emek isolate from sewage effluent.
All strains had been shown to be resistant to at least one of the following antibiotics at the concentrations shown: ampicillin (Ap, 32 μg/ml), chloramphenicol (Cm, 10 μg/ml), gentamicin (Gm, 2.5 μg/ml), kanamycin (Km 10 μg/ml), streptomycin (Sm, 25 μg/ml), spectinomycin (Sp, 50 μg/ml), sulfathiazole (Su, 550 μg/ml), tetracycline (Tc, 20 μg/ml), or trimethoprim (Tp, 50 μg/ml) using procedures described previously (1, 2). The collection included a broad range of resistance profiles with six isolates resistant to only one of these antibiotics, 18 isolates resistant to two antibiotics, nine resistant to three antibiotics, seven resistant to four, and 36 resistant to five or more (up to nine) antibiotics. Resistance to nalidixic acid (Na, 50 μg/ml) and ciprofloxacin (Cp, 2 μg/ml) was also recorded in some isolates, mainly from humans. A single SGI1-containing DT104 strain, isolated from a human and traced back to an imported food (15), was included for comparison.
PCR.
The primers used are listed in Table 1. For initial PCR screening, 5 μl of template prepared by resuspending a single colony in 1 ml of sterile Milli-Q water, heating to 100°C for 5 min, and pelleting debris by centrifugation (13,000 rpm, 10 min) was used. DNA of higher purity was isolated using a standard protocol (38), and 10 to 50 ng was used in all subsequent analyses. For detailed PCR mapping, DNA fragments were amplified in a reaction mixture (final volume of 50 μl) containing 10 mM Tris-HCl (pH 8.3), 10 mM KCl, 2 mM MgCl2 and 0.2 mM of each deoxynucleoside triphosphate with 20 pmol of each primer and 1 U of Taq DNA polymerase. Thermal cycling conditions consisted of an initial denaturation cycle (95°C, 5 min) followed by 35 cycles of denaturation (95°C, 30 s), annealing at temperatures ranging from 57 to 65°C (30 to 60 s), and extension (72°C, 60 to 120 s), and a final cycle of amplification (72°C, 5 min). Annealing temperatures for all primer pairs shown in Fig. 1 were 60°C except for groEL-F/pse-R2 (65°C), groEL-F/orfC (65°C), and L1/R1 (57°C). Amplified DNA fragments were resolved by electrophoresis using a 1 to 2% (wt/vol) agarose gel, stained with ethidium bromide (5 μg/ml), visualized with UV, and imaged using a GelDoc 1000 image analysis station (Bio-Rad, Hercules, CA).
TABLE 1.
Primers
| Primer | Sequence (5′-3′) | Location | Nucleotide positions | Accession no. | Referencea |
|---|---|---|---|---|---|
| U7-L12 | ACA CCT TGA GCA GGG CAA AG | thdF | 1-20 | AF261825.2 | 4 |
| LJ-R1 | AGT TCT AAA GGT TCG TAG TCG | int | 500-480 | AF261825.2 | 4 |
| SGI19-F3 | TTC TTC AAT GCG CGC TTT GG | S023 | 19009-19028 | AF261825.2 | This study |
| S026-FW | TCG GGT AAT CTC AGC AGA GC | S026 | 25021-25040 | AF261825.2 | 6 |
| int-RV | GGG CAT GGT GGC TGA AGG ACC | intI1 | 27266-27246 | AF261825.2 | 6 |
| L1 | GGC ATC CAA GCA GCA AGC | 5′-CS | 27892-27909, 37164-37181 | AF261825.2 | 21 |
| R1 | AAG CAG ACT TGA CCT GAT | 3′-CS | 28900-28883, 38360-38343 | AF261825.2 | 21 |
| aadA2-L | TGT TGG TTA CTG TGG CCG | aadA2 | 28146-28163 | AF261825.2 | 26 |
| aadA2-R2 | TGC TTA GCT TCA AGT AAG ACG | aadA2 | 28683-28663 | AF261825.2 | 3 |
| QS-1 | ATG AAA GGC TGG CTT TTT CTT G | qacEΔ1 | 28953-28974, 38413-38424 | AF261825.2 | 4 |
| sul1-F | GTG ACG GTG TTC GGC ATT CT | sul1 | 29297-29316, 35757-35776 | AF261825.2 | 20 |
| QS-2 | TGA GTG CAT AAC CAC CAG CC | sul1 | 29674-29655, 31934-31915 | AF261825.2 | 4 |
| MDR-7 | AAC CGT GCA TCT ATC GAG C | —b | 30094-30112 | AF261825.2 | 3 |
| StCM-L | CAC GTT GAG CCT CTA TAT GG | floR | 30640-30659 | AF261825.2 | 3 |
| StCM-R | ATG CAG AAG TAG AAC GCG AC | floR | 31527-31508 | AF261825.2 | 3 |
| F6 | TTG GAA ACA GAC GGC ATG G | tetR(G) | 31995-31977 | AF261825.2 | 3 |
| tetG-L | CAG CTT TCG GAT TCT TAC GG | tetA(G) | 32819-32838 | AF261825.2 | 26 |
| tetG-R | GAT TGG TGA GGC TCG TTA GC | tetA(G) | 33662-33643 | AF261825.2 | 26 |
| orf1-F | CGC CGA ATA TCT CAA CTT CCG | orf1 | 33927-33947 | AF261825.2 | This study |
| orf1-R | CCG CGA TGC CGA AAT CCC | orf1 | 34581-34564 | AF261825.2 | This study |
| orf2-F2 | ATG CAG TGA GAA GCC GC | orf2 | 35291-35307 | AF261825.2 | This study |
| orf2-R2 | TAC TCG AGC ACG GCT TC | orf2 | 36024-36008 | AF261825.2 | This study |
| groEL-F | ATG CCG CCC ATA CCG CCA GC | groEL | 36490-36509 | AF261825.2 | This study |
| pse-L | AAT GGC AAT CAG CGC TTC CC | blaP1 | 37495-37514 | AF261825.2 | 26 |
| pse-R2 | ACA ATC GCA TCA TTT CGC TC | blaP1 | 38147-38128 | AF261825.2 | 3 |
| sul1-R | TTT ACA GGA AGG CCA ACG GT | sul1 | 39424-39405 | AF261825.2 | 20 |
| orf5-F | AGG TTG TGC GGC TGA TGC | orf5 | 39776-39793 | AF261825.2 | This study |
| orf5-R2 | CGA GTT CTA GGC GTT CTG C | orf5 | 40213-40195 | AF261825.2 | This study |
| orf6-R | ACT ATC TTC GGC CTT CAC ACG | orf6 | 40508-40488 | AF261825.2 | This study |
| DB-T1 | TGC CAC GCT CAA TAC CGA C | IS6100 | 41120-41138 | AF261825.2 | 3 |
| IS6100-Rv2 | AAT GGT GGT TGA GCA TGC C | IS6100 | 41475-41457 | AF261825.2 | This study |
| MDR-Bc | GAA TCC GAC AGC CAA CGT TCC | S044 | 41905-41884 | AF261825.2 | 3 |
| 104-RJ | TGA CGA GCT GAA GCG AAT TG | S044 | 42373-42392 | AF261825.2 | 3 |
| C9-L2 | AGC AAG TGT GCG TAA TTT GG | Retron element | 42868-42887 | AF261825.2 | 3 |
| 104-D | ACC AGG GCA AAA CTA CAC AG | yidY | 47130-47111 | AF261825.2 | 3 |
| dfrA1-F | CGA AGA ATG GAG TTA TCG G | dfrA1 | 621-639 | X17477.1 | This study |
| dfrA1-R | TTA GAG GCG AAG TCT TGG | dfrA1 | 1029-1012 | X17477.1 | This study |
| orfC-F | CAT TAC GAA GCG AAT GCA CC | orfC | 1172-1191 | X17477.1 | This study |
| orfC-R | TCT CGA ATC AAG CAG GAA CC | orfC | 1534-1515 | X17477.1 | This study |
| orf513-F | ATG TCG CTG GCA AGG AAC G | orf513 | 3521-3539 | L06418.4 | This study |
| RH379 | ACC AGA GCA TTC GGT AAT CAA G | dfrA10 | 5530-5551 | L06418.4 | This study |
| RH380 | GCT TCA GAT AAT AAA CCA ACA CCA CC | dfrA10 | 5828-5803 | L06418.4 | This study |
References are for the primer.
MDR-7 targets the sequence preceding floR.
The sequence from AF261825.2 is GAATCCGACAGCCAACGCTTCC.
Hybridization probes for tet(G) (amplified with the tetG-L and tetG-R primers) and orf2 (amplified using orf2-F2 and orf2-R2 primers) were labeled with digoxigenin using the digoxigenin Easy Hyb kit (Roche Diagnostics Corporation, Indianapolis, Indiana) and the manufacturer's instructions. PCR cycling conditions consisted of a denaturation cycle (94°C, 5 min) followed by 30 cycles of denaturation (94°C, 30 s), annealing at 60°C for tet(G) and 64°C for orf2 (30 s), and extension (72°C, 90 s), and a final extension cycle (72°C, 10 min).
Southern hybridization.
DNA was digested with BsaI, resolved by electrophoresis through 0.7% agarose, and transferred to N+ nylon membrane (Amersham, Buckinghamshire, United Kingdom) by capillary action as described previously (38). The DNA was cross-linked to the membrane by baking at 80°C for 2 h and exposed to denatured digoxigenin-labeled tet(G) and orf2 probes overnight at 42°C. Membranes were then washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)/0.1% sodium dodecyl sulfate (SDS) for 15 min at room temperature and twice, more stringently, with 2× SSC/0.1% SDS for 15 min at 68°C. Bands were visualized fluorescently using standard protocols supplied the manufacturer (Roche Diagnostics Corporation). Digoxigenin-labeled molecular mass markers (molecular marker set II, Roche Diagnostics Corporation) were included on all membranes.
DNA sequencing.
PCR products were prepared for sequencing using the QIAquick PCR purification kit (QIAGEN, Hilden, Germany) following protocols supplied by the manufacturer. Sequencing was performed using the Big Dye Terminator reaction mix version 3.2 (Applied Biosystems, Foster City, CA). Cycle sequencing reactions (20 μl final volume) contained a PCR amplicon (30 to 90 ng), 3.2 pmol of primer, and 8 μl of reaction mix and consisted of 28 cycles of denaturation (90°C, 30 s), annealing (50°C, 15 s) and extension (60°C, 240 s).
Nucleotide sequence accession number.
The sequence of the Emek (SGI1-J) drfA1-orfC cassette array has been submitted to the GenBank database under accession no. AY963803.
RESULTS
Identification of SGI1-containing strains.
All strains were screened by PCR for the presence of SGI1 with or without the retron element using published primer pairs (3) (Table 1) that span the boundaries of SGI1 with the chromosome or with the retron element (Fig. 1A). The MR DT104 control and nine isolates representing seven serovars, Cerro, Derby, Dusseldorf, Emek, Infantis, Kiambu (two isolates), and Paratyphi B dT+ (two isolates), contained both ends of SGI1 (Table 2). The retron element was found only in the DT104 control (data not shown). These strains (Table 2) included 8 of the 39 human isolates in the collection but none originating from animals. The ninth strain was from sewage effluent.
TABLE 2.
Analysis of the boundaries of SGI1 and In104 within SGI1 in multiple-antibiotic-resistant Salmonella spp.
| Strain | Serovar | Source | Countrya | Date of isolation (day/mo/yr) | Junctionsb
|
|||
|---|---|---|---|---|---|---|---|---|
| SGI LJ | SGI RJ | In104 LJ | In104 RJ | |||||
| SRC 4 | Derby | Human | Malaysia | 20/05/99 | + | + | + | + |
| SRC 5 | Cerro | Human | Thailand | 10/09/99 | + | + | + | + |
| SRC 10c | Kiambu | Human | Australia | 01/08/99 | + | + | + | + |
| SRC 11c | Kiambu | Human | Australia | 01/04/99 | + | + | + | + |
| SRC 19e | Emek | Effluent | Australia | 15/09/99 | + | + | — | — |
| SRC 38 | Dusseldorf | Human | Malaysia | 30/08/01 | + | + | + | + |
| SRC 46 | Infantis | Human | Australia | 10/02/01 | + | + | + | + |
| SRC 49c | Paratyphi B dT+ | Human | Australia | 04/09/01 | + | + | + | + |
| SRC 50c | Paratyphi B dT+ | Human | Australia | 02/05/01 | + | + | + | + |
| DT104-2 | Typhimurium | Human | Australiaf | ??/05/01 | + | —d | + | + |
Countries listed represent regions recently visited by affected patients.
Primer pairs used (3, 4) with product sizes in brackets are SGI1-LJ, U7-L12, and LJ-R1 (0.5 kb); SGI1 RJ, 104-RJ, and 104-D (0.5 kb); In104 LJ, S026FW, and int-RV (2.245 kb); In104 RJ, DB-T1, and MDR-B (0.786 kb).
Same species isolates 10 and 11 or 49 and 50 are not known to be epidemiologically linked.
DT104 strains with SGI1 possess a retron element that disrupts the boundary spanned by primers 104-RJ/104-D.
Primers SGI19(F3) and int-RV were used to link the SGI1 backbone to In104 in SRC19.
Isolate linked to the consumption of halva (origin, Turkey) contaminated with DT104 (15).
To determine if the integron characteristic of SGI1 and its variants was present, published primer pairs (3, 6) spanning the boundaries of In104 with the SGI1 backbone (Fig. 1B) were used. PCR amplification yielded products of the sizes expected for the integron located in SGI1 in all except the serovar Emek strain, where neither boundary was detected (Table 2). However, using the outward-facing primer internal to the integron together with primers in other SGI1 backbone genes, the left or IRi end of a class 1 integron was found adjacent to part of the S023 gene. The sequence of this fragment revealed that 6,889 bp of SGI1 backbone sequence normally found adjacent to the IRi end of the integron was missing. Primers internal to the IS6100 element at the right end of In104 coupled with right-hand primers in the adjacent chromosomal yidY gene did not reveal a junction, suggesting that a large insertion may be present at this end in the serovar Emek strain.
Identification of resistance genes.
When the resistance profiles were examined (Table 3), only four of the strains, SRC49 and SRC50 (both serovar Paratyphi B dT+ isolates) and SRC10 and SRC11 (both serovar Kiambu isolates), were resistant to all antibiotics in the standard SGI1-containing DT104 set (Ap, Cm and Fl, Sm and Sp, Su, and Tc). Resistance to trimethoprim (Tp) was observed in seven strains and resistance to gentamicin (Gm) in one strain. The nine strains were also screened for the presence of each of the genes and open reading frames (ORFs) found in In104 (see Fig. 1B) using pairs of primers internal to the genes (Table 1). The standard SGI1 resistance phenotype (ApCmFlSmSpSuTc) and resistance gene complement, aadA2, blaP1, sul1, tetA(G), and floR, consistent with the presence of an integron identical to or closely related to In104, were only present in the two Paratyphi B dT+ strains (Table 3). Hence, most of the strains contained a variant of SGI1. Consistent with this conclusion, five strains lacked one or both of the cassette-associated resistance genes aadA2 (Sm and Sp) and blaP1 (Ap) (Table 3). In the serovar Infantis strain, the floR and tet(G) genes as well as orf1 and orf2 (Fig. 1B), which can be lost together if recombination between the duplicated 5′-CS or 3′-CS regions occurs (3), were also missing, indicating loss of the central region.
TABLE 3.
Properties of Salmonella spp. containing SGI1 or its variants
| Strain | Serovar | Antibiotic resistance profilea
|
Resistance genes
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ap | Fl/Cm | Sm/Sp | Su | Tc | Tp | Gm | Cp | Na | blaP1 | floR | aadA2 | sul1 | tetG | dfrA1 | dfrA10 | ||
| SRC 4 | Derby | R | R | R | R | R | R | − | + | + | + | + | + | − | |||
| SRC 5 | Cerro | R | R | R | R | R | R | R | + | + | − | + | + | + | − | ||
| SRC 10 | Kiambu | R | R | R | R | R | R | + | + | + | + | + | − | + | |||
| SRC 11 | Kiambu | R | R | R | R | R | R | R | R | + | + | + | + | + | − | + | |
| SRC 19 | Emek | R | R | R | R | R | R | − | + | − | + | + | + | − | |||
| SRC 38 | Dusseldorf | R | R | R | R | R | R | + | + | − | + | + | + | − | |||
| SRC 46 | Infantis | R | R | R | − | − | + | + | − | − | + | ||||||
| SRC 49 | Paratyphi B dT+ | R | R | R | R | R | + | + | + | + | + | − | − | ||||
| SRC 50 | Paratyphi B dT+ | R | R | R | R | R | + | + | + | + | + | − | − | ||||
| DT104-2 | Typhimurium | R | R | R | R | R | + | + | + | + | + | − | − | ||||
Antibiotics: Ap, ampicillin (32 μg/ml); Cm, chloramphenicol (10 μg/ml); Gm, gentamicin (2.5 μg/ml); Sm, streptomycin (25 μg/ml); Sp, spectinomycin (50 μg/ml); Su, sulfamethoxazole (550 μg/ml); Tc, tetracycline (20 μg/ml); Tp, trimethoprim (50 μg/ml); Cp, ciprofloxacin (2 μg/ml); Na, nalidixic acid (50 μg/ml). Florfenicol (Fl) resistance was tested by streaking isolates onto LB-agar containing 15 μg/ml of florfenicol.
Because seven of the nine strains were resistant to trimethoprim, additional screens for two trimethoprim resistance genes that have been detected previously in SGI1 variants (3, 10, 12) were also performed. These were for the dfrA1 gene, which is found in a gene cassette (35), and the dfrA10 gene (27), which is found associated with CR1 (32). The two Kiambu strains, which contain the five resistance genes of In104 but were additionally resistant to trimethoprim, also included the dfrA10 gene (Table 3) and CR1. The serovar Infantis strain also included CR1 and the dfrA10 gene. Four strains included the cassette-associated dfrA1 gene either alone or together with either the cassette-associated aadA2 or blaP1 gene, suggesting that one or both of the cassette arrays had been exchanged.
Variation in the gene cassettes.
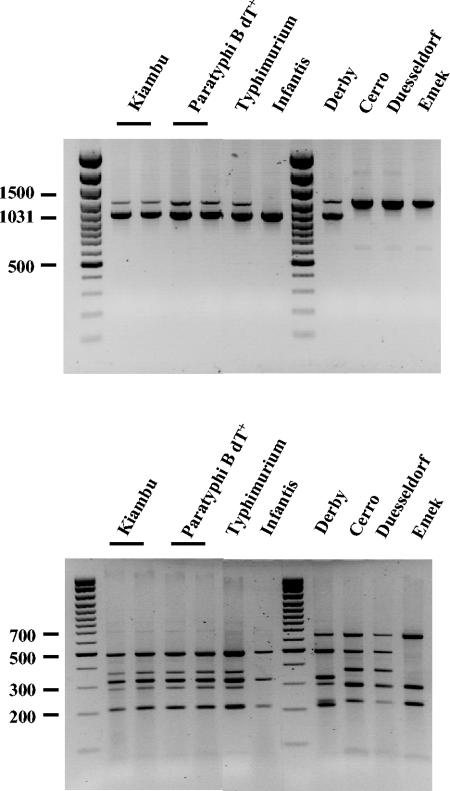
The regions containing gene cassettes were amplified using standard primers in the 5′-CS and 3′-CS of class 1 integrons (21) (Fig. 1B). The DT104 control strain as well as the serovar Kiambu and serovar Paratyphi B strains yielded the 1.0- and 1.2-kb amplicons containing, respectively, the aadA2 and blaP1 cassettes, as expected for In104 (SGI1). However, the serovar Derby strain, which does not include the blaP1 gene, gave an indistinguishable pattern (Fig. 2, top panel), indicating that the cassette amplicon sizes alone are insufficient to reliably indicate the presence of the original In104 configuration (Fig. 1).
FIG. 2.
Characterization of the cassette arrays. PCR amplification products (top panel) of cassette regions (using primers L1 and R1) and RsaI-generated restriction endonuclease fragments from them (bottom panel) were separated on agarose gels. Size markers are a 100-bp ladder.
Digestion of the PCR fragments with the restriction enzymes RsaI and Tsp5091 was therefore used to distinguish and more accurately identify the cassette arrays. For the serovar Kiambu and serovar Paratyphi strains, where both the aadA2 and blaP1 cassette had been detected (Table 3), the restriction fragment patterns were identical to that of the MR DT104 (In104 type) control (Fig. 2, lower panel). However, the serovar Derby strain yielded a different pattern, confirming that it does not contain the blaP1 cassette found in SGI1-continaing DT104. The three strains that apparently yielded a single 1.2-kb PCR product (Fig. 2, top panel) were differentiated into two types, one (serovars Cerro and Dusseldorf) with two different cassette arrays and one (serovar Emek) with one array. The fragments seen in digests from the Emek strain were also seen in the serovars Cerro, Derby, and Dusseldorf digests, where they are combined with bands corresponding to either the aadA2 amplicon (seen alone in serovar Infantis) or the blaP1 amplicon.
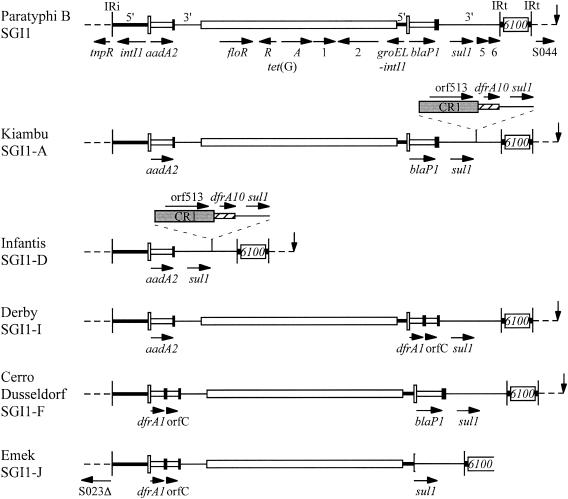
The Infantis strain yielded a single 1.0-kb product, consistent with the presence of only one attI1 site containing the aadA2 cassette in this strain, and this was confirmed by sequencing. To identify the cassette array in the isolates which contained the dfrA1 gene, the single 1.2-kb product from serovar Emek was sequenced. It contained the dfrA1 cassette with the orfC cassette and is identical to a cassette array reported in Vibrio cholerae (46) (GenBank accession no. AF455254), Escherichia coli (AB161449), and Salmonella spp. (AB186122). The restriction fragment analysis indicated that the same cassette array was also present in serovars Cerro, Derby, and Dusseldorf, and this was confirmed by PCR using a primer in each cassette. Hence, the Cerro and Dusseldorf strains include both the dfrA1-orfC and blaP1 cassette arrays and serovar Derby includes dfrA1-orfC with aadA2 (Fig. 3).
FIG. 3.
Maps of In104 variants. Features are as in Fig. 1B, and the vertical arrow indicates the right-hand boundary of SGI1. Regions associated with the additional dfrA10 genes are indicated above, with CR1 as a shaded box and the unique region including the dfrA10 gene as a hatched box. The strain or strains from which each map is derived, together with the SGI1 designation, are indicated on the left.
Location of the gene cassettes.
As there is no a priori reason why the cassette arrays amplified could not have arisen from integrons on resident plasmids rather than the integron in SGI1, further mapping was performed to confirm that the gene cassettes were part of the integron in SGI1 and to assign the cassettes to the left- or right-hand attI1 site. This was achieved by PCR using one primer within the cassette and one in a region unique to the surroundings of the left- or right-hand attI1 site (Fig. 1B). These were within S026 in the SGI1 backbone or in the floR gene for the left side and in the groEL region or orf5 for the right side (Table 1, Fig. 1B). The assignments are shown in Fig. 3. The serovar Emek strain was found to have the dfrA1-orfC cassette combination only on the left-hand side. A 1.2-kb groEL-sul1 product was generated by amplification using the groEL-F and QS-2 primers (Table 1) and sequenced to determine the structure of the right-hand side. A deletion that removes both the gene cassette and 472 bp of adjacent sequences, including part of the attI1 site and all of qacEΔ1, and is identical to a deletion found in Tn610 (23) (GenBank accession no. X53635) was present in this position.
Structure of the SGI1 integron variants.
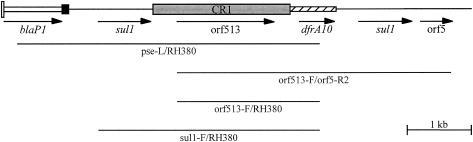
A complete map of each In104 homologue (Fig. 3) was obtained by establishing PCR linkage between most adjacent gene pairs using the forward primer of the first with the reverse primer of the second as well as longer linkages (Table 1). Linkage between orf2 and tetA(G) was established by Southern hybridization with tetA(G) and CR3 digoxigenin-labeled probes to BsaI-digested whole-cell DNA. As predicted from the DNA sequence of SGI1 (4), a 4.1-kb BsaI fragment hybridized with the tetA(G) and orf2 probes in all the SGI1-containing strains except serovar Infantis. The presence of the CR1-dfrA10 region was confirmed, and its location was determined using PCR primers in the relevant cassettes with a primer in CR1 to link it to the right-hand or left-hand cassette array (Fig. 4). The integrons detected here include the original SGI1 (4) as well as the previously identified types designated SGI1-A, SGI1-D, and SGI1-F (3, 11, 12). In addition, two new types were found and designated SGI1-I (in serovar Derby) and SGI1-J (in serovar Emek) using the next available letters.
FIG. 4.
Mapping the CR1-dfrA10 region. The primer combinations used to link genes in serovar Kiambu and serovar Infantis strains that possessed CR1 are shown. Primer sequences are listed in Table 1.
DISCUSSION
Since the discovery of genomic island SGI1 containing In104 in S. enterica DT104 strains, SGI1 or SGI1 homologues with an integron related to In104 have been found in several serovars of S. enterica. The range now includes serovar Typhimurium phage types DT104, DT120, DT12, DT1, and U302 (4, 6) and serovars Agona (3, 4, 7, 10), Paratyphi B dT+ (24, 25, 44; this study), Albany (12), Meleagridis (14), and Newport (13) as well as serovars Emek, Cerro, Derby, Dusseldorf, Infantis, and Kiambu identified in this study. This wide distribution provides a strong indication that the SGI1 genomic island is a significant determinant of multiple drug resistance in Salmonella and is clearly able to move from strain to strain.
One way in which this horizontal gene transfer could occur is by Int-mediated excision of SGI as a circular molecule, which is then packaged into a bacteriophage and transduced into another strain, and such transduction has been demonstrated (41). However, for each serovar, horizontal transfer need not necessarily have occurred on many occasions. The selective advantage conferred by the resistance genes in the many environments where antibiotics are used heavily, such as in the intensive rearing of food-producing animals, means that a single cell that had acquired SGI1 could be amplified (clonal expansion) and come to represent a significant fraction of the population of that serovar. The larger number of bacteria arising from such amplification would in turn increase the likelihood of spread of the new strain to new locations, and this is clearly what has occurred in the case of SGI1-containing DT104 (36).
SGI1-A has been shown be prevalent in poultry in Belgium (11), but whether any of the remaining serovars described here or by others are prevalent, particularly in food-producing animal populations, in the countries (including Australia) from which the isolates studied here originated remains to be established. The pairs of the same serovar isolates (Paratyphi B dT+ and Kiambu) identified here, though not known to be linked epidemiologically, could nonetheless be members of a single clonal line. In support of this, XbaI-digested chromosomal DNA from the Paratyphi B dT+ strains gave identical pulsed-field gel electrophoresis profiles (R. S. Levings and S. P. Djordjevic, unpublished observations). In addition, the properties of the multidrug resistance region in an S. enterica serovar Emek isolated in the United Kingdom (34) are consistent with its being within an SGI1 variant that is the same as SGI1-J in the serovar Emek strain studied here. Moreover, it appears that the absence of SGI1-containing DT104 in Australia does not reflect an absence of SGI1-containing strains belonging to other serovars.
Variation in the resistance gene content of the SGI1 integron observed here is likely to have arisen mostly by homologous recombination between identical segments, one within the integron in the SGI1 island and the second in a coresident plasmid or a smaller nonreplicating circular molecule. The latter can arise from a plasmid via homologous recombination as demonstrated for the CR-dfrA10-sul1 circle (32). Gene exchange by homologous recombination is likely to represent an important route for the exchange of gene cassettes in class 1 integrons because they are flanked by two regions, the 5′-CS and the 3′-CS, that are present in most class 1 integrons (16, 17, 32). This mechanism has been demonstrated experimentally (30, 31) and is likely to be responsible for the substitution of the dfrA1-orfC cassette array for either the aadA2 or the blaP1 cassette in the isolates studied here and previously.
The deletion removing part of the attI1 site found in one strain studied here (SGI1-J) could also have been acquired by this route from Tn610 (23) and, as Tn610 was originally found in Mycobacterium fortuitum, this suggests that this transposon has a very broad distribution. Previous claims of movement of gene cassettes from In104 into plasmids containing a class 1 integron via site-specific recombination (40) are unlikely to be correct. Cassette exchange most probably occurred by homologous recombination, granted that the SGI1-containing DT104 strains used are wild type and hence recombination proficient. Small nonreplicating circular molecules that have arisen by homologous recombination from a class 1 integron that has direct duplication of either the 5′-CS or 3′-CS, such as those found in In104 and those in CR1(orf513)-containing integrons (32, 42), are also likely to be important. These circles, once formed, can recombine into new locations, e.g., plasmids that contain a copy of any sequence included within them. The dfrA10 gene has been shown experimentally to move in this way (32) and is likely to have entered the In104 variants that contain it via this route. This route is also one that could lead to the movement of the floR-tet(G) region onto a plasmid that contains an integron and thence out into the broader population of Enterobacteriaceae. Indeed, such a plasmid may represent the original source of these genes in In104.
A particularly surprising finding of this study was that SGI1 or an SGI1 variant was found in 8 of the 39 strains included in the study that were isolated from humans with severe Salmonella infections. Recently, a single case of a human infection with SGI1-containing serovar Agona (11) and sporadic cases of serovar Paratyphi B dT+ infections in humans in Canada and the United Kingdom (25, 44) have also been reported. The United Kingdom Paratyphi B strains are associated with owners of tropical fish tanks, as are the Australian isolates reported here. As these serotypes are historically viewed as having low infectivity for humans, it was suggested that the presence of SGI1 may increase the likelihood of human infection (25). Our findings also support the contention that the presence of SGI1 introduces a determinant that may predispose to human infection. However, the possibility that these human infections simply reflect a widespread distribution of SGI1-containing Salmonella serovars in animal populations in the Southeast Asia-Australia region and in poultry in Belgium is still to be eliminated.
Acknowledgments
R.S.L. was supported by a University of Wollongong Postgraduate Award. This project was partly supported by a grant from the New South Wales Department of Primary Industries. S.R.P was supported by a grant (number 192108) from the National Health and Medical Research Council.
REFERENCES
- 1.Amavisit, P., P. F. Markham, D. Lighfoot, K. G. Whithear, and G. F. Browning. 2001. Molecular epidemiology of Salmonella Heidelberg in an equine hospital. Vet. Microbiol. 80:85-98. [DOI] [PubMed] [Google Scholar]
- 2.Bettelheim, K. A., M. A. Hornitzky, S. P. Djordjevic, and A. Kuzevski. 2003. Antibiotic resistance among verocytotoxigenic Escherichia coli (VTEC) and non-VTEC isolated from domestic animals and humans. J. Med. Microbiol. 52:155-162. [DOI] [PubMed] [Google Scholar]
- 3.Boyd, D., A. Cloeckaert, E. Chaslus-Dancla, and M. R. Mulvey. 2002. Characterization of variant Salmonella genomic island 1 multidrug resistance regions from serovars Typhimurium DT104 and Agona. Antimicrob. Agents Chemother. 46:1714-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, D., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistant region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briggs, C. E., and P. M. Fratamico. 1999. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob. Agents Chemother. 43:846-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carattoli, A., E. Filetici, L. Villa, A. M. Dionisi, A. Ricci, and I. Luzzi. 2002. Antibiotic resistance genes and Salmonella genomic island 1 in Salmonella enterica serovar Typhimurium isolated in Italy. Antimicrob. Agents Chemother. 46:2821-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cloeckaert, A., K. S. Boumedine, G. Flaujac, H. Imberechts, I. D'Hooge, and E. Chaslus-Dancla. 2000. Occurrence of a Salmonella enterica serovar Typhimurium DT104-like antibiotic resistance gene cluster including the floR gene in S. enterica serovar Agona. Antimicrob. Agents Chemother. 44:1359-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collis, C. M., G. Grammaticopoulos, J. Briton, H. W. Stokes, and R. M. Hall. 1993. Site-specific insertion of gene cassettes into integrons. Mol. Microbiol. 9:41-52. [DOI] [PubMed] [Google Scholar]
- 9.Davis, M. A., D. D. Hancock, and T. E. Besser. 2002. Multiresistant clones of Salmonella enterica: the importance of dissemination. J. Lab. Clin. Med. 140:135-141. [DOI] [PubMed] [Google Scholar]
- 10.Doublet, B., P. Butaye, H. Imberechts, D. Boyd, M. R. Mulvey, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Salmonella genomic island 1 multidrug resistance clusters in Salmonella enterica serovar Agona isolated in Belgium in 1992 to 2002. Antimicrob. Agents Chemother. 48:2510-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doublet, B., P. Butaye, H. Imberechts, J.-M. Collard, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Salmonella Agona harboring genomic island 1-A. Emerg. Infect. Dis. 10:756-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doublet, B., R. Lailler, D. Meunier, A. Brisabois, D. Boyd, M. R. Mulvey, E. Chaslus-Dancla, and A. Cloeckaert. 2003. Variant Salmonella genomic island 1 antibiotic resistance gene cluster in Salmonella enterica serovar Albany. Emerg. Infect. Dis. 9:585-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doublet, B., F.-X. Weill, L. Fabre, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Variant Salmonella genomic island 1 antibiotic resistance gene cluster containing a novel 3′-N-aminoglycoside acetyltransferase gene cassette, aac(3)-Id, in Salmonella enterica serovar Newport. Antimicrob. Agents Chemother. 48:3806-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebner, P., K. Garner, and A. Mathew. 2004. Class 1 integrons in various Salmonella enterica serovars isolated from animals and identification of genomic island SGI1 in Salmonella enterica var. Meleagridis. J. Antimicrob. Chemother. 53:1004-1009. [DOI] [PubMed] [Google Scholar]
- 15.Fisher, I., Y. Andersson, B. de Jong, K. O'Grady, and J. Powling. 9 August 2001. Eurosurveillance Wkly. 5. (www.eurosurveillance.org/ew/2001/010809.asp).
- 16.Hall, R. M., and C. M. Collis. 1995. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol. Microbiol. 15:593-600. [DOI] [PubMed] [Google Scholar]
- 17.Hall, R. M., and C. M. Collis. 1998. Antibiotic resistance in Gram negative bacteria: the role of gene cassettes and integrons. Drug Resist. Updates 1:109-119. [DOI] [PubMed] [Google Scholar]
- 18.Kamali-Moghaddam, M., and L. Sundström. 2000. Transposon targeting determined by resolvase. FEMS Microbiol. Lett. 186:55-59. [DOI] [PubMed] [Google Scholar]
- 19.Lawson, A. J., M. U. Dassama, L. R. Ward, and E. J. Threlfall. 2002. Multiply resistant (MR) Salmonella enterica serotype Typhimurium DT12 and DT120: A case of MR DT104 in disguise? Emerg. Infect. Dis. 8:434-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leverstein-van Hall, M. A., A. Paauw, A. T. A. Box, H. E. M. Blok, J. Verhoef, and A. C. Fluit. 2002. Presence of integron-associated resistance in the community is widespread and contributes to multidrug resistance in the hospital. J. Clin. Microbiol. 40:3038-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lévesque, C., and P. Roy. 1993. PCR analysis of integrons, p. 590-594. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.) Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, D. C.
- 22.Ling, J., P. Y. Chau, and B. Rowe. 1987. Salmonella serotypes and incidence of multiply-resistant salmonellae isolated from diarrhoeal patients in Hong Kong from 1973-82. Epidemiol. Infect. 99:295-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin, C., J. Timm, J. Rauzier, R. Gomez-Lus, J. Davies, and B. Gicquel. 1990. Transposition of an antibiotic resistance element in mycobacteria. Nature 345:739-743. [DOI] [PubMed] [Google Scholar]
- 24.Meunier, D., D. Boyd, M. R. Mulvey, S. Baucheron, C. Mammina, A. Nastasi, E. Chaslus-Dancla, and A. Cloeckaert. 2002. Salmonella enterica serotype Typhimurium DT104 antibiotic resistance genomic island 1 in serotype Paratyphi B. Emerg. Infect. Dis. 8:430-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulvey, M. R., D. Boyd, A. Cloeckaert, R. Ahmed, L.-K. Ng, and the Provincial Public Health Laboratories. 2004. Emergence of multidrug-resistant Salmonella Paratyphi B dT+, Canada. Emerg. Infect. Dis. 10:1307-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng, L., M. R. Mulvey, I. Martin, G. A. Peters, and W. Johnson. 1999. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT104. Antimicrob. Agents Chemother. 43:3018-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parsons, Y., R. M. Hall, and H. W. Stokes. 1991. A new trimethoprim resistance gene, dhfrX, in the In7 integron of plasmid pDGO100. Antimicrob. Agents Chemother. 35:2436-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Partridge, S. R., H. J. Brown, H. W. Stokes, and R. M. Hall. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Partridge, S. R., G. D. Recchia, H. W. Stokes, and R. M. Hall. 2001. Family of class 1 integrons related to In4 from Tn1696. Antimicrob. Agents Chemother. 45:3014-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Partridge, S. R., H. J. Brown, and R. M. Hall. 2002. Characterization and movement of the class 1 integron known as Tn2521 and Tn1405. Antimicrob. Agents Chemother. 46:1288-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Partridge, S. R., C. M. Collis, and R. M. Hall. 2002. Class 1 integron containing a new gene cassette, aadA10, associated with Tn1404 from R751. Antimicrob. Agents Chemother. 46:2400-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Partridge, S. R., and R. M. Hall. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob. Agents Chemother. 47:342-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Popoff, M. Y., and L. LeMinor. 2001. Antigenic formulas of the Salmonella serovars, 8th ed. W.H.O. Collaborating Centre for Reference and Research on Salmonella. Institute Pasteur, Paris, France.
- 34.Randall, L. P., S. W. Cooles, M. K. Osborn, L. J. V. Piddock, and M. J. Woodward. 2004. Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the United Kingdom. J. Antimicrob. Chemother. 53:208-216. [DOI] [PubMed] [Google Scholar]
- 35.Recchia, G. D., and R. M. Hall. 1995. Gene cassettes: a new class of mobile element. Microbiology 141:3015-3027. [DOI] [PubMed] [Google Scholar]
- 36.Ribot, E. M., R. K. Wierzba, F. J. Angulo, and T. J. Barrett. 2002. Salmonella enterica serotype Typhimurium DT104 isolated from humans, United States, 1985, 1990, and 1995. Emerg. Infect. Dis. 8:387-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ridley, A., and E. J. Threlfall. 1998. Molecular epidemiology of antibiotic resistance genes in multiresistant Salmonella typhimurium DT104. Microb. Drug Resist. 2:113-118. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Sandvang, D., F. M. Aarestrup, and L. B. Jensen. 1998. Characterisation of integron and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 160:37-41. [DOI] [PubMed] [Google Scholar]
- 40.Sandvang, D., M. Diggle, and D. J. Platt. 2002. Translocation of integron-associated resistance in a natural system: acquisition of resistance determinants by IncP and IncW plasmids from Salmonella enterica Typhimurium DT104. Microb. Drug Res. 8:151-160. [DOI] [PubMed] [Google Scholar]
- 41.Schmeiger, H., and P. Schicklmaier. 1999. Transduction of multiple drug resistance of Salmonella enterica serovar Typhimurium DT104. FEMS Microbiol. Lett. 170:251-256. [DOI] [PubMed] [Google Scholar]
- 42.Stokes, H. W., C. Tomaras, Y. Parsons, and R. M. Hall. 1993. The partial 3′-conserved segment duplications in the integrons In6 from pSa and In7 from pDGO100 have a common origin. Plasmid 30:39-50. [DOI] [PubMed] [Google Scholar]
- 43.Threlfall, E. J. 2002. Antimicrobial drug resistance in Salmonella: problems and perspectives in food- and water-borne infections. FEMS Microbiol. Rev. 26:141-148. [DOI] [PubMed] [Google Scholar]
- 44.Threlfall, J., B. Levent, K. L. Hopkins, E. de Pinna, L. Ward, and D. J. Brown. 2005. Multidrug resistant Salmonella Java. Emerg. Infect. Dis. 11:170-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Threlfall, E. J., L. R. Ward, J. A. Frost, and G. A. Willshaw. 2000. The emergence and spread of antibiotic resistance in food-borne bacteria. Int. J. Food Microbiol. 62:1-5. [DOI] [PubMed] [Google Scholar]
- 46.Thungapathra, M., A. Kislay, K. Sinha, S. R. Chaudhuri, P. Garg, T. Ramamurthy, G. B. Nair, and A. Ghosh. 2002. Occurrence of antibiotic resistance gene cassettes aac(6')-Ib, dfrA5, dfrA12, and ereA2 in class I integrons in non-O1, non-O139 Vibrio cholerae strains in India. Antimicrob. Agents Chemother. 46:2948-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]