Abstract
A quantitative real-time TaqMan PCR assay for detection of human adenoviruses (HAdV) was developed using broadly reactive consensus primers and a TaqMan probe targeting a conserved region of the hexon gene. The TaqMan assay correctly identified 56 representative adenovirus prototype strains and field isolates from all six adenovirus species (A to F). Based on infectious units, the TaqMan assay was able to detect as few as 0.4 and 0.004 infectious units of adenovirus serotype 2 (AdV2) and AdV41, respectively, with results obtained in less than 90 min. Using genomic equivalents, the broadly reactive TaqMan assay was able to detect 5 copies of AdV40 (which had zero mismatches with the PCR primers and probe), 8 copies of AdV41, and 350 copies of AdV3 (which had the most mismatches [seven] of any adenovirus serotype tested). For specific detection and identification of F species serotypes AdV40 and AdV41, a second real-time PCR assay was developed using fluorescence resonance energy transfer (FRET) probes that target the adenovirus fiber gene. The FRET-based assay had a detection limit of 3 to 5 copies of AdV40 and AdV41 standard DNA and was able to distinguish between AdV40 and AdV41 based on melting curve analysis. Both the TaqMan and FRET PCR assays were quantitative over a wide range of virus titers. Application of these assays for detection of adenoviruses and type-specific identification of AdV40 and AdV41 will be useful for identifying these viruses in environmental and clinical samples.
Adenoviruses (AdV) are grouped into six species (A to F) and 51 serotypes based on a variety of biochemical, immunological, and genetic parameters (2, 26). Adenoviruses have been identified as a cause of waterborne illnesses in the United States and in other countries (15), and several serotypes have been closely associated with acute gastroenteritis. AdV serotype 40 (AdV40) and AdV41 (species F) were second only to rotaviruses as a leading cause of acute gastroenteritis among children according to one study (3), and AdV31 (species A) and, to a lesser extent, other adenovirus serotypes of species A, B, and C have been linked to acute gastroenteritis in infants (1, 4). Recreational waters have been reported to contain species B and C adenoviruses which have been linked to outbreaks of pharyngoconjunctivitis (9, 19) and may play an important role in the transmission of respiratory diseases in recreational waters through aerosol transmission (6, 26). Other serotypes, such as AdV31 (1) and AdV51 (7), may be associated with fecal-oral transmission.
The spatial-temporal detection of waterborne viruses is of great importance to public health for prevention of illness and response to outbreaks. Although adenoviruses are thought to be common in the environment, insufficient data are available to evaluate their prevalence and distribution. Research has shown that adenoviruses can be relatively stable in water compared to other viruses (8) and can be resistant to disinfection in drinking water systems (21, 22). Adenoviruses have been detected in environmental waters and drinking water (16, 17, 19, 25) and are of sufficient concern to public health that they have been placed by the U.S. Environmental Protection Agency on its Contaminant Candidate List for drinking water (24). While it has been generally perceived that AdV40 and AdV41 are the most prevalent adenoviruses in water systems, insufficient analytical tools and research results have been available to confidently evaluate this view, especially given the fact that other adenovirus serotypes can be shed fecally and may be associated with gastrointestinal disease.
The species F adenoviruses that cause gastroenteritis (e.g., AdV40 and AdV41) are particularly difficult to isolate, in contrast to other adenovirus species (27), because they grow slowly in cell culture. Identification of adenovirus isolates can be accomplished by neutralization with type-specific antisera or by DNA restriction analysis (1). However, these methods are time-consuming and results are often difficult to interpret, and therefore, these methods are impractical for routine testing of clinical or environmental samples. A sensitive, broadly reactive test based on PCR amplification is needed for rapid screening of environmental samples for adenoviruses. Furthermore, specific identification of adenovirus species F would be useful. PCR assays can detect adenovirus DNA directly from extracts of clinical or environmental samples (14, 20), or they can be used in conjunction with cell culture to determine virus infectivity (16, 25). In addition to its speed, quantitative real-time PCR offers the additional benefit of permitting estimation of adenovirus concentrations in environmental samples. Although real-time PCR assays have been reported for detection of specific species or all species of adenoviruses (10, 11, 13), a broadly reactive assay with known sensitivity for low levels of infectious adenoviruses would be useful for screening environmental samples. No investigators have reported a real-time assay to discriminate F species (AdV40 and AdV41) from the other species. This study was designed to produce these quantitative, real-time PCR tools: a broadly reactive real-time PCR assay for detection of all human adenovirus species and a specific, discriminatory and sensitive real-time PCR assay for AdV40 and AdV41.
MATERIALS AND METHODS
Virus isolates and preparations.
Adenovirus isolates used in this study were obtained from Centers for Disease Control and Prevention archives. Fifty-six isolates previously typed by neutralization assay included AdV1 (2 strains), AdV2 (4 strains), AdV3 (4 strains), AdV4 (3 strains), AdV5 (1 strains), AdV6 (1 strains), AdV7 (7 strains), AdV8 (3 strains), AdV11 (2 strains), AdV19 (3 strains), AdV20 (1 strains), AdV23 (1 strains), AdV31 (5 strains), AdV34 (3 strains), AdV37 (2 strains), AdV40 (7 strains), and AdV41 (7 strains). AdV2 (ATCC VR-846; American Type Culture Collection, Rockville, MD) and CDC stocks of AdV40 (Dugan strain) and AdV41 (Tak strain) were cultured in A549 cells in Eagle's minimum essential medium with 10% fetal bovine serum. AdV2, AdV40, and AdV41 stocks were prepared by freezing and thawing of cultures at −70°C three times when cytopathic effect was complete, followed by removal of cell debris at 900 × g for 10 min at 4°C. The infectivity titers of these stocks, expressed in infectious units (IU), were estimated by observation of cytopathic effect in cell cultures of A549 inoculated with four replicates of serial 10-fold dilutions and calculation of the most probable number (16).
DNA extraction.
DNA was purified from 10 μl of adenovirus-infected cell culture lysate using the QIAamp viral DNA kit (QIAGEN, Valencia, CA) according to the manufacturer's instructions. DNA was eluted with 50 μl of 10 mM Tris-1 mM EDTA (pH 8.0) and stored at −70°C until use. Quantitative studies were performed using 10-fold serial dilutions of stock preparations of AdV2 (109 IU/ml) and AdV41 (5 × 106 IU/ml) in phosphate-buffered saline.
Cloning and sequencing of hexon and fiber gene regions.
The sensitivity of the TaqMan assay for AdV3, AdV40, and AdV41 was determined using genomic equivalent copies (GEC). Specified regions of the genome were amplified and cloned, and the plasmids were used as GEC. Amplification of the 380-bp region of the hexon gene of AdV3 was carried out using the forward primer 5′-TGGCCACCCCATCGATGA-3′ (positions 2 to 19) and reverse primer 5′-CTTAGGAGCGAGTGAATTGTA-3′ (positions 381 to 361) of GenBank accession no. AF542129. Amplification of the hexon region of AdV40 was carried out with the forward primer 5′-TGGCCACCCCCTCGATGA-3′ (positions 2 to 19) and reverse primer 5′-TTTGGGGGCCAGGGAGTTGTA-3′ (positions 381 to 361) of GenBank accession no. X51782. Amplification of the hexon region of AdV41 was carried out with the forward primer 5′-TGGCCACCCCCTCGATGA-3′ (positions 219 to 236) and reverse primer 5′-TTTAGGAGCCAGGGAGTTATA-3′ (positions 598 to 578) of GenBank accession no. X51783.
The sensitivity of the fluorescence resonance energy transfer (FRET) PCR assay for AdV40 and AdV41 was also determined using GEC. For this fiber gene assay, the PCR products were generated separately from AdV40 and AdV41 using the forward primer 5′-CAAAATAACGCGCTCACTCTT-3′ (positions 1606 to 1626) and reverse primer 5′-AGGGTTAAGTTTTCGTTTTCTATTTTT-3′ (positions 1946 to 1920) of GenBank accession no. M28822 to obtain a 341-bp product. The PCR products were cloned into a pDrive cloning vector using a PCR Cloningplus kit (QIAGEN, Valencia, CA). DNA sequences of the cloned products were determined with the BigDye terminator cycle sequence kit and ABI 377A sequencer (Applied Biosystems). Plasmids were purified using the Nucleobond 100 kit (Promega).
Standard curve generation.
Standard curves were generated for quantitation of assay sensitivities using stocks of adenoviruses for which IU titers had been determined and using GEC from cloned sequences of the hexon and fiber gene regions. Plasmid DNA was prepared, and its concentration was determined spectrophotometrically using the Nanodrop ND_1000 instrument according to the manufacturer's instructions (Nanodrop, Wilmington, DE). The DNA was 10-fold serially diluted in nuclease-free water containing carrier tRNA (100 ng/microliter). Standard curves were generated using 100 to 108 copies of plasmid DNA.
Prevention of PCR carryover contamination.
All standard precautions were followed to prevent any PCR contamination by adhering to strict laboratory practices. The pre-PCR manipulations (DNA isolation and PCR set-up) were performed in a clean room that was physically isolated from the real-time PCR machine and the post-PCR processing area. Dedicated pipettes and reagents were used for each location. Negative controls were run with all assays, and no indications of contamination were detected. Plasmids used for generation of standard curves were prepared in a separate room. Plasmid standards were never taken into the PCR set-up room.
Amplification and detection of adenovirus species A to F by TaqMan real-time PCR.
Hexon gene sequences obtained from GenBank for different adenovirus serotypes representing the six adenovirus species were aligned to identify sequence homologies suitable for selection of primer and probe targets. The sequences selected for the forward (JTVXF; nucleotide positions 18895 to 18915) and reverse (JTVXR; nucleotide positions 18990 to 18968) primers and probe (JTVXP) are shown in Table 1. The JTVXR primer contained deoxyinosine at position 18988 to compensate for a fourfold degeneracy at this site. The selected primers amplified a 96-bp fragment of the adenovirus hexon gene region. The TaqMan probe was labeled with a reporter dye 6-carboxyfluorescein (FAM) at the 5′ end and a quencher dye (black hole quencher) at the 3′ end. Hexon gene sequences from different adenovirus isolates, including at least two isolates from each of the six AdV species (A, B, C, D, E, and F), except for AdV species E, were obtained from GenBank for analysis (Fig. 1). Multiple alignments of the hexon sequences were performed to investigate sequence homologies and select appropriate primer and probe targets. Sequences selected for the forward primer (JTVXF), TaqMan probe (JTVXP), and reverse primer (JTVXR) are shown in Fig. 1 on the top of the alignment. Based on GenBank searches, the primers and probe do not cross-react with any published sequences of animal adenoviruses, except for a feline adenovirus.
TABLE 1.
Oligonucleotide primer/probe sequences
| Assay | Primer or probe | Name (position no.) (polarity) | Sequence (5′-3′) | Tm (°C) |
|---|---|---|---|---|
| TaqMana | ||||
| Forward primer (A-F) | JTVXF (18895-18915) (+) | GGACGCCTCGGAGTACCTGAG | 68 | |
| Reverse primer (A-F) | JTVXR (18990-18968) (−) | ACIGTGGGGTTTCTGAACTTGTT | 63 | |
| TaqMan probe (A-F) | JTVXP (18923-18944) (+) | CTGGTGCAGTTCGCCCGTGCCA | 78 | |
| FRET probeb | ||||
| Forward primer (AdV40-41) | JTVFF (619-641) (+) | AACTTTCTCTCTTAATAGACGCC | 57 | |
| Annealing probe (AdV40-41) | JTVFAP (671-652) (−) | GCGAAGAGTGCCCGTGTCAG | 69 | |
| Detection probe (AdV40-41) | JTVFDP (693-673) (−) | CAAGAGGTGCAgCACTTtGAA | 62 | |
| Reverse primer (AdV40-41) | JTVFR (736-718) (−) | AGGGGGCTAGAAAACAAAA | 62 |
Position is based on the hexon region of AdV5 of GenBank accession no. AC_000008 Tm was calculated based on the nearest-neighbor method.
Positions are based on the fiber protein gene of AdV41 of GenBank accession no. X16583.1. The detection probe was labeled with LC Red 640 at its 3′ end (693-673) and with a mismatch (small letter) at 676 (T to C) and at 682 (G to T) for AdV40.
FIG. 1.
Primers and TaqMan probe sequences aligned with hexon gene sequences of representative AdV serotypes obtained from GenBank. Corresponding accession numbers are shown in the right hand column. A to F represent the six AdV species and AdV1 to AdV41 refer to the different serotypes. The primer and probe sequences are capitalized.
The real-time TaqMan PCR assay was performed using the QuantiTect Probe PCR kit (QIAGEN, CA) in a R.A.P.I.D. real-time PCR system (Idaho Technology, Salt Lake City, UT). Amplification reaction mixtures contained 2 μl template DNA, 0.25 mM primers, and 150 nM fluorogenic probe in a final reaction volume of 20 μl in glass capillaries. The protocol took approximately 90 min to complete with the following PCR conditions: hot-start denaturation step at 95°C for 15 min, followed by 45 cycles with a 95°C denaturation for 10 s, 55°C annealing for 30 s, and 72°C elongation for 15 s (at a temperature transition rate of 20°C/s). All amplification reactions were carried out in duplicate. Amplicons were visualized by agarose gel electrophoresis and ethidium bromide staining to confirm the specificity of PCR products. PCR amplification was not improved by increasing the concentration of magnesium beyond the 3 mM concentration provided in the buffer.
Detection of species F (AdV40 and AdV41) using FRET probes.
For detection and differentiation of AdV40 and AdV41, the primers and probes were designed to detect a conserved 118-bp sequence of the fiber gene region (23) as shown in Table 1. Primers and probes were based on the formulation of a consensus sequence in the fiber region developed using four GenBank sequences (each) for AdV40 and AdV41. The accession numbers used include L19443.1 for human adenovirus type 40, M28822.1 for the mastadenovirus h40 fiber gene, X16583.1 for human enteric adenovirus type 41, and M60327 for mastadenovirus 41. The primers and probes should not detect animal adenoviruses (except, possibly, feline adenovirus) based on comparison with available sequences in GenBank.
The anchor probe (JTVFAP; positions 671 to 652) was fluorescein labeled at the 5′ end with LightCycler (LC) fluorescein and phosphorylated at the 3′ end to block extension and has sequence complementarity to both AdV40 and AdV41. The detection probe (JTVFDP; positions 692 to 673) was labeled at the 3′ end with LC Red 640 and with a mismatch at positions 676 (T to C) and 682 (G to T) for AdV40. The detection probe was designed with 100% complementarity to AdV41 but with two mismatches to AdV40, based on available GenBank sequences. Positions are based on GenBank accession no. X16583.1, the fiber protein gene of AdV41. Sequences of primers and probes are shown in Table 1.
The proximity of the LC Red 640 and fluorescein labels results in FRET, which is monitored at the end of each annealing step during PCR and continuously during melting curve analysis on channel 2. For AdV40, nucleotide mismatches at positions 676 and 682 between the detection probe and sense strand destabilizes the hybrid, resulting in a lower melting temperature during melting curve analysis. For AdV41, the detection probe forms a perfect match with the sense strand, thereby resulting in a higher melting temperature for the hybridized target-probe construct.
The FRET PCR assay was performed using the QuantiTect probe PCR kit as described above. Amplification reaction mixtures contained 100 nM concentrations of the sensor and anchor probes, 250 nM concentrations of the primers, and 2 μl of sample DNA in a final reaction volume of 20 μl. Optimal assay sensitivity was obtained with the nonamended master mix containing 3 mM magnesium. A negative control (without DNA) and positive DNA controls (AdV40 and AdV41) were included in each run. The temperature profile of the real-time FRET PCR assay included an initial denaturation step at 95°C for 15 min, followed by 45 cycles of denaturation at 95°C for 5 s (at a temperature transition rate of 20°C/s), annealing at 45°C for 10 s (at a temperature transition rate of 20°C/s), and extension at 72°C for 5 s (at a temperature transition rate of 2°C/s). Detection of the fluorescent signal was made after each cycle's annealing phase. After amplification, melting curve analysis was performed by heating the product to 95°C for 5 s, cooling it to 40°C for 15 s, and slowly heating it to 80°C (at a rate of 0.1°C/s) under continuous fluorescence monitoring.
RESULTS
Detection of adenoviruses using hexon-based real-time TaqMan assay.
The sensitivity of the TaqMan assay was evaluated in two ways: using adenovirus stocks (of AdV2 and AdV41) with a known infectious unit titer and using cloned DNA (of AdV40, AdV41, and AdV3) from the hexon gene. DNA extracted from 10-fold serial dilutions of stock cultures of AdV2 and AdV41 was analyzed, and the dilution curves were observed to be linear over a range of 8 log10 for AdV2 and 6 log10 for AdV41 (data not shown). Based on infectious units, the sensitivities of the TaqMan assay for AdV2 and AdV41 were 0.4 and 0.004 IU, respectively. The difference in sensitivity for the two types may be related to the differences in the ratios of noninfectious versus infectious particles. Investigators have reported high noninfectious particle-to-infectious particle ratios for AdV40 and AdV41 (5). Using GEC, the sensitivities of the TaqMan assay were 5 GEC for AdV40, 8 GEC for AdV41, and 350 GEC for AdV3 (Table 2). AdV40 was used as the model adenovirus for determining the maximum sensitivity of the TaqMan assay because there are no mismatches between the AdV40 hexon gene sequence and the primers and TaqMan probe. AdV3 was used as the model adenovirus for estimating the lower end of the sensitivity range for the TaqMan assay because there are seven mismatches between the AdV3 hexon gene sequence and the primers and TaqMan probe (with one mismatch being at the 3′ end of the probe). Although the sensitivity of the TaqMan assay for AdV3 was much lower than for AdV40 and AdV41, the amplification efficiency of the assay for AdV3 was very good, based on observation of an exponential increase in fluorescence (data not shown).
TABLE 2.
Quantitative analysis of plasmid DNA dilution series of each adenovirusa
| Assay | Slope | Rb | Detection limit (GEC/reaction) |
|---|---|---|---|
| TaqMan | |||
| AdV3 hexon | −3.517 | 1.0 | 350 |
| AdV40 hexon | −3.524 | 1.0 | 5 |
| AdV41 hexon | −3.661 | 1.0 | 8 |
| FRET | |||
| AdV40 fiber | −3.465 | 0.99 | 5 |
| AdV41 fiber | −3.086 | 1.00 | 3 |
Relative standard curves were derived from analysis of a dilution series of the plasmid from each strain.
R, regression coefficient.
A total of 56 adenovirus field isolates representing species A to F were tested using the real-time TaqMan assay. At least one serotype from each species was tested. All 56 isolates were detected (Table 3). AdV3, a serotype of species B, was also consistently detected despite having the most template mismatches (seven) with the PCR primers and TaqMan probe. Real-time PCR fluorescence curves for this serotype and other serotypes representing all adenovirus species showed that the TaqMan assay was efficient at producing and detecting target amplicons (data not shown). Using agarose gel electrophoresis, amplicons from positive tests were found to have the expected amplicon size of 96 bp for the TaqMan PCR assay (data not shown).
TABLE 3.
Comparison of TaqMan and FRET PCR assays with representative serotypes from different species of previously typed field isolates
| AdV species | AdV serotype | No. of isolates tested | No. positive/no. negative by:
|
|
|---|---|---|---|---|
| TaqMan (all adenoviruses) | FRET (species F) | |||
| A | 31 | 5 | 5/0 | 0/5 |
| B | 3 | 4 | 4/0 | 0/4 |
| 7 | 7 | 7/0 | 0/7 | |
| 11 | 2 | 2/0 | 0/2 | |
| 34 | 3 | 3/0 | 0/3 | |
| C | 1 | 2 | 2/0 | 0/2 |
| 2 | 4 | 4/0 | 0/4 | |
| 5 | 1 | 1/0 | 0/1 | |
| 6 | 1 | 1/0 | 0/1 | |
| D | 8 | 3 | 3/0 | 0/3 |
| 19 | 3 | 3/0 | 0/3 | |
| 20 | 1 | 1/0 | 0/1 | |
| 23 | 1 | 1/0 | 0/1 | |
| 37 | 2 | 2/0 | 0/2 | |
| E | 4 | 3 | 3/0 | 0/3 |
| F | 40 | 7 | 7/0 | 7/0 |
| 41 | 7 | 7/0 | 7/0 | |
Evaluation of FRET PCR assay for detection and differentiation of AdV40 and 41.
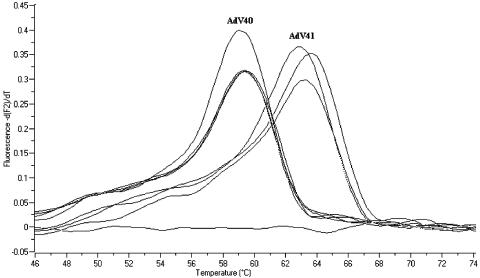
When the panel of 56 adenovirus clinical isolates was tested using the species F FRET PCR assay, all AdV40 and AdV41 isolates were consistently detected, whereas serotypes from all other adenovirus species were not (Table 3). Using gel electrophoresis, amplicons from positive tests were found to have the expected amplicon size of 118 bp for the FRET PCR assay (data not shown). In addition to detection of AdV40 and AdV41 by FRET PCR, isolates of AdV40 and AdV41 could be distinguished by melting curve analysis (Fig. 2). Four clinical isolates of AdV40 and three clinical isolates of AdV41 were correctly identified with the FRET probe assay (Fig. 2). The melting temperatures (Tm) of the FRET probes hybridized to the AdV40 and AdV41 templates ranged from 58.95 to 59.17°C and 62.57 to 63.15°C, respectively.
FIG. 2.
Melting curve (dF/dT) derivative of LC Red 640-labeled sensor probe with different samples shows qualitative detection of point mutation exploiting changes in Tm during melting curve analysis.
The sensitivity of the FRET assay was determined using cloned DNA of AdV40 and AdV41 from the fiber gene region as well as with DNA extracted from a stock of AdV41 with a known IU titer. Based on genomic equivalent copies, the sensitivity of the FRET assay was determined to be 3 GEC for AdV41 and 5 GEC for AdV40 (Table 2). Based on infectious units, the FRET PCR assay was able to detect as few as 0.004 IU of AdV41 DNA (data not shown), which was equivalent to the sensitivity of the hexon-based TaqMan assay for AdV41. The detection of 3 GEC of AdV41 and 0.004 IU of AdV41 is in agreement with the noninfectious-to-infectious particle ratio of species F viruses of 1,000 to 10,000 to 1 (5, 18).
DISCUSSION
Conventional PCR assays have been described for adenoviruses based on hexon and fiber gene sequences (2, 13, 28) but are labor intensive due to the need for post-PCR product analysis by gel electrophoresis and confirmatory hybridization assays or sequencing. The present study describes two real-time PCR assays for sensitive and specific detection of adenoviruses. The hexon gene was targeted to develop a broadly reactive TaqMan assay for detection of all adenovirus species. The adenovirus fiber gene was targeted for specific identification of AdV40 and AdV41 using FRET PCR. To our knowledge, this is the first report to describe a real-time PCR assay targeting the adenovirus fiber gene. The hexon gene has been the focus of efforts by other researchers (11) to develop a broadly reactive TaqMan assay for adenoviruses. Although additional work would need to be performed to comprehensively compare the specificity and sensitivity of the present assay with the assay of Heim et al. (11), the data from the present study suggest that the TaqMan assay provides sensitivities and specificities for detection of adenoviruses that are similar to or better than previously published general detection assays. The TaqMan assay was able to detect all 56 adenovirus field isolates representing species A to F. Using cloned plasmid copies of AdV40 and AdV3 hexon gene sequences, the present TaqMan assay was determined to have a detection limit of 5 GEC for AdV40, 8 GEC for AdV41, and 350 GEC for AdV3. The consensus sequences used for the TaqMan assay primers and probe exactly matched the target sequence of AdV40, so these data indicate that the TaqMan assay described here has a detection limit of near 5 GEC for AdV40 and other adenoviruses with few mismatches. The detection limit for AdV3 can be considered to be the worst-case sensitivity estimate for the TaqMan assay, as this species has seven mismatches with the TaqMan assay primers and probe (the most mismatches of any of the adenoviruses investigated) (Fig. 1), with one of the mismatches located near the 3′ end of the probe. The sensitivity for AdV41 was determined to be 8 GEC, although there are five mismatches for this strain, which further indicates that the location and type of mismatch is also important. Heim et al. (11) reported data indicating the sensitivity of their assay to be between 1.5 and 15 GEC for AdV2. They also reported their assay sensitivity for AdV2 based on infectivity to be 43 50% tissue culture infective doses (11). The assay we have described can detect as few as 0.4 IU of AdV2, which indicates that this assay is as sensitive as or more sensitive than other published broadly reactive adenovirus assays.
The FRET PCR assay was tested using a panel of 56 adenovirus isolates representing species A to F and was found to be specific for the detection of species F adenoviruses. Sensitivity testing of the FRET PCR assay using AdV40 and AdV41 genomic copies showed that the FRET PCR assay is highly sensitive for these viruses (with sensitivities of 3 GEC for AdV41 and 5 GEC for AdV40). The FRET PCR assay sensitivity for AdV40 was similar to the sensitivity of the TaqMan PCR assay reported here for AdV40 based on genomic equivalents. Melting curve analysis of the FRET PCR product indicated that the FRET assay can be useful for distinguishing between AdV40 and AdV41. Additional testing of more isolates of AdV40 and AdV41 will provide more information on the ability of this assay to differentiate AdV40 from AdV41. This species F-specific real-time PCR assay is expected to be a useful method for rapid, sensitive and specific detection of AdV40 and AdV41, especially when used in conjunction with a broadly reactive real-time PCR assay.
The development of a broadly reactive real-time PCR assay for human adenoviruses was challenging due to the extensive genetic diversity among the different adenovirus serotypes. TaqMan assays generally have low tolerance for mismatched sequences within the primer/probe target regions (11-13). Nevertheless, the TaqMan assay in the present study was able to detect all of the representative adenoviruses belonging to species A to F in less than 90 min. The development of a FRET PCR assay that can distinguish between AdV40 and AdV41 will be useful for epidemiological investigations of transmission routes for these serotypes. F species adenoviruses (AdV40 and AdV41) are the most recognized adenoviruses in suspected fecal-oral transmission through water and have been the prototypes for studies of gastroenteritis caused by adenoviruses. However, other adenovirus serotypes are fecally shed, thereby raising the possibility that they may also cause illness via contaminated water.
The broadly reactive TaqMan PCR assay developed in this study can be a useful tool for performing environmental surveys to determine whether any adenovirus serotypes are present in water to which humans are exposed. The FRET PCR assay for AdV40 and AdV41 can be used in conjunction with the broadly reactive TaqMan assay to determine whether adenovirus-positive samples contain these serotypes, which are known to cause gastroenteritis. Under this scenario, TaqMan-positive samples that are FRET negative could be further studied to determine what other adenovirus serotypes other than types 40 and 41 are present. Thus, these molecular tools will be useful for environmental monitoring, epidemiological investigations of disease outbreaks, and molecular epidemiological investigations of adenovirus infections and illnesses associated with waterborne exposures. Further studies are planned to determine the applicability, specificity, and detection sensitivity of the assays in environmental sample concentrates such as water.
Acknowledgments
This study was partially supported by a grant from the American Water Works Association Research Foundation (project no. 2591).
Use of trade names and commercial sources is for identification only and does not imply endorsement by the Centers for Disease Control and Prevention or the U.S. Department of Health and Human Services.
REFERENCES
- 1.Adrian, T., and R. Wigand. 1989. Genome type analysis of adenovirus 31, a potential causative agent of infants' enteritis. Arch. Virol. 105:81-87. [DOI] [PubMed] [Google Scholar]
- 2.Allard, A., B. Albinsson, and G. Wadell. 2001. Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis. J. Clin. Microbiol. 39:498-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allard, A., R. Girones, P. Juto, and G. Wadell. 1990. Polymerase chain reaction for detection of adenoviruses in stool samples. J. Clin. Microbiol. 28:2659-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, M. 1990. Laboratory identification of adenoviruses associated with gastroenteritis in Canada from 1983 to 1986. J. Clin. Microbiol. 28:1525-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, M., H. L. Wilson-Friesen, and F. Doane. 1992. A block in release of progeny virus and a high particle-to-infectious unit ratio contribute to poor growth of enteric adenovirus types 40 and 41 in cell culture. J. Virol. 66:3198-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castignolles, N., F. Petit, I. Mendel, L. Simon, L. Cattolico, and C. Buffet-Janvresse. 1998. Detection of adenovirus in the waters of the Seine River estuary by nested-PCR. Mol. Cell. Probes 12:175-180. [DOI] [PubMed] [Google Scholar]
- 7.De Jong, J. C., A. G. Wermenbol, M. W. Verweij-Uijterwaal, K. W. Slaterus, P. Wertheim-Van Dillen, G. J. Van Doornum, S. H. Khoo, and J. C. Hierholzer. 1999. Adenoviruses from human immunodeficiency virus-infected individuals, including two strains that represent new candidate serotypes Ad50 and Ad51 of species B1 and D, respectively. J. Clin. Microbiol. 37:3940-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enriquez, C. E., and C. P. Gerba. 1995. Concentration of enteric adenovirus 40 from tap, sea, and waste water. Water Res. 29:2554-2560. [Google Scholar]
- 9.Foy, H. M., M. K. Cooney, and J. B. Hatlen. 1968. Adenovirus type 3 epidemic associated with intermittent chlorination of a swimming pool. Arch. Environ. Health 17:795-802. [DOI] [PubMed] [Google Scholar]
- 10.Gu, Z., S. W. Belzer, C. S. Gibson, M. J. Bankowski, and R. T. Hayden. 2003. Multiplexed, real-time PCR for quantitative detection of human adenovirus. J. Clin. Microbiol. 41:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heim, A., C. Ebnet, G. Harste, and P. Pring-Akerblom. 2003. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J. Med. Virol. 70:228-239. [DOI] [PubMed] [Google Scholar]
- 12.Holland, P. M., R. D. Abramson, R. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houng, H. S., S. Liang, C. M. Chen, J. Keith, M. Echavarria, J. L. Sanchez, S. A. Kolavic, D. W. Vaughn, and L. N. Binn. 2002. Rapid type-specific diagnosis of adenovirus type 4 infection using a hexon-based quantitative fluorogenic PCR. Diagn. Microbiol. Infect. Dis. 42:227-236. [DOI] [PubMed] [Google Scholar]
- 14.Jiang, S., R. Noble, and W. Chu. 2001. Human adenoviruses and coliphages in urban runoff-impacted coastal waters of Southern California. Appl. Environ. Microbiol. 67:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leclerc, H., L. Schwartzbrod, and E. Dei-Cas. 2002. Microbial agents associated with waterborne diseases. Crit. Rev. Microbiol. 28:371-409. [DOI] [PubMed] [Google Scholar]
- 16.Lee, S. H., and S. J. Kim. 2002. Detection of infectious enteroviruses and adenoviruses in tap water in urban areas in Korea. Water Res. 36:248-256. [DOI] [PubMed] [Google Scholar]
- 17.Lees, D. 2000. Viruses and bivalve shellfish. Int. J. Food Microbiol. 59:81-116. [DOI] [PubMed] [Google Scholar]
- 18.Mautner, V. 1999. Methods for growth and purification of enteric adenovirus type 40, p. 282-293. In W. S. M. Wold (ed.), Methods in molecular medicine-adenovirus methods and protocols, vol. 23. Humana Press, Totowa, N.J. [DOI] [PubMed] [Google Scholar]
- 19.Papapetropoulou, M., and A. C. Vantarakis. 1998. Detection of adenovirus outbreak at a municipal swimming pool by nested PCR amplification. J. Infect. 36:101-103. [DOI] [PubMed] [Google Scholar]
- 20.Pina, S., M. Puig, F. Lucena, J. Jofre, and R. Girones. 1998. Viral pollution in the environment and in shellfish: human adenovirus detection by PCR as an index of human viruses. Appl. Environ. Microbiol. 64:3376-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thurston-Enriquez, J. A., C. N. Haas, J. Jacangelo, and C. P. Gerba. 2003. Chlorine inactivation of adenovirus type 40 and feline calicivirus. Appl. Environ. Microbiol. 69:3979-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thurston-Enriquez, J. A., C. N. Haas, J. Jacangelo, K. Riley, and C. P. Gerba. 2003. Inactivation of feline calicivirus and adenovirus type 40 by UV radiation. Appl. Environ. Microbiol. 69:577-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tiemessen, C. T., and M. J. Nel. 1996. Detection and typing of subgroup F adenoviruses using the polymerase chain reaction. J. Virol. Methods 59:73-82. [DOI] [PubMed] [Google Scholar]
- 24.U.S. Environmental Protection Agency. 1998. Announcement of the drinking water contaminant candidate list. EPA-815-Z-98-001. Fed. Regist. 63:10273-10287. [Google Scholar]
- 25.Van Heerden, J., M. M. Ehlers, W. B. Van Zyl, and W. O. Grabow. 2003. Incidence of adenoviruses in raw and treated water. Water Res. 37:3704-3708. [DOI] [PubMed] [Google Scholar]
- 26.Wadell, G. 1984. Molecular epidemiology of human adenoviruses. Curr. Top. Microbiol. Immunol. 110:191-220. [DOI] [PubMed] [Google Scholar]
- 27.Wigand, R. 1987. Pitfalls in the identification of adenoviruses. J. Virol. Methods 16:161-169. [DOI] [PubMed] [Google Scholar]
- 28.Xu, W., M. C. McDonough, and D. D. Erdman. 2000. Species-specific identification of human adenoviruses by a multiplex PCR assay. J. Clin. Microbiol. 38:4114-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]