Abstract
A two-step multiplex PCR-based method was designed for the rapid detection of 16 species of lactobacilli known to be commonly present in sourdough. The first step of multiplex PCR was developed with a mixture of group-specific primers, while the second step included three multiplex PCR assays with a mixture of species-specific primers. Primers were derived from sequences that specify the 16S rRNA, the 16S-23S rRNA intergenic spacer region, and part of the 23S rRNA gene. The primer pairs designed were shown to exclusively amplify the targeted rrn operon fragment of the corresponding species. Due to the reliability of simultaneously identifying Lactobacillus plantarum, Lactobacillus pentosus, and Lactobacillus paraplantarum, a previously described multiplex PCR method employing recA gene-derived primers was included in the multiplex PCR system. The combination of a newly developed, quick bacterial DNA extraction method from sourdough and this multiplex PCR assay allows the rapid in situ detection of several sourdough-associated lactobacilli, including the recently described species Lactobacillus rossii, and thus represents a very useful alternative to culture-based methodologies.
A successful sourdough fermentation plays a pivotal role in the development of the characteristic flavor and physical attributes of the resulting bread (28). The traditional preparation and use of a sourdough rely on repeated bacterial propagation of variable frequency, during which a selection occurs that usually leads to the establishment of one or two species of lactic acid bacteria (LAB) at numbers that are three to four orders of magnitude above those of the adventitious microbial flora (25, 26). In a mature sourdough, a mixed population of different LAB and yeasts represent the characteristic fermenting microflora (15). Typical sourdough LAB mainly belong to the genus Lactobacillus and include obligately and facultatively heterofermentative and obligately homofermentative species (27). In recent years a number of new species of lactobacilli isolated from wheat and rye sourdoughs have been described, Lactobacillus paralimentarius (7), Lactobacillus frumenti (44), Lactobacillus mindensis (17), L. rossii (11), Lactobacillus hammesii (59), and Lactobacillus acidifarinae and Lactobacillus zymae (60) being the most recent.
The identification of LAB, when exclusively based on physiological and biochemical characteristics, is intrinsically ambiguous and to a degree unreliable (30). Phenotypic analysis represents the most tedious task during the process of microorganism identification, as it requires time, skills, and, in order to avoid subjective observations, technical standardization (51). Moreover, especially for LAB, similar nutritional requirements of different species due to adaptation to a particular environment hamper identification by traditional methods (2, 24). Since the interest in using defined LAB cultures as starters in sourdough fermentation is growing (25), while also recognizing that different Lactobacillus spp. can affect the sourdough ecosystem in different ways, it is evident that a need exists for reliable and rapid detection techniques for such sourdough lactobacilli.
Genotype-based methods are useful to identify bacteria as a complement or alternative to phenotypic methods. Identification based on PCR amplification of targeted genes is a reliable technique. Several bacterial species identification methodologies using primers that target different sequences, such as the 16S rRNA-encoding gene (31, 40, 41), the 16S-23S rRNA intergenic spacer region (ISR) (4, 5, 19, 50, 52, 54, 58), the 23S rRNA-encoding DNA (35, 42), as well as the recA (9, 33, 34, 49, 57) and ldhD (43) genes, have been reported in literature.
The 16S rRNA approach is one of the most widely used standard techniques to infer phylogenetic relationships among bacteria (51) but is sometimes insufficient to distinguish closely related species (47, 57). The DNA encoding the 23S rRNA subunit represents a larger (ca. 3,300 nucleotides) source of genomic information than the 16S one (ca. 1,650 nucleotides), but due to its length, very few 23S rRNA sequences are available in databases. The 16S-23S rRNA ISR sequence is much more variable than that of the 16S rRNA structural gene, both in size and sequence, even within closely related taxonomic groups (23), which makes it a suitable target for typing microbial populations using species-specific primers (3). Length variability is mainly due to the presence (not universal) of important units such as tRNA genes (number and type may vary among species), RNase III gene, or boxA, characterized by a very low degree of nucleotide changes (19).
Monoplex PCR strategies are not suitable for the study of complex flora consisting of multiple bacterial species. To rapidly detect multiple bacteria in a single reaction, simultaneous amplification of more than one locus is required, a methodology referred to as multiplex PCR (8). Multiplex PCR targeting different genes has been successfully applied to identify different groups of bacteria (6, 29, 36, 37, 39, 43, 46, 48, 61, 62), including lactobacilli (38, 45, 55, 57, 63). Recently, the identification tools for lactobacilli of cheeses and other dairy products have been reviewed by Coeuret et al. (10), and it has become clear that multiplex PCR represents an extremely useful tool to quickly obtain reliable information on composition and population dynamics of a complex microbial community. In the particular case of sourdough fermentation, multiplex PCR may be used to trace key LAB which had been added as starter or those that emerge naturally during sourdough fermentation.
The objectives of this study were to develop a two-step multiplex PCR strategy for the rapid identification of 16 Lactobacillus spp. commonly associated with sourdough fermentation; develop a rapid and simple method to extract total microbial DNA from sourdough; and detect sourdough lactobacilli by means of a culture-independent strategy based on the above multiplex PCR assays.
MATERIALS AND METHODS
Strains and growth conditions.
The 16 reference strains of the genus Lactobacillus reported in Table 1 represent Lactobacillus species commonly found in sourdough. Twelve Lactobacillus strains isolated from sourdoughs and identified previously (L. rossii CI35, CR20, CD76, and CF51, Lactobacillus brevis 5Z, L. plantarum 20, 3DM, and DC400, and Lactobacillus sanfranciscensis E17, A15, 7A, and 5D) (11, 12, 13, 22) plus 25 unidentified isolates, all belonging to the Culture Collection of the Department of Food Science-Section of Food Technology and Biotechnology-Laboratory of Microbiology (University of Perugia, Italy), and eight Lactobacillus strains available at the DSMZ Culture Collection (Lactobacillus alimentarius DSM 20181, Lactobacillus farciminis DSM 20180, Lactobacillus fermentum DSM 20391, Lactobacillus fructivorans DSM 20607, Lactobacillus hilgardii DSM 20051, L. paraplantarum DSM 10641, L. pentosus DSM 20199, and Lactobacillus pontis DSM 8476) were included to validate the multiplex PCR system. Lactobacillus acidophilus DSM 20079T, Lactobacillus amylolyticus DSM 11664T, Lactobacillus amylovorus DSM 20531T, Lactobacillus crispatus DSM 20584T, Lactobacillus delbrueckii subsp. delbrueckii DSM 20074T, Lactococcus lactis subsp. lactis DSM 20481T, Leuconostoc mesenteroides DSM 20343T and Weissella confusa DSM 20196T were included with the aim of investigating the specificity of the two-step multiplex PCR system. Lactobacillus strains obtained from the DSMZ were propagated as indicated by the culture collection for 24 h. Sourdough LAB were propagated in sourdough bacteria broth (32) at 30°C for 24 h.
TABLE 1.
List of reference strains used in this study representing Lactobacillus species commonly found in sourdough and the nucleotide sequence accession numbers
| Reference strain | Other reference strain no. | Accession no.
|
|
|---|---|---|---|
| 16S rRNA | 16S-23S rRNA ISR and partial 23S rRNA | ||
| L. alimentarius LMG 9187T | DSM 20249T | M58804 | AJ616015 |
| L. brevis ATCC 14869T | M58810 | AJ616223 | |
| L. farciminis DSM 20184T | AJ417499 | AJ616017 | |
| L. fermentum ATCC 14931T | JCM 1173T | M58819 | AF182720 |
| L. fructivorans DSM 20203T | X76330 | AJ616220 | |
| L. frumenti DSM 13145T | AJ250074 | AJ616011 | |
| L. hilgardii DSM 20176T | M58821 | AJ616222 | |
| L. mindensis DSM 14500T | AJ313530 | AJ616016 | |
| L. panis DSM 6035T | X94230 | AJ616012 | |
| L. paralimentarius DSM 13238T | AJ417500 | AJ616014 | |
| L. paraplantarum DSM 10667T | AJ306297 | AJ616224 | |
| L. pentosus ATCC 8041T | JCM 1558T | D79211 | AJ635329 |
| L. plantarum ATCC 14917T | NCDO 1752T; JCM 1149T | X52653 | AF182722 |
| L. pontis DSM 8475T | LMG 14187T | AJ422032 | AJ616013 |
| L. rossii DSM 15814T | AJ564009 | AJ635330 | |
| L. sanfranciscensis DSM 20451T | M58830 | AJ616221 | |
DNA isolation.
Genomic DNAs from broth cultures were extracted as reported by De Los Reyes-Gavilán et al. (16) from 2-ml samples of overnight cultures grown in the appropriate medium and incubated at the optimal growth temperature. The final concentration of lysozyme used for cell lysis was 2 mg/ml. The concentration and purity of DNA was assessed by determining the optical densities at 260 and 280 nm, as described by Sambrook et al. (53). To extract genomic DNA directly from colonies of lactobacilli isolated from sourdough, a single colony was suspended in 20 μl of microLYSIS buffer (Labogen, Terrazzano di Rho, Italy) and subjected to a heating-cooling treatment (5 min at 65°C, 2 min at 96°C, 4 min at 65°C, 1 min at 96°C, 1 min at 65°C, and 30 s at 96°C) using a thermal cycler (GeneAmp PCR System 9700: PE Applied Biosystems, Norwalk, CT), as indicated by the manufacturer.
Total DNA from doughs was extracted according to the following protocol: 10 g of each dough was suspended in 90 ml of sterile distilled H2O and homogenized (Classic Blender, PBI International, Milan, Italy); 500 μl of the suspension was subjected to centrifugation (10,000 × g for 5 min), the supernatant was discarded, and the resulting pellet was resuspended in 500 μl of 0.1 M NaOH. After 1 h of incubation at room temperature, the suspension was subjected to centrifugation at 10,000 × g for 5 min and the resulting pellet was resuspended in 500 μl of sterile double-distilled H2O, from which 1.5 μl was used for the PCR amplification reaction.
Isolation of the 16S rRNA/16S-23S rRNA ISR/flanking 23S rRNA gene sequences.
The 16S rRNA/16S-23S rRNA ISR/flanking 23S rRNA gene sequences for the 16 reference strains listed in Table 1 were obtained from EMBL-GenBank-DDBJ nucleotide sequence databases or, if not available, determined in this study. Primers (Life Technologies, Milan, Italy) MulISRF/MulISRR, MulISRF/ISRmulR, and 16 (5)/23-10C (23) (see Table 2 for the corresponding positions) were used to amplify the 16S-23S rRNA ISR and corresponding flanking 23S rRNA gene of L. pentosus ATCC 8041T, L. rossii DSM 15814T and all the other Lactobacillus strains, respectively.
TABLE 2.
Oligonucleotide primer sequences and their locations
| Primer | Oligonucleotide sequence (5′ → 3′) | Position (accession no.) | Reference |
|---|---|---|---|
| 16 | GCTGGATCACCTCCTTTC | 5 | |
| 23-10C | CCTTTCCCTCACGGTACTG | 23 | |
| MulISRF | CACCGCCCGTCACACCATG | 1419-1437 (X52653) | This study |
| MulISRR | GCCTTGSGAGATGGTCCTC | 657-675 (AJ616224) reversed | This study |
| ISRMulR | CCGTACTCAGGATHCTGAA | 601-619 (AJ616016) reversed | This study |
| Gru1′′′F | GCAGTTTTAATCAACTGTTACC | 335-356 (AJ616224) | This study |
| Gru2′′F | AGGGTTGTAGGACTGATGTTG | 492-512 (AJ616016) | This study |
| Gru3F | GGGGAACTGAAACATCTMAGTACCC | 408-432 (AJ616221) | This study |
| Gru4F | CGCAACGAGCGCAACCCTTGTTAC | 1058-1081 (AJ422032) | This study |
| LrosF | GTATCTGAGAGTAACTGTTCAGA | 11 | |
| LrosR | AGGGAACGTCCGATCTCTCG | 11 | |
| paraF | GTCACAGGCATTACGAAAAC | 57 | |
| pentF | CAGTGGCGCGGTTGATATC | 57 | |
| planF | CCGTTTATGCGGAACACCTA | 57 | |
| pREV | TCGGGATTACCAAACATCAC | 57 | |
| LaliF | TAGTTGAGATAGCTGAACAGC | 529-549 (AJ616015) | This study |
| LfarF | CTACTTTCACATGATCGTAGC | 194-214 (AJ417499) | This study |
| LminF | GGTAGGATGATGCGTAAGCAT | 1543-19 (AJ313530-AJ616016) | This study |
| LpalF | GACGAAAGTCATGGCAAATTG | 163-183 (AJ616014) | This study |
| LbreF | TTTGACGATCACGAAGTGACCG | 196-217 (AJ616223) | This study |
| LfrucF | CGACAGTGAATTCATAGCTGTCG | 375-397 (AJ616220) | This study |
| LhilF | CGGAAACCTACACAATGTCG | 8-27 (AJ616222) | This study |
| Lsan′F | TGAAGTAGTTGGGAAGCTACA | 557-577 (AJ616221) | This study |
| LferF | AAGAATCAGGTAGTCGAAGTG | 527-547 (AF182720) | This study |
| Lfru′F | CACCGCGTTATTTTGAGTTGT | 126-146 (AJ616011) | This study |
| Lpan′′F | CCAACTTAGTCGTTGGTTATC | 336-356 (AJ616012) | This study |
| LponF | TCAAAACCACATGGTTTTGATTTC | 136-159 (AJ422032) | This study |
| LacbF | TGCCTAATACATGCAAGT | 12 | |
| LacbR | CTTGTTACGACTTCACCC | 12 | |
| LABR | GATCCGCGATTACTAGCG | 1352-1369 (X54268) reversed | This study |
Each reaction mixture contained 200 μM of each 2′-deoxynucleoside 5′-triphosphate, 1 μM of both forward and reverse primers, 2 mM MgCl2, 1 U of Taq DNA polymerase (Life Technologies), 2.5 μl of 10× PCR buffer, ca. 25 ng of DNA, and enough sterile double-distilled H2O to bring the final volume to 25 μl. PCR amplification was performed using the GeneAmp PCR System 9700 thermal cycler (Applied Biosystems). Amplification consisted of 30 cycles of denaturation for 45 s at 94°C, annealing for 45 s at 49°C for primer pair 16/23 and 10C, at 58°C for primer pair MulISRF and MulISRR, or at 55°C for primer pair MulISRF and ISRmulR, and extension for 1 min at 72°C. The first cycle was preceded by an initial template denaturation step for 4 min at 94°C, and the last cycle was followed by a final extension step for 7 min at 72°C.
PCR products were separated by electrophoresis on 1.5% (wt/vol) agarose gel (Gibco BRL, Cergy Pontoise, France) containing ethidium bromide (0.5 μg/ml), and the DNA was detected by UV transillumination. The expected amplicons of about 700, 750, and 800 bp (after amplification with primer pairs 16/23 and 10C, MulISRF and ISRmulR, and MulISRF and MulISRR, respectively) were excised from the gel and purified by the GFX PCR DNA and gel band purification kit (Amersham Biosciences, Piscataway, NJ). DNA sequencing reactions were performed by MWG Biotech AG (Ebersberg, Germany).
Analysis of sequence data and phylogenetic grouping.
The identities of the sequences obtained after analysis of amplified PCR products were verified by a BlastN (1) search against the NCBI nonredundant sequence database located at http://www.ncbi.nlm.nih.gov. The ClustalW program (56) was used for nucleotide sequence alignment of the 16S-23S rRNA ISR genes and of the 16S rRNA/16S-23S rRNA ISR/flanking 23S rRNA genes. Sequence alignments were analyzed and adjusted by GeneDoc program version 2.5.000 (K. B. Nicholas and H. B. Nicholas, unpublished data), and phylogenetic grouping was obtained using programs in the PHYLIP (Phylogeny Inference Package) software version 3.5c (18).
Two-step multiplex PCR conditions.
Based on the obtained 16S-23S rRNA ISR sequences, the 16 Lactobacillus strains were first typed by a multiplex PCR assay named Grouping-multiplex PCR. Subsequently, four multiplex PCR assays, named Group1-, Group2-, Group3-, and Group4-multiplex PCRs, were performed in order to discriminate Lactobacillus representatives of each group at the species level. PCR mixtures contained from four up to seven primers.
Oligonucleotide primer (Life Technologies) sequences and their locations are reported in Table 2. Amplification reactions, except for group 1-multiplex PCR, were carried out in a total volume of 15 μl of a solution containing 10% (vol/vol) of 10× PCR buffer, 2.5 mM MgCl2, 200 μM of each 2′-deoxynucleoside 5′-triphosphate, 0.6 U of Taq DNA polymerase (Life Technologies), approximately 15 ng of template DNA, and different primer concentrations, depending on the multiplex PCR assay: 0.25 μM of primer Gru3F, 0.50 μM of primer Gru2′′F, 0.60 μM of primer Gru1‴F, 0.75 μM of primer MulISRR, and 1 μM of the other primers involved in Grouping-multiplex PCR; 0.5 μM of each Group2-multiplex PCR primer; 1 μM of primer Lsan′F and 0.5 μM of all other Group3-multiplex PCR primers; 0.15 μM of primer Lfru′F, 0.5 μM of primers LferF, Lpan′′F, and MulISRR, and 1.5 μM of primer LponF for Group4-multiplex PCR. Sterile double-distilled H2O was added to bring the mixture to the final reaction volume. The applied PCR conditions for Grouping-, Group2-, Group3-, and Group4-multiplex PCR comprised an initial template denaturation step for 4 min at 94°C followed by 30 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 65°C, and elongation for 1 min at 72°C. The final extension step was for 7 min at 72°C. PCR mixture composition (20 μl) and PCR conditions reported by Torriani et al. (57), used for Group1-multiplex PCR, were modified as follows: 2 mM MgCl2, 200 μM of each 2′-deoxynucleoside 5′-triphosphate, 0.8 U of Taq DNA polymerase (Life Technologies), ca. 20 ng of template DNA, 0.5 μM of primers paraF, pentF, and pREV, and 0.25 μM of primer planF. The PCR conditions included 30 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 55°C, and elongation for 45 s at 72°C.
DNA accessibility of strains used as negative controls.
The presence of DNA in the negative controls was verified by PCR employing the LacbF/LABR primer pair. Amplifications were performed using the above reported thermal cycler with reaction mixtures containing 200 μM of each 2′-deoxynucleoside 5′-triphosphate, 1 μM of both forward and reverse primer, 2 mM MgCl2, 0.6 U of Taq DNA polymerase (Life Technologies), 1.5 μl of 10× PCR buffer, ca. 15 ng of DNA, and sterile double-distilled H2O to bring the final volume to 15 μl. Amplification conditions consisted of an initial template denaturation at 94°C for 4 min and 30 cycles of denaturation for 45 s at 94°C, annealing for 45 s at 57°C, and elongation for 1 min at 72°C. The final cycle was followed by a final extension step for 7 min at 72°C.
Genotypic identification of sourdough lactobacilli.
In order to perform 16S rRNA gene sequence analysis of sourdough-isolated lactobacilli, a large part of the 16S rRNA gene from 25 strains was amplified using the LacbF/LacbR primer pair employing PCR conditions reported by Corsetti et al. (12). PCR products (ca. 1,400 nucleotides) were purified by the GFX PCR DNA and gel band purification kit (Amersham Biosciences). DNA sequencing reactions were performed by MWG Biotech AG.
Dough production and characteristics.
Doughs were prepared in order to extract total DNA to be analyzed by the two-step multiplex PCR system described above in order to evaluate if this method could be applied for in situ detection of sourdough lactobacilli. Lactobacilli used as dough starters were cultured as reported above. Cells were harvested by centrifugation at 8,000 × g for 10 min, washed twice with sterile distilled H2O, and then suspended in sterile distilled H2O; the cell suspension, diluted 1:10, gave an optical density at 620 nm of 1.25. Wheat flour contained 12.5% moisture; protein (N × 5.70), 11% of dry matter; fat, 1.82% of dry matter; and ash, 0.60% of dry matter. Wheat flour (12.5 g), cellular suspension (1.1 ml for each strain) and tap H2O (enough to bring the volume of the cellular suspension to 7.5 ml) were used to produce six doughs with a dough yield (weight of the dough/weight of the flour × 100) of 160 and with a cellular concentration of about 108 CFU/g.
To reflect the various Lactobacillus groups as outlined in Fig. 2, doughs were produced that contained various combinations of strains as indicated in Table 3. Moreover, a dough (6, Table 3) containing one strain of each representative group was produced. Doughs (20 g) were immediately analyzed by plate count: 10 g of each dough was suspended in 90 ml of sterile distilled H2O and homogenized (Classic Blender, PBI International). A reference dough (R) without starter was produced and treated as described above.
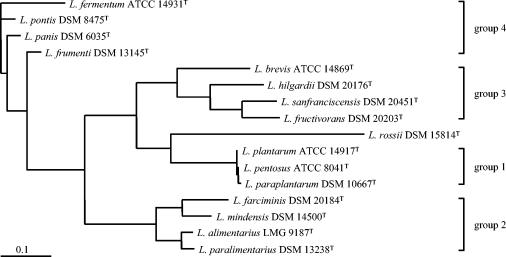
FIG. 2.
Phylogenetic tree showing the relative positions of 16 Lactobacillus reference strains representing sourdough-associated species based on 16S-23S rRNA ISR gene sequences. Bar, 0.1 nucleotide substitution per site.
TABLE 3.
Lactobacilli contained in doughs and their detection by two-step multiplex PCRa
| Dough no. | Starter | First-step reliability | Second-step reliability |
|---|---|---|---|
| 1 | L. plantarum ATCC 14917T | + | + |
| L. pentosus ATCC 8041T | + | + | |
| L. paraplantarum DSM 10667T | + | + | |
| 2 | L. farciminis DSM 20184T | + | + |
| L. mindensis DSM 14500T | + | + | |
| L. alimentarius LMG 9187T | + | + | |
| L. paralimentarius DSM 13238T | + | + | |
| 3 | L. brevis ATCC 14869T | + | + |
| L. hilgardii DSM 20176T | + | + | |
| L. sanfranciscensis DSM 20451T | + | + | |
| L. fructivorans DSM 20203T | + | + | |
| 4 | L. fermentum ATCC 14931T | + | + |
| L. pontis DSM 8475T | + | + | |
| L. panis DSM 6035T | + | + | |
| L. frumenti DSM 13145T | + | + | |
| 5 | L. rossii DSM 15814T | + | nn |
| 6 | L. plantarum ATCC 14917T | + | − |
| L. farciminis DSM 20184T | + | − | |
| L. sanfranciscensis DSM 20451T | + | − | |
| L. fermentum ATCC 14931T | + | − | |
| L. rossii DSM 15814T | + | nn | |
| R | None | + | − |
Symbols: +, reliable; −, not reliable; nn, not needed.
Nucleotide sequence accession number.
The 16S-23S ISR sequences were submitted to the EMBL-GenBank-DDBJ nucleotide sequence databases under the accession numbers reported in Table 1.
RESULTS
Sequence analysis of the 16S-23S rRNA ISR and phylogenetic grouping.
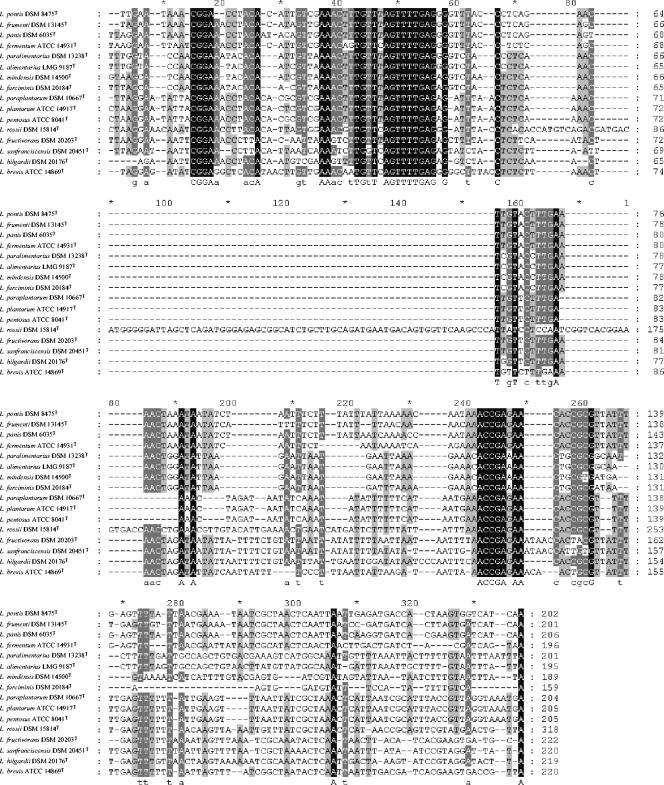
The alignment of the 16S-23S rRNA ISR sequences from the 16 reference strains of lactobacilli used in this study is presented in Fig. 1. ISR sequences showed a high heterogeneity while varying in length from 159 (L. farciminis DSM 20184T) to 318 bp (L. rossii DSM 15814T). It is worth noting that for 14 out of the 16 Lactobacillus strains the ISR length is between 189 and 222 bp, making the L. rossii DSM 15814T ISR sequence almost 100 bp longer. The phylogram inferred from the above 16S-23S rRNA ISR alignment is reported in Fig. 2.
FIG. 1.
Alignment of the 16S-23S rRNA ISR gene sequences of 16 reference strains representing Lactobacillus species commonly found in sourdough.
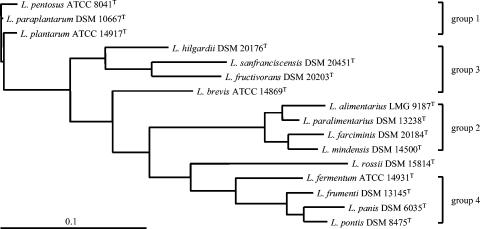
A phylogram was also constructed using the 16S rRNA/16S-23S rRNA ISR/flanking 23S rRNA gene sequences of the 16 reference strains after manual adjustment of the nucleotide sequence alignment (Fig. 3).
FIG. 3.
Phylogenetic tree showing the relative positions of 16 Lactobacillus reference strains representing sourdough-associated species based on 16S rRNA/16S-23S rRNA ISR/partial 23S rRNA gene sequences. Bar, 0.1 nucleotide substitution per site.
The phylogram constructed from ISR alignment (Fig. 2) clearly distinguishes four phylogenetic groups, whereas, when also 16S rRNA and partial 23S rRNA genes are taken into account (Fig. 3), three out of the four previous groups were still clearly recognizable (group 1, group 2, and group 4). In both cases L. rossii DSM 15814T was not included in any group.
Primer design.
Aligned 16S rRNA/16S-23S rRNA ISR/partial 23S rRNA nucleotide sequences of the 16 Lactobacillus reference strains were analyzed to design group- and species-specific primers to be used in a two-step multiplex PCR assay. Group-specific primers were chosen on regions showing sequence homogeneity among strains grouped together (see Fig. 2) but characterized by a high level of heterogeneity among groups. Species-specific primers were designed on regions with high sequence heterogeneity, paying particular attention to their specificity against lactobacilli belonging to the same group (Fig. 2). The group- and species-specific primers are listed in Table 2, while their locations relative to the rRNA-encoding region, together with the sizes of their expected amplicon, are schematically presented in Fig. 4 (see Grouping-multiplex PCR and Group2- to Group4-multiplex PCR, respectively).
FIG. 4.
Schematic alignment of primer location on the rrn operon fragment.
Multiplex PCR for grouping and discrimination of lactobacilli at the species level.
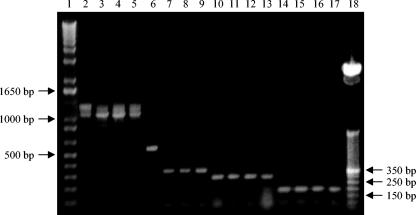
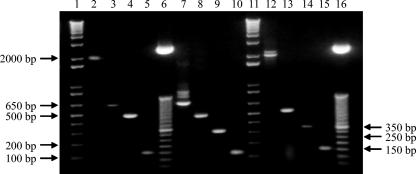
Grouping-multiplex PCR generated the following expected (Fig. 4 and 5) major amplicons for each group of lactobacilli reported in Fig. 2: ca. 1,100 bp for group 4, ca. 340 bp for group 1, ca. 280 bp for group 3, and ca. 180 for group 2 (Fig. 5). L. rossii DSM 15814T was characterized by a 575-bp DNA amplification product. For lactobacilli belonging to group 4, some unexpected additional amplicons were produced with the Gru4F/MulISRR primer pair (Fig. 5, lanes 2 to 5), which, however, did not affect the specificity of the Grouping-multiplex PCR assay, as their observed size was different from that of any of the expected amplified products.
FIG. 5.
Grouping-multiplex PCR (first step) assay. Lanes: 1, 1-kb DNA molecular size markers (Invitrogen); 2, L. fermentum ATCC 14931T; 3, L. frumenti DSM 13145T; 4, L. panis DSM 6035T; 5, L. pontis DSM 8475T; 6, L. rossii DSM 15814T; 7, L. pentosus ATCC 8041T; 8, L. plantarum ATCC 14917T; 9, L. paraplantarum DSM 10667T; 10, L. brevis ATCC 14869T; 11, L. fructivorans DSM 20203T; 12, L. hilgardii DSM 20176T; 13, L. sanfranciscensis DSM 20451T; 14, L. alimentarius LMG 9187T; 15, L. farciminis DSM 20184T; 16, L. mindensis DSM 14500T; 17, L. paralimentarius DSM 13238T; 18, 50-bp DNA molecular size markers (Invitrogen).
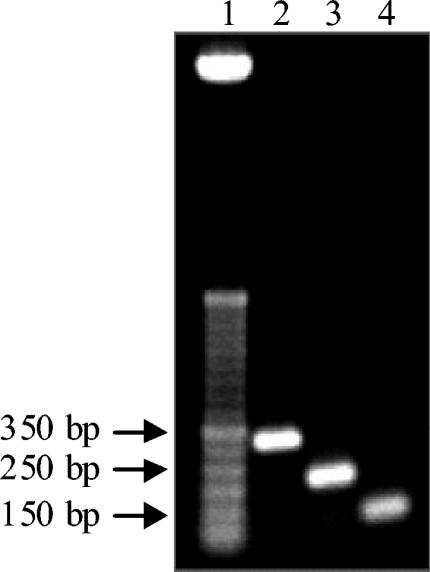
Individual Lactobacillus strains belonging to group 2, group 3, or group 4 were successfully detected and discriminated at the species level by the second step of the two-step multiplex PCR developed in this work. Group2-multiplex PCR was shown to distinguish L. farciminis DSM 20184T, L. mindensis DSM 14500T, L. paralimentarius DSM 13238T and L. alimentarius LMG 9187T (Fig. 6, lanes 2 to 5). Furthermore, L. hilgardii DSM 20176T, L. brevis ATCC 14869T, L. fructivorans DSM 20203T and L. sanfranciscensis DSM 20451T were detected and individually distinguished by Group3-multiplex PCR (Fig. 6, lanes 7 to 10), whereas Group4-multiplex PCR enabled the detection and distinction of L. pontis DSM 8475T, L. frumenti DSM 13145T, Lactobacillus panis DSM 6035T and L. fermentum ATCC 14931T (Fig. 6, lanes 12 to 15). Unexpected bands were obtained with the LhilF/MulISRR and LponF/MulISRR primer pairs (Fig. 6, lanes 7 and 12), but these additional bands did not diminish the resolving power of Group3- and Group4-multiplex PCR. As previously stated by Torriani et al. (57), Group1-multiplex PCR enables the identification of Lactobacillus strains belonging to group 1, yielding the following amplicons: 318 bp for L. plantarum ATCC 14917T, 218 bp for L. pentosus ATCC 8041T and 107 bp for L. paraplantarum DSM 10667T (Fig. 7).
FIG. 6.
Group2-, Group3-, and Group4-multiplex PCR (second step) assays. Lanes: 1 and 11, 1-kb DNA molecular size markers (Invitrogen); 2, L. farciminis DSM 20184T; 3, L. mindensis DSM 14500T; 4, L. paralimentarius DSM 13238T; 5, L. alimentarius LMG 9187T; 6 and 16, 50 bp DNA molecular weight marker (Invitrogen); 7, L. hilgardii DSM 20176T; 8, L. brevis ATCC 14869T; 9, L. fructivorans DSM 20203T; 10, L. sanfranciscensis DSM 20451T; 12, L. pontis DSM 8475T; 13, L. frumenti DSM 13145T; 14, L. panis DSM 6035T; 15, L. fermentum ATCC 14931T.
FIG. 7.
Group1-multiplex PCR (second step) assay. Lanes: 1, 50-bp DNA molecular size markers (Invitrogen); 2, L. plantarum ATCC 14917T; 3, L. pentosus ATCC 8041T; 4, L. paraplantarum DSM 10667T.
In order to use the system for in situ detection of lactobacilli at the species level, the specificity of species-specific primers of each Group-multiplex PCR assay was further checked using all other Lactobacillus reference strains as negative controls: nonspecific bands were obtained, probably due to cross amplification reactions. For example, a band of 134 bp, identifying L. sanfranciscensis, was obtained from the DNA of L. farciminis, L. rossii, and L. plantarum when subjected to Group3-multiplex PCR.
Multiplex PCR system specificity check and validation.
The specificity of Grouping-multiplex PCR was tested against species representing microorganisms isolated from sourdough with lower frequency than the above reference strains: L. acidophilus DSM 20079T, L. amylolyticus DSM 11664T, L. amylovorus DSM 20531T, L. crispatus DSM 20584T, L. delbrueckii subsp. delbrueckii DSM 20074T, Lactococcus lactis subsp. lactis DSM 20481T, Leuconostoc mesenteroides DSM 20343T, and Weissella confusa DSM 20196T. No PCR products were obtained with any of the Lactobacillus strains or L. mesenteroides DSM 20343T, while a 180-bp and a 280-bp band, characteristic of groups 2 and 3, respectively, were obtained with W. confusa DSM 20196T and Lactococcus lactis subsp. lactis DSM 20481T, respectively (results not shown). However, the latter two strains did not generate a false positive result when analyzed by the Group2- and Group3-multiplex PCR assays.
Twelve strains isolated from sourdough and identified previously (11, 12, 13, 22) and eight nontype strains obtained from an official culture collection were used to validate the established two-step multiplex PCR system. The system correctly attributed each strain to the expected species.
According to the results of the 16S rRNA gene sequencing, the multiplex PCR results showed that 25 previously unidentified sourdough isolates were two strains of L. brevis, one strain of L. fermentum, five strains of L. paralimentarius, two strains of L. paraplantarum, three strains of L. plantarum, six strains of L. rossii, and six strains of L. sanfranciscensis.
In situ detection of lactobacilli.
In order to verify the reliability of the PCR system developed in this study to detect sourdough Lactobacillus species in situ, Lactobacillus reference strains were used as starters to produce six doughs from which total DNA was extracted. The strains were inoculated at a final concentration of ca. 108 CFU/g, a number which roughly corresponds to that of a prevailing Lactobacillus species present in a fermented sourdough, and DNA was extracted from the doughs using a rapid extraction method with a 0.1 M NaOH solution. Less than 4 h was needed from dough sample collection to the reading of the results through the following steps: DNA extraction (ca. 1 h), PCR amplification (ca. 2 h), and electrophoretic separation (ca. 45 min).
The reliability of the in situ two-step multiplex PCR system is reported in Table 3, which shows that both steps of the analysis (group- and species-specific PCR) generated the expected bands for doughs 1 to 5, while no bands were obtained from the reference dough (R) produced without starters. In the case of dough 6, containing a representative strain for each group, the group-multiplex PCR allowed the direct detection of L. rossii, while the second step was not performed, having been proven to be unreliable at the species level.
DISCUSSION
Sourdough is a very complex ecosystem hosting a multitude of microorganisms, among which a relatively large number of different Lactobacillus species may be present. The application of sourdough-derived LAB as starter cultures for controlled fermentation requires evaluation of their persistence during sourdough propagation, necessitating a simple, rapid, and reliable detection method. A way to rapidly identify different microbial species at a trustworthy level is by multiplex PCR, a molecular method for simultaneous bacterial detection. Such a genotype-based approach has the attractive advantage of being reliable, because based on amplification of conserved regions on DNA sequences, while at the same time being less time-consuming than phenotype-based methods.
With regard to sourdough fermentation, the multiplex PCR assay has previously been successfully applied for the detection of L. pontis and two related species (45), whereas the maximum numbers of different species distinguished by this method is 11 human intestinal Lactobacillus spp. (55). In the present study, a two-step multiplex PCR system (using group- and species-specific primers) for grouping and subsequent discrimination of 16 Lactobacillus species, commonly isolated from sourdoughs of different origins and including the new species L. rossii (11), was developed. The group of reference strains was based on comparative sequence analysis of the 16S-23S rRNA ISR, and 16S rRNA/16S-23S rRNA ISR/partial 23S rRNA sequences (ca. 2,100 bp). Based on 16S rRNA/16S-23S rRNA ISR/partial 23S rRNA gene-derived grouping, L. rossii DSM 15814T was assigned to group 4 (Fig. 3), but its relatively high sequence dissimilarity with its closest related species used in this work, only 92% identity of 16S rRNA genes (11), did not allow its inclusion into any multiplex PCR group and no group-specific primers could be designed for L. rossii DSM 15814T. This allowed the identification of L. rossii directly by the first step of the multiplex PCR system.
Group - and species-specific primers were designed based on the nucleotide sequences of the 16S rRNA/16S-23S rRNA ISR/partial 23S rRNA genes. These sequences, in particular the 16S-23S rRNA ISR, allowed the generation of differently sized amplicons allowing convenient identification by agarose gel electrophoresis. Except for L. rossii 15814T, the 15 other species of Lactobacillus did not require a specific reverse primer to be identified at the species level, and thus the same reverse primer (MulISRR) was used for each multiplex PCR assay developed (except group 1), making it possible to reduce the number of primers to be used in a single multiplex reaction.
A total of five multiplex PCR assays were necessary to discriminate the 16 Lactobacillus species considered in our study. Since species of group 1 (L. pentosus, L. plantarum, and L. paraplantarum) were characterized by a high identity value on 16S rRNA (57) and on 16S-23S rRNA ISR gene sequences (Fig. 1), their distinction was based on a modification of the multiplex PCR assay (reported in the present study as group 1-multiplex PCR) with the recA gene-derived primers developed by Torriani et al. (57).
The specificity of Grouping-multiplex PCR was ensured by using eight strains belonging to four genera of LAB representing Lactobacillus and non-Lactobacillus species associated with sourdough fermentation but isolated at lower frequency compared to the 16 species used as a reference: W. confusa DSM 20196T and Lactococcus lactis subsp. lactis DSM 20481T were assigned to group 2 and group 3, respectively. DNAs from W. confusa DSM 20196T and Lactococcus lactis subsp. lactis DSM 20481T were further analyzed by group 2- and group 3-multiplex PCR, respectively, yielding no PCR amplification products. False negative results were avoided by checking the DNA accessibility of negative controls with the LacbF/LABR primer pair.
The different species used as negative controls and the high annealing temperature of 65°C, except for Group1-multiplex PCR, for which the specificity is ensured by recA gene sequences, make this system highly specific for the rapid detection of and distinction among commonly prevailing sourdough lactobacilli. The two-step multiplex PCR system was also validated using sourdough lactobacilli identified previously or obtained from a culture collection. The usefulness of the system was tested by the identification of 25 strains of undetermined species previously isolated from sourdough, which, compared by 16S rRNA analysis, showed that the system managed to simultaneously and reliably identify to the species level, following isolation and propagation, 16 species of lactobacilli (L. alimentarius, L. brevis, L. farciminis, L. fermentum, L. fructivorans, L. frumenti, L. hilgardii, L. mindensis, L. panis, L. paralimentarius, L. paraplantarum, L. pentosus, L. plantarum, L. pontis, L. rossii, and L. sanfranciscensis) commonly found in sourdough.
The system was also capable of rapid identification of lactobacilli at species level using DNA extracted directly from colonies, avoiding time-consuming DNA isolation procedures based on enzymatic lysis. Nevertheless, for monitoring the Lactobacillus community without prior cultivation, direct isolation of total bacterial DNA from dough with a subsequent specific PCR can be a valuable system (45). With this in mind, a fast and simple extraction protocol of DNA from dough was developed which made it possible to have target DNA accessible for PCR assays in about 1 h. The handling of samples and solutions is reduced to a minimum as just 0.1 M NaOH is needed for DNA extraction.
For the above reasons, the system could even be adapted for rapid sourdough microbial characterization on-line with a reliable multiplex PCR system. The group-specific primer combinations allow an unequivocal Lactobacillus group attribution when the first multiplex PCR is applied to total DNA extracted from dough and, by only one-step PCR, to L. rossii. As reported above, species-specific primers were designed to be specific for each group, and faulty results were avoided by the combination of both steps of the multiplex PCR system. The appearance of false positive reactions using species-specific primers designed for one group of lactobacilli with species belonging to other groups limits the multiplex PCR approach developed here to the in situ detection of each group of lactobacilli. Nevertheless, in case the strains of a group represent a mixed starter to be used for sourdough fermentation, or if they are added as a single culture, the multiplex PCR system developed in this work can be suitable to track in situ, at the species level, each LAB contained in the mixture. The system may in particular be suitable to investigate the microbial composition of those commercial sourdough starter preparations which are continuously propagated without disturbance of the production process and in which the dominant microflora is represented by only a few species (e.g., L. sanfranciscensis and L. pontis) over a long time (25).
Moreover, Grouping-multiplex PCR (first step of the developed system) enabled the detection of L. rossii in situ without the need of colony isolation and cultivation. It is worth noting that the new sourdough associated species L. rossii, which can erroneously be identified as L. brevis by classical phenotypic methods (e.g., API 50CHL system), seems to have a wide distribution in Italian sourdoughs. This species belongs to the group of obligately heterofermentative lactobacilli (11), the group of sourdough microorganisms with the most interesting characteristics for sourdough baked products (14, 20, 21). At present 30 strains, isolated in our laboratory from sourdoughs of different origin (South and Central Italy), have been identified by means of 16S rRNA sequence analysis as belonging to L. rossii species (data not shown). The interest to study this species, in particular with regards to its impact on sourdough microbial ecology and final product characteristics, is increasing. The combined application of multiplex PCR and fast DNA extraction as described here, will allow routine, high through-put analysis of sourdoughs for the presence of such potentially commercially interesting species, as the combined system ensures the reading of the results obtained by Grouping-multiplex PCR by means of agarose gel electrophoresis in less than 4 h from the time of dough sampling.
Work is in progress to evaluate the real distribution of such new Lactobacillus species in Italian sourdoughs of different geographical origins. Moreover, work is being prepared to establish the sensitivity of Grouping-multiplex PCR for the in situ detection of sourdough lactobacilli and the combination of such a system with other tools, such as denaturing gradient gel electrophoresis, for rapid detection of lactobacilli belonging to each group up to the species level using total sourdough bacterial DNA.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Grapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ampe, F., N. ben Omar, and J.-P. Guyot. 1999. Culture-independent quantification of physiologically active microbial groups in fermented foods using rRNA-targeted oligonucleotide probes: application to pozol, a Mexican lactic acid fermented maize dough. J. Appl. Microbiol. 87:131-140. [DOI] [PubMed] [Google Scholar]
- 3.Barry, T., G. Colleran, M. Glennon, L. K. Dunnican, and F. Gannon. 1991. The 16S/23S ribosomial spacer region as a target for DNA probes to identify eubacteria. PCR Methods Appl. 1:51-56. [DOI] [PubMed] [Google Scholar]
- 4.Berridge, B. R., H. Bercovier, and P. F. Frelier. 2001. Streptococcus agalactiae and Streptococcus difficile 16S-23S intergenic rDNA: genetic homogeneity and species-specific PCR. Vet. Microbiol. 78:165-173. [DOI] [PubMed] [Google Scholar]
- 5.Berthier, F., and S. D. Ehrlich. 1998. Rapid species identification within two groups of closely related lactobacilli using PCR primers that target the 16S/23S rRNA spacer region. FEMS Microbiol. Lett. 161:97-106. [DOI] [PubMed] [Google Scholar]
- 6.Bonjock, X., E. Ballesté, and A. R. Blanch. 2004. Multiplex PCR with 16S rRNA gene-tergeted primers of Bifidobacterium spp. to identify sources of fecal pollution. Appl. Environ. Microbiol. 70:3171-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, Y., H. Okada, H. Mori, Y. Benno, and T. Nakase. 1999. Lactobacillus paralimentarius sp. nov., isolated from sourdough. Int. J. Syst. Evol. Microbiol. 49:1451-1455. [DOI] [PubMed] [Google Scholar]
- 8.Chamberlain, J. S., R. A. Gibbs, J. E. Ranier, P. N. Nguyen, and C. T. Caskey. 1988. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res. 16:11141-11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan, C. H., D. E. Stead, and R. H. A. Coutts. 2003. Development of a species-specific recA-based PCR test for Burkholderia fungorum. FEMS Microbiol. Lett. 224:133-138. [DOI] [PubMed] [Google Scholar]
- 10.Coeuret, V., S. Dubernet, M. Bernardeau, M. Gueguen, and J. P. Vernoux. 2003. Isolation, characterisation and identification of lactobacilli focusing mainly on cheeses and other dairy products. Lait 83:269-306. [Google Scholar]
- 11.Corsetti, A., L. Settanni, D. Van Sinderen, G. E. Felis, F. Dellaglio, and M. Gobbetti. 2005. Lactobacillus rossii sp. nov. isolated from wheat sourdough. Int. J. Syst. Evol. Microbiol. 55:35-40. [DOI] [PubMed] [Google Scholar]
- 12.Corsetti, A., L. Settanni, and D. Van Sinderen. 2004. Characterization of bacteriocin-like inhibitory substances (BLIS) from sourdough lactic acid bacteria and evaluation of their in vitro and in situ detection. J. Appl. Microbiol. 96:521-534. [DOI] [PubMed] [Google Scholar]
- 13.Corsetti, A., P. Lavermicocca, M. Morea, F. Baruzzi, N. Tosti, and M. Gobbetti. 2001. Phenotypic and molecular identification and clustering of lactic acid bacteria and yeasts from wheat (species Triticum durum and Triticum aestivum) sourdoughs of Southern Italy. Int. J. Food Microbiol. 64:95-104. [DOI] [PubMed] [Google Scholar]
- 14.Corsetti, A., M. Gobbetti, J. Rossi, and P. Damiani. 1998. Antimould activity of sourdough lactic acid bacteria: identification of a mixture of organic acids produced by Lactobacillus sanfrancisco CB1. Appl. Microbiol. Biotechnol. 50:253-256. [DOI] [PubMed] [Google Scholar]
- 15.Corsetti, A., M. Gobbetti, and E. Smacchi. 1996. Antibacterial activity of sourdough lactic acid bacteria: isolation of a bacteriocin-like inhibitory substance from Lactobacillus sanfrancisco C57. Food Microbiol. 13:447-456. [Google Scholar]
- 16.De Los Reyes-Gavilán, C. G., G. K. Y. Limsowtin, P. Tailliez, L. Séchaud, and J.-P. Accolas. 1992. A Lactobacillus helveticus-specific DNA probe detects restriction fragment length polymorphisms in this species. Appl. Environ. Microbiol. 58:3429-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ehrmann, M. A., M. R. Müller, and R. F. Vogel. 2003. Molecular analysis of sourdough reveals Lactobacillus mindensis sp. nov. Int. J. Syst. Evol. Microbiol. 53:7-13. [DOI] [PubMed] [Google Scholar]
- 18.Felsenstein, J. 1993. PHYLIP (phylogeny inference package), version 3.5c. Distributed by the author. Department of Genetics, University of Washington, Seattle.
- 19.García-Martínez, J., S. G. Acinas, A. I. Antón, and F. Rodríguez-Valera. 1999. Use of the 16S-23S ribosomal genes spacer region in studies of prokaryotic diversity. J. Microbiol. Methods 36:55-64. [DOI] [PubMed] [Google Scholar]
- 20.Gobbetti, M., and A. Corsetti. 1997. Lactobacillus sanfrancisco a key sourdough lactic acid bacterium: a review. Food Microbiol. 14:175-187. [Google Scholar]
- 21.Gobbetti, M., A. Corsetti, and S. De Vincenzi. 1995. The sourdough microflora. Characterization of heterofermentative lactic acid bacteria based on acidification kinetics and impedance test. Ital. J. Food Sci. 2:103-111. [Google Scholar]
- 22.Gobbetti, M., A. Corsetti, J. Rossi, F. La Rosa, and S. De Vincenzi. 1994. Identification and clustering of lactic acid bacteria and yeasts from wheat sourdoughs of Central Italy. Ital. J. Food Sci. 1:85-94. [Google Scholar]
- 23.Gürtler, V., and V. A. Stanisich. 1996. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology 142:3-16. [DOI] [PubMed] [Google Scholar]
- 24.Hamad, S. H., M. C. Dieng, M. A. Ehrmann, and R. F. Vogel. 1997. Characterization of the bacterial flora of Sudanese sorghum sourdough. J. Appl. Microbiol. 83:764-770. [DOI] [PubMed] [Google Scholar]
- 25.Hammes, W. P., and M. G. Gänzle. 1998. Sourdough breads and related products, p. 199-216. In B. J. B. Wood (ed.), Microbiology of fermented foods, vol. 1. Blackie Academic and Professional, London, United Kingdom. [Google Scholar]
- 26.Hammes, W. P., P. Stolz, and M. G. Gänzle. 1996. Metabolism of lactobacilli in traditional sourdoughs. Adv. Food Sci. 18:176-184. [Google Scholar]
- 27.Hammes, W. P., and R. F. Vogel. 1995. The genus Lactobacillus, p. 19-54. In B. J. B. Wood and W. H. Holzapfel (ed.), The lactic acid bacteria, : the genera of lactic acid bacteria. Blackie Academic and Professionalvol. 2, London, United Kingdom. [Google Scholar]
- 28.Hansen, A., and B. Hansen. 1996. Flavour of sourdough wheat bread crumb. Z. Lebensm. Unters. Forsch. 202:244-249. [Google Scholar]
- 29.Houf, K., A. Tutenel, L. De Zutter, J. Van Hoof, and P. Vandamme. 2000. Development of a multiplex PCR assay for the simultaneous detection and identification of Arcobacter butzleri, Arcobacter cryaerophilus and Arcobacter skirrowii. FEMS Microbiol. Lett. 193:89-94. [DOI] [PubMed] [Google Scholar]
- 30.Kandler, O., and N. Weiss. 1986. Genus Lactobacillus Beijerinck 1901, p. 1209-1234. In P. H. A. Sneath, N. S. Mairm, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams and Wilkins, Baltimore, Md. [Google Scholar]
- 31.Kaufmann, P., A. Pfefferkorn, M. Teuber, and L. Meile. 1997. Identification and quantification of Bifidobacterium species isolated from food with genus-specific 16S rRNA-targeted probes by colony hybridization and PCR. Appl. Environ. Microbiol. 63:1268-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kline, L., and T. F. Sugihara. 1971. Microorganisms of the San Francisco sour dough bread process. II. Isolation and characterization of undescribed bacterial species responsible for the souring activity. Appl. Microbiol. 21:459-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krawczyk, B., K. Lewandowski, and J. Kur. 2002. Comparative studies of the Acinetobacter genus and the species identification method based on the recA sequences. Mol. Cell. Probes 16:1-11. [DOI] [PubMed] [Google Scholar]
- 34.Kullen, M. J., L. J. Brady, and D. J. O'Sullivan. 1997. Evaluation of using a short region of the recA gene for rapid and sensitive speciation of dominant bifidobacteria in the human large intestine. FEMS Microbiol. Lett. 154:377-383. [DOI] [PubMed] [Google Scholar]
- 35.Kurabachew, M., R. A. Sandaa, Ø. Enger, and B. Bjorvatn. 2003. Sequence analysis in the 23S rDNA region of Mycobacterium tuberculosis and related species. J. Microbiol. Methods 54:373-380. [DOI] [PubMed] [Google Scholar]
- 36.Lee, H. J., S. Y. Park, and J. Kim. 2000. Multiplex PCR-based detection and identification of Leuconostoc species. FEMS Microbiol. Lett. 193:243-247. [DOI] [PubMed] [Google Scholar]
- 37.Liu, C., Y. Song, M. McTeague, A. W. Vu, H. Wexler, and S. M. Finegold. 2003. Rapid identification of the species of the Bacteroides fragilis group by multiplex PCR assays using group- and species-specific primers. FEMS Microbiol. Lett. 222:9-16. [DOI] [PubMed] [Google Scholar]
- 38.Lucchini, F., V. Kmet, C. Cesena, L. Coppi, V. Bottazzi, and L. Morelli. 1998. Specific detection of a probiotic Lactobacillus strain in faecal samples by using multiplex PCR. FEMS Microbiol. Lett. 158:273-278. [DOI] [PubMed] [Google Scholar]
- 39.Mata, A. I., A. Gibello, A. Casamayor, M. M. Blanco, L. Domínguez, and J. F. Fernández-Garayzábal. 2004. Multiplex PCR assay for detection of bacterial pathogens associated with warm-water streptococcosis in fish. Appl. Environ. Microbiol. 70:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuki, T., K. Watanabe, R. Tanaka, M. Fukuda, and H. Oyaizu. 1999. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl. Environ. Microbiol. 65:4506-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsuki, T., K. Watanabe, R. Tanaka, and H. Oyaizu. 1998. Rapid identification of human intestinal bifidobacteria by 16S rRNA-targeted species- and group-specific primers. FEMS Microbiol. Lett. 167:113-121. [DOI] [PubMed] [Google Scholar]
- 42.Minnick, M. F., S. J. Mitchell, S. J. McAllister, and J. M. Battisti. 1995. Nucleotide sequence analysis of the 23S ribosomal RNA-encoding gene of Bartonella bacilliformis. Gene 162:75-79. [DOI] [PubMed] [Google Scholar]
- 43.Mora, D., M. G. Fortina, C. Parini, and P. L. Manachini. 1997. Identification of Pediococcus acidilactici and Pediococcus pentosaceus based on 16S rRNA and ldhD gene-targeted multiplex PCR analysis. FEMS Microbiol. Lett. 151:231-236. [DOI] [PubMed] [Google Scholar]
- 44.Müller, M. R. A., M. A. Ehrmann, and R. F. Vogel. 2000. Lactobacillus frumenti sp. nov., a new lactic acid bacterium isolated from rye-bran fermentations with a long fermentation period. Int. J. Syst. Evol. Microbiol. 50:2127-2133. [DOI] [PubMed] [Google Scholar]
- 45.Müller, M. R. A., M. A. Ehrmann, and R. F. Vogel. 2000. Multiplex PCR for the detection of Lactobacillus pontis and two related species in a sourdough fermentation. Appl. Environ. Microbiol. 66:2113-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mullié, C., M. F. Odou, E. Singer, M. B. Romond, and D. Izard. 2003. Mutliplex PCR using 16S rRNA gene-targeted primers for the identification of bifidobacteria from human origin. FEMS Microbiol. Lett. 222:129-136. [DOI] [PubMed] [Google Scholar]
- 47.Normand, P., C. Ponsonnet, X. Nesme, M. Neyra, and P. Simonet. 1996. Molecular microbial ecology manual, p. 1-12. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 48.Nowak, A., and J. Kur. 1996. Differentiation of seventeen genospecies of Acinetobacter by multiplex polymerase chain reaction and restriction fragment length polymorphism analysis. Mol. Cell. Probes 10:405-411. [DOI] [PubMed] [Google Scholar]
- 49.Nowak, A., and J. Kur. 1995. Genomic species typing of acinetobacters by polymerase chain reaction amplification of the recA gene. FEMS Microbiol. Lett. 130:327-332. [DOI] [PubMed] [Google Scholar]
- 50.Rachman, C. N., P. Kabadjova, H. Prévost, and X. Dousset. 2003. Identification of Lactobacillus alimentarius and Lactobacillus farciminis with 16S-23S rDNA intergenic spacer region polymorphism and PCR amplification using species-specific oligonucleotide. J. Appl. Microbiol. 95:1207-1216. [DOI] [PubMed] [Google Scholar]
- 51.Rosselló-Mora, R., and R. Amann. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25:39-67. [DOI] [PubMed] [Google Scholar]
- 52.Rossi, F., R. Tofalo, S. Torriani, and G. Suzzi. 2001. Identification by 16S-23S rDNA intergenic region amplification, genotypic and phenotypic clustering of Staphylococcus xylosus strains from dry sausages. J. Appl. Microbiol. 90:365-371. [DOI] [PubMed] [Google Scholar]
- 53.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 54.Schmid, M., S. Schmitz-Esser, M. Jetten, and M. Wagner. 2001. 16S-23S rDNA intergenic spacer and 23S rDNA of anaerobic ammonium-oxidizing bacteria: implications for phylogeny and in situ detection. Environ. Microbiol. 3:450-459. [DOI] [PubMed] [Google Scholar]
- 55.Song, Y.-L., N. Kato, C.-X. Liu, Y. Matsumiya, H. Kato, and K. Watanabe. 2000. Rapid identification of 11 human intestinal Lactobacillus species by multiplex PCR assays using group- and species-specific primers derived from the 16S-23S rRNA intergenic spacer region and its flanking 23S rRNA. FEMS Microbiol. Lett. 187:167-173. [DOI] [PubMed] [Google Scholar]
- 56.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Torriani, S., G. E. Felis, and F. Dellaglio. 2001. Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene-derived primers. Appl. Environ. Microbiol. 67:3450-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trcek, J., and M. Teuber. 2002. Genetic and restriction analysis of the 16S-23S rDNA internal transcribed spacer regions of the acetic acid bacteria. FEMS Microbiol. Lett. 208:69-75. [DOI] [PubMed] [Google Scholar]
- 59.Valcheva, R., M. Korakli, B. Onno, H. Prévost, I. Ivanova, M. A. Ehrmann, X. Dousset, M. G. Gänzle, and R. F. Vogel. 2005. Lactobacillus hammesii sp. nov., isolated from French sourdough. Int. J. Syst. Evol. Microbiol. 55:763-767. [DOI] [PubMed] [Google Scholar]
- 60.Vancanneyt, M., P. Neysens, M. Dewachter, K. Engelbeen, C. Snauwaert, I. Cleenwerck, R. Van der Meulen, B. Hoste, E. Tsakalidou, L. De Vuyst, and J. Swings. 2005. Lactobacillus acidifarinae sp. nov. and Lactobacillus zymae sp. nov., from wheat sourdoughs. Int. J. Syst. Evol. Microbiol. 55:615-620. [DOI] [PubMed] [Google Scholar]
- 61.Ventura, M., R. Reniero, and R. Zink. 2001. Specific identification and targeted characterization of Bifidobacterium lactis from different environmental isolated by a combined multiplex-PCR approach. Appl. Environ. Microbiol. 67:2760-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wisselink, H. J., J. J. Joosten, and H. E. Smith. 2002. Multiplex PCR assays for simultaneous detection of six major serotypes and two virulence-associated phenotypes of Streptococcus suis in tonsillar specimens from pigs. J. Clin. Microbiol. 40:2922-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yost, C. K., and F. M. Nattress. 2000. The use of multiplex PCR reactions to characterize populations of lactic acid bacteria associated with meat spoilage. Lett. Appl. Microbiol. 31:129-133. [DOI] [PubMed] [Google Scholar]