Abstract
Campylobacter jejuni is a predominant cause of food-borne bacterial gastroenteritis in the developed world. We have investigated the importance of a homologue of the periplasmic HtrA protease in C. jejuni stress tolerance. A C. jejuni htrA mutant was constructed and compared to the parental strain, and we found that growth of the mutant was severely impaired both at 44°C and in the presence of the tRNA analogue puromycin. Under both conditions, the level of misfolded protein is known to increase, and we propose that the heat-sensitive phenotype of the htrA mutant is caused by an accumulation of misfolded protein in the periplasm. Interestingly, we observed that the level of the molecular chaperones DnaK and ClpB was increased in the htrA mutant, suggesting that accumulation of nonnative proteins in the periplasm induces the expression of cytoplasmic chaperones. While lack of HtrA reduces the oxygen tolerance of C. jejuni, the htrA mutant was not sensitive to compounds that increase the formation of oxygen radicals, such as paraquat, cumene hydroperoxide, and H2O2. Using tissue cultures of human epithelial cells (INT407), we found that the htrA mutant adhered to and invaded human epithelial cells with a decreased frequency compared to the wild-type strain. This defect may be a consequence of the observed altered morphology of the htrA mutant. Thus, our results suggest that in C. jejuni, HtrA is important for growth during stressful conditions and has an impact on virulence.
Campylobacter jejuni is a gram-negative human pathogen and is recognized as a major cause of bacterial food-borne infections worldwide. The symptoms of campylobacteriosis are malaise, fever, severe abdominal pain, and diarrhea. In rare cases, C. jejuni triggers the Guillain-Barré syndrome, which is an autoimmune disorder of the peripheral nervous system (32). C. jejuni is a common and efficient gut colonizer of many animals, and fecal contamination of poultry meat during slaughtering is considered to be the major source of Campylobacter organisms that cause campylobacteriosis in humans. The optimum growth temperature of C. jejuni is 42°C, which may reflect adaptation to the core temperature of the avian host. C. jejuni does not normally multiply on food products (35) because most strains grow only between 31 and 45°C and require reduced oxygen tension as well as 5 to 10% carbon dioxide for growth. In contrast, C. jejuni survives well in food products at low temperature (5, 26), and due to a low infective dose (6), even few surviving Campylobacter organisms can present a health risk to humans.
During transmission from the intestinal tract of poultry to the human gut, C. jejuni experiences unfavorable conditions which may lead to protein misfolding and loss of protein function. Since misfolded proteins tend to form potentially harmful aggregates, removal of nonnative proteins is required in all cellular compartments. The HtrA protease was first identified in Escherichia coli as being required for growth at 42°C (28) and for degradation of misfolded protein in the periplasm (49). Later it was shown that HtrA degrades heat-denatured proteins in vivo and in vitro (25), and because only a few native proteins have been demonstrated to be in vivo substrates for HtrA proteolytic activity (2, 8, 20), it is suggested that the major biological role of HtrA is the removal of nonnative misfolded protein from the cellular envelope. In E. coli, the HtrA protease is located at the periplasmic side of the inner membrane (29, 43) and is derived from a pre-HtrA precursor protein by cleavage of the 26 N-terminal amino acids during export (29). The HtrA protein contains a catalytic triad (His105-Asp135-Ser210) that is required for proteolytic activity (44) and two PDZ domains that are responsible for oligomerization of the protein complex, substrate recognition, and binding (42, 46). In addition to the proteolytic activity, E. coli HtrA also possesses chaperone activity in vitro at low temperatures (47), where a conformational change of the protein masks the proteolytic residues (24). At elevated temperatures, the catalytic residues are accessible and the proteolytic activity of HtrA predominates (24, 47).
HtrA homologues are found in most bacteria, and even though the protein is well conserved in evolution, the impact on bacterial physiology differs among the gram-negative bacteria. In contrast to E. coli, HtrA is not essential for growth at high temperatures in Salmonella enterica serovar Typhimurium (19). Interestingly, the S. enterica serovar Typhimurium htrA mutant showed a reduced virulence in the murine salmonellosis model (9) and reduced survival in macrophages (19). Phenotypic characterization of the S. enterica serovar Typhimurium htrA mutant furthermore revealed a decreased tolerance to oxidative stress, which may explain the reduced survival in macrophages, where reactive oxygen intermediates are released during the oxidative burst (19). htrA mutants of other gram-negative pathogenic bacteria, such as Yersinia enterocolitica, Klebsiella pneumoniae, and Brucella abortus, are sensitive both to high temperatures and oxidative stress (10, 11, 27, 58). Furthermore, htrA mutants of Y. enterocolitica and B. abortus show reduced virulence in murine models (12, 58). It is unclear why htrA mutants of gram-negative bacteria are attenuated in virulence. However, since they are more susceptible to stress than the parental strains, the mutants may also be less viable in host tissues. In addition, it has been speculated that the chaperone and protein processing functions of HtrA are needed for the folding of secreted proteins or that HtrA might be involved in the oligomerization and export of virulence factors (36).
In general, bacteria that are exposed to stressful conditions such as heat shock induce synthesis of proteases and chaperones, which are involved in degradation and refolding of unfolded protein substrates, respectively. Using a whole-genome DNA microarray, it was shown that transcription of some protease genes (lon, clpB, and hslU) and several chaperone genes (groEL, groES, grpE, dnaK, and dnaJ) of C. jejuni NCTC11168 were up-regulated in response to a temperature up-shift from 37°C (human body temperature) to 42°C (temperature of the chicken core) (48). In addition, transcription of the lon, clpB, and dnaK genes as well as the groELS operon was found to be induced by heat shock (48°C) (50-53), and recently, we have shown that clpB, dnaK, and groELS are negatively regulated by HspR (1). Experiments suggest that heat shock proteins play a role in vivo, since a C. jejuni dnaJ mutant is unable to colonize chickens (22). In contrast, the HtrA protease does not seem to be important in the ability of C. jejuni to colonize chickens (7). The aim of the present study was to examine the role of HtrA in C. jejuni stress tolerance by analyzing growth and survival of a constructed htrA mutant. In addition, the importance of HtrA for interaction between C. jejuni and the human host has been investigated.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. jejuni NCTC11168 (National Collection of Type Cultures) and C. jejuni NCTC11168 ΔhtrA::cat (LB1281; this study) were routinely grown on blood agar baseII (Oxoid) supplemented with 5% calf blood (base II) or in brain heart infusion (BHI) broth (Difco) in a microaerobic environment provided by CampyGen (Oxoid). E. coli DH5α [φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1] was grown in Luria-Bertani broth or on Luria-Bertani agar (Difco). When appropriate, media were supplemented with ampicillin (100 μg/ml), chloramphenicol (20 μg/ml), or kanamycin (50 μg/ml).
Transformation.
C. jejuni NCTC11168 was transformed by electroporation essentially as described by Wassenaar and coworkers (57). To produce competent cells, C. jejuni NCTC11168 was harvested from overnight-incubated agar plates with 2 ml ice-cold wash buffer (272 mM sucrose, 15% glycerol) and subjected to four repeated centrifugations (4°C, 10,000 rpm, 10 min) and wash steps. After the last centrifugation, the cells were resuspended in 1/10 volume wash buffer, resulting in a concentration of 109 CFU/ml. Cell volumes of 50 μl were electroporated (1.80 kV, 200 Ω, 25 F) with 1 to 5 μl plasmid DNA. Immediately after electroporation, 1 ml of recovery broth (10% glycerol, Mueller-Hinton broth [2/3], brucella broth [1/3]) was added, and cells were plated on nonselective plates. After incubation overnight at 37°C under microaerobic conditions, the cells were harvested with recovery broth and plated on selective plates. E. coli DH5α was transformed by electroporation using standard methods.
Primers used in this study.
The primers used in this study were as follows (PstI restriction enzyme sites are underlined, and bases complementary to Ht2 are indicated in bold): Ht1, 5′-AATTTAATGGTTTCGCCTTG-3′; Ht2Ps, 5′-CTGCAGTGCAGCAAATAAAGCACTTGC-3′; Ht3Ps, 5′-GCTTTATTTGCTGCACTGCAGGAATTTACCAAAGTTTGGGT-3′; Ht4, 5′-AGCTTATAACCTATTCCACG-3′; HtrA-up, 5′-CATCATCTTCTCTTGTGTAA-3′; HtrA-down, 5′-TTCACTGATAACTCCTGC-3′; htrA-start, 5′-GCTAACTCCAAGAGTTTCG-3′; htrA-end, 5′-GAATATTTGTCATAGTTTTCC-3′; htrA-intF, 5′-GTAAAGATCCAAAAACAGATT-3′; htrA-intR, 5′-ACTAAATCTCCACGCTTAACG-3′.
Construction of a C. jejuni htrA mutant.
Chromosomal DNA from C. jejuni NCTC11168 was used as a template for amplification of DNA fragments containing either the 5′ end of the htrA gene and upstream sequences or the 3′ end of the htrA gene and downstream sequences. For the upstream fragment, primers Ht1 and Ht2Ps (731 bp) were used, while primers Ht3Ps and Ht4 (799 bp) were used for the downstream fragment. In a second round of PCR, the htrA fragments were joined by using the splicing-by-overlap-extension PCR method (16), creating a PCR fragment containing an in-frame deletion of 1,308 bp in htrA and introducing a PstI site between the upstream and downstream fragments. The ΔhtrA PCR fragment was cloned in the TOPO TA cloning vector pCR2.1 (Invitrogen), resulting in plasmid pLB217. Subsequently, pLB225 was constructed by cloning the ΔhtrA PCR fragment from pLB217 into the EcoRI site of pGEM-7Zf(+) (Promega). Finally, the cat gene (obtained from pRY109) was cloned into the PstI site of pLB225, resulting in pLB229. The cat gene was transcribed in the same direction as the htrA gene.
C. jejuni NCTC11168 was transformed with pLB229, and several chloramphenicol-resistant colonies were isolated. Chromosomal DNA was isolated from four different chloramphenicol-resistant colonies and used as templates in PCRs. By using primers htrA-up and htrA-down, which anneal to chromosomal sequences upstream and downstream of the region cloned in pLB229, respectively, it was verified that the ΔhtrA::cat mutation was transferred to the chromosome of C. jejuni NCTC11168 in all four isolates. Furthermore, PCR using primers htrA-up and htrA-down demonstrated that no wild-type htrA allele was present in the genome, showing that a double-crossover event had occurred. In the present report, the C. jejuni NCTC11168 htrA::cat mutant is named LB1281.
Complementation of an E. coli htrA mutant.
The htrA gene of C. jejuni NCTC11168, including 231-bp upstream sequences, was amplified in a PCR using chromosomal DNA extracted from C. jejuni NCTC11168 as a template and the primers htrA-start and htrA-end. The 1,740-bp fragment was cloned in the TOPO TA cloning vector pCR2.1 (Invitrogen), resulting in plasmid pLB262. E. coli htrA20 (BL20) and E. coli wild-type BL78 (28) were transformed with pLB262 and the parental plasmid pCR2.1. By selecting for kanamycin resistance, colonies that carried the plasmids were isolated. Single colonies of each strain were streaked on three different Luria-Bertani agar plates containing kanamycin, and after incubation overnight at 30, 39, or 42°C, the plates were inspected for growth. The experiment was repeated three times.
Adherence and invasion assay.
Adherence and invasion assays were performed with INT407 cell monolayers growing in Eagle's minimal essential medium (EMEM) supplemented with 10% fetal bovine serum at 37°C in a humidified atmosphere containing 5% CO2. Approximately 108 bacterial cells in EMEM were added to a monolayer consisting of 5 × 105 epithelial cells (multiplicity of infection, 200) and incubated for 2 h. For determination of adherence, the monolayers were washed three times with 10 mM phosphate-buffered saline (PBS), pH 7.2, and epithelial cells were lysed by adding 0.1% Triton X-100. The adhered bacteria were enumerated by plate count. For determination of invasion, the monolayers were subsequently incubated with EMEM containing gentamicin (250 μg/ml) for 2 h at 37°C in 5% CO2 to kill all extracellular bacteria. The monolayers were washed three times with PBS, epithelial cells were lysed by adding 0.1% Triton X-100, and the internalized bacteria were enumerated by plate count. The values represent the mean counts ± standard deviations derived from four wells. One representative experiment of four is shown.
Motility assay.
C. jejuni strains were grown overnight on base II agar plates at 37°C under microaerobic conditions. Bacterial cells were harvested using BHI broth, and the optical density at 600 nm (OD600) was adjusted to 0.1. One microliter of the cell suspension was deposited at the center of each heart infusion broth (Difco) plate containing 0.25% agar. After 48 h of microaerophilic incubation at 37°C, the ability of the strain to move in the soft agar was evaluated. Three independent experiments were performed.
Autoagglutination assay.
The ability to autoagglutinate was investigated essentially as described by Misawa and Blaser (30). Bacterial cells were harvested from base II agar plates with MilliQ water and washed once before the OD600 was adjusted to 1 in 10 mM PBS, pH 7.2. Cell suspensions of 4 ml were incubated at 25°C for 24 h, and the OD600 was measured by carefully removing 1 ml from the top phase. Bacterial cells which have autoagglutinated fall to the bottom of the tube, and the OD600 diminishes. The percentage decrease in OD600 was calculated, and the values represent the mean counts ± standard deviations derived from three independent assays.
Growth on plates.
C. jejuni NCTC11168 and LB1281 were grown overnight on base II agar plates at 37°C under microaerobic conditions. Bacterial cells were harvested using BHI broth, and the OD600 was adjusted to 0.1. Serial dilutions were made, and 10-μl volumes of the 10−1, 10−2, 10−3, 10−4, 10−5, and 10−6 dilutions were spotted onto four base II agar plates, which were incubated under microaerobic conditions using CampyGen (Oxoid) for 3 days at 37, 42, or 44°C or at 42°C in the presence of a lit candle. Alternatively, 10-μl volumes of the 10−1, 10−2, 10−3, 10−4, 10−5, and 10−6 dilutions were spotted onto a base II agar plate containing 5 μg/ml puromycin and incubated under microaerobic conditions for 4 days at 37°C. Experiments were repeated at least three times.
Protein analysis.
C. jejuni NCTC11168 and LB1281 were grown in BHI broth at 37°C or 42°C under microaerobic conditions. Cells (50 ml of a culture with an OD600 of 0.3) were harvested, washed with 0.9% NaCl, and resuspended in TM buffer (5 mM MgCl2,10 mM Tris-HCl [pH 7.5]) to the same protein concentration. A NuPage 4 to 12% Bis-Tris gel (Invitrogen) was used to analyze samples of protein extracts, followed by Coomassie staining with Safe Stain (Invitrogen). To detect DnaK immunologically, proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Invitrogen) and probed with rabbit polyclonal anti-DnaK (E. coli) (Abcam). Bound antibody was detected with the WesternBreeze chemiluminiscent anti-rabbit antibody kit (Invitrogen).
Electron microscopy.
C. jejuni NCTC11168 and LB1281 were grown overnight in sterile filtered Mueller-Hinton broth (Oxoid) under microaerobic conditions at 37°C. Bacteria were negatively stained with saturated aqueous uranyl acetate and photographed in a JEOL EX/B transmission microscope at 80 kV.
Northern blotting.
C. jejuni NCTC11168 was grown under microaerobic conditions in BHI broth to an OD600 of approximately 0.3 at 37°C; cells were cooled on ice and harvested by centrifugation for 5 min at 10,000 rpm at 4°C, and the pellet was resuspended in Tri agent (Sigma). Another portion of the cells was exposed to 46°C for 15 min, before the cells were harvested. RNA was isolated as described by Andersen and coworkers (1). Four micrograms of RNA was denatured at 75°C for 10 min and subsequently separated on 1% agarose gels in 10 mM sodium phosphate buffer as described by Pelle and Murphy (39). RNA was blotted onto PVDF membranes, and finally the RNA was fixed to the membrane by exposure to UV light. An internal 536-bp htrA fragment (obtained by PCR using primers htrA-intF and htrA-intR) was used as a probe. The probe was labeled with [α-32P]dCTP and Ready-To-Go labeling beads from Amersham Biosciences, and unincorporated nucleotides were subsequently removed by using ProbeQuant G-50 micro columns. Hybridization was performed overnight at 65°C in hybridization buffer (0.5 M sodium phosphate buffer, pH 7.2, 7% [wt/vol] sodium dodecyl sulfate [SDS]) after 1 h of prehybridization under the same conditions. To remove unspecific binding of the probes, the membranes were washed three times (15, 10, and 10 min) with 20 mM sodium phosphate buffer (pH 7.2)-1% (wt/vol) SDS. Membranes were autoradiographed at −80°C.
RESULTS
HtrA is expressed from a monocistronic transcript.
The open reading frame cj1228c of the C. jejuni NCTC11168 genome (accession number NP_002163) encodes an HtrA homologue. The amino acid sequence of the C. jejuni HtrA protein has the highest degree of identity to HtrA of H. pylori (49% identity) but also shows homology to HtrA proteins of other bacteria, e.g., E. coli (38% identity). By using the program SignalP V1.1 (34), we predicted that the C. jejuni HtrA protein possesses a 16-amino-acid leader peptide, suggesting a periplasmic location similar to that of E. coli HtrA (29, 43). The htrA gene (1,416 bp) of C. jejuni NCTC11168 is preceded by a Shine-Dalgarno sequence (AAGGAA) and two putative C. jejuni promoters that are located 27 and 82 bp upstream of the translational start site (data not shown). Because the upstream gene (cbpA) is divergently transcribed and a putative rho-independent terminator (ATAACGGAAGGATTTTCCTTTCGTTAT; ΔG = −13 kcal/mol) is located downstream of the htrA gene, the htrA gene is suggested to be not in an operon with the neighboring genes. This possibility was verified by Northern blot analysis using an internal htrA fragment as a probe, as the only hybridization found was to a transcript that corresponded in size to an mRNA containing htrA (data not shown).
HtrA is required for growth under conditions that increase the level of nonnative protein.
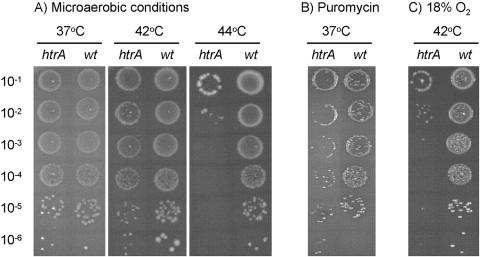
With the aim of studying the role of HtrA in stress tolerance and virulence in C. jejuni, we constructed an htrA mutant in which the main part of the htrA (cj1228c) gene was replaced with a selective marker. A total of 436 amino acids, including the catalytic triad (His120-Asp151-Ser225) and the two PDZ domains, were removed (for details, see Materials and Methods). Previous studies of HtrA homologues in various bacterial species have pointed to a role of HtrA in heat tolerance in some organisms but not in others (10, 11, 19, 27, 28, 58). In C. jejuni we found that while the htrA mutant formed colonies with the same frequency as the wild type did at 37°C and 42°C, the ability of the htrA mutant to form colonies at 44°C was greatly reduced (104-fold) (Fig. 1A). In contrast, the wild type formed colonies with the same frequency at all three temperatures (Fig. 1A). To determine if HtrA is required generally for growth under conditions in which the nonnative proteins accumulate, we compared the abilities of the wild type and the htrA mutant to form colonies in the presence of puromycin. This antibiotic increases the level of misfolded proteins by competing with charged tRNAs for the ribosomal A-site, thus leading to premature termination of protein synthesis (14). When growth was investigated using plates containing 5 μg/ml puromycin, we found that the htrA mutant formed fewer and smaller colonies than the wild type (Fig. 1B). Thus, our results suggest that in C. jejuni, HtrA is required for growth under conditions in which misfolded proteins accumulate.
FIG. 1.
Effect of temperature, oxygen concentration, and puromycin on growth of the C. jejuni htrA mutant on solid surfaces. Serial dilutions (10−1, 10−2, 10−3, 10−4, 10−5, and 10−6) of C. jejuni NCTC11168 (wt) and LB1281 (htrA) cultures with an OD600 of 0.1 were spotted in 10-μl volumes onto base II agar plates. The plates were incubated at 37, 42, or 44°C under microaerobic conditions (A), at 37°C under microaerobic conditions on a base II agar plate containing 5 μg/ml puromycin (B), or at 42°C in a candle jar providing 17 to 18% oxygen (C). One representative of three independent experiments is presented.
Complementation of an E. coli htrA mutant.
Our results suggest that HtrA of C. jejuni may retain the same function as E. coli HtrA, since an E. coli htrA mutant is also temperature and puromycin sensitive (28). To investigate whether C. jejuni HtrA was able to complement an E. coli htrA mutant, we supplied a fragment containing the C. jejuni htrA gene and the putative promoter on a plasmid (pLB262) and examined growth at 30, 39, and 42°C (Fig. 2). While the E. coli htrA mutant containing the vector plasmid pCR2.1 was able to form colonies only at 30°C, we found that the sequences present in pLB262 were able to complement the temperature-sensitive phenotype of the E. coli htrA mutant at 39°C but not at 42°C (Fig. 2). The incomplete suppression may be due to an inappropriate expression level of the C. jejuni htrA gene or that the HtrA protein of C. jejuni, to some extent, is different from E. coli HtrA (38% identity). However, the partial suppression of the temperature-sensitive phenotype of the E. coli htrA mutant indicates that htrA of C. jejuni is a functional homologue of the htrA gene from E. coli.
FIG. 2.
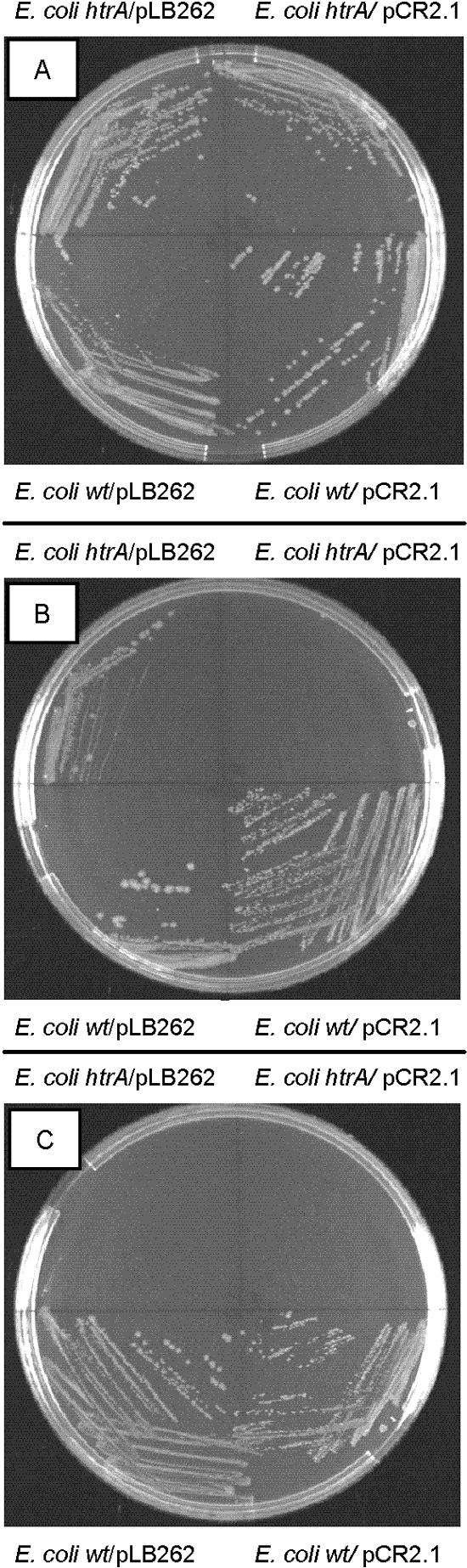
Complementation of an E. coli htrA mutant. E. coli strains (wt or htrA21 mutant) containing pLB262 (C. jejuni htrA) or pCR2.1 (vector) were streaked on Luria-Bertani agar plates containing kanamycin (50 μg/ml) and incubated at 30°C (A), 39°C (B), or 42°C (C) overnight.
The cytoplasmic heat shock response is induced in the absence of HtrA.
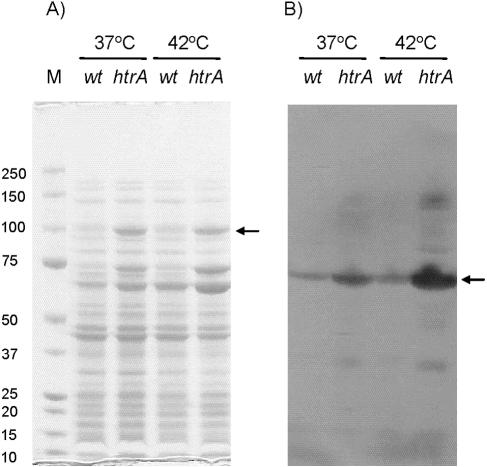
During our initial characterization of the C. jejuni htrA mutant, we observed that the pattern of proteins extracted from htrA mutant cells was different from that of wild-type cells. In particular, protein bands with approximate sizes of 100, 70, and 60 kDa were more intense in cells lacking HtrA than in the wild-type cells (Fig. 3A). Protein sequence analysis revealed that the 100-kDa protein corresponds to the ClpB chaperone, which in E. coli is required for the resolution of protein aggregates (31). The 70-kDa band specifically reacted with DnaK antibodies raised against E. coli DnaK, thus identifying the 70-kDa band as DnaK (Fig. 3B). DnaK is a chaperone, which in E. coli refolds nonnative proteins and, in cooperation with ClpB, disaggregates protein aggregates (3). The predicted molecular sizes of C. jejuni ClpB and DnaK are 95.6 and 67.4 kDa, and these sizes correspond well to the observed sizes. Furthermore, the Western blot revealed that in wild-type cells, equal amounts of DnaK were present at 37 and 42°C (Fig. 3B, lanes 1 and 3). In contrast, growth at 42°C resulted in an increase in the amount of DnaK in the htrA mutant compared to that at 37°C (Fig. 3B, lanes 1 and 3). The protein present in the 60-kDa protein band was not identified; however, the size corresponds to the predicted size of the C. jejuni groEL protein product (58 kDa) which encodes a third chaperone.
FIG. 3.
Effect of the htrA mutation on protein composition of C. jejuni. Proteins were extracted from C. jejuni NCTC11168 (wt) and LB1281 (htrA) cells growing in exponential phase (OD600 = 0.3) at 37 or 42°C. (A) Extracted proteins were separated on a SDS-polyacrylamide gel electrophoresis gel, and the 100-kDa band (arrow) was cut out and identified by mass spectrometry as ClpB. Lane M, molecular weight markers. (B) In a Western blot, proteins were transferred to a PVDF membrane and probed with rabbit polyclonal anti-DnaK (E. coli). An arrow indicates the DnaK band.
The htrA mutant is sensitive to oxygen but not to H2O2, paraquat, or cumene hydroperoxide.
In previous studies, HtrA has been shown to be important for oxidative stress tolerance in several gram-negative pathogenic bacteria (10, 12, 19, 27, 58). Therefore we compared the sensitivities of the C. jejuni htrA mutant and wild-type cells to oxidative stress inducers by using a disk diffusion assay. When exposed to H2O2, cumene hydroperoxide, or paraquat, the mutant and wild-type cells were equally sensitive, and this finding was confirmed by survival assays (data not shown).
Since reactive oxygen species may also be produced from oxygen via metabolism, we investigated oxygen sensitivity by incubation of htrA and wild-type cells in an atmosphere with a lit candle, in which the oxygen concentration is around 17 to 18% (55). After 3 days of incubation at 42°C, we found that the htrA mutant formed colonies at a lower frequency than the wild type did (Fig. 1C), suggesting that the htrA mutant is oxygen sensitive. In addition, the viability of the htrA mutant was reduced approximately 500-fold compared to that of the wild type when liquid cultures were exposed to vigorous shaking in atmospheric air for 6 h, thus confirming that the htrA mutant has a reduced oxygen tolerance. Since iron catalyzes reactions that lead to formation of reactive oxygen species, we investigated whether the reduced oxygen tolerance was dependent on iron. However, the presence of the iron chelator desferal did not eliminate the inhibitory effect of oxygen, nor did additional iron decrease the oxygen tolerance further (data not shown). These results suggest that the toxic effect of oxygen on the htrA mutant is mediated mainly via an iron-independent reaction.
HtrA is important for adherence to and invasion of INT407 epithelial cells.
Although C. jejuni is considered to be primarily an extracellular pathogen, the ability to adhere to and invade epithelial cells is also believed to be part of its pathogenesis (23). With the aim of determining whether HtrA plays a role in the interaction between C. jejuni and human epithelial cells, we compared the abilities of htrA mutant cells and wild-type cells to adhere to and invade the human epithelial cell line INT407. We found that when HtrA was absent, both adhesion to and invasion of the epithelial cells were reduced compared to those of the wild-type strain of C. jejuni (Fig. 4A and B). The sensitivity to gentamicin and Triton-X of the htrA mutant was not significantly affected compared to that of the wild type (data not shown). Even though the reduced adherence and invasion of the htrA mutant may be an indirect effect of C. jejuni lacking HtrA, our results show that HtrA is important for both adherence to and invasion of INT407 epithelial cells.
FIG. 4.
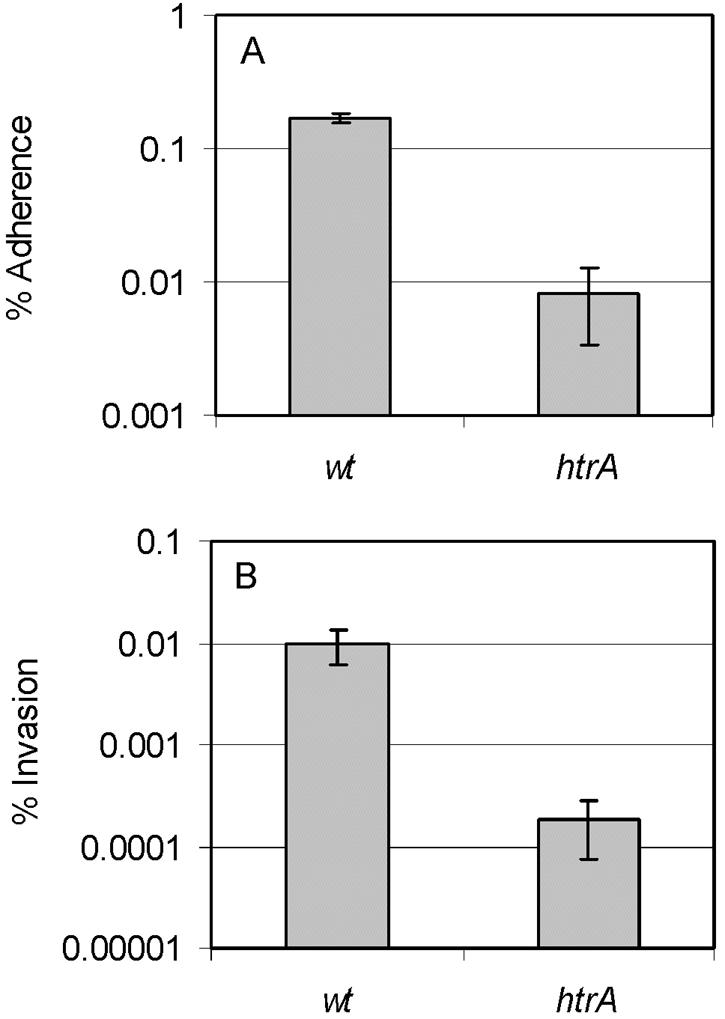
Effect of the htrA mutation on the ability of C. jejuni to adhere to and invade cultured INT407 intestinal cells. Adhered (A) and invaded (B) cells are expressed as percentages of those in inocula. The values represent the means (grey bar) and standard deviations (thin lines) of results for four wells. One representative of three independent experiments is presented.
To investigate whether the lack of HtrA affects the human response mediated by the complement system, we measured the viabilities of the htrA mutant and the wild type after incubation in human serum. The complement system present in human serum is part of the human defense system against infection by gram-negative bacteria, which are killed through interaction with specific components of the complement. While a Klebsiella pneumoniae htrA mutant has decreased resistance to human serum (10), we found that the viability of the htrA mutant in human serum was not significantly different from that of the wild-type C. jejuni strain NCTC11168, regardless of whether the complement was inactivated by heat treatment (data not shown).
HtrA is not required for autoagglutination and motility, but lack of HtrA alters the morphology of C. jejuni.
Since the ability to self-associate or autoagglutinate has been associated with virulence in some gram-negative pathogenic bacteria such as enteropathogenic E. coli (4) and Y. enterocolitica (41), we determined the abilities of the C. jejuni wild type and htrA mutant to autoagglutinate. Because bacterial cells in suspension fall to the bottom of the tube when they autoagglutinate, the reduction in OD600 in a suspension of cells adjusted to an OD600 of 1 after 24 h of static incubation at 25°C is a measure of autoagglutination. However, a lack of HtrA did not affect the ability of C. jejuni to autoagglutinate, since the OD600 of the htrA mutant was reduced to 9.1% ± 0.4%, a result similar to that of the wild type (9.7% ± 1.7%).
Previous studies have shown that lack of the major flagellar subunit FlaA of C. jejuni reduces adherence and invasion of C. jejuni into INT407 epithelial cells, suggesting that motility is associated with virulence (56, 59). When investigating motility in a soft agar assay, we found that the htrA mutant and wild-type cells were equally motile, indicating that neither motility nor chemotaxis is affected by deletion of htrA (data not shown). This notion was confirmed by phase-contrast microscopy, in which the htrA mutant and wild-type C. jejuni appeared equally motile (data not shown). However, when we examined the morphology of the htrA mutant cells by using electron microscopy, we repeatedly observed that the htrA mutant cells were longer and less curved than the wild-type cells but carried flagella resembling those of the wild type (data not shown).
DISCUSSION
The food-borne pathogen C. jejuni continues to be a public health problem all over the world. The persistence of C. jejuni in the different environments that the bacterium experiences during the contamination cycle may be a consequence of a significant ability to adapt to and survive these conditions. In the present study, we have investigated the role of the HtrA protease in the stress tolerance of C. jejuni. We showed that the HtrA protease is required for both heat and oxygen tolerance, and our results furthermore indicate that HtrA is important for interaction with human epithelial cells, suggesting that HtrA contributes to the successful survival of C. jejuni during the contamination cycle. In contrast to data presented here, it was previously noted that a C. jejuni htrA mutant did not display any obvious phenotype (7, 36). However, since there are no details available concerning construction of this mutant and the physiological conditions used, it is not possible to pinpoint the reason for this apparent discrepancy between our results and those of previous studies.
In the present communication, we show that in C. jejuni, HtrA is required for growth at high temperatures and that the htrA mutant is impaired in growth in the presence of puromycin. Since we predict that HtrA of C. jejuni is located in the periplasmic space, we propose that the temperature sensitivity of the htrA mutant is a result of the accumulation of misfolded protein in the periplasm and that C. jejuni HtrA, like E. coli HtrA (29, 43, 49), is involved in the degradation of nonnative proteins in this cellular compartment. Thus, we suggest that HtrA of C. jejuni is a functional homologue of HtrA in E. coli, which is supported by the complementation of the temperature-sensitive phenotype of an E. coli htrA mutant. Interestingly, we found that a lack of HtrA increased the cellular content of the cytoplasmic chaperones ClpB and DnaK, which are conserved chaperones involved in the refolding of heat-denatured proteins and solubilization of larger protein aggregates by the expenditure of ATP in E. coli (3, 31). In general, the heat shock response is induced in response to an unfolded protein signal, and the increased levels of the ClpB and DnaK chaperones in the htrA mutant thus indicate that a nonnative protein signal is elicited in the absence of HtrA in C. jejuni. In contrast, increased amounts of ClpB and DnaK are not, to our knowledge, found in an E. coli htrA mutant, suggesting that the heat shock response is not induced. While C. jejuni lacks homologues of the heat shock sigma factor and the sigma factor responding to envelope stress (37), the genome of C. jejuni encodes two homologues of the negative heat shock regulatory proteins HrcA and HspR that are found mostly in gram-positive bacteria (33). Analysis of a C. jejuni hspR mutant has revealed that the expression of dnaK, groEL, and clpB is controlled by HspR (1), while binding sites for HrcA are found only upstream of dnaK and groEL, suggesting that the nonnative protein signal elicited in the absence of HtrA might be communicated to the heat shock genes via HspR.
Questions arise as to whether the accumulation of misfolded proteins in the periplasm increases the level of cytoplasmic chaperones and how exactly the signal is transmitted from periplasm to cytoplasm. The fact that a clpB mutation could not be transduced to an E. coli htrA mutant stresses the biological requirement for ClpB in the absence of HtrA (25). In E. coli, blocking of protein export leads to an intracellular accumulation of precursor molecules of the envelope proteins, which was found to induce heat shock proteins (17). Similarly, when C. jejuni lacks HtrA, nonnative proteins that accumulate in the periplasm may inhibit the protein translocation systems located in the inner membrane, leading to stalling of unfolded precursor proteins in the cytoplasm and induction of the heat shock proteins, including ClpB and DnaK. Alternatively, the accumulation of misfolded proteins in the periplasm is sensed and communicated via specific signal transduction systems to the cytoplasm of C. jejuni. In E. coli, the presence of misfolded proteins in the periplasm is sensed by the two-component systems Cpx or Bae or by the RseA/DegS system (40, 60). However, examination of the C. jejuni genome failed to reveal homologues of RseA or the HtrA-like protease DegS. In addition, we were unable to find close homologues of the Cpx or Bae two-component systems. Thus, further work to identify the molecular mechanisms by which the nonnative protein signal is transmitted from periplasm to cytoplasm will be needed.
In contrast to the growth and survival of bacterial htrA mutants of other pathogenic gram-negative bacteria (11, 19, 27, 58), those of the C. jejuni htrA mutant were not reduced by oxidative stress inducers. Conversely, the C. jejuni htrA mutant showed reduced oxygen tolerance, a phenotype that, to our knowledge, has not been previously associated with bacterial htrA mutants. Even though C. jejuni is a microaerobic organism, it is able to perform aerobic respiration (45), and an increase in oxygen tension might result in the accumulation of oxidatively damaged proteins in the periplasm that require HtrA for removal. Also a C. jejuni strain with a mutation in the fdxA gene, encoding a putative ferredoxin that promotes one-electron transfer from a highly negative potential, shows reduced oxygen tolerance but no sensitivity towards H2O2 or cumene hydroperoxide (54). A lack of ferredoxin may interfere with the respiration process, as the small iron-sulfur proteins may be involved in electron transfer in the respiratory pathways.
By the use of tissue cultures of human epithelial cells (INT407), we demonstrated that both adherence to and invasion of the htrA mutant were severely reduced compared to those of the wild-type C. jejuni, showing that HtrA has an impact on the interaction with human epithelial cells in vitro. Although we did not include a C. jejuni mutant containing a cat gene in a neutral position in the assay, it is our experience (based on other cat mutants) that the impact on invasion and adherence is not likely an effect of the cat gene itself. In C. jejuni, several factors have been shown to be important for in vitro adherence and invasion. C. jejuni mutants lacking the major flagellar subunit FlaA are less motile and show a reduced ability to adhere to and invade intestinal cells in vitro (15, 18, 56). However, the impaired adherence and invasion of the htrA mutant cannot be explained by reduced motility, since the htrA mutation does not affect these parameters. But surface components other than flagella, for example, the outer membrane proteins PebA1 and CadF, seem to be involved in C. jejuni adherence to epithelial cells (21, 38). Although we did not observe differences in outer membrane-associated proteins between the htrA mutant and the wild-type strain on an SDS-polyacrylamide gel electrophoresis gel, the surface components of the htrA mutant may be different (data not shown). Recent results indicate that the morphology of C. jejuni affects invasion in INT-407 epithelial cells, as a straight variant invades with a lower frequency than one with a spiral shape (13); thus, the changed morphology of the htrA mutant may result in reduced adherence to and invasion of the human epithelial cells.
Acknowledgments
We thank Dorte Frees and Line Elnif Thomsen for critical reading of the manuscript. Knut Buettner, Department of Microbiology and Molecular Biology, Ernst-Moritz-Arndt University, Greifswald, Germany, is thanked for identifying ClpB. Patricia Guerry is thanked for providing plasmid pRY109.
The Danish Directorate for Food, Fisheries and Agro Business and The Royal Veterinary and Agricultural University, Denmark, supported this work.
REFERENCES
- 1.Andersen, M. T., L. Brøndsted, B. Pearson, F. Mulholland, M. Parker, C. Pin, J. Wells, and H. Ingmer. 2004. Diverse roles for HspR in Campylobacter jejeuni revealed by the proteome, transcriptome and phenotypic characterisation of an hspR mutant. Microbiology 151:905-915. [DOI] [PubMed]
- 2.Bakker, D., C. E. Vader, B. Roosendaal, F. R. Mooi, B. Oudega, and F. K. de Graaf. 1991. Structure and function of periplasmic chaperone-like proteins involved in the biosynthesis of K88 and K99 fimbriae in enterotoxigenic Escherichia coli. Mol. Microbiol. 5:875-886. [DOI] [PubMed] [Google Scholar]
- 3.Ben Zvi, A. P., and P. Goloubinoff. 2001. Review: mechanisms of disaggregation and refolding of stable protein aggregates by molecular chaperones. J. Struct. Biol. 135:84-93. [DOI] [PubMed] [Google Scholar]
- 4.Bieber, D., S. W. Ramer, C. Y. Wu, W. J. Murray, T. Tobe, R. Fernandez, and G. K. Schoolnik. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114-2118. [DOI] [PubMed] [Google Scholar]
- 5.Birk, T., H. Ingmer, M. T. Andersen, K. Jørgensen, and L. Brøndsted. 2004. Chicken juice, a food-based model system suitable to study survival of Campylobacter jejuni. Lett. Appl. Microbiol. 38:66-71. [DOI] [PubMed] [Google Scholar]
- 6.Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157:472-479. [DOI] [PubMed] [Google Scholar]
- 7.Boer, P., J. A. Wagenaar, R. P. Achterberg, J. P. Putten, L. M. Schouls, and B. Duim. 2002. Generation of Campylobacter jejuni genetic diversity in vivo. Mol. Microbiol. 44:351-359. [DOI] [PubMed] [Google Scholar]
- 8.Cavard, D., C. Lazdunski, and S. P. Howard. 1989. The acylated precursor form of the colicin A lysis protein is a natural substrate of the DegP protease. J. Bacteriol. 171:6316-6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatfield, S. N., K. Strahan, D. Pickard, I. G. Charles, C. E. Hormaeche, and G. Dougan. 1992. Evaluation of Salmonella typhimurium strains harbouring defined mutations in htrA and aroA in the murine salmonellosis model. Microb. Pathog. 12:145-151. [DOI] [PubMed] [Google Scholar]
- 10.Cortes, G., B. de Astorza, V. J. Benedi, and S. Alberti. 2002. Role of the htrA gene in Klebsiella pneumoniae virulence. Infect. Immun. 70:4772-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elzer, P. H., R. W. Phillips, M. E. Kovach, K. M. Peterson, and R. M. Roop. 1994. Characterization and genetic complementation of a Brucella abortus high-temperature-requirement A (htrA) deletion mutant. Infect. Immun. 62:4135-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elzer, P. H., R. W. Phillips, G. T. Robertson, and R. M. Roop. 1996. The HtrA stress response protease contributes to resistance of Brucella abortus to killing by murine phagocytes. Infect. Immun. 64:4838-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaynor, E. C., S. Cawthraw, G. Manning, J. K. MacKichan, S. Falkow, and D. G. Newell. 2004. The genome-sequenced variant of Campylobacter jejuni NCTC 11168 and the original clonal clinical isolate differ markedly in colonization, gene expression, and virulence-associated phenotypes. J. Bacteriol. 186:503-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldberg, A. L. 1972. Correlation between rates of degradation of bacterial proteins in vivo and their sensitivity to proteases. Proc. Natl. Acad. Sci. USA 69:2640-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guerry, P., R. A. Alm, M. E. Power, S. M. Logan, and T. J. Trust. 1991. Role of two flagellin genes in Campylobacter motility. J. Bacteriol. 173:4757-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horton, R. M., S. N. Ho, J. K. Pullen, H. D. Hunt, Z. Cai, and L. R. Pease. 1993. Gene splicing by overlap extension. Methods Enzymol. 217:270-279. [DOI] [PubMed] [Google Scholar]
- 17.Ito, K., Y. Akiyama, T. Yura, and K. Shiba. 1986. Diverse effects of the MalE-LacZ hybrid protein on Escherichia coli cell physiology. J. Bacteriol. 167:201-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jagannathan, A., C. Constantinidou, and C. W. Penn. 2001. Roles of rpoN, fliA, and flgR in expression of flagella in Campylobacter jejuni. J. Bacteriol. 183:2937-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, K., I. Charles, G. Dougan, D. Pickard, P. O'Gaora, G. Costa, T. Ali, I. Miller, and C. Hormaeche. 1991. The role of a stress-response protein in Salmonella typhimurium virulence. Mol. Microbiol. 5:401-407. [DOI] [PubMed] [Google Scholar]
- 20.Jones, C. H., P. N. Danese, J. S. Pinkner, T. J. Silhavy, and S. J. Hultgren. 1997. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J. 16:6394-6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konkel, M. E., S. G. Garvis, S. L. Tipton, D. E. Anderson, Jr., and W. Cieplak, Jr. 1997. Identification and molecular cloning of a gene encoding a fibronectin-binding protein (CadF) from Campylobacter jejuni. Mol. Microbiol. 24:953-963. [DOI] [PubMed] [Google Scholar]
- 22.Konkel, M. E., B. J. Kim, J. D. Klena, C. R. Young, and R. Ziprin. 1998. Characterization of the thermal stress response of Campylobacter jejuni. Infect. Immun. 66:3666-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konkel, M. E., M. R. Monteville, V. Rivera-Amill, and L. A. Joens. 2001. The pathogenesis of Campylobacter jejuni-mediated enteritis. Curr. Issues Intest. Microbiol. 2:55-71. [PubMed] [Google Scholar]
- 24.Krojer, T., M. Garrido-Franco, R. Huber, M. Ehrmann, and T. Clausen. 2002. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416:455-459. [DOI] [PubMed] [Google Scholar]
- 25.Laskowska, E., D. Kuczynska-Wisnik, J. Skorko-Glonek, and A. Taylor. 1996. Degradation by proteases Lon, Clp and HtrA, of Escherichia coli proteins aggregated in vivo by heat shock; HtrA protease action in vivo and in vitro. Mol. Microbiol. 22:555-571. [DOI] [PubMed] [Google Scholar]
- 26.Lee, A., S. C. Smith, and P. J. Coloe. 1998. Survival and growth of Campylobacter jejuni after artificial inoculation onto chicken skin as a function of temperature and packaging conditions. J. Food Prot. 61:1609-1614. [DOI] [PubMed] [Google Scholar]
- 27.Li, S. R., N. Dorrell, P. H. Everest, G. Dougan, and B. W. Wren. 1996. Construction and characterization of a Yersinia enterocolitica O:8 high-temperature requirement (htrA) isogenic mutant. Infect. Immun. 64:2088-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lipinska, B., O. Fayet, L. Baird, and C. Georgopoulos. 1989. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J. Bacteriol. 171:1574-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipinska, B., M. Zylicz, and C. Georgopoulos. 1990. The HtrA (DegP) protein, essential for Escherichia coli survival at high temperatures, is an endopeptidase. J. Bacteriol. 172:1791-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Misawa, N., and M. J. Blaser. 2000. Detection and characterization of autoagglutination activity by Campylobacter jejuni. Infect. Immun. 68:6168-6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mogk, A., and B. Bukau. 2004. Molecular chaperones: structure of a protein disaggregase. Curr. Biol. 14:R78-R80. [DOI] [PubMed] [Google Scholar]
- 32.Nachamkin, I., B. M. Allos, and T. Ho. 1998. Campylobacter species and Guillain-Barré syndrome. Clin. Microbiol. Rev. 11:555-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narberhaus, F. 1999. Negative regulation of bacterial heat shock genes. Mol. Microbiol. 31:1-8. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst. 8:581-599. [DOI] [PubMed] [Google Scholar]
- 35.Notermans, S., and A. Hoogenboom-Verdegaal. 1992. Existing and emerging foodborne diseases. Int. J. Food Microbiol. 15:197-205. [DOI] [PubMed] [Google Scholar]
- 36.Pallen, M. J., and B. W. Wren. 1997. The HtrA family of serine proteases. Mol. Microbiol. 26:209-221. [DOI] [PubMed] [Google Scholar]
- 37.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 38.Pei, Z., C. Burucoa, B. Grignon, S. Baqar, X. Z. Huang, D. J. Kopecko, A. L. Bourgeois, J. L. Fauchere, and M. J. Blaser. 1998. Mutation in the peb1A locus of Campylobacter jejuni reduces interactions with epithelial cells and intestinal colonization of mice. Infect. Immun. 66:938-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pelle, R., and N. B. Murphy. 1993. Northern hybridization: rapid and simple electrophoretic conditions. Nucleic Acids Res. 21:2783-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raffa, R. G., and T. L. Raivio. 2002. A third envelope stress signal transduction pathway in Escherichia coli. Mol. Microbiol. 45:1599-1611. [DOI] [PubMed] [Google Scholar]
- 41.Roggenkamp, A., H. R. Neuberger, A. Flugel, T. Schmoll, and J. Heesemann. 1995. Substitution of two histidine residues in YadA protein of Yersinia enterocolitica abrogates collagen binding, cell adherence and mouse virulence. Mol. Microbiol. 16:1207-1219. [DOI] [PubMed] [Google Scholar]
- 42.Sassoon, N., J. P. Arie, and J. M. Betton. 1999. PDZ domains determine the native oligomeric structure of the DegP (HtrA) protease. Mol. Microbiol. 33:583-589. [DOI] [PubMed] [Google Scholar]
- 43.Skorko-Glonek, J., B. Lipinska, K. Krzewski, G. Zolese, E. Bertoli, and F. Tanfani. 1997. HtrA heat shock protease interacts with phospholipid membranes and undergoes conformational changes. J. Biol. Chem. 272:8974-8982. [DOI] [PubMed] [Google Scholar]
- 44.Skorko-Glonek, J., A. Wawrzynow, K. Krzewski, K. Kurpierz, and B. Lipinska. 1995. Site-directed mutagenesis of the HtrA (DegP) serine protease, whose proteolytic activity is indispensable for Escherichia coli survival at elevated temperatures. Gene 163:47-52. [DOI] [PubMed] [Google Scholar]
- 45.Smith, M. A., M. Finel, V. Korolik, and G. L. Mendz. 2000. Characteristics of the aerobic respiratory chains of the microaerophiles Campylobacter jejuni and Helicobacter pylori. Arch. Microbiol. 174:1-10. [DOI] [PubMed] [Google Scholar]
- 46.Spiers, A., H. K. Lamb, S. Cocklin, K. A. Wheeler, J. Budworth, A. L. Dodds, M. J. Pallen, D. J. Maskell, I. G. Charles, and A. R. Hawkins. 2002. PDZ domains facilitate binding of high temperature requirement protease A (HtrA) and tail-specific protease (Tsp) to heterologous substrates through recognition of the small stable RNA A (ssrA)-encoded peptide. J. Biol. Chem. 277:39443-39449. [DOI] [PubMed] [Google Scholar]
- 47.Spiess, C., A. Beil, and M. Ehrmann. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339-347. [DOI] [PubMed] [Google Scholar]
- 48.Stintzi, A. 2003. Gene expression profile of Campylobacter jejuni in response to growth temperature variation. J. Bacteriol. 185:2009-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strauch, K. L., K. Johnson, and J. Beckwith. 1989. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J. Bacteriol. 171:2689-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thies, F. L., H. P. Hartung, and G. Giegerich. 1998. Cloning and expression of the Campylobacter jejuni lon gene detected by RNA arbitrarily primed PCR. FEMS Microbiol. Lett. 165:329-334. [DOI] [PubMed] [Google Scholar]
- 51.Thies, F. L., H. Karch, H. P. Hartung, and G. Giegerich. 1999. Cloning and expression of the dnaK gene of Campylobacter jejuni and antigenicity of heat shock protein 70. Infect. Immun. 67:1194-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thies, F. L., H. Karch, H. P. Hartung, and G. Giegerich. 1999. The ClpB protein from Campylobacter jejuni: molecular characterization of the encoding gene and antigenicity of the recombinant protein. Gene 230:61-67. [DOI] [PubMed] [Google Scholar]
- 53.Thies, F. L., A. Weishaupt, H. Karch, H. P. Hartung, and G. Giegerich. 1999. Cloning, sequencing and molecular analysis of the Campylobacter jejuni groESL bicistronic operon. Microbiology 145:89-98. [DOI] [PubMed] [Google Scholar]
- 54.van Vliet, A. H., M. A. Baillon, C. W. Penn, and J. M. Ketley. 2001. The iron-induced ferredoxin FdxA of Campylobacter jejuni is involved in aerotolerance. FEMS Microbiol. Lett. 196:189-193. [DOI] [PubMed] [Google Scholar]
- 55.Wang, W. L., N. W. Luechtefeld, M. J. Blaser, and L. B. Reller. 1983. Effect of incubation atmosphere and temperature on isolation of Campylobacter jejuni from human stools. Can. J. Microbiol. 29:468-470. [DOI] [PubMed] [Google Scholar]
- 56.Wassenaar, T. M., N. M. Bleumink-Pluym, and B. A. van der Zeijst. 1991. Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. EMBO J. 10:2055-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wassenaar, T. M., B. N. Fry, and B. A. van der Zeijst. 1993. Genetic manipulation of Campylobacter: evaluation of natural transformation and electro-transformation. Gene 132:131-135. [DOI] [PubMed] [Google Scholar]
- 58.Yamamoto, T., T. Hanawa, S. Ogata, and S. Kamiya. 1996. Identification and characterization of the Yersinia enterocolitica gsrA gene, which protectively responds to intracellular stress induced by macrophage phagocytosis and to extracellular environmental stress. Infect. Immun. 64:2980-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao, R., D. H. Burr, P. Doig, T. J. Trust, H. Niu, and P. Guerry. 1994. Isolation of motile and non-motile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol. Microbiol. 14:883-893. [DOI] [PubMed] [Google Scholar]
- 60.Young, J. C., and F. U. Hartl. 2003. A stress sensor for the bacterial periplasm. Cell 113:1-2. [DOI] [PubMed] [Google Scholar]