Abstract
Sardinian wine strains of Saccharomyces cerevisiae used to make sherry-like wines form a biofilm at the air-liquid interface at the end of ethanolic fermentation, when grape sugar is depleted and further growth becomes dependent on access to oxygen. Here, we show that FLO11, which encodes a hydrophobic cell wall glycoprotein, is required for the air-liquid interfacial biofilm and that biofilm cells have a buoyant density greater than the suspending medium. We propose a model for biofilm formation based on an increase in cell surface hydrophobicity occurring at the diauxic shift. This increase leads to formation of multicellular aggregates that effectively entrap carbon dioxide, providing buoyancy. A visible biofilm appears when a sufficient number of hydrophobic cell aggregates are carried to and grow on the liquid surface.
Flor or velum formation by certain wine strains of Saccharomyces cerevisiae (flor strains) is a form of cellular aggregation that manifests as an air-liquid interfacial biofilm at the end of alcoholic fermentation. Increased cell buoyancy and the resultant biofilm that forms on the wine surface appear to be an adaptive mechanism because the biofilm assures access to oxygen and therefore permits continued growth on nonfermentable ethanol. In general, nonbuoyant cells cease growth at the end of completed wine fermentations not for lack of carbon, but for lack of oxygen. In contrast to other microbial biofilms, those formed by flor strains appear to consist of a layer of buoyant cells without a suspending extracellular polysaccharide or protein matrix, as no evidence for such extracellular material has been reported. Biofilm cells have been found to have an elevated and/or altered lipid content and an increased surface hydrophobicity (7, 9, 15, 16, 24). Recently, Zara et al. (35) found that the small heat shock protein Hsp12 is required for biofilm formation in a Sardinian flor strain. Reynolds and Fink (28) reported that a laboratory strain of S. cerevisiae could be induced to form a biofilm at a liquid-hydrophobic solid interface and that such formation was dependent on FLO11. In addition, flo11Δ mutants were reported to be less hydrophobic than the wild type.
FLO11 has an open reading frame (ORF) of 4,104 bp, which encodes a hydrolase belonging to the glycosylphosphatidylinositol-anchored class of cell wall proteins rich in serine and threonine. The central domain of Flo11 is similar to that of the flocculins Flo1, Flo5, and Flo10 (33). The FLO11 promoter is at least 2,800 bp (22) and is complex, consisting of four upstream activating sequences and at least nine upstream repressing sequences, the activities of which depend upon growth stage and nutritional conditions (30). In the present study, we demonstrate that FLO11 is required for yeast biofilm formation at an air-liquid interface and that the biofilm cells are not less dense than the suspending medium, and we propose a model to explain the role of FLO11 in biofilm formation.
MATERIALS AND METHODS
Yeast strains, media, and genetic methods.
Yeast strains are listed in Table 1. Strain 3238-32 was derived by crossing a homothallic spore from the wild-type Sardinian biofilm-forming Arvisionadu wine strain A9 with the laboratory strain YPH499. The resultant diploid was sporulated and backcrossed to another A9 spore. A total of four successive backcrosses was performed, yielding heterothallic, auxotrophic, biofilm-forming segregants, one of which was designated 3238-32. Standard growth and sporulation media were used (6). Spore-to-cell matings (2), other crosses, and tetrad dissections were performed by standard procedures (6). Yeast transformations were performed as previously described (11). Flor medium (5) is yeast nitrogen base (Difco, Detroit, Mich.) containing 4% ethanol as a sole carbon source, supplemented when necessary with bases and amino acids at standard concentrations (6).
TABLE 1.
Yeast strains
| Strain | Genotype | Phenotypea | Source |
|---|---|---|---|
| A9 | MATa/MATαHO/HO | FLOR+ | DISAABAb |
| 3238-32 | MATα leu2-Δ1 lys2-801 ura3-52 | FLOR+ | This study |
| 3238-32Δflo11 | MATα leu2-Δ1 lys2-801 flo11Δ::URA3 ura3-52 | FLOR− | This study |
| 3238-4 | MATalys2-801 ura3-52 FLOR+ | FLOR+ | This study |
| 10560-23C | MATα ura3-52 his3::hisG leu2::hisG | FLOR− | G. Fink |
FLOR+ indicates the ability to form an air-liquid interfacial biofilm during growth on ethanol.
DISAABA, Dipartimento di Scienze Ambientali Agrarie e Biotecnologie Agroalimentari.
Measurement of buoyant density.
An estimate of cell buoyant density was made by equilibrium sedimentation with a Percoll (Amersham Biosciences, Piscataway, N.J.) gradient (3). Briefly, cells were grown at 30°C in static culture in yeast extract-peptone-dextrose medium (YEPD) and harvested in log phase or were grown statically for 3 days at 30°C in flor medium, where they formed an air-liquid interfacial biofilm. Cells from both cultures were concentrated and fixed in 3.7% formaldehyde by the addition of 1 part of concentrated formaldehyde (37%) with 9 parts of culture (27). Fixed cells were sonicated on ice for 10 s (setting no. 5 in a model 60 Sonic Dismembrator; Fisher Scientific, Pittsburgh, Pa.) to disperse multicellular aggregates at a dose found not to affect the viability of unfixed cells. Cells were then pelleted and resuspended in 60% Percoll. A step gradient of 60, 70, 80, and 90% Percoll (in 1× YEPD) was prepared by layering 0.5 ml of each solution into centrifuge tubes. The density of each of the solutions was determined by weighing measured volumes. About 5 μl of concentrated cell suspensions was carefully added to the top of the gradients, and the tubes were centrifuged to equilibrium at 500 × g, requiring 5 to 10 min.
Construction and confirmation of FLO11 deletion allele.
A start-to-stop codon FLO11 deletion allele was constructed by PCR (6) using plasmid pRS416 as a template to create a URA3 gene flanked by 60 bp of FLO11 sequence immediately upstream of the start codon and 60 bp immediately downstream of the stop codon. The PCR was performed using newflo11:ura UP and newflo11:ura LO primers (Table 2). Temperature cycling parameters were as follows: an initial hold at 95°C for 2 min and 45 s; 3 cycles, each at 94°C for 45 s, 51°C for 1 min,and 72°C for 2 min; 30 cycles, each at 94°C for 45 s, 62°C for 1 min, and 72°C for 3 min; and a final elongation at 72°C for 8 min, performed with a Robocycler 96 (Stratagene, La Jolla, Calif.). A 1.3-kb PCR product was gel isolated (Qiaquick gel extraction kit; QIAGEN, Inc., Valencia, Calif.) and used to transform strain 3238-32. Confirmation of the deletion was determined by PCR using the “Flo11 UP 45 and Flo11 LO 737” pair of primers (700-bp product), and the Flo11 UP 45 and URA3 down primer pair (1.3-kb product).
TABLE 2.
DNA primers
| Primer | Sequence (5′→3′) |
|---|---|
| newflo11:ura UP | ATCCCTCGTCATGTTGTGGTTCTAATTAAAATATACTTTTGTAGGCCTCAAAAATCCATAAGATTGTACTGAGAGTGCAC |
| newflo11:ura LO | TATCTTAATTTAAGAATGAAAACATCGTAATGAAGAAACGAACATTTGGAATTGTATCACTGTGCGGTATTTCACACCG |
| Flo11 UP 45 | AACCCTAAAAGTGCCTGCTC |
| Flo11 LO 737 | GTGCCAATTGAAGTCTAAGT |
| URA3 down | CCTGCTTCAAACCGCTAACAATACCTG |
| Up prom flo 82 | CCAACTAAATCTGAATAACAA |
| Lo prom flo 3022 | AAGCGAAAGGACCAAATAAGC |
| Seq prom flo 134 | GCAATGATTATGTGGTAT |
| Seq prom flo 584 | AGCTGAAAAGTCCATCTA |
| Seq prom flo 1071 | ACATCTTTGCTCCCTTAC |
| Seq prom flo 1538 | GGTGAGATTTGTTTTATG |
| Seq prom flo 1837 | AATGTCGCCCAAAGAGTT |
| Seq prom flo 2022 | TGCGACAGTGGCTTCAAA |
| Seq prom flo 2549 | TGTGGGTCATCTTTTTAG |
Air-liquid interfacial biofilm formation and invasive growth assays.
Formation of an air-liquid interfacial biofilm was performed as follows: strains were grown in 2 ml of YEPD overnight at 30°C in an incubator-shaker, recovered by centrifugation, washed once in sterile distilled water, and resuspended in 2 ml flor medium in test tubes. Samples were incubated at 30°C for 3 to 5 days under static conditions.
The invasive growth assay was performed as previously described (29). Briefly, strains were patched onto YEPD plates with toothpicks, with care being taken to avoid scratching the agar surface, and allowed to grow for 3 to 5 days at 30°C. Plates were then washed with sterile distilled water to remove cells from the agar surface, leaving subsurface cells that had effectively invaded the agar. Plates were subsequently observed microscopically to confirm the invasive growth phenotype (data not shown).
Colony morphology on soft agar.
Strains were inoculated onto YPD soft agar plates (0.3% agar) with a toothpick 1 to 2 days after the plates were poured, as previously described (28). The plates were wrapped with parafilm, incubated at 25°C, and photographed.
DNA sequencing.
The FLO11 promoter from strain 3238-32 was amplified by PCR using primers Up prom flo 82 and Lo prom flo 3022. The PCR product was gel isolated (Qiaquick gel extraction kit; QIAGEN, Inc.), and sequenced by the dideoxy-dye terminator method with an ABI model 373A sequencer (Applied Biosystems, Foster City, Calif.) with the primers listed in Table 2, at the CRIBI DNA sequencing service, University of Padova, Padova, Italy. DNA and protein homology searches were performed using the BLAST algorithm and the Saccharomyces Genome Database (http://www.yeastgenome.org/) and through the National Center for Biotechnology Information.
RESULTS
FLO11 is essential for an air-liquid interfacial biofilm.
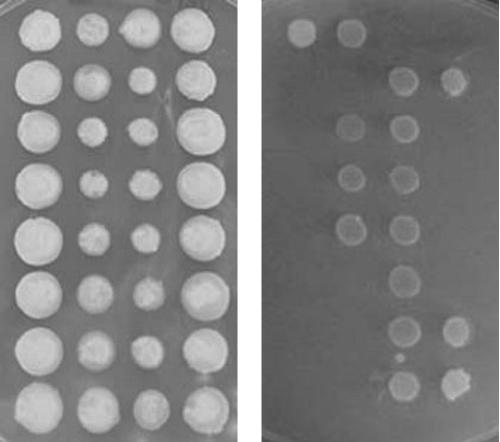
Based on the observation that FLO11 is required for yeast biofilm formation on a hydrophobic solid surface (28), we reasoned that it would also be required for biofilm formation at the air-liquid interface, as gas-liquid interfaces are excellent models for hydrophobic surface-liquid interfaces (14). A null allele of FLO11 was constructed by replacing the entire ORF with the selectable marker URA3 (6) in strain 3238-32, derived from the biofilm-forming Sardinian wine strain A9. The resultant FLO11 deletant strain, 3238-32Δflo11, was found to be unable to form an air-liquid interfacial biofilm during growth in flor medium, even when incubated for 7 days, while 2 days were sufficient for the parent strain to form a visible film (Fig. 1). Thus, FLO11 is essential for air-liquid interfacial biofilm formation. While deletion of FLO11 was confirmed by PCR (data not shown), a confirmatory cross was performed between 3238-32Δflo11 and a congenic FLO11 strain of opposite mating type, 3238-4. The diploid formed by this cross, the two haploid parent strains, and progeny from 24 dissected tetrads were scored for air-liquid interfacial biofilm formation in flor medium, agar invasivity, a known FLO11-dependent phenotype, and Ura prototrophy. As expected, the diploid and FLO11 parent, 3238-4, were found to be biofilm positive, invasive, and Ura−; 3238-32Δflo11 was found to be biofilm negative, noninvasive, and Ura+. Cosegregation (2+:2−) of the two FLO11-associated phenotypes with Ura− was observed among 24 dissected progeny, of which seven tetrads segregating 2+:2− for agar invasivity are shown in Fig. 2. This genetic evidence confirms that FLO11 is essential for interfacial biofilm formation in an A9 background.
FIG. 1.

Biofilm formation in flor medium (yeast nitrogen base plus 4% ethanol). Cells were grown in 2 ml of YEPD overnight at 30°C in an incubator-shaker, recovered by centrifugation, washed once with sterile water, and resuspended in 2 ml flor medium. The tubes were photographed after 3 days of static incubation at 25°C. Left panel, 3238-32; right panel, 3238-32 Δflo11.
FIG. 2.
Segregation for agar invasivity, a FLO11-dependent phenotype, among tetrads produced by a cross between 3238-32 (Δflo11) and 3238-4 (FLO11) after 5 days at 30°C on YEPD. The colonies were photographed before (left panel) and after (right panel) cells were washed from the plate surface.
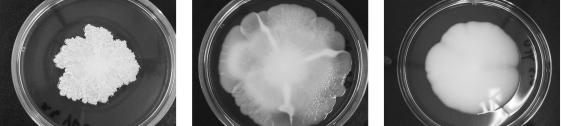
Disruption of FLO11 was also found to interfere with another FLO11-dependent phenotype, mat formation on soft agar (28). While mat formation was observed in strain 3238-32, it was absent in 3238-32Δflo11, and was present but differed morphologically in the presumably unrelated Σ1278b derivative strain 10560-23C (Fig. 3).
FIG. 3.
Comparison of mat formation on soft agar by 10560-23C (derived from Σ1278b) (left panel), 3238-32 (center panel), and 3238-32 Δflo11 (right panel) following incubation for 12 days at 25°C.
No evidence for gas vesicles.
In bacteria, gas vesicles that increase cell buoyancy appear as refractile bodies by phase-contrast microscopy and can be lysed under pressure, i.e., centrifugation in a microfuge (20). In our examination of the A9 strain, no such refractile bodies were seen in buoyant biofilm cells, nor did centrifugation at 12,000 × g for 30 min change the appearance of the cells (data not shown). While not definitive, these negative data suggest that bacterial-type gas vesicles are not present in the A9 strain. These findings are consistent with our observations that physical disruption of the air-liquid interfacial biofilm by gentle mixing resulted in rapid sedimentation of the cells. In such a case, a visible biofilm reformed over a prolonged period of many hours to days, depending on the rate of growth of the culture and size of the growth vessel.
Buoyant density of biofilm cells is greater than that of the suspending medium.
One explanation for the ability of the Sardinian flor strain to form a biofilm at the air-liquid interface during growth on ethanol is that the cells growing on ethanol are less dense than the suspending medium. To test this hypothesis, an estimate of cell density of the wild-type A9 strain was made by equilibrium sedimentation in a Percoll gradient (3). Sonicated or unsonicated cells grown on ethanol banded at the 80 to 90% Percoll interface, corresponding to a density between 1.11 and 1.10 g/cm3. Biofilm cells were found to consist mostly of large multicellular aggregates which sonication efficiently disrupted (photomicrograph not shown). Cells grown on glucose, which do not form an air-liquid interfacial biofilm, yielded a major band at the same 80 to 90% Percoll interface and a minor band at the 70 to 80% Percoll interface. The latter band corresponded to a density between 1.06 and 1.07 g/cm3. The density of both YEPD and flor medium was found to be 1.01 g/cm3. Thus, a decrease in density of the biofilm-forming A9 cells cannot explain their buoyancy.
FLO11 promoter sequence in flor strain not different than that of S288C.
While we found that FLO11 is essential for formation of an air-liquid interfacial biofilm in a Sardinian wine strain, laboratory strains of S. cerevisiae also have FLO11 but do not form a biofilm during growth on ethanol as sole carbon source, i.e., following the diauxic shift. One explanation is that FLO11 is regulated differently in the Sardinian strain and may have a unique promoter sequence. The FLO11 promoter is at least 2,800 bp long, and it is one of the longest and most complex promoters in the S. cerevisiae genome as it has specific binding sites for several activating and repressing elements (30). Sequence analysis revealed that the 3238-32 FLO11 promoter (GenBank accession no. AY618269) shares 97% homology with that of the standard laboratory strain S288C (http://www.yeastgenome.org/) and that no differences were found in previously identified sequence elements with known functional significance. In particular, the predicted binding sites for the transcription factors Tec1 (23), Ste12 (23), and Flo8 (18) are identical in the two strains, and occur in the 3238-32 promoter sequence at positions −706 to −702, −728 to −722, and −1,447 to −1,423, respectively. Relative to the S288C sequence, a total of 71 base pair substitutions were found throughout the 2,933-bp 3238-32 FLO11 promoter, amounting to 2.4% of the total sequence. Of the 71 differences, 43 are transitions, 12 are single-base-pair deletions, one is a deletion of 2 adjacent base pairs, and the remaining 14 are transversions.
DISCUSSION
In the present study, we demonstrate that FLO11 is necessary for formation of an air-liquid interfacial biofilm during growth of a Sardinian wine strain of S. cerevisiae on ethanol. While the sequence of the wine strain promoter was not found to differ significantly from that of laboratory strains, the functional significance of these minor differences awaits experimental evaluation. Although the extensive analysis of Rupp et al. (30) revealed regions containing activating and repressing sequences along the entire length of the promoter, specific sequence elements were not identified in their study. Differential activity or regulation of Flo11 cannot be ruled out because the wine strain FLO11 ORF has not yet been sequenced, nor do we know what sequence differences exist in wine strain alleles of the many genes known to regulate FLO11 expression (12, 19, 30). The extent of Flo11 mannosylation may also differ between laboratory and flor strains of S. cerevisiae. In Candida albicans, the acid-labile mannan component of cell wall-associated glycoproteins has been found to correlate with cell surface hydrophobicity (25, 26).
A model for air-liquid interfacial biofilm formation.
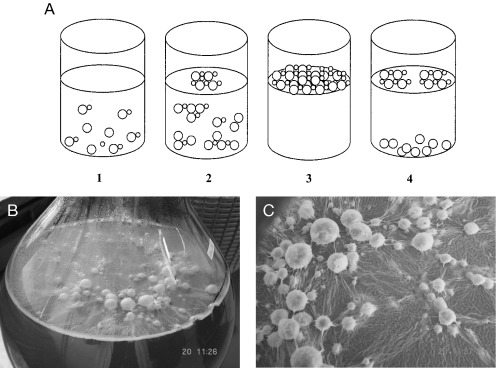
Based on what is known about FLO11 and the air-liquid interfacial biofilm formed by Sardinian and related flor strains of S. cerevisiae, we propose the following general model (Fig. 4A), based in part on a mechanism proposed by Martínez et al. (24).
FIG. 4.
Air-wine interfacial biofilm formation by flor strains of S. cerevisiae. (A) Model for biofilm formation. (1) Budded and unbudded cells ferment sugar while in suspension or at the bottom of the vessel and do not form a visible biofilm. (2) At the diauxic shift, when sugar levels decrease to about 0.2%, cells begin to aggregate into multicellular flocs due to an increase in cell surface hydrophobicity. The flocs entrap CO2 (bubbles not depicted), which continues to evolve from metabolism of the residual sugar, and are carried to the liquid surface. (3) A visible biofilm forms on the liquid surface. Direct access of the uppermost layer of biofilm cells to air provides a growth advantage when sugar is completely depleted as respiration of ethanol is strictly dependent on oxygen. (4) The biofilm begins to break apart and collapse as cells die due to depletion of available nutrients, i.e., nitrogen, vitamins, or are killed by prolonged exposure to high ethanol and acetaldehyde levels. (B) Biofilm-covered carbon dioxide bubbles produced by a spore segregant of Sardinian flor yeast (M25) 11 days postinoculation of Vernaccia grape juice. (C) A portion of the fermentation depicted in panel B is shown at 24× magnification.
Increased FLO11 expression at the diauxic shift significantly increases cell surface hydrophobicity, which in turn leads to formation of multicellular aggregates. The hydrophobic aggregates entrap carbon dioxide produced by fermentation of residual sugar (<0.2%), and the resulting bubbles carry them to the liquid surface, leading to formation of a visible biofilm. The following observations are consistent with the model. FLO11 confers significant hydrophobicity to the yeast cell surface (28), and in laboratory strains, FLO11 expression is known to be repressed by glucose (8, 19, 30). Hydrophobicity contributes to the related phenomenon of flocculence (32), which leads to the rising of ale yeasts or the rapid sedimentation of lager yeasts at the end of beer fermentation (34). Air-liquid interfacial biofilm-forming (9, 15) or foam-forming (31) strains of S. cerevisiae are hydrophobic. Rising gas bubbles are a recognized mechanism for concentrating hydrophobic chemicals, particles, and microorganisms at air-liquid interfaces (4, 10). Figures 4B and C illustrate how evolution of carbon dioxide bubbles produced by a spore segregant of Sardinian flor strain M25 can carry biofilm cells to the wine surface towards the end of a wine fermentation.
While our model does not rule out sources of cell surface hydrophobicity other than FLO11, it does suggest that FLO11 expression in biofilm-forming strain 3238-32, derived from strain A9, must differ significantly from that of laboratory strains which express FLO11 but do not form air-liquid interfacial biofilms. One recognized difference is that the FLO11 transcriptional activator FLO8 harbors a nonsense mutation in S288C-derived laboratory strains (21). It is of interest that Shimoi et al. (31) have reported that foam-formation by a sake strain of S. cerevisiae is dependent on a different hydrophobic cell surface protein, Awa1, that is absent in the laboratory strain S288C. Similarly, Alexandre et al. (1), described an unidentified but abundant 49-kDa cell wall-associated mannoprotein with relative hydrophobic character, present in the film-forming wine yeast P3 isolated from a French sherry-like wine (vin jaune).
The physical chemistry of how Flo11 confers hydrophobicity to the yeast cell surface is unknown. While Flo11 is known to be mannosylated, the number and type of mannan linkages and the degree of polymerization have not been determined precisely in any S. cerevisiae strain. With respect to flocculation, addition of mannose and other monosaccharides and proteolytic treatment have all been observed to be disruptive (13, 32). Martínez et al. (24) found that proteolytic treatment of a yeast biofilm on the surface of sherry had a similarly disruptive effect. It has been suggested that a reduction in the amount of phosphodiester-linked β-1,2-oligomannosyl branches of cell surface mannoproteins, rather than a reduction in total glycosylation, is sufficient to decrease the hydrophobicity of Candida albicans (26).
ADDENDUM
Ishigami et al. (17) recently reported that FLO11 is essential for flor formation in S. cerevisiae.
Acknowledgments
We thank Jeff Greenwood for use of the sonicator and Mike Penner for useful discussions and for critically reviewing the manuscript. We also thank Piersisinnio Asunis for his assistance and Davide Orro for providing photographs of Fig. 4.
This research was supported by PON-Misura 1.3: progetto AgriEchnos.
REFERENCES
- 1.Alexandre, H., S. Blanchet, and C. Charpentier. 2000. Identification of a 49-kDa hydrophobic cell wall mannoprotein present in velum yeast which may be implicated in velum formation. FEMS Microbiol. Lett. 185:147-150. [DOI] [PubMed] [Google Scholar]
- 2.Bakalinsky, A. T., and R. Snow. 1990. Conversion of homothallic wine strains of Saccharomyces cerevisiae to heterothallism. Appl. Environ. Microbiol. 56:849-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldwin, W. W., and H. E. Kubitschek. 1984. Buoyant density variation during the cell cycle of Saccharomyces cerevisiae. J. Bacteriol. 158:701-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchard, D. C., and L. Syzdek. 1970. Mechanism for the water-to-air transfer and concentration of bacteria. Science 170:626-628. [DOI] [PubMed] [Google Scholar]
- 5.Budroni, M. 1994. Ph.D. thesis. University of Sassari, Sassari, Italy.
- 6.Burke, D. A., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 7.Cantarelli, C., and A. Martini. 1969. On the pellicle formation by “flor” yeast. Antoine Leeuwenhoek 35:F35-F36. [PubMed] [Google Scholar]
- 8.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 9.Farris, G. A., M. Sinigaglia, M. Budroni, and M. E. Guerzoni. 1993. Cellular fatty acid composition in film-forming strains of two physiological races of Saccharomyces cerevisiae. Lett. Appl. Microbiol. 17:215-219. [Google Scholar]
- 10.Frew, N. M. 1997. The role of organic films in air-sea gas exchange, p. 121-172. In P. S. Liss and R. A. Duce (ed.), The sea surface and global change. Cambridge University Press, Cambridge, United Kingdom.
- 11.Gietz, R. D., and R. H. Schiestl. 1995. Transforming yeast with DNA. Methods Mol. Biol. 5:255-269. [Google Scholar]
- 12.Halme, A., S. Bumgarner, C. Styles, and G. R. Fink. 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116:405-415. [DOI] [PubMed] [Google Scholar]
- 13.Hammond, J. R. M. 1993. Brewer's yeast, p. 7-67. In A. H. Rose and J. S. Harrison (ed.), The yeasts, vol. 5. Yeast technology. Academic Press, N.Y. [Google Scholar]
- 14.Hunter, J. R., P. K. Kilpatrick, and R. G. Carbonell. 1990. Lysozyme adsorption at the air/water interface. J. Colloid Interface Sci. 137:462-482. [Google Scholar]
- 15.Iimura, Y., S. Hara, and K. Otsuka. 1980. Cell surface hydrophobicity as a pellicle formation factor in a film strain of Saccharomyces. Agric. Biol. Chem. 44:1215-1222. [Google Scholar]
- 16.Iimura, Y., S. Hara, and K. Otsuka. 1980. Fatty acids as hydrophobic substances on the cell surface of a film strain of Saccharomyces. Agric. Biol. Chem. 44:1223-1229. [Google Scholar]
- 17.Ishigami, M., Y. Nakagawa, M. Harakawa, and Y. Iimura. 2004. FLO11 is essential for flor formation caused by the C-terminal deletion of NRG1 in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 237:425-430. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi, O., H. Yoshimoto, and H. Sone. 1999. Analysis of the genes activated by the FLO8 gene in Saccharomyces cerevisiae. Curr. Genet. 36:256-261. [DOI] [PubMed] [Google Scholar]
- 19.Kuchin, S., V. K. Vyas, and M. Carlson. 2002. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 22:3994-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, N., and M. C. Cannon. 1998. Gas vesicle genes identified in Bacillus megaterium and functional expression in Escherichia coli. J. Bacteriol. 180:2450-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, H., C. A. Styles, and G. R. Fink. 1996. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics 144:967-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo, W. S., and A. M. Dranginis. 1996. FLO11, a yeast gene related to the STA genes, encodes a novel cell surface flocculin. J. Bacteriol. 178:7144-7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo, W. S., and A. M. Dranginis. 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martínez, P., L. Perez, and T. Benítez. 1997. Mechanism of velum formation by flor yeast isolated from sherry wine. Am. J. Enol. Vitic. 48:55-62. [Google Scholar]
- 25.Masuoka, J., and K. C. Hazen. 1997. Cell wall protein mannosylation determines Candida albicans cell surface hydrophobicity. Microbiology 143:3015-3021. [DOI] [PubMed] [Google Scholar]
- 26.Masuoka, J., and K. C. Hazen. 1999. Differences in the acid-labile component of Candida albicans mannan from hydrophobic and hydrophilic cells. Glycobiology 9:1281-1286. [DOI] [PubMed] [Google Scholar]
- 27.Pringle, J. R., and J. R. Mor. 1975. Methods for monitoring the growth of yeast cultures and for dealing with the clumping problem, p. 131-168. In D. M. Prescott (ed.), Methods in cell biology, vol. XI. Yeast cells. Academic Press, New York, N.Y. [DOI] [PubMed] [Google Scholar]
- 28.Reynolds, T. B., and G. R. Fink. 2001. Bakers yeast, a model for fungal biofilm formation. Science 291:878-881. [DOI] [PubMed] [Google Scholar]
- 29.Roberts, R. L., and G. R. Fink. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8:2974-2985. [DOI] [PubMed] [Google Scholar]
- 30.Rupp, S., E. Summers, H. J. Lo, H. Madhani, and G. R. Fink. 1999. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 18:1257-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimoi, H., K. Sakamoto, M. Okuda, R. Atthi, K. Iwashita, and K. Ito. 2002. The AWA1 gene is required for the foam-forming phenotype and cell surface hydrophobicity of sake yeast. Appl. Environ. Microbiol. 68:2018-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smit, G. M. H. Straver, B. J. J. Lugtenberg, and J. W. Kijne. 1992. Flocculence of Saccharomyces cerevisiae cells is induced by nutrient limitation, with cell surface hydrophobicity as a major determinant. Appl. Environ. Microbiol. 58:3709-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teunissen, A. W. R. H., E. Holub, J. Van der Hucht, J. A. Van den Berg, and H. Y. Steensma. 1993. Sequence of the open reading frame of the FLO1 gene from Saccharomyces cerevisiae. Yeast 9:423-427. [DOI] [PubMed] [Google Scholar]
- 34.Verstrepen, K. J., G. Derdelinckx, H. Verachtert, F. R. Delvaux. 2003. Yeast flocculation: what brewers should know. Appl. Microbiol. Biotechnol. 61:197-205. [DOI] [PubMed] [Google Scholar]
- 35.Zara, S., G. A. Farris, M. Budroni, and A. T. Bakalinsky. 2002. HSP12 is essential for biofilm formation by a Sardinian sherry strain of S. cerevisiae. Yeast 19:269-276. [DOI] [PubMed] [Google Scholar]