Abstract
Bacterial communication signals, acylated homoserine lactones (AHLs), were extracted from samples of commercial bean sprouts undergoing soft-rot spoilage. Bean sprouts produced in the laboratory did not undergo soft-rot spoilage and did not contain AHLs or AHL-producing bacteria, although the bacterial population reached levels similar to those in the commercial sprouts, 108 to 109 CFU/g. AHL-producing bacteria (Enterobacteriaceae and pseudomonads) were isolated from commercial sprouts, and strains that were both proteolytic and pectinolytic were capable of causing soft-rot spoilage in bean sprouts. Thin-layer chromatography and liquid chromatography-high-resolution mass spectrometry revealed the presence of N-3-oxo-hexanoyl-l-homoserine lactone in spoiled bean sprouts and in extracts from pure cultures of bacteria. During normal spoilage, the pH of the sprouts increased due to proteolytic activity, and the higher pH probably facilitated the activity of pectate lyase. The AHL synthetase gene (I gene) from a spoilage Pectobacterium was cloned, sequenced, and inactivated in the parent strain. The predicted amino acid sequence showed 97% homology to HslI and CarI in Erwinia carotovora. Spoilage of laboratory bean sprouts inoculated with the AHL-negative mutant was delayed compared to sprouts inoculated with the wild type, and the AHL-negative mutant did not cause the pH to rise. Compared to the wild-type strain, the AHL-negative mutant had significantly reduced protease and pectinase activities and was negative in an iron chelation (siderophore) assay. This is the first study demonstrating AHL regulation of iron chelation in Enterobacteriaceae. The present study clearly demonstrates that the bacterial spoilage of some food products is influenced by quorum-sensing-regulated phenotypes, and understanding these processes may be useful in the development of novel food preservation additives that specifically block the quorum-sensing systems.
Many gram-negative bacteria use chemical signal molecules, acylated homoserine lactones (AHLs), to regulate the expression of particular phenotypes as a function of cell density. This phenomenon, known as quorum sensing (QS), is involved in the expression of virulence factors in several pathogenic bacteria and in the expression of factors important for the bacterial colonization of higher organisms (35, 52, 59, 62, 63). The growth and activity of gram-negative bacteria are also important in food microbiology, where spoilage of fresh foods, such as fish, meat, milk, and vegetables, is mostly a consequence of the degradative activity of gram-negative bacteria growing to high densities (108 to 109 CFU per gram). Food spoilage is a major economic and societal problem, and large amounts of foods are lost due to bacterial growth and spoilage (9). Understanding of the microbial processes leading to spoilage would facilitate development of novel preservation techniques and reduce loss of food. We hypothesized that bacterial food-degradative processes could be regulated by QS, and if QS systems are involved in the regulation of bacterial spoilage, targeted inhibition of the communication underlying the QS systems could reduce or prevent the spoilage reactions.
AHLs have been detected in a variety of different spoiled commercial food products, such as cold-smoked salmon (29), fish fillet, minced fish, turkey meat, vacuum-packed beef, and bean sprouts (12, 30), and many gram-negative bacteria involved in food spoilage are capable of producing AHLs (53). The involvement of AHL-dependent QS in bacterial food spoilage is, however, not resolved. QS may play an important role in the spoilage of, e.g., milk (15), while in other products, such as meat, QS is of little or no importance for the microbial spoilage process (12).
Microbial food spoilage may be caused by turnover of low-molecular-weight compounds (amino acids and other N-containing compounds and sugars), producing offensive off odors (20, 34), or by degradation of polymers, such as proteins or pectins (50). Proteolytic and pectinolytic activities in some gram-negative bacteria are regulated by QS, and the proteolytic activities of, for instance, Pseudomonas aeruginosa (51), Serratia liquefaciens (23), and Vibrio anguillarum (19) are expressed in a quorum-dependent manner. The extracellular enzymes pectate lyase, pectin lyase, polygalacturonase, cellulase, and protease are regulated by N-3-oxohexanoyl-l-homoserine lactone (3-oxo-C6-HSL)-dependent QS in the plant pathogen Erwinia carotovora subsp. carotovora (35, 52). Hence, we hypothesized that QS could be an important feature of vegetable spoilage, since this process typically involves pectinolytic degradation (43). Vegetables, in particular, as ready-to-eat products (e.g., salads), are food commodities of increasing popularity worldwide. Their shelf life is short, and knowledge of factors controlling spoilage would enable further expansion of this type of product.
We chose bean sprouts as a model for vegetables, since they can be produced under controlled conditions in the laboratory. Commercially available bean sprouts usually have very high bacterial counts, reaching 108 CFU/g during 2 days of sprouting (47, 60). Bacterial spoilage of bean sprouts is characterized by soft rot, in which maceration of plant tissue by bacterial enzymes results in collapse of the cell wall structure. The most important bacteria causing soft rot of vegetables and fruits are the gram-negative E. carotovora and pectinolytic strains of Pseudomonas fluorescens (43). Pectate lyase, which cleaves polygalacturonic acid by β-trans-elimination, is believed to be the principal enzyme responsible for soft rot (44, 48).
The purpose of this study was to investigate the role of AHL-based QS in spoilage of bean sprouts. Bean sprouts in this context were considered a model for food products in which (i) the bacterial count is high (>107 CFU/g) at the time of spoilage, (ii) the spoilage flora is dominated by gram-negative bacteria, and (iii) spoilage is characterized by enzymatic degradative activity.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Supernatants from bacterial cultures, as well as extracts of sprouts or pure bacterial cultures, were tested for the presence of AHLs using the bacterial AHL monitor strains Agrobacterium tumefaciens strain NT1 with pZLR4 (14), Chromobacterium violaceum strain CV026 (46, 65), and Escherichia coli JB525 (4). The growth media were supplemented with 20 μg/ml of gentamicin and kanamycin for A. tumefaciens and C. violaceum, respectively. Strains used for positive and negative controls in phenotypic tests and enzyme assays are listed in Table 1. E. coli S17-1/λpir[pBSL180] NCCB3397 (2) was used for construction of an AHL-negative mutant.
TABLE 1.
Strains used as positive and negative controls in phenotypic tests and enzyme assays
| Assay | Positive control
|
Negative control
|
||
|---|---|---|---|---|
| Strain | Reference | Strain | Reference | |
| Pectinase | E. carotovora GS101 | 8 | E. coli MG1655 | 32 |
| Deoxyribonuclease | S. liquefaciens MG1 | 27 | E. coli MT102 | 4 |
| S. liquefaciens PL10 | 42 | |||
| Maltose assimilation | S. liquefaciens MG1 | 27 | E. carotovora GS101 | 8 |
| Lipase | P. aeruginosa ATCC 27853 | E. coli MG1655 | 32 | |
| Cellulase | E. coli MG1655 | 32 | ||
| Carbapenem | E. carotovora GS101 | 8 | ||
All strains were routinely grown in LB5 broth (Luria-Bertani broth [10] using 5 g/liter of NaCl instead of 10 g/liter). When appropriate, the broth was supplemented with 1.2% (wt/vol) agar. A. tumefaciens was grown in AB medium (16) supplemented with 2.5 mg/liter thiamine (ABT), 0.5% (wt/wt) Casamino Acids, and 0.5% (wt/wt) glucose (ABTG) for replica plating, thin-layer chromatography (TLC), and estimation of levels of AHLs.
Bean sprouts.
Several samples of commercial bean sprouts (from two processors) were purchased on several occasions in nearby grocery stores, and the presence of AHLs and AHL-producing bacteria was determined as described below. Bean sprouts were produced in the laboratory by soaking 100 g of mung beans (Rømer, Silkeborg, Denmark) in 500 ml tap water in a 2-liter beaker. After 24 h at 25°C, the water was carefully discarded, and the bean sprouts were kept at 25°C in the dark with daily soaking in tap water for 30 min. A circular metal plate placed 1 cm above the beaker bottom drained excess water from the beans. The beans were stored at 5°C after a total of 5 days of sprouting. When bacterial cultures were added to the laboratory-produced bean sprouts, it was done by adding 1 ml of an overnight culture to the 500 ml tap water used for the initial soaking.
Determination of pH in bean sprouts.
Five grams of bean sprouts was homogenized with 5 ml of distilled water using an Ultra-Turrax homogenizer (T 25 basic; IKA-WERKE, Staufen, Germany). pH was measured with a PHM95 pH meter equipped with a GK2401c electrode (Radiometer, Copenhagen, Denmark).
Determination of aerobic bacterial count in bean sprouts.
Ten grams of bean sprouts was homogenized with 90 ml of peptone saline water (8 g/liter NaCl and 1 g/liter Bacto Peptone [211677; Becton Dickinson Co., Sparks, MD]) for 60 s at high speed in a stomacher (Stomacher 400; Seward, London, United Kingdom). Tenfold dilutions were made, and 0.1 ml of appropriate dilutions was spread on LB5 agar and incubated at 25°C for 2 days.
Isolation and characterization of AHL-producing bacterial strains from bean sprouts.
AHL-producing bacterial strains were isolated by replica plating LB5 agar plates with 5 to 50 colonies onto ABTG or LB5 agar plates cast with A. tumefaciens or C. violaceum, respectively (12). The replica plates were incubated at 25°C for 24 h, and AHL-producing bacteria (colonies) were detected by the presence of blue (with A. tumefaciens) and violet (for C. violaceum) zones around the colonies. Bacteria assumed to produce AHLs were isolated from the LB5 master plate, and AHL production was verified in the pure cultures by testing bacterial supernatants in a well diffusion assay with A. tumefaciens or C. violaceum as the AHL monitor (53). By this procedure, a total of 32 AHL-producing bacterial strains were isolated from six samples of commercial bean sprouts.
Preliminary identification of the isolated strains was based on Gram reaction (3% KOH [31]), oxidase reaction (BBL DrySlide; Becton Dickinson Co., Sparks, MD), and catalase reaction (3% H2O2 [38]); cell shape and motility, as observed by phase-contrast microscopy; and metabolism of glucose in Hugh and Leifson medium (Merck OF-basal medium; catalog no. 1.10282). Spoilage potential was determined by inoculating the strains in the soaking water from laboratory-produced bean sprouts and inspecting the sprouts for signs (e.g., soft rot) of spoilage. The presence of pectinolytic activity was evaluated by spotting 10 μl of an overnight culture on agar plates containing polygalacturonic acid (Sigma; P-3850) (11). Pectinolytic activity was detected as a depression in the agar around the colonies. The agar turned yellow around some colonies, presumably caused by a decrease in the pH of the medium. When the water in the agar was replaced by HEPES buffer (0.05 M; pH 7), the agar retained its original red color and still allowed depressions to be formed. Qualitative evaluation of proteolytic activity was performed on ABTG agar plates supplemented with 10% (vol/vol) sterile skimmed milk (105°C for 30 min). Wells (6 mm) were punched in the solidified agar, and 60 μl of sterile filtered supernatant from an overnight culture was added to each well. The plates were incubated at 25°C for 3 days. The ability to degrade DNA was evaluated on DNase Test Agar plates (Difco catalog no. 211179) incubated at 30°C for 48 h. Clearing zones were detected by flooding the plates with 5 ml of 1 N HCl. The ability to assimilate maltose was evaluated on ABT minimal medium supplemented with 0.2% (wt/wt) maltose and solidified with 2% (wt/wt) Bacto Agar (Becton Dickinson; 214030). Following 72 h of incubation at 30°C, the ABT-maltose plate was visually inspected for the formation of colonies. Lipolytic activity was assayed on tributyrin-containing agar plates (2.5 g/liter Bacto Peptone [Becton Dickinson; 211672], 2.5 g/liter tryptone [Becton Dickinson; 211705], 3.0 g/liter yeast extract [Becton Dickinson; 212750], 12 g/liter agar [Bie & Berntsen, Rødovre, Denmark; BBB10030], and 10 ml/liter tributyrin [Merck catalog no. 1.01958.0100]). Ten microliters of an overnight culture was spotted on the agar plates, and the plates were incubated overnight at 25°C. Clear zones around the colonies were evaluated as positive for lipolytic activity. Further phenotypic characterization was carried out on API 20E strips (BioMérieux, Marcy-l'Etoile, France) used according to the manufacturer's instructions. The strips were incubated at 25°C for 24 h. Carbapenem production was assayed on LB5 agar containing E. coli ESS (a β-lactam-supersensitive indicator strain) (8) and evaluated for the presence of a zone of antibiosis after 48 h of incubation at 25°C. One strain of Enterobacteriaceae, A2JM, caused soft-rot spoilage of bean sprouts and was selected for further studies of AHL regulation.
Extraction of AHLs.
AHLs were extracted from bean sprouts by homogenizing 10 g of bean sprouts with 30 ml ethyl acetate (acidified by supplementation with 0.5% formic acid) with an Ultra-Turrax homogenizer (T 25 basic; IKA-WERKE, Staufen, Germany). Large debris was removed by the use of a Bodum French press coffee maker, after which the samples were filtered through a Whatman no. 4 filter (catalog no. 1004 150; Maidstone, England). The filtrate was evaporated under nitrogen flow to dryness, redissolved in 1 ml of acidified ethyl acetate, transferred to a high-performance liquid chromatography vial, and stored at −20°C for further analysis. Extracts of bacterial cultures were prepared by extracting sterile filtered culture supernatants three times with equal volumes of acidified ethyl acetate, evaporated, and redissolved as described above.
Estimation of level of AHLs.
The level of AHLs was estimated in a semiquantitative well diffusion assay with A. tumefaciens NT1(pZLR4) as the indicator organism (53) using 20 μg/ml gentamicin and 3-oxo-C6-HSL (Sigma-Aldrich 143537-62-6) as a standard (53).
Identification of AHLs.
AHLs were profiled by TLC (14, 46, 53, 58). In brief, 1 to 100 μl of AHL extracts or culture supernatants was applied to a TLC plate (TLC aluminum sheet; 20 by 20 cm2; RP-18 F254 S plate; catalog no. 1.05559 [Merck, Darmstadt, Germany]). The plates were developed in 10 ml 60:40 (vol/vol) methanol-Millipore water. The plates were overlaid with agar containing either A. tumefaciens or C. violaceum. Synthetic 3-oxo-C6-HSL, N-hexanoyl-l-homoserine lactone (C6-HSL; Sigma-Aldrich 106983-28-2), and N-octanoyl-l-homoserine lactone (C8-HSL; obtained from Protein Chemistry, Novozymes, Bagsvaerd, Denmark) were used as reference standards.
LC-HR-MS.
Culture extracts were extracted and analyzed by liquid chromatography-high-resolution mass spectrometry (LC-HR-MS) as described by Bruhn et al. (12).
Bean sprouts (15 g) were freeze-dried for 2 days at 20 kPa and extracted with 25 ml methanol-dichloromethane-ethyl acetate (1:2:3) for 2 h, and the solvent was decanted and evaporated in vacuo. The samples were dissolved in 2 × 100 μl water-methanol (19:1) and applied to 60-mg Strata-X SPE cartridges (Phenomenex, Torrance, CA), which had been activated by 1 ml methanol and then 1 ml water containing 1% formic acid. The cartridges were washed with 1 ml water-methanol (19:1) containing 1% formic acid, and the column was eluted with 1 ml water-methanol (1:9) containing 1% formic acid. The eluates were then evaporated in vacuo and redissolved in 100 μl methanol-water (1:4), and 3 μl was injected and analyzed by LC-HR-MS on an LCT orthogonal time-of-flight mass spectrometer (Micromass, Manchester, United Kingdom) by the same method as the culture extracts (12).
DNA manipulation.
Standard techniques for DNA manipulation were used (55). Plasmid DNA was prepared using the QIAGEN minispin prep kit (QIAGEN, Germany), and chromosomal DNA was extracted using the QIAGEN Genomic DNA Tip 100/G system (QIAGEN, Germany). Transformation of E. coli was done by electroporation in a 0.2-cm cuvette using a GenePulser apparatus (Bio-Rad) set to 25 μF, 400 Ω, and 2.5 kV/cm. PCR was carried out using the Expand High Fidelity PCR system (Boehringer, Mannheim, Germany), and a GFX PCR DNA and gel band purification kit (Amersham Pharmacia Biotech) was employed for purification of DNA fragments.
PCR amplification and sequencing of 16S rRNA and the rpoB gene of A2JM.
The 16S rRNA gene of strain A2JM was sequenced by Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH according to their standard protocol. The resulting 1,496-bp 16S rRNA sequence was deposited in GenBank. PCR-amplified fragments of rpoB were purified and cycle sequenced on an ABI 377 automatic sequencer (Chemistry Guide; Applied Biosystems, Foster City, CA) as described recently (37). DNA sequencing resulted in 518 bp of the rpoB gene, and the sequence was deposited in GenBank. Searches for DNA sequences were performed in GenBank by BLAST (3). Pairwise similarities were calculated by BESTFIT (Wisconsin Sequence Analysis Package; Genomics Computer Group, Madison, WI). Representative sequences of organisms belonging to Enterobacteriaceae were aligned using the ClustalX (version 1.83) algorithm (64), and a phylogenetic tree was constructed by the neighbor-joining method. Bootstrap analysis was performed in ClustalX with 1,000 replicates. Positions with gaps were excluded, and the analysis was corrected for multiple substitutions.
DNA nucleotide sequencing of I and R genes.
The oligonucleotides used in this study are listed in Table 2. DNA sequencing was performed at a commercial facility (GATC Biotech AG, Germany). DNA flanking the Tn10-nptII transposon in the A2JM luxI mutant was sequenced using the primers Tn10-nptII(C), Tn10-nptII(N), a2jmR(C), and I1(up) as sequencing primers and pJBA232 as the template DNA. The PCR-amplified luxIR region of A2JM was sequenced using pJBA270 as the template DNA and the primers a2jmI(−250), I1(up), a2jmI(596), a2jmR(669), a2jmR(C), and a2jmR(−30) as sequencing primers.
TABLE 2.
Primers used for gene sequencing in the present study
| Primer | Sequence |
|---|---|
| Tn10nptII(N) | 5′-TGCAATCCATCTTGTTCAATCAT-3′ |
| Tn10nptII(C) | 5′-CTTCCTCGTGCTTTACGGTAT-3′ |
| A2jmR(C) | 5′-CAAATATCTTGGCCAACTGC-3′ |
| I1(up) | 5′-TAAACCTTGCTCGACCACGC-3′ |
| A2jmR(−30) | 5′-TTTTTAAGCTTGTAGGTATATACTTTT CTATTACAA-3′ |
| A2jmI(−250) | 5′-ATATAAGGATCCGGAATCGTCT-3′ |
| A2jmI(596) | 5′-CAACGAACTGAAGCAGTGG-3′ |
| A2jmR(669) | 5′-AATGCCAAGCATGCCATCAG-3′ |
Protein sequence analysis.
A Blastp homology search (http://www.ncbi.nlm.nih.gov/BLAST) was performed searching for amino acid sequences in the nonredundant peptide sequence database and conserved domains in the CDD v1.62-11088PSSMs protein database.
Construction of AHL-deficient mutant of A2JM.
A mutant library consisting of approximately 15,000 potentially independent Tn10 transposon insertion mutants of A2JM was constructed by diparental mating between A2JM and E. coli S17-1/λpir[pBSL180] NCCB3397 (2). A2JM was incubated for 1 h at 42°C prior to mating, and 1 ml of culture of A2JM and NCCB3397 was harvested, washed twice in 1 ml of 0.9% NaCl, and resuspended in 100 μl of 0.9% NaCl. The strains were mixed in a 1:1 ratio and spotted on a prewarmed (42°C) LB plate, and the mating plate was incubated at 42°C for 3 h, followed by 20 h of incubation at 30°C. The resulting mating spot was suspended in 0.9% NaCl, and the conjugation mixture was spread onto ABT plates containing 2% agar (Difco; catalog no. 214010), 20 mM citrate, and 150 μg/ml of kanamycin to create an A2JM mutant library. The mutant library was replicated onto indicator plates containing C. violaceum CV026 seeded in LB agar and 50 μg/ml of kanamycin, and one putative AHL-deficient mutant (hereafter referred to as the A2JM luxI mutant) that failed to induce violacein production was picked for further analysis.
Characterization of AHL-deficient mutant.
To map the insertion site of the Tn10-nptII transposon in the A2JM luxI mutant, chromosomal DNA was restricted with StuI, and the resulting StuI fragments were ligated to an SmaI-restricted plow1 vector (33) and electroporated into E. coli strain MT102 (4). Transformants were selected on LB plates supplemented with 25 μg/ml of kanamycin, and a restriction analysis of plasmid DNA prepared from eight of these transformants showed that the Tn10-nptII transposon from the A2JM luxI mutant had been cloned in plow1 on a 12-kb chromosomal StuI fragment. The insertion site of the Tn10-nptII transposon in one such plasmid (pJBA232) was determined by DNA sequencing using primers annealing to each end of the Tn10-nptII transposon [primers Tn10nptII(N) and Tn10nptII(C) (Table 2)]. New primers were designed from these initial sequences and used for progressive sequencing of neighboring DNA, and the resulting sequence covered 887 bp upstream and 1,010 bp downstream of the Tn10 transposon. A BlastN and a BlastX (http://www.ncbi.nlm.nih.gov/BLAST) search performed with the sequence revealed that the Tn10 transposon had been inserted at position 563 in a 651-bp open reading frame exhibiting homology to genes belonging to the luxI family. The BLAST searches also revealed that this luxI homologue shared 17 bp at its 3′ terminus with a 729-bp convergently transcribed luxR homologue. These luxI and luxR homologues were subsequently named luxI and luxR. To confirm that the AHL-deficient strain was derived from A2JM, a PCR amplification was carried out using the luxRI flanking primers a2jmI(−250) and a2jmR(−30) (Table 2) and chromosomal DNA of A2JM as a template. The resulting 1.7-kb fragment was cut with BamHI and HindIII and ligated to the BamHI- and HindIII-restricted plow1 vector (33) to give the plasmid pJBA270. To confirm that the 1.7-kb PCR fragment derived from A2JM contained the expected luxIR region, the fragment was sequenced and compared to the luxIR region of the A2JM luxI mutant.
AHL-regulated phenotypes.
Quantitative evaluation of pectinase activity was modified from Collmer et al. (17) using 850 μl of the substrate solution and 150 μl of sample (or sample diluted in water). Samples were either sterile filtered supernatants from bacterial cultures or sterile filtered samples of bean sprout soaking water. Absorbance at 232 nm was measured with a UV-visible-light recording spectrophotometer (UV-160A; Shimadzu). A quantitative protease assay was performed on the sprouting water and sterile filtered supernatants from pure bacterial cultures (22, 67). In brief, 100 μl of sample was incubated with 100 μl azocasein (sulfanilamide-azocasein; A-2765; Sigma) (5 mg/ml dissolved in 50 mM Tris-HCl, pH 8) at 30°C for 90 min. The reaction was terminated by addition of 400 μl trichloroacetic acid (10% [wt/vol]). The precipitated protein was removed by centrifugation (16,000 × g; 3 min). Five hundred microliters of the supernatant was transferred to 700 μl of 525 mM NaOH, and the absorbance was measured at 442 nm on a Novaspec II visible-light spectrophotometer (Pharmacia Biotech). Each sample was assayed in triplicate with two controls, which were vials to which the sample was added after trichloroacetic acid. A qualitative assay of cellulase activity was modified from the method of Andro et al. (6), using ABTG in the agar instead of M9. Ten microliters of an overnight culture was spotted on the agar plates, and the plates were incubated overnight at 25°C before being stained with a 0.1% (wt/vol) Congo red solution and washed with a 1 M NaCl solution. The presence of clear zones around the colonies was evaluated as positive for cellulase activity. Siderophore production was detected by spotting overnight cultures of A2JM and its mutants (luxI and luxR mutants) grown at 25°C in LB on siderophore-indicative plates with and without 1 μM 3-oxo-C6-HSL. AB minimal medium without FeCl3 and supplemented with 1.2% agar was autoclaved, and the molten agar was supplemented with 0.01 mM FeCl3, 0.5% glucose, 0.5% Casamino Acids, and 10% CAS solution (57). The pH was adjusted to approximately 6.5 using pH paper (Merck 1.09547.0001) and visually inspected for color. The plates were incubated for 2 days at 25°C before being read.
Construction of an AHL receptor-negative mutant.
The luxR gene present on pJBA270 was inactivated by introducing the aacC1 gene (conferring resistance to gentamicin) of pUCGm (56) as a 0.85-kb SmaI fragment into the NdeI site located at nucleotide 465 in the coding sequence of luxR. From the resulting plasmid, named pJBA271, the 2.55-kb NotI fragment including a functional luxI gene and an accC1-interrupted luxR gene was isolated and cloned into the unique NotI site of the delivery vector pCK318 (36) to give pJBA275. As pJBA275 failed to replicate in A2JM (replication was dependent on the lambda pir protein), and as it also includes a sacB gene located outside of the luxI-luxR::aacC1 cassette, pJBA275 was employed for construction of a luxR mutant of A2JM by homologous recombination. pJBA275 was introduced into A2JM by diparental mating between A2JM and S17-1(λ-pir) (21) harboring pJBA275 (donor) as described above for A2JM and NCCB3397, and transconjugants were selected on ABT plates (16) containing 2% agar (Difco), 20 mM citrate, and 10 μg/ml of gentamicin (ABTC-Gm10 plates). Transconjugants were replicated to ABTC-Gm10 plates and ABTC-Gm10 plates supplemented with 7% (wt/wt) sucrose, and gentamicin-resistant transconjugants resulting from double crossover of the luxI-luxR::aacC1 cassette from pJBA275 into the chromosome of A2JM were scored as transconjugants capable of growing in the presence of 7% sucrose (SacB selection). Finally, the genotype of one such luxR mutant, named JB1159, was verified by PCR using the primer set A2jmI(−250)-A2jmR(−30) (Table 2) and chromosomal DNA of JB1159 as a template.
Nucleotide sequence accession numbers.
The 1,496-bp 16S rRNA gene sequence of strain A2JM was deposited in GenBank under accession no. AY338233. The 518-bp sequence of the rpoB gene was deposited in GenBank under AY842256. luxI and luxR were assigned GenBank accession number AY323227.
RESULTS
Quality changes in commercial and laboratory-produced sprouts.
The spoilage of commercial bean sprouts was often associated with soft rot, and the pH varied between 5.8 and 6.4. Bacterial counts were high (108 to 109 CFU/g), and AHLs could be extracted from five out of six samples investigated (Table 3), as detected with the sensors A. tumefaciens and C. violaceum. AHL-producing bacteria were detected at a level of 2 × 107 to 8 × 107 CFU/g. Bean sprouts produced in the laboratory did not undergo soft-rot spoilage, even though the bacterial count was on the same order of magnitude as the commercial bean sprouts (108 to 109 CFU/g) (Table 3). This indicates that the bacteria causing spoilage of the commercial bean sprouts do not necessarily originate from the mung beans but could be present in the production environment. AHLs were not detected in extracts from the laboratory bean sprouts, and no AHL-producing bacteria appeared on replica plates. The laboratory bean sprouts therefore qualified as a model system to study the effect of AHL-producing bacteria on the spoilage of bean sprouts.
TABLE 3.
Bacterial counts and AHLs extracted from commercial and laboratory-produced bean sproutsa
| Origin | Code | CFU g−1 | AHL-producing (CFU g−1) | 3-oxo-C6-HSL equivalents (nmol kg−1) | Presumed AHLs by TLC analysisb |
|---|---|---|---|---|---|
| Commercial | A | 7.6 × 108 | 8 × 107 | 63 | 3-oxo-C6-HSL |
| B | 1.4 × 108 | NTc | 11 | NId | |
| C | 4.5 × 108 | 3 × 107 | 5 | NDe | |
| D | 3.1 × 108 | 4 × 107 | 8 | 3-oxo-C6-HSL | |
| E | 2.9 × 108 | 2 × 107 | ND | ND | |
| F | 9.4 × 108 | 4 × 107 | 35 | 3-oxo-C6-HSL and NI | |
| Laboratory | G | 2.4 × 108 | <107 | ND | ND |
| H | 4.0 × 108 | <107 | ND | ND |
Aerobic bacterial counts were determined on LB5 agar. The commercial bean sprouts were from two different producers and were sampled on three different occasions.
Based on TLC profiling with Agrobacterium tumefaciens and Chromobacterium violaceum as monitor strains.
NT, not tested.
NI, not identified (spots on TLC plates did not correspond to any known standard).
ND, not detected.
Isolation and identification of AHL-producing bacteria.
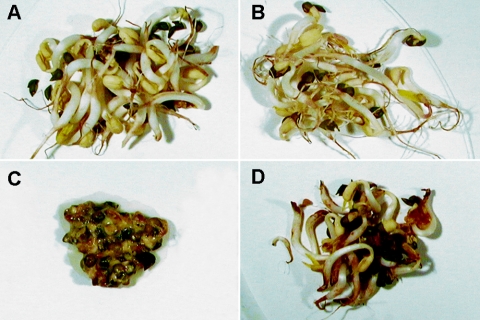
A total of 32 AHL-producing bacterial isolates were obtained from different batches of commercial bean sprouts (Table 4). All of the isolates were gram-negative motile rods exhibiting a positive catalase reaction. Fermentative strains with negative oxidase reactions were grouped as Enterobacteriaceae, and nonfermentative strains with positive oxidase reactions were grouped as Pseudomonas spp. Fermentative, oxidase-positive strains were grouped as Vibrionaceae. A total of seven isolates caused softening of the vegetable tissue when inoculated on bean sprouts, but most of the isolates did not cause any change in sprout texture (Fig. 1A, B, and C). The pH of freshly produced sprouts was between 6.3 and 6.4, but following inoculation with bacteria causing tissue softening, the pH increased to 8.0 to 8.4. The pH of the sprouts did not change or decreased slightly when the sprouts were inoculated with strains that did not cause soft rot. All isolates that caused soft rot were both pectinolytic and proteolytic (Table 4). Two bacterial strains (MRa40 and C1JM) were pectinolytic but were unable to degrade protein (MRa40) or displayed only a vague reaction on casein agar (C1JM). These strains were unable to spoil the bean sprouts. Three strains (C3-2JM, MRa42, and MRa53) were proteolytic but unable to degrade pectin. None of these strains was able to spoil bean sprouts.
TABLE 4.
Characterization of AHL-producing bacterial isolates from commercial bean sprouts
| Isolate | Identification | Presumed AHLs by TLCa | AHLs identified by LC-HR-MS | Pectinolytic activityb | Proteolytic activityb | Ability to spoil bean sproutsc |
|---|---|---|---|---|---|---|
| A2JM | Enterobacteriaceae | 3-oxo-C6-HSL | 3-oxo-C6-HSL | + | + | +++ |
| A4JM | Enterobacteriaceae | 3-oxo-C6-HSL + C6-HSL + NId | NDe | − | − | − |
| A7JM | Enterobacteriaceae | NI | ND | − | − | − |
| A9JM | Enterobacteriaceae | 3-oxo-C6-HSL | 3-oxo-C6-HSL | + | + | ++ |
| A10JM | Enterobacteriaceae | 3-oxo-C6-HSL | 3-oxo-C6-HSL | + | + | ++ |
| B4JM | Enterobacteriaceae | 3-oxo-C6-HSL | 3-oxo-C6-HSL | + | + | + |
| C2JM | Enterobacteriaceae | 3-oxo-C6-HSL | 3-oxo-C6-HSL, (C6-HSL) | + | + | + |
| C3-1JM | Enterobacteriaceae | NI | 3-hydroxy-C10-HSL | − | − | − |
| MRa40 | Enterobacteriaceae | NI | ND | + | − | − |
| MRa43 | Enterobacteriaceae | C6-HSL+NI | ND | − | (−) | − |
| MRa44-1 | Enterobacteriaceae | C6-HSL+NI | ND | − | − | − |
| MRa44-2 | Enterobacteriaceae | NI | ND | − | − | − |
| MRa45 | Enterobacteriaceae | 3-oxo-C6-HSL | 3-oxo-C6-HSL, (C6-HSL) | + | + | +++ |
| MRa46-1 | Enterobacteriaceae | NI | C4-HSL | − | − | − |
| MRa46-2 | Enterobacteriaceae | 3-oxo-C6-HSL+NI | C6-HSL | − | − | − |
| MRa47 | Enterobacteriaceae | C6-HSL+NI | ND | − | − | − |
| MRa48 | Enterobacteriaceae | 3-oxo-C6-HSL | C6-HSL | + | + | + |
| MRa49 | Enterobacteriaceae | 3-oxo-C6-HSL | C6-HSL | − | − | − |
| MRa50 | Enterobacteriaceae | 3-oxo-C6-HSL | C6-HSL, 3-hydroxy-C10-HSL | − | − | − |
| MRa51 | Enterobacteriaceae | 3-oxo-C6-HSL | 3-oxo-C6-HSL, C6-HSL | − | (+) | − |
| MRa54 | Enterobacteriaceae | 3-oxo-C6-HSL | 3-oxo-C6-HSL | − | − | − |
| A5JM | Pseudomonas | NI | ND | − | − | − |
| A6JM | Pseudomonas | NI | ND | − | − | − |
| B2JM | Pseudomonas | NI | ND | − | − | − |
| C1JM | Pseudomonas | 3-oxo-C6-HSL | C10-HSL, 3-oxo-C6-HSL | + | (+) | − |
| C3-2JM | Pseudomonas | NI | ND | − | (+) | − |
| MRa52 | Pseudomonas | NI | ND | − | − | − |
| MRa55 | Pseudomonas | NI | ND | − | − | − |
| B6JM | Vibrionaceae | NI | ND | − | + | − |
| MRa39 | Vibrionaceae | NI | C10-HSL, C8-HSL, 3-oxo-C10-HSL, 3-hydroxy-C10-HSL | − | + | − |
| MRa42 | Vibrionaceae | NI | C4-HSL | − | + | − |
| MRa53 | Vibrionaceae | NI | ND | − | + | − |
Based on TLC profiling with Agrobacterium tumefaciens and Chromobacterium violaceum as monitor strains.
−, (+), and +, negative, weakly positive, and positive reactions, respectively, in qualitative assays.
−, no difference compared to the uninoculated control (Fig. 1B); +, visible signs of spoilage; ++, clear signs of spoilage; +++, spoilage with total collapse of bean sprouts (Fig. 1C).
NI, not identified (spots on TLC plates did not correspond to any known standard).
ND, not detected.
FIG. 1.
Appearance of bean sprouts, where the soaking water has been inoculated with different bacterial strains to determine their spoilage potentials. (A) Control (uninoculated); (B) bean sprouts inoculated with the nonspoiling strain C1JM; (C) bean sprouts inoculated with the spoiling strain Pectobacterium sp. strain A2JM; and (D) bean sprouts inoculated with the AHL-deficient A2JM luxI mutant.
AHLs in bean sprouts and pure cultures of bacteria.
TLC profiling of extracts from the 32 isolates revealed that 14 isolates produced an AHL with an Rf value and shape similar to that of 3-oxo-C6-HSL, and 12 of them produced this molecule as the only AHL compound (Table 4). Four isolates produced an AHL with an Rf value and shape similar to that of C6-HSL. The isolates produced the same types of AHLs after inoculation on bean sprouts. AHLs from bacterial extracts were also identified chemically by LC-HR-MS. In general, there was good agreement between the presumptive identification of AHLs by TLC and the chemical identification. Minor differences were probably caused by differences in sensitivity between the two methods. The presumed main AHL compound produced by a selected spoilage strain (A2JM; see below) was 3-oxo-C6-HSL, as determined by TLC (Table 4). This was confirmed by LC-HR-MS, in which trace amounts of C6-HSL were also detected. Furthermore, 3-oxo-C6-HSL was detected by LC-HR-MS in bean sprouts spoiled by A2JM (data not shown).
Characterization of the bean sprout spoilage isolate A2JM.
Based on its ability to spoil bean sprouts, strain A2JM was chosen for further studies. Preliminary identification based on phenotypic tests placed A2JM as a member of the Enterobacteriaceae. The highest similarities of the 16S rRNA gene sequence of strain A2JM were found to the type strain of Pectobacterium atrosepticum (accession no. Z96090) with 97.6% and to the type strain of Serratia marcescens (accession no. M59160) with 98.1%.
A phylogenetic analysis based on a partial 16S rRNA sequence confirmed the family classification of A2JM, and from its position in a phylogenetic tree reconstructed with sequences from a subset of species within this family, A2JM clustered with Serratia spp. Bootstrap analysis revealed that this position in the phylogenetic tree was not reliable (data not shown), and as pectinolytic activity has never been reported to be associated with members of Serratia spp., A2JM was submitted to additional biochemical analysis. Although the ability to degrade DNA and the ability to use maltose as the only carbon source are common traits within Serratia spp., A2JM lacked DNase activity, which virtually all Serratia species produce (24), and it could not metabolize maltose. When characterized by the API 20E system, A2JM did not decarboxylate lysine and ornithine but was able to utilize sodium citrate. The negative lysine and ornithine tests indicate that A2JM is a Pectobacterium (previously Erwinia) strain, whereas the ability to use sodium citrate indicates that it is a Serratia strain. A2JM was not lipolytic, whereas Serratia spp. are positive on tributyrin agar and Pectobacterium spp. are negative. A2JM did not produce carbapenem, which is an antibiotic produced by a number of Erwinia carotovora strains. The highest similarity of the deduced partial rpoB sequence of A2JM was found to strain SCRT1043 of Pectobacterium atrosepticum (accession no. CAG73142), with 98% (two amino acids difference). The highest similarity to other bacteria, such as Enterobacter, Morganella, and Serratia, was no more than 94%. The phenotypic characteristics of A2JM, as well as the high similarity of the 16S rRNA, the housekeeping gene rpoB, and luxI and luxR homologues (see below), lead to the conclusion that A2JM is a Pectobacterium strain.
Construction of an AHL-negative mutant.
An AHL-deficient mutant of A2JM (an A2JM luxI homologue mutant) was isolated after random Tn10 transposon mutagenesis. The transposon had been inserted at position 563 in a 651-bp open reading frame designated luxI. This gene was 92% identical to the hslI gene of E. carotovora, 91% identical to the carI gene of E. carotovora, and 89% identical to the ecbI gene of Erwinia carotovorum subsp. betavasculorum, indicating that luxI encodes a 216-amino-acid putative AHL synthetase. LuxI was similar to the I proteins of several Enterobacteriaceae (Table 5). A search for lux box-like regulatory elements (inverted repeats) in the 300-bp region preceding the start codon of luxI proved futile, implying that luxI belongs to the still-growing group of luxI homologues that lack a consensus lux box. Further analysis of the DNA flanking the transposon revealed that luxI shared 17 bp at its 3′ end with a 729-bp convergently transcribed luxR homologue (see below).
TABLE 5.
Similarity of I and R proteins of Enterobacteriaceae strain A2JM to I and R proteins of other strains
| Protein type | Strain | Protein | Similarity (%) | GenBank accession no. |
|---|---|---|---|---|
| I | ||||
| E. carotovora | HslI | 97 | AAA62483 | |
| E. carotovora | Carl | 97 | P33880 | |
| E. carotovorum subsp. betavasculorum | Ecbl | 96 | AAB69645 | |
| E. carotovora | ExpI | 88 | P33882 | |
| E. chrysanthemi | EchI | 78 | Q46968 | |
| S. liquefaciens | SwrI | 78 | P52989 | |
| E. chrysanthemi | AhlI | 77 | AAM46699 | |
| Serratia sp. strain ATCC 39006 | SmaI | 76 | CAB92553 | |
| Yersinia pestis | YpeI | 73 | AAF21290 | |
| S. marcescens | SpnI | 67 | AAN52498 | |
| Serratia proteomaculans | SprI | 65 | AAK76733 | |
| R | ||||
| E. carotovorum subsp. betavasculorum | EcbR | 99 | AAB69464 | |
| E. carotovora | ExpR | 81 | CAA56646 | |
| E. chrysanthemi | EchR | 79 | Q46967 | |
| E. chrysanthemi | ExpR | 78 | Q47188 | |
| Serratia sp. strain ATCC39006 | SmaR | 75 | CAB92554 | |
| Hafnia alvei | HalR | 73 | AAP30848 | |
| Yersinia pestis | YspR | 72 | NP_404602 | |
| S. proteomaculans | SprR | 71 | AAK76734 | |
| S. marcescens | SpnR | 69 | AAN52499 | |
| E. carotovora | CarI | 61 | Q46751 |
Prior to further analysis of the A2JM luxI mutant, its identity as an A2JM mutant was confirmed by PCR amplification of 100% identical luxI and luxR genes from chromosomal DNA of A2JM. When this luxIR region of wild-type A2JM was introduced into the AHL-deficient E. coli strain MT102, transformants acquired the ability to synthesize 3-oxo-C6-HSL (data not shown), supporting the notion that luxI encodes an AHL synthetase.
AHL-regulated phenotypes in A2JM.
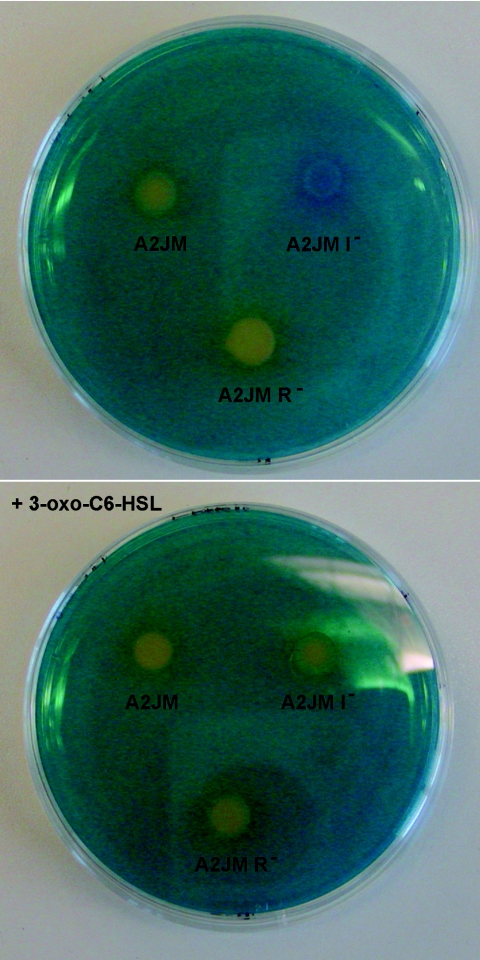
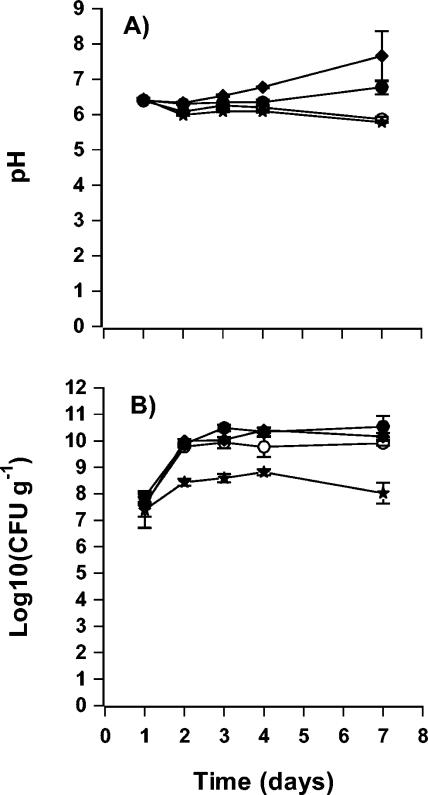
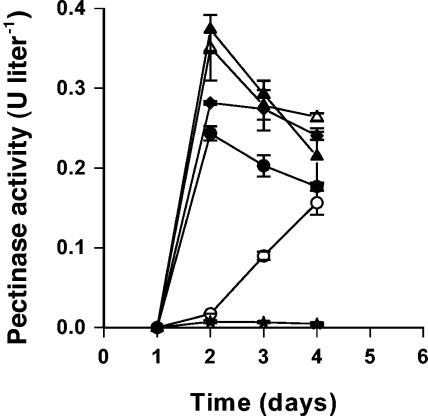
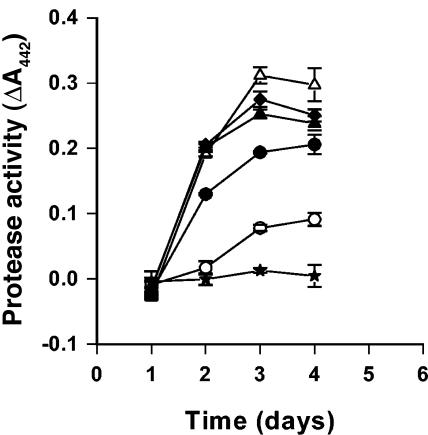
Phenotypes which could be regulated by AHL were tested both in laboratory media and in bean sprouts. The AHL-negative mutant of A2JM was impaired in pectinase, protease, cellulase (data not shown), and siderophore activities (Fig. 2). The A2JM luxI mutant grew as well as the wild-type strain on bean sprouts (Fig. 3B); however, spoilage was less pronounced (Fig. 1C and D). Furthermore, the pH of sprouts inoculated with the AHL-negative mutant during sprouting (Fig. 3A) remained low, whereas the pH increased in sprouts inoculated with the wild type. The level of pectinase or protease activity in sterile filtered samples of the soaking water from sprouts inoculated with the mutant was significantly lower than in sprouts inoculated with the wild-type A2JM (Fig. 4 and 5). When the soaking water was supplemented with 1 μM 3-oxo-C6-HSL, the A2JM luxI mutant expressed wild-type levels of pectinase and protease activities (Fig. 4 and 5).
FIG. 2.
Siderophore-indicative plates spotted with Pectobacterium sp. strain A2JM, the A2JM luxI mutant (I−), and the A2JM luxR mutant (R−). The bottom plate was complemented with 1 μM 3-oxo-C6-HSL.
FIG. 3.
pH (A) and bacterial counts (B) of bean sprouts during sprouting. Sprouts inoculated with Pectobacterium sp. strain A2JM (♦), the A2JM luxI mutant (○), the A2JM luxI mutant complemented with 3-oxo-C6-HSL (•), and the control (★) are shown. The values are the means and standard deviations of duplicate determinations of the production of bean sprouts.
FIG. 4.
Pectinase activity in the soaking water from the bean sprouts inoculated with Pectobacterium sp. strain A2JM (♦), the A2JM luxI mutant (○), the A2JM luxI mutant complemented with 3-oxo-C6-HSL (•), the A2JM luxR mutant (▵), the A2JM luxR mutant complemented with 3-oxo-C6-HSL (▴), and the control (★). The values are the means and standard deviations of triplicate determinations.
FIG. 5.
Protease activity in the soaking water from the bean sprouts inoculated with Pectobacterium sp. strain A2JM (♦), the A2JM luxI mutant (○), the A2JM luxI mutant complemented with 3-oxo-C6-HSL (•), the A2JM luxR mutant (▵), the A2JM luxR mutant complemented with 3-oxo-C6-HSL (▴), and the control (★). The values are the means and standard deviations of triplicate determinations.
Construction and characterization of an R-negative mutant.
The luxR homologue in A2JM was designated luxR and was 89% identical to the ecbR gene of E. carotovorum subsp. betavasculorum (GenBank accession no. AF001050), strongly suggesting that luxR encodes a 242-amino-acid putative transcriptional regulator. Other luxR homologues showed only localized identity to the luxR gene, and this identity was primarily restricted to the first 250 nucleotides of the aligned luxR homologues. LuxR showed its highest degree of similarity (99%) to EcbR of E. carotovorum subsp. betavasculorum (Table 5). For further support of its function as a LuxR homologue, a search for conserved domains using RPS-BLAST revealed the existence of an N-terminal AHL binding domain and a C-terminal helix-turn-helix motif.
The R mutant had similar or slightly elevated expression of pectinase (Fig. 4), protease (Fig. 5), cellulase (data not shown), and siderophore activity (Fig. 2), and it spoiled bean sprouts as well as the wild-type strain (data not shown).
DISCUSSION
The present study demonstrates that bacterial QS can play an important role in spoilage of food products in which gram-negative bacteria grow to high cell densities. We base this conclusion on the following observations: (i) the complete lack of soft-rot spoilage in sprouts which contained high (normal) levels of non-AHL-producing bacteria, (ii) a delay in the spoilage process of an AHL-negative mutant compared to its AHL-producing parent, and (iii) the ability of exogenous 3-oxo-C6-HSL to restore the spoilage process of the AHL-negative mutant. At least four phenotypes were regulated by the QS system: pectinase, protease, and cellulase activities and siderophore-mediated iron chelation. All these phenotypic traits are likely to be important for the ability of the bacteria to establish themselves and spoil this food product.
Bean sprouts typically have very high bacterial counts (47, 60), due to the sprouting period at high temperatures, and our finding of approximately 108 CFU/g is similar to other studies. Approximately 10% of the microorganisms produced AHLs and were identified as Enterobacteriaceae and pseudomonads, which are common on bean sprouts (43). Spoilage of bean sprouts was easily determined by sensory evaluation (soft rot), but the spoilage process was also accompanied by an increase in pH, as has been observed in spoiled spinach (7). The increase in pH was less pronounced for the commercial sprouts than for sprouts inoculated with the Pectobacterium strain A2JM; however, the degradation caused by A2JM was more severe than that of the naturally spoiled sprouts. Pectinolytic activity alone was not sufficient to enable a bacterial strain to spoil bean sprouts, but spoilage required the simultaneous expression of proteolytic and pectinolytic activities. Marits et al. (45) also found that protease activity was necessary for the normal progression of the disease symptoms caused by E. carotovora subsp. carotovora in living plants. However, it contrasts with an observation by Mount et al. (48), in which “tissue maceration” was seen when pure pectate lyase devoid of protease was tested. It is likely that the proteolytic activity is required for initial degradation of plant material and that the main purpose is to increase the pH, since the pectate lyase has a pH optimum of 8.5 (41). Therefore, although the pectinolytic activity alone will eventually lead to soft rot, it is enhanced and accelerated by accompanying proteolytic activity.
AHLs were readily extracted from commercial bean sprouts, and 3-oxo-C6-HSL was the dominant molecule. Despite the increase in pH, which may result in degradation of AHLs (13, 25), we found high levels of 3-oxo-C6-HSL equivalents in bean sprouts, in the range of 5 to 63 nmol per kg, as determined by a semiquantitative well diffusion assay.
One spoilage Pectobacterium strain, A2JM, which was both pectinolytic and proteolytic, produced 3-oxo-C6-HSL as its main AHL compound. AHLs have been detected in many species of Enterobacteriaceae (8, 23, 61), and 3-oxo-C6-HSL is commonly produced by strains of Enterobacteriaceae associated with food spoilage (29, 53). The A2JM luxI mutant, a 3-oxo-C6-HSL-deficient mutant of A2JM, revealed the existence of a 651-bp luxI homologue (designated luxI), which shared 17 bp at its 3′ end with a 729-bp convergently transcribed luxR homologue (designated luxR). Such a genetic arrangement has previously been described in members of the Enterobacteriaceae, including E. carotovorum subsp. betavasculorum (18), E. carotovora (5), Erwinia chrysanthemi (49) and S. marcescens (47). At the nucleotide level, the highest degrees of identity to the luxIR genes were found among the luxIR homologues of Erwinia spp. No gaps were observed throughout the alignment of the luxIR region with the ecbRI region of E. caratovorum subsp. betavasculorum (18), and these regions proved to be 89% identical. Although LuxI and LuxR protein homologues of Serratia spp. and Yersinia spp. were 76% similar to the A2JM LuxI and LuxR proteins, the highest degrees of similarity were found to EcbI and EcbR of E. carotovorum subsp. betavasculorum, indicating that these proteins have identical functions in A2JM and E. carotovorum subsp. betavasculorum. In support of this notion, functional A2JM LuxI was required for the synthesis of 3-oxo-C6-HSL, as is EcbI in E. carotovorum subsp. betavasculorum (18). Despite the strong homology of luxIR to ecbIR of E. carotovorum subsp. betavasculorum, a partial 16S rRNA identified A2JM as being phylogenetically affiliated with S. marcescens and Serratia odorifera. Strain A2JM did, however, express pectinolytic activity, a trait not previously observed within Serratia spp. but quite common among Pectobacterium spp. Other phenotypic traits (DNase activity, maltose catabolism, lysine decarboxylase, ornithine decarboxylase, and lipolytic activity on tributyrin agar) indicated that A2JM is a Pectobacterium strain. The uncertainty of the identification by the 16S rRNA gene could be caused by the lack of resolving power at the species level for 16S rRNA (54).
To test the spoilage abilities of bacterial strains and to study the role of AHLs, we cultured bean sprouts devoid of AHLs and AHL-producing bacteria. These sprouts had counts of 108 to 109 CFU/g, similar to commercial sprouts, but did not undergo soft rot. Hence, the spoilage caused by A2JM (and other bacterial cultures) is not caused by higher bacterial densities per se but by the metabolic activities of the strains. Similarly, the reduced spoilage activity of the A2JM luxI mutant was not a consequence of lower bacterial densities.
The AHL-negative mutant had reduced spoilage ability and had a lower protease activity than the wild type, but protease activity increased when exogenous 3-oxo-C6-HSL was added. In fact, all the defects observed in the luxI mutant turned out to be complemented upon exogenous addition of 3-oxo-C6-HSL, demonstrating that the insertion of the Tn10 transposon in the luxI mutant exclusively affects the synthesis of the signal molecule (3-oxo-C6-HSL). Consequently, we concluded that any potential polar effects of the Tn10 transposon-mediated mutation on neighboring genes were marginal, if they existed at all. Exoprotease activity is commonly regulated by AHLs. This is seen in organisms such as P. aeruginosa (51), S. liquefaciens (23), Aeromonas hydrophila (62), and V. anguillarum (19). Pectinase activity in E. carotovora has been linked to AHL regulation (52), similar to our findings in the bean sprout model system. It was recently reported that rot of onions caused by Burkholderia cepacia was regulated by quorum sensing. Both protease and polygalacturonase activities were regulated via the quorum-sensing system (1). Our study is the first to demonstrate a clear link between siderophore and AHL production in Enterobacteriaceae (Fig. 2). In some bacteria, the ability to grow under iron limitation is hampered in presumed quorum-sensing mutants (26). In B. cepacia, siderophore production does not appear to be systematically linked to AHL regulation (28), although ornibactin synthesis in one strain, K56-2, is regulated by the CepR-CepI system (40). Interestingly, the luxI-negative mutants hyperproduced the siderophore ornibactin (39).
The A2JM luxR mutant expressed the phenotypes (pectinase, protease, cellulase, and siderophore production) of the wild type and had slightly higher pectinase and protease activities than the wild type (Fig. 4 and 5). One may hypothesize that the R protein serves as a transcriptional repressor and that AHL production results in alleviation of the repression. Similarly, in B. cepacia strain K56-2, CepR is a negative regulator of siderophore (ornibactin) production (40). Alternatively, a second R gene may be present but not yet found in the strain (5, 66).
In conclusion, the present work has demonstrated that quorum sensing can be important in controlling bacterial food spoilage. We have demonstrated this using bean sprouts as a model food system; however, we anticipate that similar regulation of spoilage is found in other food products with similar spoilage characteristics. The fact that quorum sensing regulates food spoilage raises the interesting option of using quorum-sensing inhibitors as a novel targeted food preservation strategy.
Acknowledgments
This work was supported by the Danish Technical Research Council.
The Agrobacterium tumefaciens monitor was obtained from Clay Fuqua, and the Escherichia coli ESS strain was from Paul Williams. We thank Adam C. Martiny for the alignment and preliminary phylogenetic analysis of our 16S rRNA sequence and Peter Sestoft for assistance in alignment and bootstrap analyses. The technical assistance of Linda Stabell, Jette Melchiorsen, and Tina Nørgaard is greatly appreciated.
REFERENCES
- 1.Aguilar, C., I. Bertani, and V. Venturi. 2003. Quorum-sensing system and stationary-phase sigma factor (rpoS) of the onion pathogen Burkholderia cepacia genomovar I type strain, ATCC 25416. Appl. Environ. Microbiol. 69:1739-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexeyev, M. F., and I. N. Shokolenko. 1995. Mini-Tn10 transposon derivatives for insertion mutagenesis and gene delivery into the chromosome of Gram-negative bacteria. Gene 160:59-62. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen, J. B., A. Heydorn, M. Hentzer, L. Eberl, O. Geisenberger, B. B. Christensen, S. Molin, and M. Givskov. 2001. gfp based N-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl. Environ. Microbiol. 67:575-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersson, R. A., A. R. Eriksson, R. Heikinheimo, A. Mae, M. Pirhonen, V. Koiv, H. Hyytianen, A. Tuikkala, and E. T. Palva. 2000. Quorum sensing in the plant pathogen Erwinia caratovora subsp. carotovora: the role of expREcc. Mol. Plant-Microbe Interact. 13:384-393. [DOI] [PubMed] [Google Scholar]
- 6.Andro, T., J. P. Chambost, A. Kotoujansky, J. Cattaneo, Y. Bertheau, F. Barras, F. Van Gijsengem, and A. Coleno. 1984. Mutants of Erwinia chrysanthemi defective in secretion of pectinase and cellulase. J. Bacteriol. 160:1199-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babic, I., S. Roy, A. E. Watada, and W. P. Wergin. 1996. Changes in microbial populations on fresh cut spinach. Int. J. Food Microbiol. 31:107-119. [DOI] [PubMed] [Google Scholar]
- 8.Bainton, N. J., P. Stead, S. R. Chhabra, B. W. Bycroft, G. P. C. Salmond, G. S. A. B. Stewart, and P. Williams. 1992. N-(3-Oxohexanoyl)-l-homoserine lactone regulates carbapenem antibiotic production in Erwinia carotovora. Biochem. J. 288:997-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baird-Parker, T. C. 2000. The production of microbiologically safe and stable foods, p. 3-18. In B. M. Lund, T. C. Baird-Parker, and G. W. Gould (ed.), The microbiological safety and quality of food. Aspen Publishers, Gaithersburg, Md.
- 10.Bertani, G. 1951. Studies on lysogenesis. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowen, J. H., and S. D. Kominos. 1979. Evaluation of a pectin agar medium for isolation of Yersinia enterocolitica within 48 hours. Am. J. Clin. Pathol. 72:586-590. [DOI] [PubMed] [Google Scholar]
- 12.Bruhn, J. B., A. B. Christensen, L. R. Flodgaard, K. F. Nielsen, T. O. Larsen, M. Givskov, and L. Gram. 2004. Presence of acylated homoserine lactones (AHLs) and AHL-producing bacteria in meat and potential role of AHL in spoilage of meat. Appl. Environ. Microbiol. 70:4293-4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Byers, J. T., C. Lucas, G. P. C. Salmond, and M. Welch. 2002. Nonenzymatic turnover of an Erwinia carotovora quorum-sensing signaling molecule. J. Bacteriol. 184:1163-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cha, C., P. Gao, Y. C. Chen, P. D. Shaw, and S. K. Farrand. 1998. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant-Microbe Interact. 11:1119-1129. [DOI] [PubMed] [Google Scholar]
- 15.Christensen, A. B., K. Riedel, L. Eberl, L. R. Flodgaard, S. Molin, L. Gram, and M. Givskov. 2003. Quorum sensing directed protein expression in Serratia proteamaculans B5a. Microbiology 149:471-483. [DOI] [PubMed] [Google Scholar]
- 16.Clark, D. J., and O. Maaløe. 1967. DNA replication and division cycle in Escherichia coli. J. Mol. Biol. 23:99-112. [Google Scholar]
- 17.Collmer, A., J. L. Ried, and M. S. Mount. 1988. Assay methods for pectic enzymes. Methods Enzymol. 161:329-335. [Google Scholar]
- 18.Costa, J. M., and J. E. Loper. 1997. EcbI and EcbR: homologs of LuxI and LuxR affecting antibiotic and exoenzyme production by Erwinia carotovora subsp. betavasculorum. Can. J. Microbiol. 43:1164-1171. [DOI] [PubMed] [Google Scholar]
- 19.Croxatto, A., V. J. Chalker, J. Lauritz, J. Jass, A. Hardman, P. Williams, M. Camara, and D. L. Milton. 2002. VanT, a homologue of Vibrio harveyi LuxR, regulates serine, metalloprotease, pigment, and biofilm production in Vibrio anguillarum. J. Bacteriol. 184:1617-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dainty, R. H., and B. M. Mackey. 1992. The relationship between the phenotypic properties of bacteria from chill-stored meat and spoilage processes. Soc. Appl. Bacteriol. Symp. Ser. 21:103S-114S. [DOI] [PubMed] [Google Scholar]
- 21.de Lorenzo, V., L. Eltis, B. Kessler, and K. N. Timmis. 1993. Analysis of Pseudomonas gene products using lacI q/ Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123:17-24. [DOI] [PubMed] [Google Scholar]
- 22.Denkin, S. M., and D. R. Nelson. 1999. Induction of protease activity in Vibrio anguillarum by gastrointestinal mucus. Appl. Environ. Microbiol. 65:3555-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eberl, L., M. K. Winson, C. Sternberg, G. S. A. B. Stewart, G. Christiansen, S. R. Chhabra, B. Bycroft, P. Williams, S. Molin, and M. Givskov. 1996. Involvement of N-acyl-l-homoserine lactone autoinducers in controlling the multicellular behaviour of Serratia liquefaciens. Mol. Microbiol. 20:127-136. [DOI] [PubMed] [Google Scholar]
- 24.Farmer, J. J., F. Silva, and D. R. Williams. 1973. Isolation of Serratia marcescens on deoxyribonuclease-toluidine blue-cephalothin agar. Appl. Microbiol. 25:151-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flodgaard, L. R., A. B. Christensen, S. Molin, M. Givskov, and L. Gram. 2003. Influence of food preservation parameters and associated microbiota on production rate, profile and stability of acylated homoserine lactones from food-derived Enterobacteriaceae. Int. J. Food Microbiol. 84:145-156. [DOI] [PubMed] [Google Scholar]
- 26.Fong, K. P., L. Gao, and D. R. Demuth. 2003. luxS and arcB control aerobic growth of Actinobacillus actinomycetemcomitans under iron limitation. Infect. Immun. 71:298-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Givskov, M., M. Olsen, and S. Molin. 1988. Cloning and expression in Escherichia coli of the gene for extracellular phospholipase A1 from Serratia liquefaciens. J. Bacteriol. 170:5855-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gotschlich, A., B. Huber, O. Geisenberger, A. Togl, A. Steidle, K. Riedel, P. Hill, B. Tummler, P. Vandamme, B. Middleton, M. Camara, P. Williams, A. Hardman, and L. Eberl. 2001. Synthesis of multiple N-acylhomoserine lactones is wide-spread among the members of the Burkholderia cepacia complex. Syst. Appl. Microbiol. 24:1-14. [DOI] [PubMed] [Google Scholar]
- 29.Gram, L., A. B. Christensen, L. Ravn, S. Molin, and M. Givskov. 1999. Production of acylated homoserine lactones by psychrotrophic members of the Enterobacteriaceae isolated from foods. Appl. Environ. Microbiol. 65:3458-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gram, L., L. Ravn, M. Rasch, J. B. Bruhn, A. B. Christensen, and M. Givskov. 2002. Food spoilage—interactions between food spoilage bacteria. Int. J. Food Microbiol. 78:79-97. [DOI] [PubMed] [Google Scholar]
- 31.Gregersen, T. 1978. Rapid method for distinction of Gram-negative and Gram-positive bacteria. Eur. J. Appl. Microbiol. 5:123-127. [Google Scholar]
- 32.Guyer, M. S., R. R. Reed, J. A. Steitz, and K. B. Low. 1980. Identification of a sex-factor-affinity site in Escherichia coli as gamma-delta. Cold Spring Harbor Symp. Quant. Biol. 45:135-140. [DOI] [PubMed] [Google Scholar]
- 33.Hansen, L. H., S. J. Sørensen, and L. B. Jensen. 1997. Chromosomal insertion of the entire Escherichia coli lactose operon, into two strains of Pseudomonas, using a modified mini-Tn5 delivery system. Gene 186:167-173. [DOI] [PubMed] [Google Scholar]
- 34.Herbert, H. A., M. A. Hendire, D. M. Gibson, and J. M. Shewan. 1971. Bacteria active in the spoilage of certain seafoods. J. Appl. Bacteriol. 34:41-50. [DOI] [PubMed] [Google Scholar]
- 35.Jones, S., B. Yu, N. J. Bainton, M. Birdsall, B. W. Bycroft, S. R. Chhabra, A. R. J. Cox, P. Golby, P. J. Reeves, S. Stephens, M. K. Winson, G. P. C. Salmond, G. S. A. B. Stewart, and P. Williams. 1993. The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa. EMBO J. 12:2477-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aaes-Jørgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48:1511-1524. [DOI] [PubMed] [Google Scholar]
- 37.Korczak, B., H. Christensen, S. Emler, J. Frey, and P. Kuhnert. 2004. Phylogeny of the family Pasteurellaceae based on rpoB sequences. Int. J. Syst. Evol. Microbiol. 54:1393-1399. [DOI] [PubMed] [Google Scholar]
- 38.Lauderdale, T.-L., K. C. Chapin, and P. R. Murray. 1999. Reagents, p. 1665-1673. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 39.Lewenza, S., B. Conway, E. P. Greenberg, and P. A. Sokol. 1999. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181:748-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewenza, S., and P. A. Sokol. 2001. Regulation of ornibactin biosynthesis and N-acyl-l-homoserine lactone production by CepR in Burkholderia cepacia. J. Bacteriol. 183:2212-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao, C.-H., J. Sullivan, J. Grady, and L.-J. C. Wong. 1997. Biochemical characterization of pectate lyases produced by fluorescent pseudomonads associated with spoilage of fresh fruit and vegetables. J. Appl. Microbiol. 83:10-16. [Google Scholar]
- 42.Lindum, P. W., U. Anthoni, C. Christophersen, L. Eberl, S. Molin, and M. Givskov. 1998. N-Acyl-l-homoserine lactone autoinducers control production of an extracellular lipopeptide biosurfactant required for swarming motility of Serratia liquefaciens MG1. J. Bacteriol. 180:6384-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lund, B. M. 1982. The effect of bacteria on post-harvest quality of vegetables and fruits, with particular reference to spoilage. Soc. Appl. Bacteriol. Symp. Ser. 10:133-153. [Google Scholar]
- 44.Lund, B. M. 1983. Bacterial spoilage, p. 219-257. In C. Dennis (ed.), Post harvest pathology of fruits and vegetables. Academic Press, London, United Kingdom.
- 45.Marits, R., V. Koiv, E. Laasik, and A. Mae. 1999. Isolation of an extracellular protease gene of Erwinia carotovora subsp. carotovora strain SCC3193 by transposon mutagenesis and the role of protease in phytopathogenicity. Microbiology 145:1959-1966. [DOI] [PubMed] [Google Scholar]
- 46.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. S. A. B. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703-3711. [DOI] [PubMed] [Google Scholar]
- 47.Michard, J., N. Jardy, and J. L. Gey. 1993. Bacteriologie des pousses de “soja” (Vigna radiata). Microbiol. Aliments Nutr. 11:135-146. [Google Scholar]
- 48.Mount, M. S., D. F. Bateman, and H. G. Basham. 1970. Induction of electrolyte loss, tissue maceration, and cellular death of potato tissue by an endopolygalacturonate trans-eliminase. Phytopathology 60:924-931. [Google Scholar]
- 49.Nasser, W., M. L. Bouillant, G. Salmond, and S. Reverchon. 1998. Characterization of the Erwinia chrysanthemi expl-expR locus directing the synthesis of two N-acyl-homoserine lactone signal molecules. Mol. Microbiol. 29:1391-1405. [DOI] [PubMed] [Google Scholar]
- 50.Nguyen-The, C., and F. Carlin. 2000. Fresh and processed vegetables, p. 620-684. In B. M. Lund, T. C. Baird-Parker, and G. W. Gould (ed.), The microbiological safety and quality of food. Aspen Publishers, Gaithersburg, Md.
- 51.Passador, L., J. M. Cook, M. J. Gambello, L. Rust, and B. H. Iglewski. 1993. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260:1127-1130. [DOI] [PubMed] [Google Scholar]
- 52.Pirhonen, M., D. Flego, R. Heikinheimo, and E. T. Palva. 1993. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 12:2467-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ravn, L., A. B. Christensen, S. Molin, M. Givskov, and L. Gram. 2001. Methods for detecting acylated homoserine lactones produced by Gram-negative bacteria and their application in studies of AHL-production kinetics. J. Microbiol. Methods 44:239-251. [DOI] [PubMed] [Google Scholar]
- 54.Roselló-Mora, R., and R. Amann. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25:39-67. [DOI] [PubMed] [Google Scholar]
- 55.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 56.Schweizer, H. P. 1993. Small broad-host-range gentamicin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques 15:831-834. [PubMed] [Google Scholar]
- 57.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 58.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. Cronan, K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 94:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith, R. S., S. G. Harris, R. Phipps, and B. Iglewski. 2002. The Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)-homoserine lactone contributes to virulence and induces inflammation in vivo. J. Bacteriol. 184:1132-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Splittstoesser, D. F., D. T. Queale, and B. W. Andaloro. 1983. The microbiology of vegetable sprouts during commercial production. J. Food Saf. 5:79-86. [Google Scholar]
- 61.Swift, S., M. K. Winson, P. F. Chan, N. J. Bainton, M. Birdsall, P. J. Reeves, C. E. D. Rees, S. R. Chhabra, P. J. Hill, J. P Throup, B. W. Bycroft, G. P. C. Salmond, P. Williams, and G. S. A. B. Stewart. 1993. A novel strategy for the isolation of luxI homologues: evidence for the widespread distribution of a LuxR:LuxI superfamily in enteric bacteria. Mol. Microbiol. 10:511-520. [DOI] [PubMed] [Google Scholar]
- 62.Swift, S., M. J. Lynch, L. Fish, D. F. Kirke, J. M. Tomas, G. S. A. B. Stewart, and P. Williams. 1999. Quorum sensing-dependent regulation and blockade of exoprotease production in Aeromonas hydrophila. Infect. Immun. 67:5192-5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Telford, G., D. Wheeler, P. Williams, P. T. Tomkins, P. Appleby, H. Sewell, G. S. A. B. Stewart, B. W. Bycroft, and D. I. Pritchard. 1998. The Pseudomonas aeruginosa quorum-sensing signal molecule N-(3-oxododecanoyl)-l-homoserine lactone has immunomodulatory activity. Infect. Immun. 66:36-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thompson, J. D., T. J. Gibson, F. Plewniak F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Throup, J., N. J. Bainton, B. W. Bycroft, P. Williams, and G. S. A. B. Stewart. 1995. Signalling in bacteria beyond luminescence, p. 89-92. In A. K. Cambell, L. J. Kricka, and P. E. Stanley (ed.), Bioluminescence and chemiluminescence: fundamental and applied aspects. Wiley, Chichester, United Kingdom.
- 66.Whitehead, N. A., J. T. Byers, P. Commander, M. J. Corbett, S. J. Coulthurst, L. Everson, A. K. P. Harris, C. L. Pemberton, N. J. L. Simpson, H. Slater, D. S. Smith, M. Welch, N. Williamson, and G. P. C. Salmond. 2002. The regulation of virulence in phytopathogenic Erwinia species: quorum sensing, antibiotics and ecological considerations. Antonie Leeuwenhoek 81:223-231. [DOI] [PubMed] [Google Scholar]
- 67.Windle, H. J. P., and D. Kelleher. 1997. Identification and characterization of a metalloprotease activity from Helicobacter pylori. Infect. Immun. 65:3132-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]