Abstract
The characterization of microbial assemblages within solid gas hydrate, especially those that may be physiologically active under in situ hydrate conditions, is essential to gain a better understanding of the effects and contributions of microbial activities in Gulf of Mexico (GoM) hydrate ecosystems. In this study, the composition of the Bacteria and Archaea communities was determined by 16S rRNA phylogenetic analyses of clone libraries derived from RNA and DNA extracted from sediment-entrained hydrate (SEH) and interior hydrate (IH). The hydrate was recovered from an exposed mound located in the northern GoM continental slope with a hydrate chipper designed for use on the manned-submersible Johnson Sea Link (water depth, 550 m). Previous geochemical analyses indicated that there was increased metabolic activity in the SEH compared to the IH layer (B. N. Orcutt, A. Boetius, S. K. Lugo, I. R. Macdonald, V. A. Samarkin, and S. Joye, Chem. Geol. 205:239-251). Phylogenetic analysis of RNA- and DNA-derived clones indicated that there was greater diversity in the SEH libraries than in the IH libraries. A majority of the clones obtained from the metabolically active fraction of the microbial community were most closely related to putative sulfate-reducing bacteria and anaerobic methane-oxidizing archaea. Several novel bacterial and archaeal phylotypes for which there were no previously identified closely related cultured isolates were detected in the RNA- and DNA-derived clone libraries. This study was the first phylogenetic analysis of the metabolically active fraction of the microbial community extant in the distinct SEH and IH layers of GoM gas hydrate.
Marine gas hydrates, which are ice-like crystalline solids, are composed of rigid water molecules with trapped gas molecules, primarily methane and other hydrocarbons. Gas hydrate reservoirs, which are distributed in the sediments of active and passive continental slope margins, as well as in terrestrial (i.e., permafrost) regions (38), are a proposed fossil fuel energy source (10). Additionally, the estimated global volume of submarine methane hydrates exceeds 1016 m3 (7, 10), highlighting the impact of hydrates on global carbon cycling, climate conditions, and seafloor stability (16, 18, 28, 31, 35). The formation of gas hydrates is dependent upon suitable gas, temperature, and pressure conditions (reviewed in reference 38). Geological and chemical conditions in the northern continental slope of the Gulf of Mexico (GoM) promote the formation of gas hydrates where seepage of hydrocarbon gases forms extensive surface-breaching mounds on the seafloor, as well as vast vein-filling hydrates in hemipelagic sediments (27).
Geochemical characteristics, including gas composition and isotopic ratios of surface breaching hydrate, in the GoM have been well documented (19, 33, 34, 38). Growth and dissolution of GoM hydrate mounds have also been observed, with changes in mound size and shape evident over a period of months (19). Such hydrate growth patterns increase fluid and solid (i.e., sediment) inclusions and also increase the frequency of interconnecting flaws and fissures (43). Thus, Sassen et al. (35) have proposed that rapidly growing hydrate crystals in the outer layers of hydrate mounds, such as those found in the GoM (32), can be colonized by microorganisms. While rate measurements indicate that active microbial populations are present in the distinct layers of solid gas hydrate (26), there is no information regarding the composition of the corresponding metabolically active fractions of the microbial communities extant in these hydrate environments. Thus, characterization of microbial assemblages within solid hydrate, especially those that may be physiologically active under in situ hydrate conditions, is essential to gain a better understanding of the effects and contributions of microbial activities in GoM hydrate ecosystems.
In the present study, nucleic acids (DNA and RNA) were extracted from samples representing two distinct layers of a gas hydrate environment. One layer, designated the interior hydrate (IH), included samples collected from the interior portion of solid gas hydrate (>5 cm from the outside surface) devoid of sediment particles. The second layer, designated the sediment-entrained hydrate (SEH), included samples collected at the interface between the interior portion of the hydrate (IH) and the sediment that was directly in contact with hydrate. SEH samples were composed mainly of solid gas hydrate, and less than 5% of the mixture was composed of sediment particles. The primary objective of this study was to characterize the metabolically active fraction of the microbial communities present in these distinct hydrate layers. Total rRNA was extracted from the IH and SEH layers and subjected to reverse transcription-PCR with primers specific for the domains Bacteria and Archaea. Coextracted DNA was also amplified with the domains Bacteria and Archaea so that comparisons to RNA-derived 16S rRNA gene libraries could be made. This study was the first phylogenetic analysis of interior hydrate-associated microbial communities from surface-breaching GoM gas hydrate mounds and the first study of the metabolically active fractions of the IH- and SEH-associated microbial communities.
MATERIALS AND METHODS
Gulf of Mexico site description and sample collection.
The study site, GC234 (depth, 575 m), is located in the Green Canyon area in the northern GoM continental slope province at 27°44′N, 91°13′W. A detailed site description has recently been published by Orcutt et al. (26). This area contains oil and gas seepage along with extensive (i.e., several meters thick) surface-breaching gas hydrate mounds and displaced sediments colonized by chemosynthetic tube worms, mussels, and polychaete ice worms (2, 8). Samples of solid gas hydrate with overlying sediment drape were collected from visible gas hydrate mounds during multiple dives of the Johnson Sea Link manned submersible in July 2002. The recovery, preservation, and storage of solid gas hydrate samples and shipboard manipulations have been described in detail by Mills et al. (22). As gas hydrate samples were transported to the surface under in situ pressure and temperature conditions, little or no dissociation was observed. Solid gas hydrate with overlying, entrained sediment was aseptically divided by cutting the solid hydrate with sterilized scalpels and cutting tools into (i) SEH and (ii) IH. SEH samples were composed of solid gas hydrate, and generally no more than 5% of the mixture was sediment particles. IH samples were composed of interior solid hydrate devoid of any sediment and were acquired by aseptically cutting and paring away the outer sediment-entrained layers of intact solid gas hydrate (diameter, 12 cm). Multiple aliquots (150 to 250 g) of SEH and IH were subsequently stored in liquid N2. Direct cell counting was performed with unfrozen aliquots (0.5 g, wet weight) of displaced, overlying sediment, SEH, and IH samples as previously described (30). Replicate samples were processed for geochemical analyses (analyses of gas composition and anion concentrations) and measurement of microbial rates of sulfate reduction (SR) and methane oxidation as described by Orcutt et al. (26). Relevant geochemical and rate data are shown in Table 1, and a description of the (geo)chemistry of the hydrate samples has been reported previously (26).
TABLE 1.
Details of three distinct GoM hydrate layers
| Sample type | Cells (g−1) | RNA (μg g−1) | DNA (μg g−1) | RNA/DNA ratio | Sulfate (mM)a | AOM (nmol cm−3 day−1)a | SR (nmol cm − 3 day−1)a |
|---|---|---|---|---|---|---|---|
| Sediment overlying hydrate | 4.3 × 108 ± 4.8 × 107b | NDc | ND | ND | 12.3 ± 1.4 | 0.60 ± 0.2 | 76.2 ± 20.8 |
| Sediment-entrained hydrate | 2.0 × 107 ± 4.2 × 106 | 11.4 | 11.5 | 0.99 | 9.5 ± 1.0 | 0.13 ± 0.1 | 23.0 ± 1.0 |
| Interior hydrate | 4.3 × 106 ± 2.1 × 106 | 0.9 | 3.6 | 0.25 | 3.2 ± 2.1 | 0.28 ± 0.3 | 3.2 ± 4.1 |
Preparation of reagents and materials used for RNA extraction.
Prior to nucleic acid extraction, RNases were removed from solutions and solids by treating stock solutions and water with 0.1% diethyl pyrocarbonate overnight at 37°C and autoclaving. All glassware and nonplastics were baked at 250°C for 24 h. All surfaces and plastics were cleaned with RNase Erase (ICN, Aurora, OH) to remove contaminating RNases during shipboard and laboratory manipulations.
RNA and DNA isolation.
Total nucleic acids were extracted from triplicate samples of SEH and IH aliquots (50 to 100 g) as described by Hurt et al. (14). In brief, SEH and IH samples stored in liquid N2 were repeatedly thawed by physical grinding with sterilized 0.1-mm-diameter zirconium beads in the presence of a denaturing solution (4 M guanidine isothiocyanate, 10 mM Tris-HCl [pH 7.0], 1 mM EDTA, 0.5% 2-mercaptoethanol) and refrozen by immersion in liquid N2. The SEH and IH samples were incubated for 30 min at 65°C in pH 7.0 extraction buffer (100 mM sodium phosphate [pH 7.0], 100 mM Tris-HCl [pH 7.0], 100 mM EDTA [pH 8.0], 1.5 M NaCl, 1% hexadecyltrimethylammonium bromide, and 2% sodium dodecyl sulfate) and centrifuged (1,800 × g for 10 min). The supernatants from three separate extractions were pooled, extracted with 24:1 (vol/vol) chloroform-isoamyl alcohol, and centrifuged (1,800 × g for 20 min). The nucleic acids were precipitated at room temperature with isopropanol (30 min), pelleted by centrifugation (16,000 × g for 20 min), resuspended in diethyl pyrocarbonate-treated water, and subsequently purified by ion-exchange chromatography (14, 39) to obtain aliquots containing only DNA and aliquots containing only RNA.
Reverse transcription of rRNA.
Aliquots of rRNA were reverse transcribed with Moloney murine leukemia virus reverse transcriptase used according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Purified RNA was initially denatured by heating (65°C) for 10 min. The reverse transcription reaction mixture consisted of 5 μM 16S rRNA reverse primer DXR518 (5′-CGTATTACCGCGGCTGCTGG-3′) amplifying domain-specific Bacteria sequences (25) or 5 μM 16S rRNA reverse primer Ar958r (5′-YCCGGCGTTGAMTCCAATTT-3′) amplifying domain-specific Archaea sequences (6), 50 to 100 ng of denatured RNA, and 200 μM deoxynucleoside triphosphates. The mixture was incubated for 5 min at 65°C and for 2 min at 4°C, and this was followed by addition of 1× first-strand buffer (50 mM Tris-HCl [pH 8.3], 75 mM KCl, 3 mM MgCl2) and 75 U RNase inhibitor and heating at 37°C for 2 min. Moloney murine leukemia virus (200 U) was added prior to a 50-min incubation at 37°C that resulted in transcription of the RNA into complementary ribosomal DNA (crDNA). The crDNA end product was used as the template for a standard PCR. The RNA and DNA concentrations were routinely monitored visually (by gel electrophoresis) and spectrophotometrically (by determining absorbance at 260 nm and 280 nm). DNA contamination of RNA templates was routinely monitored by PCR amplification of aliquots of RNA that were not reverse transcribed. No contaminating DNA was detected in any of these reactions. The primers used for PCR amplification (a maximum of 25 to 30 cycles) included the reverse primers described above and 16S rRNA gene forward domain-specific primers (for Bacteria, 27F [5′-AGAGTTTGATCCTGGCTCAG-3′]; and for Archaea, A341f [5′-CCTAIGGGGIGCAICAG-3′]) (44). The PCR mixture contained 10 to 50 ng crDNA, 1× PCR buffer (Stratagene, La Jolla, CA), 1.5 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 200 μM, 1 pmol of each forward and reverse primer, and 0.025 U μl−1TaKaRa Taq. Amplicons were analyzed by electrophoresis on 1.0% agarose gels with Tris-borate-EDTA buffer that were stained with ethidium bromide and UV illuminated.
Environmental clone library construction.
Aliquots of purified DNA (1 μl, corresponding to the DNA recovered from an approximately 1.0-g sample) were PCR amplified as previously described (22). 16S amplicons, derived from SEH and IH DNA (e.g., 16S rRNA gene) and RNA (e.g., 16S crDNA) samples, were subsequently pooled from three to five reactions, purified with a Qiaquick gel extraction kit (QIAGEN, Valencia, CA), and cloned into the TOPO TA cloning vector pCR2.1 according to manufacturer's instructions (Invitrogen, Carlsbad, CA). Cloned inserts were amplified from lysed colonies with primers specific for the vector (M13F [5′-GTAAAACGACGGCCAG-3′] and M13R [5′-CAGGAAACAGCTATGAC-3′]) or with primers specific for the Archaea amplicons (A341f [44] and Ar958r [6]). The M13F and M13R primers were used to amplify inserts from bacterial clones to prevent amplification of the Escherichia coli host 16S rRNA gene. PCR products were digested (2 h, 37°C) with MspI and HhaI (bacterial clones) or with HhaI and RsaI (archaeal clones). Clones were grouped according to their restriction fragment length polymorphism (RFLP) patterns, and representative clones were sequenced as previously described (22). Briefly, restriction digestion profiles (RFLP patterns) of clones were visualized on 1 to 2% agarose gels, photographed, and grouped according to the number and size of their restriction fragments. Representative clones from all phylotypes in each library, with the exception of the DNA-derived bacterial library constructed from the SEH samples, were sequenced. Phylotypes from the SEH DNA library having more than one clone member and a random selection of phylotypes with a single clone representative were sequenced. Sequencing was performed at the Georgia Institute of Technology core DNA facility using a BigDye Terminator v3.1 cycle sequencing kit with an automated capillary sequencer (model 3100 gene analyzer; Applied Biosystems). Inserts were sequenced multiple times on the sense and antisense strands. Prior to comparative sequence analysis, vector sequences flanking the bacterial 16S rRNA gene and crDNA inserts were manually removed. A total of 97 sequences representing 374 Bacteria and Archaea clones were obtained in this study.
Phylogenetic and statistical analyses.
Sequence analysis was performed as previously described by Mills et al. (22, 23). Multiple sequences of individual inserts were initially aligned using the program BLAST 2 Sequences (41) available through the National Center for Biotechnology Information and were assembled with the program BioEdit v5.0.9 (11). Sequences were checked for chimeras using Chimera Check from Ribosomal Database Project II (20). Sequences from this study and reference sequences, as determined by BLAST analysis, were subsequently aligned using CLUSTALX v1.81 (42). An average of 500 (Bacteria clones) to 600 (Archaea clones) nucleotides were included in the final phylogenetic analyses. Neighbor-joining trees were created from the shortened sequence alignments. The bootstrap data represented 1,000 samplings. The final trees were viewed using NJPlot (29) and TreeView v1.6.6 (available at http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). Rarefaction analysis was performed using equations as described by Heck et al. (12). Standard calculations were used to produce the rarefaction curve using the total number of clones obtained compared to the number of clones representing each unique RFLP pattern. Sorensen's index and the Shannon-Weiner index were calculated using standard equations. Species richness was determined by EstimateS (1, 3, 4). Additional statistical estimators, including gene (24) and nucleotide (24, 40) diversity, θ(π) (40), FST (37), and P tests (21), were calculated using Arlequin (36). Lineage-per-time plots were constructed from TreePuzzle (http://www.tree-puzzle.de) pairwise alignments assuming a molecular clock.
Nucleotide sequence accession numbers.
The 97 16S rRNA gene and 16S crDNA nucleotide sequences have been deposited in the GenBank database under accession numbers AY542171 to AY542267.
RESULTS
The composition of the Bacteria and Archaea community in a gas hydrate environment in the GoM was determined by 16S rRNA phylogenetic analyses of clone libraries derived from RNA and DNA extracted from SEH and IH. The purified RNA was of sufficient quality and quantity to be reverse transcribed. The concentration of recovered RNA and DNA and the corresponding RNA/DNA ratios were significantly higher in the SEH layer than in the IH layer (Table 1). Quantification of microbial cell numbers in the overlying sediment revealed a 1- to 2-order-of-magnitude-higher cell count than the cell count in either the SEH or IH layer (Table 1). A similar trend was observed in SR rates; the highest rates were measured in the overlying sediment, and the lowest rates were measured in the IH layer (Table 1). Higher rates of anaerobic oxidation of methane (AOM) were detected in the equivalent IH layer than the SEH (26) (Table 1). The highest rate of AOM (0.60 ± 0.2 nmol cm−3 day−1) was measured in the overlying sediment (26).
RFLP and statistical analyses of 16S rRNA libraries.
Four different 16S rRNA libraries were constructed, representing a total of 99 Bacteria 16S rRNA gene clones (DNA derived), 97 Bacteria 16S crDNA clones (RNA derived), 100 Archaea 16S rRNA gene clones, and 97 Archaea 16S crDNA clones from the IH and SEH layers. All clones were grouped according to the RFLP patterns and subjected to rarefaction and percent coverage analysis to determine if a sufficient number of clones from each of the libraries was sampled to estimate library diversity (9). The curves reached saturation for Archaea clones obtained from either DNA or RNA (data not shown). The percent coverage for the Archaea clone libraries was greater than 92%, with the exception of the DNA-derived library from the SEH (82%) (Table 2). All Archaea clone libraries had significant (P < 0.05) FST and P tests (data not shown), while lineage-per-time plots were similar to plots indicative of constant births and deaths (data not shown) (21). Greater gene and nucleotide diversity and θ(π) values were observed in IH DNA-derived libraries, whereas these indices were higher in the SEH layer RNA-derived libraries (Table 2).
TABLE 2.
Statistical analyses of Bacteria and Archaea 16S rRNA clone libraries using standard ecological and molecular estimates of sequence diversity
| Domain | Nucleic acid sampled | Sample layer | No. of clones screened | No. of operational taxonomic units | % Coverage | Species richness | Sorensen's index | Shannon- Weiner index | Gene diversity | Nucleotide diversity | θ(π) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria | DNA | SEH | 34 | 35 | 42.9 | 69 (57, 81)a | 2.785 | 0.95 ± 0.02b | 0.22 ± 0.11b | 85.0 ± 41.7b | |
| IH | 46 | 26 | 64.0 | 45 (38, 53) | 2.762 | 0.93 ± 0.02 | 0.24 ± 0.12 | 97.5 ± 47.4 | |||
| Total | 80 | 55 | 60.6 | 77 (71, 82) | 0.238 | 3.168 | 0.95 ± 0.01 | 0.23 ± 0.11 | 96.0 ± 46.2 | ||
| RNA | SEH | 48 | 18 | 81.3 | 37 (14, 60) | 2.591 | 0.93 ± 0.02 | 0.17 ± 0.08 | 82.0 ± 39.9 | ||
| IH | 49 | 22 | 75.5 | 39 (30, 47) | 2.811 | 0.94 ± 0.02 | 0.17 ± 0.08 | 76.8 ± 37.3 | |||
| Total | 97 | 30 | 83.5 | 50 (44, 57) | 0.500 | 2.946 | 0.94 ± 0.01 | 0.17 ± 0.08 | 83.0 ± 39.9 | ||
| Archaea | DNA | SEH | 50 | 12 | 82.0 | 33 (22, 45) | 1.675 | 0.72 ± 0.05 | 0.08 ± 0.04 | 23.3 ± 11.6 | |
| IH | 50 | 14 | 92.0 | 15 (14, 16) | 2.375 | 0.91 ± 0.02 | 0.21 ± 0.10 | 58.9 ± 28.7 | |||
| Total | 100 | 20 | 94.0 | 24 (22, 27) | 0.519 | 2.527 | 0.90 ± 0.02 | 0.17 ± 0.08 | 46.4 ± 22.5 | ||
| RNA | SEH | 49 | 6 | 95.9 | 7 (4, 10) | 1.301 | 0.69 ± 0.04 | 0.08 ± 0.04 | 44.7 ± 21.9 | ||
| IH | 48 | 4 | 97.9 | 4 (4, 4) | 0.897 | 0.49 ± 0.07 | 0.04 ± 0.02 | 23.3 ± 11.6 | |||
| Total | 97 | 7 | 96.9 | 9 (4, 14) | 0.600 | 1.293 | 0.69 ± 0.02 | 0.07 ± 0.03 | 38.3 ± 18.6 |
The numbers in parentheses are 95% confidence intervals.
Mean ± standard deviation.
Rarefaction curves generated for Bacteria 16S rRNA gene and crDNA clones (a determination for the sampled community only) did not indicate saturation (data not shown). The percent coverage for the RNA-derived clones libraries ranged from 76% for the IH to 81% for the SEH layer. Both of these DNA-derived Bacteria libraries had a percent coverage of less than 64% (Table 2). FST and P tests were insignificant (P > 0.05) for all Bacteria libraries (data not shown). In addition, little difference was observed for gene and nucleotide diversity, Shannon-Weiner indices, and θ(π) values calculated for the four Bacteria libraries (Table 2). In contrast to the Archaea libraries, Bacteria lineage-per-time plots were indicative of populations with an excess of highly divergent lineages (data not shown) (21).
Bacteria community structure based on 16S rRNA gene sequence analyses.
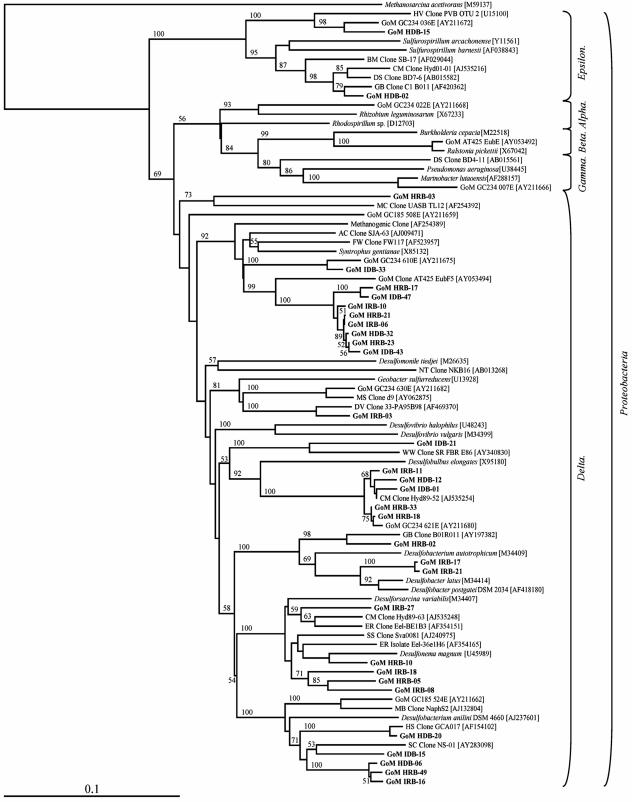
Analysis of the Bacteria 16S rRNA gene clones obtained from the SEH and IH layers revealed the greatest phylogenetic diversity compared to the other clone libraries (Table 2). The vast majority of Bacteria clones obtained were most closely related to uncultured lineages (Table 3). A total of 53 RFLP patterns (data not shown) representing eight distinct lineages and 55 phylotypes were detected. Representatives of 37 phylotypes (13 phylotypes comprising more than one clone and 24 phylotypes comprising a single clone) were sequenced and analyzed. All frequency calculations were based on the number of clones represented by phylotypes that were sequenced (n = 80). No one lineage was numerically dominant in these DNA-derived clone libraries (Table 3). Five of the eight lineages detected (δ - and ɛ-Proteobacteria, Chloroflexi, Firmicutes, and hydrocarbon-associated bacteria [Fig. 1 and 2 ]) comprised between 15 and 23% of the total 16S rRNA gene library (Table 3). A majority of the proteobacterial clones (18 of 30 clones) were most similar to the δ-Proteobacteria and grouped into 10 different phylotypes (Table 3). The phylotype designated GoM IDB-15 was one of five δ-Proteobacteria-related phylotypes detected only in the SEH layer (Table 3). Although only 1 of the 10 δ-Proteobacteria-related phylotypes, GoM IDB-47, was detected in both hydrate layers, comparable percentages of δ-Proteobacteria-related clones were observed in the SEH-derived (27%) and IH-derived (20%) libraries (Fig. 3). The remaining 12 Proteobacteria-related clones, which accounted for 15% of the total Bacteria 16S rRNA gene library, were related to the ɛ-Proteobacteria (Table 3 and Fig. 1). One phylotype, GoM HDB-02, represented the majority of the ɛ-Proteobacteria-related sequences detected (Table 3).
TABLE 3.
Summary of 16S rRNA gene sequences from SEH and IH Bacteria clone libraries
| Phylogenetic group | Clone | Nearest relative | % Similarity | No. of related clones
|
||
|---|---|---|---|---|---|---|
| Total | SEH | IH | ||||
| δ-Proteobacteria | GoM IDB-15 | HS clone GCA017 | 95 | 4 | 4 | 0 |
| GoM IDB-01 | CM clone Hyd89-52 | 98 | 1 | 1 | 0 | |
| GoM IDB-43 | GoM clone AT425 EubF5 | 91 | 1 | 1 | 0 | |
| GoM IDB-21 | WW clone SR FBR E86 | 89 | 1 | 1 | 0 | |
| GoM IDB-33 | GoM GC234 610E | 97 | 1 | 1 | 0 | |
| GoM HDB-20 | HS clone GCA017 | 99 | 3 | 0 | 3 | |
| GoM HDB-32 | GoM clone AT425 EubF5 | 93 | 2 | 0 | 2 | |
| GoM HDB-06 | MB clone NaphS2 | 92 | 2 | 0 | 2 | |
| GoM HDB-12 | CM clone Hyd89-52 | 98 | 1 | 0 | 1 | |
| GoM IDB-47 | FW clone FW117 | 90 | 2 | 1 | 1 | |
| ɛ-Proteobacteria | GoM HDB-15 | GoM GC185 036E | 96 | 1 | 0 | 1 |
| GoM HDB-02 | GB clone C1 B011 | 98 | 11 | 4 | 7 | |
| Chloroflexi | GoM IDB-24 | GB clone C1 B004 | 97 | 2 | 2 | 0 |
| GoM IDB-09 | HV clone P. palm A11 | 85 | 1 | 1 | 0 | |
| GoM IDB-50 | TS clone OlaA2 | 90 | 1 | 1 | 0 | |
| GoM IDB-35 | MC clone Eub 6 | 87 | 1 | 1 | 0 | |
| GoM HDB-07 | GoM GC185 546E | 99 | 3 | 0 | 3 | |
| GoM HDB-03 | HS clone GCA112 | 94 | 2 | 0 | 2 | |
| GoM HDB-37 | DSS clone t0.6.f | 90 | 1 | 0 | 1 | |
| GoM HDB-23 | WW clone CARB ER2.5 | 89 | 1 | 0 | 1 | |
| GoM HDB-31 | GoM GC185 546E | 99 | 1 | 0 | 1 | |
| Firmicutes | GoM IDB-40 | GoM GC234 604E | 96 | 1 | 1 | 0 |
| GoM IDB-10 | GoM GC234 604E | 95 | 1 | 1 | 0 | |
| GoM IDB-30 | GoM GC234 604E | 95 | 1 | 1 | 0 | |
| GoM HDB-46 | GoM GC234 604E | 95 | 1 | 0 | 1 | |
| GoM HDB-18 | GoM GC234 604E | 88 | 1 | 0 | 1 | |
| GoM HDB-21 | GoM GC234 604E | 97 | 7 | 2 | 5 | |
| GoM IDB-03 | Clostridium sp. | 87 | 2 | 1 | 1 | |
| Verrucomicrobiales | GoM HDB-19 | RC isolate DSM 44180 | 89 | 1 | 0 | 1 |
| GoM HDB-48 | Verrucomicrobia clone LD1-PA26 | 86 | 1 | 0 | 1 | |
| Actinomycetales | GoM HDB-08 | SO clone OHKB 16.85 | 94 | 1 | 0 | 1 |
| Spirochaetales | GoM IDB-04 | TCE clone ccslm2126 | 83 | 1 | 1 | 0 |
| Hydrocarbon-associated bacteria | GoM IDB-08 | HS clone GCA018 | 98 | 3 | 3 | 0 |
| GoM HDB-09 | GoM GC185 546E | 99 | 1 | 0 | 1 | |
| GoM HDB-04 | HS clone GCA025 | 99 | 13 | 5 | 8 | |
| Unclassified Bacteria group 1 | GoM HDB-22 | CM clone Hyd24-12 | 95 | 1 | 0 | 1 |
| Unclassified Bacteria group 2 | GoM IDB-27 | RP clone P2D6 | 94 | 1 | 1 | 0 |
FIG. 1.
Phylum Proteobacteria phylogenetic tree of relationships of 16S rRNA gene and 16S crDNA bacterial clone sequences, as determined by distance Jukes-Cantor analysis, from GoM GC234 SEH and IH samples (indicated by boldface type) to selected cultured isolates and environmental clones. The numbers in brackets are GenBank accession numbers. Clones whose designations include HDB and IDB represent sequences derived from rRNA genes extracted from SEH and IH samples, respectively. Clones whose designations include HRB and IRB represent 16S crDNA sequences derived from rRNA extracted from SEH and IH samples, respectively. One thousand bootstrap analyses were conducted, and percentages greater than 50% are indicated at the nodes. Methanosarcina acetivorans was used as the outgroup. Scale bar = 0.1 change per nucleotide position.
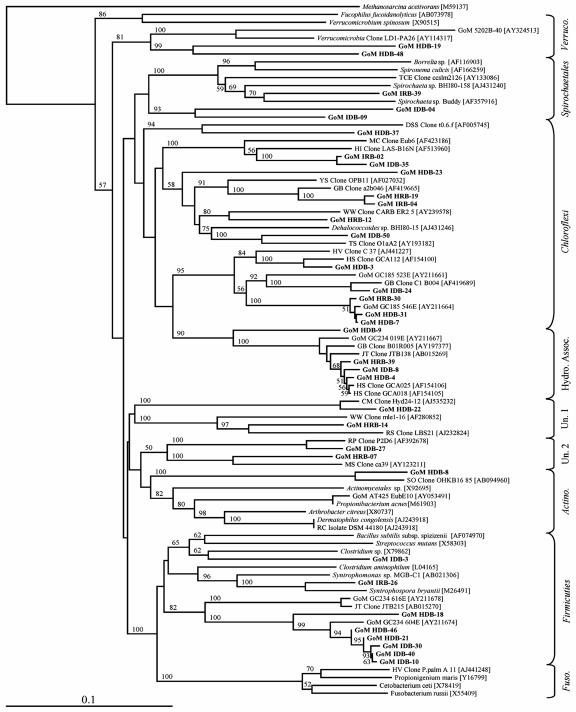
FIG. 2.
Non-Proteobacteria phylogenetic tree of relationships of 16S rRNA gene and 16S crDNA bacterial clone sequences, as determined by distance Jukes-Cantor analysis, from GoM GC234 SEH and IH samples (indicated by boldface type) to selected cultured isolates and environmental clones. The numbers in brackets are GenBank accession numbers. Clones whose designations include HDB and IDB represent sequences derived from rRNA genes extracted from SEH and IH samples, respectively. Clones whose designations include HRB and IRB represent 16S crDNA sequences derived from rRNA extracted from SEH and IH samples, respectively. One thousand bootstrap analyses were conducted, and percentages greater than 50% are indicated at the nodes. Methanosarcina acetivorans was used as the outgroup. Scale bar = 0.1 change per nucleotide position.
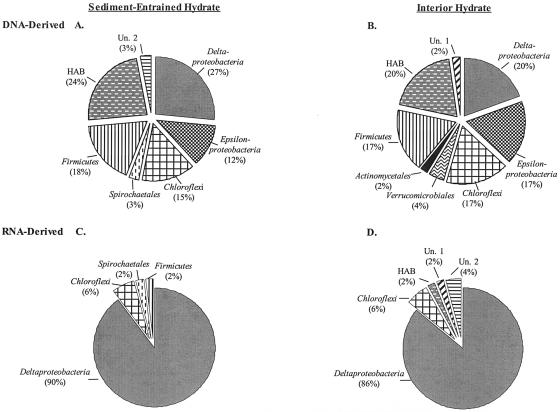
FIG. 3.
Frequencies of bacterial phylogenetic lineages detected in 16S rRNA gene and 16S crDNA clone libraries derived SEH and IH samples. Calculations were made based on the total number of clones associated with phylotypes from which a representative clone had been sequenced. (A and B) DNA-derived clone libraries. (C and D) RNA-derived clone libraries. HAB, hydrocarbon-associated bacteria; Un.1, unclassified Bacteria group 1; Un.2, unclassified Bacteria group 2.
A majority of the Bacteria 16S rRNA gene clones (63%) (Table 3) were related to four non-Proteobacteria lineages, the Chloroflexi, Firmicutes, Verrucomicrobiales, and Spirochaetales, and three uncharacterized (i.e., potentially novel) groups designated the hydrocarbon-associated bacteria, unclassified Bacteria group 1, and unclassified Bacteria group 2 (Fig. 2). The second most frequently detected group of phylotypes (21% of all rRNA gene clones [Table 3]) was most closely related to the hydrocarbon-associated bacteria, a distinct clade within the phylum Chloroflexi (Fig. 2) (15). The hydrocarbon-associated bacterium-related phylotype GoM HDB-04 was the most numerically abundant phylotype in the Bacteria library (16%) (Table 3). Nine distinct Chloroflexi-related phylotypes were detected (Table 3); however, none of these phylotypes occurred in both the SEH and IH layers. With the exception of the Firmicutes-related phylotype GoM HDB-21, which was detected 2.5-fold more frequently in the IH layer, the majority of Firmicutes-related clones exhibited a similar spatial pattern (Table 3). In contrast, the Firmicutes-related clones as a group, which represented 18% of the total DNA-derived Bacteria library, were detected at similar percentages in the two hydrate layers (Fig. 3). Comparable observations were not possible for the Spirochaetales and Actinomycetales lineages as each of these taxonomic groups was represented by a single phylotype (i.e., one clone each) (Table 3).
Determination of the metabolically active fraction of the Bacteria community.
Analysis of the 95 16S crDNA Bacteria clones revealed greater diversity than that in the Archaea clone libraries (Table 2), and these clones predominately included sequences most closely related to uncultured bacterial lineages (Table 4). A total of 30 distinct RFLP patterns were detected for clones representing five distinct phylogenetic lineages (Table 4 and Fig. 1 and 2). A considerable majority of the Bacteria crDNA clones from the SEH (90%) and IH (86%) layers were related to the δ-Proteobacteria (Fig. 3). However, only 3 of the 20 δ-Proteobacteria-related phylotypes, represented by clones GoM IRB-06, GoM HRB-10, and GoM IRB-21 (Table 4), comprised more than 10% of the total RNA-derived library. Interestingly, while nine phylotypes contained clones isolated from both the SEH and IH layers (Table 4), the δ-Proteobacteria-related phylotype GoM IRB-16 was the only phylotype with more than two members exclusively isolated from one hydrate layer. In addition, phylotypes represented by clones GoM IRB-06 and GoM HRB-10 appeared at greater frequencies in a single hydrate layer (i.e., they were two- and threefold more prevalent in the SEH and IH layers, respectively) (Table 4).
TABLE 4.
Summary of 16S crDNA sequences from SEH and IH Bacteria clone libraries
| Phylogenetic group | Clone | Nearest relative | % Similarity | No. of related clones
|
||
|---|---|---|---|---|---|---|
| Total | SEH | IH | ||||
| δ-Proteobacteria | GoM IRB-16 | SC clone NS-01 | 88 | 8 | 8 | 0 |
| GoM IRB-08 | ER isolate Eel-36e1H6 | 91 | 2 | 2 | 0 | |
| GoM IRB-03 | DV clone 33-PA95B98 | 96 | 1 | 1 | 0 | |
| GoM IRB-18 | SS clone Sva0081 | 92 | 1 | 1 | 0 | |
| GoM HRB-23 | GoM clone AT425 EubF5 | 89 | 2 | 0 | 2 | |
| GoM HRB-18 | CM clone Hyd89-52 | 98 | 1 | 0 | 1 | |
| GoM HRB-32 | CM clone Hyd89-52 | 98 | 1 | 0 | 1 | |
| GoM HRB-21 | GoM clone AT425 EubF5 | 89 | 1 | 0 | 1 | |
| GoM HRB-03 | MC clone UASE TL12 | 92 | 1 | 0 | 1 | |
| GoM HRB-05 | CM clone Hyd89-63 | 91 | 1 | 0 | 1 | |
| GoM HRB-49 | SC clone NS-01 | 88 | 1 | 0 | 1 | |
| GoM IRB-06 | GoM clone AT425 EubF5 | 89 | 12 | 8 | 4 | |
| GoM HRB-10 | ER isolate Eel-36e1H6 | 92 | 12 | 3 | 9 | |
| GoM IRB-21 | Desulfobacter postgatei DSM 2034 | 94 | 10 | 5 | 5 | |
| GoM IRB-11 | CM clone Hyd89-52 | 99 | 8 | 4 | 4 | |
| GoM IRB-27 | ER clone Eel-BE1B3 | 91 | 6 | 3 | 3 | |
| GoM IRB-17 | Desulfobacter postgatei DSM 2034 | 94 | 6 | 3 | 3 | |
| GoM IRB-10 | GoM clone AT425 EubF5 | 89 | 5 | 3 | 2 | |
| GoM HRB-16 | AC clone SJA-63 | 89 | 4 | 1 | 3 | |
| GoM HRB-02 | GB clone B01R011 | 96 | 2 | 1 | 1 | |
| Chloroflexi | GoM IRB-02 | HI clone LAS-B16N | 93 | 1 | 1 | 0 |
| GoM IRB-04 | GB clone a2b046 | 94 | 1 | 1 | 0 | |
| GoM HRB-19 | GB clone a2b046 | 95 | 1 | 0 | 1 | |
| GoM HRB-30 | GoM GC185 546E | 98 | 1 | 0 | 1 | |
| GoM HRB-12 | YS clone OPB11 | 91 | 2 | 1 | 1 | |
| Spirochaetales | GoM IRB-39 | Spirocaeta sp. strain BHI80-158 | 85 | 1 | 1 | 0 |
| Firmicutes | GoM IRB-26 | Syntrophospora bryantii | 90 | 1 | 1 | 0 |
| Hydrocarbon-associated bacteria | GoM HRB-39 | HS clone GCA025 | 99 | 1 | 0 | 1 |
| Unclassified Bacteria group 1 | GoM HRB-14 | RS clone LBS21 | 88 | 1 | 0 | 1 |
| Unclassified Bacteria group 2 | GoM HRB-07 | MS clone ca39 | 89 | 2 | 0 | 2 |
The remaining 12 16S crDNA clone sequences were most closely related to non-Proteobacteria lineages, including the Chloroflexi, Firmicutes, Spirochaetales, and groups that included the hydrocarbon-associated bacteria and unclassified Bacteria groups 1 and 2 (Table 4 and Fig. 2). A total of five Chloroflexi-related phylotypes (six clones) were identified and were detected at similar frequencies in the two hydrate layers (Fig. 3). Each of the remaining two lineages (Firmicutes and Spirochaetales) and three other phylogenetic groups (hydrocarbon-associated bacteria and unclassified Bacteria groups 1 and 2) were represented by a single phylotype (Table 4). Of these, unclassified Bacteria group 2 was the only phylotype that contained more than one clone (n = 2) (Table 4).
Archaea community structure based on 16S rRNA gene sequence analyses.
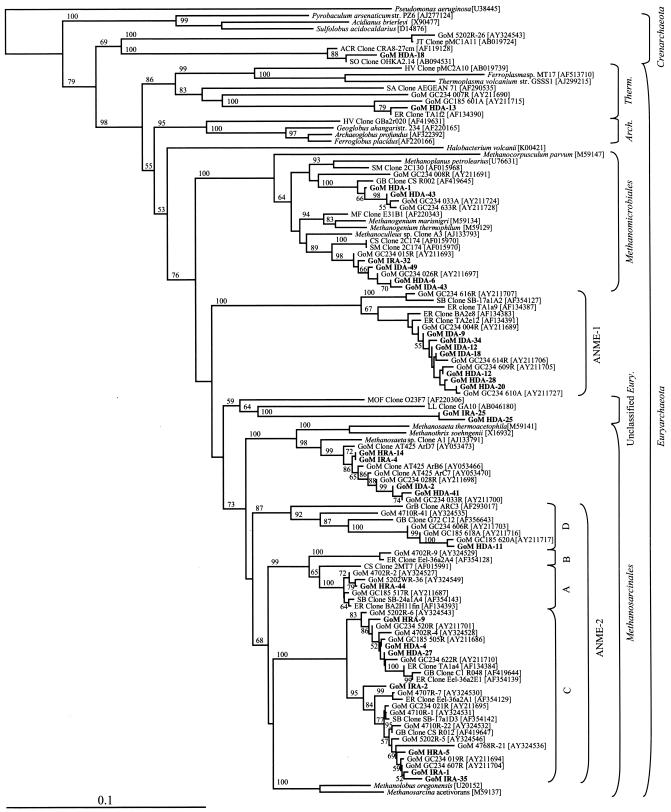
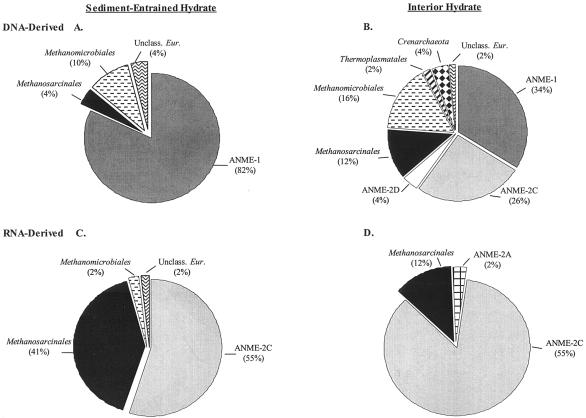
A total of 100 Archaea 16S rRNA gene clones obtained from the SEH and IH layers were grouped into 20 distinct RFLP patterns (data not shown). The 20 phylotypes were most closely related to Crenarchaeota and six lineages of Euryarchaeota, including Methanosarcinales, Methanomicrobiales, Thermoplasmatales, ANME-1, ANME-2, and one putatively novel clade designated unclassified Euryarchaeota (Fig. 4). Sixteen of the 20 phylotypes were most closely related to cloned sequences previously identified from the archaeal community extant in sediments overlying gas hydrate (22) (Table 5 and Fig. 4). The majority of the Archaea 16S rRNA gene clones (58% of the total clones) (Table 5) isolated from the SEH and IH layers were most closely related (>99% similar) to uncultured ANME-1 clones. The ANME-1-related phylotype represented by clone GoM IDA-34 was the numerically dominant phylotype for this library (n = 24) and was predominately isolated from the SEH layer (23 of 24 clones) (Table 5). The second most numerically dominant clone type in this DNA-derived library, the ANME-1-related phylotype GoM IDA-09, was detected only in the SEH (Table 5). As a group, the ANME-1-related clones were detected nearly threefold less frequently in the IH layer (Fig. 5). However, phylotype GoM HDA-28, comprising 16% of the ANME-1-related clones, was detected only in the IH (Table 5). In addition, the ANME-2C- and ANME-2D-related phylotypes were also detected only in the IH layer (Fig. 5) and were most similar to previously identified uncultured GoM clones (22) (Fig. 4).
FIG.4.
Phylogenetic tree of relationships of 16S rRNA gene and 16S crDNA archaeal clone sequences, as determined by distance Jukes-Cantor analysis, from GoM GC234 SEH and IH samples (indicated by boldface type) to selected cultured isolates and environmental clones. Pseudomonas aeruginosa was used as the outgroup. The numbers in brackets are GenBank accession numbers. Clones whose designations include HDB and IDB represent sequences derived from rRNA genes extracted from SEH and IH samples, respectively. Clones whose designations include HRB and IRB represent 16S crDNA sequences derived from rRNA extracted from SEH and IH samples, respectively. One thousand bootstrap analyses were conducted, and percentages greater than 50% are indicated at the nodes. Scale bar = 0.1 change per nucleotide position.
TABLE 5.
Summary of 16S rRNA gene sequences from SEH and IH Archaea clone libraries
| Phylogenetic group | Clone | Nearest relative | % Similarity | No. of related clones
|
||
|---|---|---|---|---|---|---|
| Total | SEH | IH | ||||
| ANME-1 | GoM IDA-09 | GoM GC234 610A | 99 | 14 | 14 | 0 |
| GoM IDA-18 | GoM GC234 610A | 99 | 1 | 1 | 0 | |
| GoM IDA-12 | GoM GC234 614R | 99 | 1 | 1 | 0 | |
| GoM HDA-28 | GoM GC234 609R | 99 | 9 | 0 | 9 | |
| GoM HDA-20 | GoM GC234 610A | 99 | 2 | 0 | 2 | |
| GoM IDA-34 | GoM GC234 609R | 99 | 24 | 23 | 1 | |
| GoM HDA-12 | GoM GC234 609R | 99 | 7 | 2 | 5 | |
| ANME-2C | GoM HDA-27 | GoM GC234 622R | 99 | 8 | 0 | 8 |
| GoM HDA-04 | GoM GC234 622R | 99 | 5 | 0 | 5 | |
| ANME-2D | GoM HDA-11 | GoM GC234 606R | 99 | 2 | 0 | 2 |
| Methanosarcinales | GoM IDA-02 | GoM GC234 033R | 99 | 1 | 1 | 0 |
| GoM HDA-41 | GoM GC234 033R | 99 | 7 | 1 | 6 | |
| Methanomicrobiales | GoM IDA-49 | GoM GC234 026R | 99 | 1 | 1 | 0 |
| GoM IDA-43 | GoM GC234 026R | 99 | 1 | 1 | 0 | |
| GoM HDA-06 | GoM GC234 026R | 99 | 6 | 1 | 5 | |
| GoM HDA-43 | GoM GC234 633A | 99 | 3 | 1 | 2 | |
| GoM HDA-01 | GB clone CS R002 | 98 | 2 | 1 | 1 | |
| Thermoplasmatales | GoM HDA-13 | ER clone TA1f2 | 99 | 1 | 0 | 1 |
| Crenarchaeota | GoM HDA-18 | SO clone CHKA2.14 | 96 | 2 | 0 | 2 |
| Unclassified Euryarchaeota | GoM HDA-25 | LL clone GA10 | 83 | 3 | 2 | 1 |
FIG. 5.
Frequencies of archaeal phylogenetic lineages detected in 16S rRNA gene and 16S crDNA clone libraries derived from SEH and IH samples. Calculations were made based on the total number of clones associated with phylotypes from which a representative clone had been sequenced. (A and B) DNA-derived clone libraries. (C and D) RNA-derived clone libraries. Unclass. Eur., unclassified Euryarchaeota.
The remaining 27% of the Archaea 16S rRNA gene library consisted of 10 phylotypes related to two methanogenic Euryarchaeota lineages (Methanosarcinales and Methanomicrobiales), two nonmethanogenic Euryarchaeota lineages (Thermoplasmatales and an unclassified group), and one Crenarchaeota lineage (Fig. 4). Although for each of the five Methanomicrobiales-related phylotypes at least one clone was isolated from the SEH layer (Table 5), the three numerically dominant Methanomicrobiales-related phylotypes (GoM HDA-06, GoM HDA-43, and GoM HDA-01) were more frequently detected in the IH (8 of 11 clones) (Table 5). Similarly, the Methanosarcinales-, Thermoplasmatales-, and Crenarchaeota-related phylotypes occurred more frequently in the IH library (Fig. 5). Phylotype GoM HDA-25 may represent a novel lineage, designated unclassified Euryarchaeota, due to low similarity to previously sequenced clones (83%) (Table 5) and deep phylogenetic branching (Fig. 4).
Determination of the metabolically active fraction of the Archaea community.
A total of 97 Archaea 16S crDNA clones obtained from the SEH and IH layers were grouped into seven distinct RFLP patterns. Clones grouped into five phylogenetic lineages, including Methanomicrobiales, Methanosarcinales, two groups of ANME-2, and one putatively novel clade designated unclassified Euryarchaeota (Table 6 and Fig. 4). The majority of the 16S crDNA Archaea clones (70%) were related to ANME-2C (Table 6). A large percentage of the ANME-2C clones (59%) grouped into a phylotype designated GoM HRA-9 and were most closely related (99% similar) (Table 6) to an environmental clone originally isolated from the overlying sediment on a GoM gas hydrate mound (22). Although this phylotype was almost fivefold more numerically abundant in the IH library (Fig. 5), a second ANME-2C-related phylotype, represented by clone GoM HRA-05, was over twofold more numerically abundant in the SEH library (Fig. 5).
TABLE 6.
Summary of 16S crDNA sequences from SEH and IH Archaea clone libraries
| Phylogenetic group | Clone | Nearest relative | % Similarity | No. of related clones
|
||
|---|---|---|---|---|---|---|
| Total | SEH | IH | ||||
| ANME-2A | GoM HRA-44 | GoM 5210WR-36 | 99 | 1 | 0 | 1 |
| ANME-2C | GoM IRA-35 | GoM GC234 607R | 99 | 2 | 2 | 0 |
| GoM HRA-09 | GoM GC185 505R | 99 | 40 | 7 | 33 | |
| GoM HRA-05 | GoM GC234 607R | 99 | 26 | 18 | 8 | |
| Methanosarcinales | GoM IRA-04 | GoM GC234 028R | 99 | 26 | 20 | 6 |
| Methanomicrobiales | GoM IRA-32 | GoM GC234 026R | 99 | 1 | 1 | 0 |
| Unclassified Euryarchaeota | GoM IRA-25 | MF clone 023F7 | 85 | 1 | 1 | 0 |
The remaining 28 clones were most closely related to Methanosarcinales and Methanomicrobiales lineages and one uncharacterized group. A single Methanosarcinales-related phylotype, designated GoM IRA-4, comprised 26 of the 28 clones which were detected threefold more frequently in the SEH library (Fig. 5). A single Methanomicrobiales-related clone, GoM IRA-32, isolated from the SEH, was most closely related (99%) to a previously sequenced GoM GC234 clone from the overlying sediment (22) (Fig. 4). Interestingly, one additional phylotype isolated from the SEH, designated unclassified Euryarchaeota GoM IRA-25, was the only phylotype not closely related to a clone previously identified from the GoM.
DISCUSSION
Microbial communities residing in cold seep environments have been the focus of numerous characterization studies (13, 17, 22, 27). To date, fluorescent in situ hybridization has been one of the most widely used techniques to identify the presumptively metabolically active fraction of the microbial community. This technique, however, does not provide in-depth characterization of the overall microbial diversity, nor can it detect novel lineages without prior clone sequence information. The present study was the first study to extract RNA directly from gas hydrate and to delineate the metabolically active Bacteria and Archaea fraction of the gas hydrate microbial community. In addition, DNA-derived libraries were created in order to compare these two different molecular approaches for community analyses. In this study, several putatively novel microbial (Bacteria and Archaea) lineages were detected in the different hydrate layers. Geochemical and rate measurement data for sample layers taken from the same bulk hydrate sample that have been reported previously (26) provide a link to gas hydrate microbial community structure and function.
Detection of metabolically active microbes associated with GoM gas hydrates.
Discrete sampling of GoM gas hydrate (i.e., the SEH and IH layers) and subsequent isolation of rRNA enable characterization of the presumptively metabolically active fraction of microbial assemblages. Gas hydrate samples can be collected with a manned submersible or by shipboard deployment of piston and gravity cores. Our method of collection, by manned submersible, provided an unprecedented opportunity to accurately sample the distinct GoM hydrate ecosystem. Thus, sufficient amounts of hydrate materials were retrieved to provide the necessary volumes that yielded suitable concentrations (quantity and quality) of RNA needed for molecular analyses.
To date, few studies have demonstrated the presence of metabolically active microbial communities within gas hydrates. One line of evidence for the presence of active microbial fractions in GoM gas hydrates is our finding of 4- and 10-fold-higher RNA/DNA ratios and total RNA concentrations (5), respectively, across the distinct hydrate layers. Phylogenetic analysis of RNA-derived clones revealed a total of 30 distinct Bacteria and 7 distinct Archaea phylotypes in the SEH and IH libraries. Rarefaction data and percent coverage calculations suggested that the bacterial 16S rRNA gene libraries in both layers did not reach saturation. This was in contrast to data obtained for the archaeal libraries. Although additional sampling of bacterial clones would be needed to determine the full extent of the Bacteria community diversity, we did obtain several numerically dominant lineages. We theorized that additional sampling of the clone libraries would not likely alter the bacterial phylotype percent distribution patterns reported here.
Geochemical data for subsamples taken from the same hydrate sample used in this study provide a second line of evidence for the presence of metabolically active microbial populations (26). Although AOM and SR rates were higher in the overlying sediment (0.60 and 76.2 nmol cm−3 day−1, respectively), active anaerobic methane-oxidizing and sulfate-reducing populations were detected in the IH layer (0.28 and 3.2 nmol cm−3 day−1, respectively) (26). Interestingly, within the hydrate layers, the highest SR rates did not occur in the same layer in which the highest rates of AOM were detected. Orcutt et al. (26) described this perhaps surprising finding as a result of a “loose coupling between SR and AOM.” Regardless, the geochemical data and RNA-based phylogenetic identification of microbial populations within intact gas hydrate and across boundary layers suggest that there is active microbial habitation of this extreme environment by diverse lineages of Bacteria and Archaea.
Putative phylotype niche specificity.
Although similar distributions of phylotypes were observed for the SEH and IH Bacteria and Archaea clone libraries, we hypothesized that some phylotypes may be restricted (or only detected) in a single hydrate layer. As SEH and IH samples were collected from the same piece of solid gas hydrate, the potential for heterogeneity effects was potentially minimized. The geochemistry and rate measurements (26) differed by severalfold for the SEH and IH layers even though the sampled layers were in close proximity to each other (∼2 to 4 cm apart). Moreover, several phylotypes, especially those related to the Chloroflexi, δ-Proteobacteria, ANME-1, and ANME-2C lineages, were detected at higher frequencies in only one layer. The differences in chemistry, metabolic activity, and clone frequency suggest the presence of two distinct hydrate-associated habitats.
While statistical determinations of the Archaea clone sequences supported the hypothesis that there are two hydrate habitats, the same analyses of Bacteria sequences did not. Specifically, 10 different statistical indices were applied to the eight DNA- and RNA-derived clone libraries constructed from the SEH and IH layers. Although the Sorensen's index indicated little overlap between Bacteria phylotypes from the SEH and IH, all other indices revealed few differences between these two layers. Insignificant FST and P tests suggested that the Bacteria sequences from the SEH and IH were from similar lineage distributions and were indistinguishable from the combined communities (21). Therefore, although differences in individual phylotype distributions were observed between the two layers, these differences were not convincing enough to indicate that two distinct bacterial populations were present. Lineage-per-time plots indicated an excess of highly divergent lineages, suggesting that selection factors affecting both hydrate layers maintain high diversity in the bacterial community (21). In contrast, plots that show an excess of closely related lineages are indicative of a recent selection event resulting in the speciation of a few surviving or newly colonizing species (21). In this study the Archaea libraries had lineage-per-time plot lines in close proximity to the plot lines indicative of constant lineage births and deaths (21). This result suggests that compared to the Bacteria, the Archaea are less divergent and may possibly have experienced a more recent selection event. Additionally, the statistical indices applied to the Archaea libraries, including FST and P tests, suggested that there is a significant difference between the SEH and IH Archaea communities. Mills et al. (23) reported a similar trend in Archaea community phylotype frequency variances across a depth profile in sediments associated with GoM microbial mats, while Bacteria phylotype frequencies were relatively less depth specific. Therefore, it is tempting to speculate that the archaeal populations are more specialized for specific environmental conditions and perhaps less able to tolerate environmental fluctuations than the bacterial populations.
Differences in lineage frequency were detected between the RNA- and DNA-derived libraries within individual hydrate layers. The utilization of PCR techniques to construct clone libraries is inherently biased by the concentration of template available for amplification. DNA-derived clone libraries are based on a constant number of 16S rRNA genes per cell and can potentially detect dead or quiescent cells. In contrast, the concentration of rRNA present in a cell is thought to be proportional to the metabolic activity of the cell and therefore can alter the detection frequency of phylotypes relative to the DNA-derived clone libraries (5). Whereas absolute quantification of microbial populations was not possible by analyzing the DNA- and RNA-derived clone libraries in this study, differences in phylotype distribution patterns between the two clone libraries were detected. For example, ɛ-Proteobacteria- and ANME-1-related lineages may have been present in the hydrate-associated samples but either were not metabolically active or were active at a level below the level of detection by clone library analyses. In contrast, δ-Proteobacteria- and ANME-2C-related lineages were the most numerical abundant lineages detected in libraries derived from the active metabolic fraction of the community, but they were less abundant in the corresponding DNA-derived libraries. Such discrepancies between 16S rRNA gene and 16S crDNA libraries demonstrate the necessity of generating DNA- and RNA-derived libraries when possible to perform a more comprehensive analysis of the extant microbial communities.
Detection of novel microbial lineages.
Molecular characterization of microbial communities from extreme environments has provided evidence for numerous new taxonomic groups having no known closely related culturable isolates (13, 22, 27). In this study, several potentially novel clones grouped into four distinct clades (three clades from the Bacteria library and one clade from the Archaea library) that were distantly related to cultured isolates. Recent reports have indicated that there is a new clade in the Chloroflexi lineage specific to hydrocarbon seeps (15). This clade, designated the hydrocarbon-associated bacteria, lacks a culturable isolate, and therefore, the specific physiology is unknown. However, clones related to this clade, along with a closely related second branch within the Chloroflexi lineage, were identified in our RNA-derived Bacteria clone library from the IH layer, which provided the first described environment in which this lineage appears to be metabolically active.
Two additional clades, designated unclassified Bacteria group 1 and unclassified Bacteria group 2, were less than 90% similar to the nearest culturable relative. Interestingly, both groups contained RNA- and DNA-derived clones, which provided evidence not only that this group is present but also that it is metabolically active in hydrate-associated habitats. Similarly, the novel Archaea cluster, designated in this study the unclassified Euryarchaeota group, was detected in the 16S rRNA gene and 16S crDNA libraries. The data presented here provide further evidence of the potential habitat or niche of each of these novel lineages, as well as other previously characterized lineages. However, we readily acknowledge that the present study lacked a temporal component. Thus, there is a need to confirm that similar results can be obtained in future GoM hydrate sampling. Additionally, attempts will be made to cultivate these and other microbes identified in these GoM clone libraries to determine their specific physiology and metabolic traits.
Acknowledgments
This work was supported by National Science Foundation LExEn grant OCE-0085549. The U.S. Department of Energy and the National Undersea Research Program provided support for submersible operations.
We thank C. Fisher for invaluable sampling and study site information.
REFERENCES
- 1.Chao, A. 1987. Estimating the population size for capture-recapture data with unequal catchability. Biometrics 43:783-791. [PubMed] [Google Scholar]
- 2.Clifford, S. M., D. Crisp, D. A. Fisher, K. E. Herkenhoff, S. E. Smrekar, P. C. Thomas, D. D. Wynn-Williams, R. W. Zurek, J. R. Barnes, B. G. Bills, E. W. Blake, W. M. Calvin, J. M. Cameron, M. H. Carr, P. R. Christensen, B. C. Clark, G. D. Clow, J. A. Cutts, D. Dahl-Jensen, W. B. Durham, F. P. Fanale, J. D. Farmer, F. Forget, K. Gotto-Azuma, R. Grard, R. M. Haberle, W. Harrison, R. Harvey, A. D. Howard, A. P. Ingersoll, P. B. James, J. S. Kargel, H. H. Kieffer, J. Larsen, K. Lepper, M. C. Malin, D. J. McCleese, B. Murray, J. F. Nye, D. A. Paige, S. R. Platt, J. J. Plaut, N. Reeh, J. W. Rice, D. E. Smith, C. R. Stoker, K. L. Tanaka, E. Mosley-Thompson, T. Thorsteinsson, S. E. Wood, A. Zent, M. T. Zuber, and H. J. Zwally. 2000. The state and future of Mars polar science and exploration. Icarus 144:210-242. [DOI] [PubMed] [Google Scholar]
- 3.Colwell, R. K. 1997. EstimateS: statistical estimation of species richness and shared species from samples, version 5. User's guide and application. [Online.] http://viceroy.eeb.uconn.edu/estimates.
- 4.Colwell, R. K., and J. A. Coddington. 1994. Estimating terrestrial biodiversity through extrapolation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 345:101-118. [DOI] [PubMed] [Google Scholar]
- 5.Dell'anno, A., M. Fabiano, G. C. A. Duineveld, A. Kok, and R. Danovaro. 1998. Nucleic acid (DNA, RNA) quantification and RNA/DNA ratio determination in marine sediment: comparison of spectophotometric, fluorometric, and high-performance liquid chromatography methods and estimation of detrital DNA. Appl. Environ. Microbiol. 64:3238-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobrynin, V. M., Y. P. Korotajev, and D. V. Plyuschev. 1981. Gas hydrates—a possible energy resource, p. 727-729. In R. F. Meyer and J. C. Olson (ed.), Long-term energy resources. Pitman, Boston, Mass.
- 8.Freytag, J. K., P. R. Girguis, D. C. Bergquist, J. P. Andras, J. J. Childress, and C. R. Fisher. 2001. A paradox resolved: sulfide acquisition by roots of seep tubeworms sustains net chemoautotrophy. Proc. Natl. Acad. Sci. USA 98:13408-13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Good, I. J. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237-264. [Google Scholar]
- 10.Grauls, D. 2001. Gas hydrates: importance and applications in petroleum exploration. Mar. Petroleum Geol. 18:519-523.
- 11.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 12.Heck, K. L., G. V. Belle, and D. Simberloff. 1975. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 56:1459-1461. [Google Scholar]
- 13.Hinrichs, K. U., J. M. Hayes, S. P. Sylva, P. G. Brewer, and E. F. DeLong. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802-805. [DOI] [PubMed] [Google Scholar]
- 14.Hurt, R. A., X. Y. Qiu, L. Y. Wu, Y. Roh, A. V. Palumbo, J. M. Tiedje, and J. H. Zhou. 2001. Simultaneous recovery of RNA and DNA from soils and sediments. Appl. Environ. Microbiol. 67:4495-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inagaki, F., M. Suzuki, K. Takai, H. Oida, T. Sakamoto, K. Aoki, K. H. Nealson, and K. Horikoshi. 2003. Microbial communities associated with geological horizons in coastal subseafloor sediments from the Sea of Okhotsk. Appl. Environ. Microbiol. 69:7224-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kvenvolden, K. A. 1999. Potential effects of gas hydrate on human welfare. Proc. Natl. Acad. Sci. USA 96:3420-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lanoil, B. D., R. Sassen, M. T. La Duc, S. T. Sweet, and K. H. Nealson. 2001. Bacteria and archaea physically associated with Gulf of Mexico gas hydrates. Appl. Environ. Microbiol. 67:5143-5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lashof, D. A., and D. R. Ahuja. 1990. Relative contributions of greenhouse gas emissions to global warming. Nature 344:529-531. [Google Scholar]
- 19.MacDonald, I. R., N. L. Guinasso, R. Sassen, J. M. Brooks, L. Lee, and K. T. Scott. 1994. Gas hydrate that breaches the sea-floor on the continental slope of the Gulf of Mexico. Geology 22:699-702. [Google Scholar]
- 20.Maidak, B. L., J. R. Cole, C. T. Parker, Jr., G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin, A. P. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 68:3673-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mills, H. J., C. Hodges, K. Wilson, I. R. MacDonald, and P. A. Sobecky. 2003. Microbial diversity in sediments associated with surface-breaching gas hydrate mounds in the Gulf of Mexico. FEMS Microbiol. Ecol. 46:39-52. [DOI] [PubMed] [Google Scholar]
- 23.Mills, H. J., R. M. Martinez, S. Story, and P. Sobecky. 2004. Identification of members of the metabolically active microbial populations associated with Beggiatoa species mat communities from Gulf of Mexico cold-seep sediments. Appl. Environ. Microbiol. 70:5447-5458. [DOI] [PMC free article] [PubMed]
- 24.Nei, M. 1987. Molecular evolutionary genetics. Columbia University Press, New York, N.Y.
- 25.Nogales, B., E. R. B. Moore, W.-R. Abraham, and K. N. Timmis. 1999. Identification of the metabolically active members of a bacterial community in a polychlorinated biphenyl-polluted moorland soil. Environ. Microbiol. 1:199-212. [DOI] [PubMed] [Google Scholar]
- 26.Orcutt, B. N., A. Boetius, S. K. Lugo, I. R. Macdonald, V. A. Samarkin, and S. Joye. 2004. Life at the edge of methane ice: microbial cycling of carbon and sulfur in Gulf of Mexico gas hydrates. Chem. Geol. 205:239-251. [Google Scholar]
- 27.Orphan, V. J., K. U. Hinrichs, W. Ussler III, C. K. Paull, L. T. Taylor, S. P. Sylva, J. M. Hayes, and E. F. Delong. 2001. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67:1922-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paull, C. K., W. J. Buelow, W. Ussler, and W. S. Borowski. 1996. Increased continental-margin slumping frequency during sea-level lowstands above gas hydrate-bearing sediments. Geology 24:143-146. [Google Scholar]
- 29.Perriere, G., and M. Gouy. 1996. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78:364-369. [DOI] [PubMed] [Google Scholar]
- 30.Powers, L. G., H. J. Mills, A. V. Palumbo, C. L. Zhang, K. Delaney, and P. Sobecky. 2002. Introduction of a plasmid-encoded phoA gene for constitutive overproduction of alkaline phosphatase in three subsurface Pseudomonas isolates. FEMS Microbiol. Ecol. 41:115-123. [DOI] [PubMed] [Google Scholar]
- 31.Sassen, R., S. L. Losh, L. Cathles III, H. H. Roberts, J. K. Whelan, A. V. Milkov, S. T. Sweet, and D. A. DeFreitas. 2001. Massive vein-filling hydrate: relation to ongoing gas migration from the deep subsurface in the Gulf of Mexico. Mar. Pet. Geol. 18:551-560. [Google Scholar]
- 32.Sassen, R., and I. R. MacDonald. 1997. Hydrocarbons of experimental and natural gas hydrates, Gulf of Mexico continental slope. Org. Geochem. 26:289-293. [Google Scholar]
- 33.Sassen, R., I. R. MacDonald, N. L. Guinasso, S. Joye, A. G. Requejo, S. T. Sweet, J. Alcala-Herrera, D. DeFreitas, and D. R. Schink. 1998. Bacterial methane oxidation in sea-floor gas hydrate: significance to life in extreme environments. Geology 26:851-854. [Google Scholar]
- 34.Sassen, R., I. R. Macdonald, A. G. Requejo, N. L. Guinasso, M. C. Kennicutt, S. T. Sweet, and J. M. Brooks. 1984. Organic geochemistry of sediments from chemosythetic communities, Gulf of Mexico slope. Geo-Mar. Lett. 14:110-119. [Google Scholar]
- 35.Sassen, R., S. T. Sweet, A. V. Milkov, D. A. DeFreitas, G. G. Salata, and E. C. McDade. 1999. Geology and geochemistry of gas hydrates, central Gulf of Mexico continental slope. Trans. Gulf Coast Assoc. Geol. Soc. 49:462-468. [Google Scholar]
- 36.Schneider, S., D. Roessli, and L. Excoffier. 2000. Arlequin ver. 2.000: a software for population genetics data analysis. Genetics and Biometry Laboratory: University of Geneva, Geneva, Switzerland.
- 37.Slatkin, M. 1991. Inbreeding coefficients and coalescence times. Genet. Res. 58:167-175. [DOI] [PubMed] [Google Scholar]
- 38.Sloan, E. D. 1990. Clathrate hydrates of natural gases. Dekker, New York, N.Y.
- 39.Sobecky, P. A., T. J. Mincer, M. C. Chang, and D. R. Helinski. 1997. Plasmids isolated from marine sediment microbial communities contain replication and incompatibility regions unrelated to those of known plasmid groups. Appl. Environ. Microbiol. 63:888-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tajima, F. 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105:437-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tatusova, T. A., and T. L. Madden. 1999. BLAST 2 SEQUENCES, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 174:247-250. (Erratum, 177: 187-188.) [DOI] [PubMed] [Google Scholar]
- 42.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Hook, A. 1961. Crystallization: theory and practice. Reinhold, New York, N.Y.
- 44.Watanabe, K., Y. Kodama, N. Hamamura, and N. Kaku. 2002. Diversity, abundance, and activity of archaeal populations in oil-contaminated groundwater accumulated at the bottom of an underground crude oil storage cavity. Appl. Environ. Microbiol. 68:3899-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]