Abstract
The ability to identify sources of fecal pollution plays a key role in the analysis of human health risk and the implementation of water resource management strategies. One approach to this problem involves the identification of bacterial lineages or gene sequences that are found exclusively in a particular host species or group. We used subtractive hybridization to enrich for target host-specific fecal Bacteroidales rRNA gene fragments that were different from those of very closely related reference (subtracter) host sources. Target host rRNA gene fragments were hybridized to subtracter rRNA gene fragments immobilized in a microplate well, and target sequences that did not hybridize were cloned and sequenced for PCR primer design. The use of microplates for DNA immobilization resulted in a one-step subtractive hybridization in which the products could be directly amplified with PCR. The new host-specific primers designed from subtracted target fragments differentiated among very closely related Bacteroidales rRNA gene sequences and distinguished between similar fecal sources, such as elk and cow or human and domestic pet (dog).
Aquatic fecal pollution is associated with human health risk, economic loss, and closure of recreational beaches. More than 12,000 closures and advisories occurred at U.S. ocean and freshwater beaches in 2002, the second highest number in over a decade (10). Eighty-seven percent of the closures were due to the presence of bacteria associated with fecal contamination, and 62% of these instances could not be attributed to a source. Current coliform standards for fecal pollution do not distinguish among sources, and this deficit creates conflict among public health agencies, farmers, environmentalists, and the shellfish industry.
This study addresses two specific fecal pollution issues requiring source identification. The Tillamook Bay watershed on the Oregon coast is home to a large dairy industry, but it also contains a growing population of wildlife, including elk herds. Fecal pollution has had an impact on the economy of the local shellfish industry. Appropriate assessment and abatement strategies and the cooperation of local farmers depend on the ability to distinguish host sources of fecal pollution. A second issue involves runoff from urban storm drains that carry rainwater into rivers and streams. This may include fecal pollution from leaking septic systems or the excrement of domestic pets. Efficient source identification will help to evaluate human health risk and expedite water quality management procedures.
Fecal members of the order Bacteroidales are abundant in the feces of warm-blooded animals (9, 15, 18, 19, 25), and some have host species or group-specific distributions (2, 18). Using terminal restriction fragment length polymorphism and clone library analysis, Bernhard and Field (3) developed PCR primers that distinguish between human and ruminant fecal pollution based on differences in Bacteroidales partial 16S rRNA gene sequences. The primers have been used successfully in trials and field studies in both freshwater and saltwater environments (4, 5, 12, 13). Similar analyses were used to develop markers for pig and horse fecal pollution (9a). However, the clone library analysis involving eight host sources showed that humans, cats, and dogs share very closely related Bacteroidales sequences, as do cows and elk. We were unsuccessful in developing markers to differentiate cow and elk feces based on sequence data from clone libraries. In addition, although two previously developed human-specific Bacteroidales PCR primers did not amplify dog fecal DNAs (3), clone library sequences did not support design of a dog-specific primer.
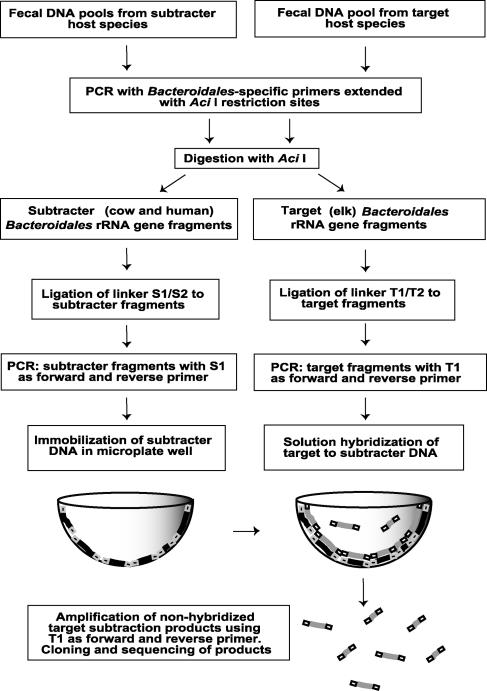
Determining microbial diversity in a complex community requires an extensive analysis of large clone libraries. In a typical library of 100 to 300 clones, often only one or two sequences are identical at the species level, indicating a significant lack of coverage (11, 16). Terminal restriction fragment length polymorphism analysis to identify unique restriction patterns can reduce the number of clones sequenced, but pattern comparison involves an additional element of subjectivity that also results in error and incomplete sequence information. These gaps in sequence data are especially detrimental when the goal is to identify unique markers among very closely related sequences. The aim of this study was to identify host-specific Bacteroidales rRNA gene markers by comparing the genes empirically rather than by relying on sequence data alone. We used a technique based on subtractive hybridization, whereby genetic differences between closely related genomes are amplified to generate unique fragments. Subtractive hybridization has been used to compare bacterial genomes for identification of virulence factors (7, 8, 24), to define regions present in a sequenced genome but absent in an unsequenced relative (1), and more recently to design primers that identify individual members of microbial communities (22). Based on previously published methods (22, 26, 27), this report describes a modified subtractive hybridization for selective enrichment of Bacteroidales rRNA gene sequences. We compared Bacteroidales rRNA genes from a source of interest for primer design (target) with those of one or more reference sources (subtracters) in a solution hybridization according to the procedure described by Zwirglmaier and colleagues (27). An experimental overview is given in Fig. 1. Each hybridization took place in a single microplate well. We used unique unhybridized sequences to design primers for fecal source identification.
FIG. 1.
Overview of subtractive hybridization in microplate wells, adapted from the technique of Zwirglmaier et al. (27).
MATERIALS AND METHODS
Target and subtracter DNA.
The first of two experiments involved elk feces as the target source and cattle and human feces as subtracters. A positive control using only human feces as a subtracter was expected to provide greater variability for experimental validation. In a second experiment, dog feces was the target source and human and cat feces were subtracters.
Sample collection and DNA preparation.
Fecal samples were collected by hunters, farmers, and colleagues and were also acquired from animal shelters. Samples were stored in guanidine isothiocyanate buffer (5 M guanidine isothiocyanate, 100 mM EDTA, pH 8, 0.5% Sarkosyl) at −80°C. The FastDNA kit for soils (Q-Biogene, Carlsbad, CA) was used for DNA isolation. DNAs from 10 to 30 individual fecal samples were mixed in approximately equal concentrations (3 ng/μl) to create a pool of genomic DNA for each host species. All DNA quantifications were done by a PicoGreen assay (Molecular Probes, Inc., Eugene, OR).
PCR amplification and restriction digestion.
Target and subtracter genomic DNA pools were amplified using PCR primers extended with AciI restriction sites. The Bacteroidales-specific 16S rRNA primer AciBac32F (3) and the universal 23S rRNA primer Aci422R (21) amplified approximately 2,400 bp, including most of the Bacteroidales 16S rRNA gene, the intergenic spacer region (internal transcribed spacer), and a portion of the 23S rRNA gene. The oligonucleotides used as primers and linkers in this study are listed in Table 1. Each 50-μl PCR mixture contained 1× Taq polymerase buffer, each primer at a concentration of 10 μM, each deoxynucleoside triphosphate at a concentration of 200 μM, 0.06% bovine serum albumin, 2 mM MgCl2, and 1.25 U of the proofreading Taq polymerase TaKaRa Ex Taq (Takara Bio, Inc., Shiga, Japan). Cycling parameters were as follows: 30 cycles of 94°C for 1 min, 68°C for 1 min, and 72°C for 2.5 min, followed by a final extension at 72°C for 7 min. PCR products were purified with a GeneClean II kit (Q-Biogene) and digested with AciI (New England Biolabs, Inc., Beverly, MA).
TABLE 1.
Primers and linkers used in this study
| Primer or linker | Sequence (5′-3′)a | Reference or source |
|---|---|---|
| AciBac32F | AATATAAACCGCAACGCTAGCTACAGGCTT | 2 |
| Aci422R | AATATAAACCGCWSTCAGGAGTATTTAGCCTT | 21 |
| Linker S1 | CGCCAGGGAACACCCAGTCACGAC | 27 |
| Linker S2 | CGGTCGTGACTGGGTGTTCCCTGGCG | 27 |
| Linker T1 | AGGGGATAACCAATTCACACACCA | 27 |
| Linker T2 | CGTGGTGTGTGAATTGGTTATCCCCT | 27 |
| EF447F | AATAACACCATCTACGTGTAGA | This study |
| EF990R | GCCTGTCCAGTGCAATTTAA | This study |
| DF475F | CGCTTGTATGTACCGGTACG | This study |
| Bac708R | CAATCGGAGTTCTTCGTG | 2 |
Restriction site extensions and overhangs are underlined.
Preparation of linkers and ligation and amplification of target and subtracter DNA.
Separate linkers for subtracter and target DNA (S1/S2 and T1/T2) were obtained from Invitrogen Corp. (Carlsbad, CA). We diluted 8 μg of each linker pair in 40 μl 10 mM Tris-HCl, pH 8.5. They were heated to 65°C and cooled to 20°C gradually to allow hybridization of double-stranded linkers. The hybridized linkers contained AciI-compatible 5′ overhangs but were otherwise the same as those used by Zwirglmaier et al. (27). Target and subtracter restriction fragments were ligated to their respective linkers using T4 DNA ligase (New England Biolabs). Excess linkers were removed using a QIAGEN column (QIAGEN, Valencia, CA). T1 and S1 oligonucleotides were used as PCR primers to amplify target and subtracter ligation products, respectively. The following cycling parameters were used: 30 cycles of 94°C for 1 min, 55°C for 45 s, and 72°C for 1 min.
Immobilization of subtracter in microplate well.
Equal amounts of the subtracter DNA from the two host sources were mixed (1 μg total) and diluted in 50 μl phosphate-buffered saline buffer (8.0 mM Na2HPO4, 1.5 mM KH2PO4, 137 mM NaCl, 27 mM KCl, [pH 7.2]; and MgCl2 added to a final concentration of 100 mM). The mixture was heated to denature the DNAs and immediately placed into a MaxiSorp microplate well (Nalge Nunc, Naperville, IL). The plate was incubated at 37°C for 1 h, the buffer was removed, and the plate was dried for 2 h at 70°C.
Solution hybridization.
Target DNA (20 ng) was diluted in 40 μl hybridization buffer (2.5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 1% blocking reagent [Roche Applied Science, Indianapolis, IN], 0.01% sodium dodecyl sulfate, 0.05% N-lauroylsarcosine, and formamide). A gradient of formamide concentrations (29%, 35%, and 41%) provided hybridizations in three wells at a range of effective temperatures for stringency optimization. Target DNA was heat denatured, iced, and added to the microplate wells. Hybridization was carried out at 70°C for 2 h.
Amplification of subtraction products, cloning, sequencing, and primer design.
Two microliters of subtracted, unhybridized target DNA was removed from the supernatant and amplified with primer T1 using the cycling parameters described above for this primer. PCR fragments were gel extracted (QiaQuick gel purification kit; QIAGEN) following dilution and reamplification and cloned into TOPO TA vectors (Invitrogen Corp.). Ten individual clones were randomly selected for bidirectional sequencing on an ABI 3100 capillary sequencer. Sequences were aligned using ARB (20), either with related GenBank sequences (NCBI BLAST) or with our clone library sequences if they matched the rRNA region for which we had sequence data (16S rRNA; E. coli positions 32 to 708). The short, subtracted (SH) target sequences were added to a neighbor-joining tree of full-length sequences using the parsimony insertion tool of ARB. New primers DF475F and EF990R were designed using the Probe Design and Probe Match functions of ARB.
Primer specificity and sensitivity.
Primers were tested for cross-reactivity against host pools of fecal DNAs representing 4 to 30 individual hosts from each species. Primer specificity was optimized by manipulation of annealing temperature, MgCl2 concentration, and cycle number on a PCR Express thermocycler (Thermo Hybaid, Middlesex, United Kingdom). Primer sensitivity was estimated in serial dilutions of plasmid-inserted templates of known copy numbers. Theoretical detection limits were determined in pure water, creek water (from Beaver Creek, Alsea, OR), and seawater (from 5 miles off the central Oregon coast). Each reaction mixture contained 3 ng total DNA from the natural water sources.
Nucleotide sequence accession numbers.
The partial 16S rRNA gene sequences have been deposited in the GenBank database under accession numbers AY70144475 through AY70144492.
RESULTS
We used subtractive hybridization in microplate wells to identify Bacteroidales 16S rRNA gene fragments found in target host species but not in subtracter species (Fig. 1). These fragments were sequenced and used to design PCR primers for fecal source identification. Table 1 lists primers and linkers used in or designed during this study.
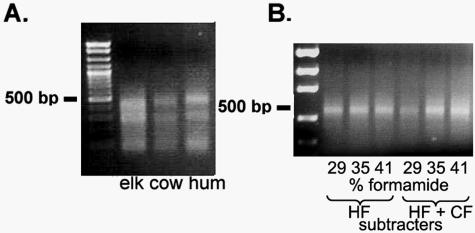
The restriction digests of target and subtracter DNAs resulted in 100- to 500-bp fragments, visualized by gel electrophoresis and shown for the elk experiment in Fig. 2A. Subtracted target fragments of 300 to 500 bp were recovered in both the elk and dog experiments following hybridization and PCR amplification with primer T1. A gel image of the subtracted fragments from the elk experiment is shown in Fig. 2B. Band intensity appeared to increase with increasing stringency (formamide concentration) in fragments from the elk wells versus those from the human and cow wells but not in the human subtracter-only controls. However, since the PCR may not have been quantitative, no conclusions could be drawn from this observation. Subtracted target fragments from the 35% formamide wells were cloned and sequenced. Products in the control wells were not sequenced.
FIG. 2.
(A) AciI restriction digest of target (elk fecal) and subtracter (cow and human [hum] fecal) rRNA genes following PCR amplification with Bacteroidales-specific primers. (B) Elk fecal subtraction products amplified with target-specific linker T1 as PCR primer. HF, human fecal; CF, cow fecal.
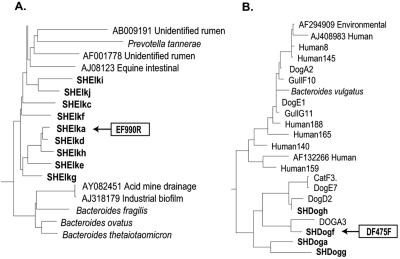
The ARB database to which subtraction clones were compared contained sequences from eight host clone libraries (human, cattle, dog, cat, elk, pig, gull, and horse) and from GenBank (NCBI BLAST). All 10 of the cloned, elk feces-specific fragments were located between E. coli positions 940 and 1370. Because our clone libraries were made from the 5′ half of the 16S rRNA gene (3), we had no sequence data for reference hosts from this region. Three clones formed a unique cluster from which primer EF990R was designed (Fig. 3A). It was paired with EF447F, a primer designed from clone library sequence data.
FIG. 3.
Relationships of subtracted target fragments to database or host-specific clone library sequences. (A) Target elk feces-derived sequences clustered separately from related database sequences. (B) Target dog feces-derived sequences clustered with dog and cat fecal Bacteroidales sequences from an earlier clone library analysis (L. K. Dick et al., unpublished data).
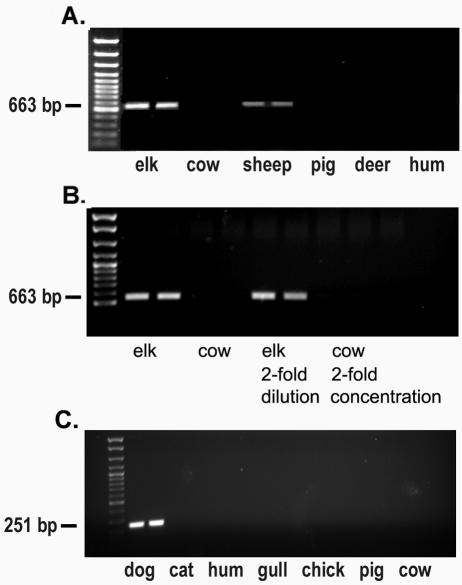
Primer EF990R did not match database sequences of fecal origin when up to five mismatches were chosen by the ARB Probe Match program. Primer specificity was further established using pools of reference (subtracter) host fecal DNAs and host sources not used in the hybridization (Fig. 4A). The new elk-specific primer successfully distinguished elk (target) from cow and human feces, the subtracters used in the experiment. Primer EF990R distinguished elk from cow feces even when the pool of total genomic DNA from cows was in fourfold excess of that of total DNA from elks (Fig. 4B). The specificity extended to some host sources not used in the experiment (pig and deer) but not to other hosts (sheep). DNAs from nine of the 10 individual elk fecal samples used in the hybridization amplified with the primer set. The theoretical detection limit for EF990 was 100 copies and did not change when DNA extracted from creek water was added to the PCRs (data not shown).
FIG. 4.
Host DNA pools PCR amplified with new source-specific primers derived from products of subtractive hybridization. (A) Primer EF990R distinguishes elk fecal DNA from cow and human reference sources, as well as from pig and deer sources. (B) Primer EF990R distinguishes elk from cow feces even when total genomic DNA in the cow fecal pool is in fourfold excess to total DNA in elk fecal pool. (C) Primer DF475F distinguishes dog fecal DNA from human and cat fecal DNAs as well as from other host sources.
Three of 10 dog feces-specific subtracted fragments aligned with a region of the 16S rRNA gene for which we had sequence data from the clone libraries, at E. coli positions 256 to 661. A fourth fragment aligned with E. coli 16S rRNA positions 940 to 1370. Three fragments aligned with the 23S rRNA gene, and three fragments that appeared to be chimeras were not used in the analysis. Clone SHDFf clustered with dog and cat clones from our clone libraries (Fig. 3B). Primer DF475F was designed from this group, and it did not match other ARB database or host clone library sequences. The primer was paired with Bacteroidales-specific Bac708R (3). The primer was tested with pools of reference host fecal DNAs (human and cat), as well as pools from sources not used in the experiment. The specificity of the primer was not limited to sources used in the hybridization. The new primer did not amplify fecal DNAs from human, cat, cow, pig, chicken, or gull sources (Fig. 4C). It amplified fecal DNAs from 17 of the 20 individual dog fecal samples used in the hybridization and from samples from seven individual dogs not used in the original experiment (data not shown). The detection limit for DF475 was 100 gene copies, and the limit was unchanged when DNA extracted from creek water was added to the reaction mixtures (data not shown). Optimized conditions for primers DF475F and EF990R were 2 mM MgCl2 and 30 cycles of 94°C for 1 min, 62°C for 45 s, and 72°C for 1 min, followed by a final 7-min extension at 72°C.
Primer DF475F was used successfully in a national comparative study of fecal-source tracking methods (12, 14). Unidentified aqueous samples containing mixtures of four fecal sources or sewage were prepared in three different matrices: distilled water, salt water, and humic acids. DF475F correctly identified the marker in all samples containing dog feces, including a sample containing humic acids.
DISCUSSION
Subtractive hybridization is a valuable, relatively inexpensive molecular tool, but it can require many complex steps to achieve adequate enrichment for target-specific DNA. The use of microplate wells to immobilize subtracter fragments eliminated the need to separate target from subtracter following hybridization. Subtracted target DNA was PCR amplified directly from the supernatant, and one round of hybridization provided sufficient enrichment for target-specific fragments. Sequencing of only 10 clones from each experiment provided enough information for primer design. Had it not done so, additional clones could have been sequenced.
Maxisorp microplates (Nalge Nunc) contain a polystyrene surface that binds DNA noncovalently. Since immobilized DNA is stable for at least 6 to 12 h at 68°C (27), we expected that the 70°C, 2-h hybridization would occur without significant loss of subtracter DNA to the solution. A large excess of subtracter over target DNA was used to allow for any loss of subtracter due to reannealing.
Three formamide concentrations were chosen based on Na2+ concentration, GC content, and fragment sizes. Formamide is a denaturant and has the effect of decreasing the melting temperature of the DNA hybrid (23). Overly stringent conditions result in hybridization of perfect matches only, and subtraction products could lack the variability necessary for primer design. If too relaxed, the conditions result in no product at all. Since all three formamide concentrations resulted in similar bands, products of the 35% concentration were chosen for sequencing because this concentration produced a sufficient amount of unhybridized DNA for cloning. Ultimately, the design of primers that distinguish target from subtracter host Bacteroidales indicated that an appropriate stringency range was attained. Future experiments might include a greater range of stringencies or sequencing of products from more than one stringency for comparison, but because our primer design objectives were met, we did not pursue this further.
No fragments of less than 300 bp were recovered in either of the experiments, for which there are several possible explanations. Some of the restriction fragments ligated to each other, forming chimeric sequences; some small fragments may have been lost in this way. PCR bias may have resulted in preferential amplification of the most-abundant larger restriction fragments. Alternatively, the inverted terminal repeats created by ligation of identical linkers to either end of the fragments may have resulted in the formation of panhandle structures that prevented primer binding and amplification (1). This would be more kinetically favorable with smaller fragment sizes. Inouye and Hondo found that hybridization efficiency declined as fragment size fell below 227 bp (17). These observations suggest the use of a restriction enzyme that produces fragments larger than 200 bp.
The intent in using the entire 16S rRNA gene, internal transcribed spacer, and part of the 23S rRNA gene was to obtain as much sequence information as possible for primer design. Three of 20 total subtracted fragments aligned with the 23S rRNA gene, but the small number of database sequences available for comparison made it difficult to use these fragments for primer design. Fourteen of the 20 subtraction products were from the 16S rRNA gene, and all 10 of the elk feces-specific clones were from a region known to be hypervariable (E. coli positions 940 to 1370) (6).
Primer DF475F was the first and only dog-specific primer designed and tested, and it did not require stringent optimization to exclude amplification of other host fecal DNAs. This illustrates the ability of this method to enrich for regions of variability. Primer EF990R was the third elk-specific primer tested and the first to successfully distinguish cow and elk fecal Bacteroidales sequences. It did not distinguish elk and sheep Bacteroidales sequences, which was not the intent of this experiment, but that may be accomplished with another subtractive hybridization. Alternatively, it may be possible to design a primer based on the 5′ and 3′ ends of a target fragment. Any cross-reacting DNA could be sequenced, allowing more directed primer design.
Primer EF990R amplified elk fecal DNAs and did not amplify cow fecal DNAs, even when four times more cow fecal DNA was added. A previous study using blind samples (12, 14) demonstrated that Bacteroidales source-specific primers identified fecal sources correctly when the sources comprised as little as 1% of the total fecal contamination in the sample.
Previous studies reported the immobilization of up to seven genomic subtracters in one well (26, 27). An initial attempt to use seven subtracters with an elk-specific target resulted in subtraction products without enough variability for primer design. Future experiments may determine what limitations exist and what adjustments can be made in the use of multiple subtracters.
Microplate subtractive hybridization was successfully employed to generate a unique source-specific marker for dog fecal pollution and a marker that differentiates elk and cow fecal pollution. The results demonstrate the method's ability to enrich for variable regions of the 16S rRNA gene. In addition, the method could readily be adapted for other gene targets. The capacity to characterize markers that distinguish sources of fecal pollution without obtaining large numbers of clones for each new host will expedite the addition of new source markers for fecal pollution.
Acknowledgments
This work was supported in part by U.S. Environmental Protection Agency Star Program grant no. R82-7639, by grant NA16RG1039 (project R/SD-07) from the National Oceanic and Atmospheric Administration to the Oregon State University Sea Grant College Program, by U.S. Department of Agriculture award no. 00-S1130-9818, and by the Nucleic Acids and Protein Service Core of the Environmental Health Sciences Center, Oregon State University (grant no. P30 ES00210 from the National Institute of Environmental Health Sciences, National Institutes of Health).
We thank Katrin Zwirglmaier for her correspondence and advice and Sarah Walters for helpful discussions and support.
REFERENCES
- 1.Agron, P. G., M. Macht, L. Radnedge, E. W. Skowronski, W. Miller, and G. L. Andersen. 2002. Use of subtractive hybridization for comprehensive surveys of prokaryotic genome differences. FEMS Microbiol. Lett. 211:175-182. [DOI] [PubMed] [Google Scholar]
- 2.Bernhard, A. E., and K. G. Field. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66:1587-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernhard, A. E., and K. G. Field. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernhard, A. E., T. Goyard, M. Simonich, and K. G. Field. 2003. Application of a rapid method for identifying fecal pollution sources in a multi-use estuary. Water Res. 37:909-913. [DOI] [PubMed] [Google Scholar]
- 5.Boehm, A. B., J. A. Fuhrman, R. D. Mrše, and S. B. Grant. 2003. Tiered approach for identification of a human fecal pollution source at a recreational beach: case study at Avalon Bay, Catalina Island, California. Environ. Sci. Technol. 37:673-680. [DOI] [PubMed] [Google Scholar]
- 6.Brosius, J. J., L. Palmer, H. P. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calia, K. E., M. K. Waldor, and S. B. Calderwood. 1998. Use of representational difference analysis to identify genomic differences between pathogenic strains of Vibrio cholerae. Infect. Immun. 66:849-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, J. Y., C. D. Sifri, B. C. Goumnerov, L. G. Rahme, F. M. Ausubel, and S. B. Calderwood. 2002. Identification of virulence genes in a pathogenic strain of Pseudomonas aeruginosa by representational difference analysis. J. Bacteriol. 184:952-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daly, K., C. S. Stewart, H. J. Flint, and S. P. Shirazi-Beechey. 2001. Bacterial diversity within the equine large intestine as revealed by molecular analysis of cloned 16S rRNA genes. FEMS Microbiol. Ecol. 38:141-151. [Google Scholar]
- 9a.Dick, L. K., A. E. Bernhard, T. J. Brodeur, J. W. Santo Domingo, J. M. Simpson, S. P. Walters, and K. G. Field. 2005. Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification. Appl. Environ. Microbiol. 71:3184-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorfman, M. 2003. Testing the waters 2003: a guide to water quality at vacation beaches. Natural Resources Defense Council, New York, N.Y.
- 11.Dunbar, J., S. M. Barns, L. O. Ticknor, and C. R. Kuske. 2002. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 68:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Field, K. G., E. C. Chern, L. K. Dick, J. Fuhrman, J. Griffith, P. A. Holden, G. LaMontagne, J. Le, B. Olson, and M. T. Simonich. 2003. A comparative study of culture-independent, library-independent genotypic methods of fecal source tracking. J. Water Health 1:181-194. [PubMed] [Google Scholar]
- 13.Gilpin, B., T. James, F. Nourozi, D. Saunders, P. Scholes, and M. Savill. 2003. The use of chemical and molecular microbial indicators for faecal source identification. Water Sci. Technol. 47:39-43. [PubMed] [Google Scholar]
- 14.Griffith, J. F., S. B. Weisberg, and C. D. McGee. 2003. Evaluation of microbial source tracking methods using mixed fecal sources in aqueous test samples. J. Water Health 1:141-151. [PubMed] [Google Scholar]
- 15.Hold, G. L., S. E. Pryde, V. J. Russell, E. Furrie, and H. J. Flint. 2002. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol. Ecol. 39:33-39. [DOI] [PubMed] [Google Scholar]
- 16.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inouye, S., and R. Hondo. 1990. Microplate hybridization of amplified viral DNA segment. J. Clin. Microbiol. 28:1469-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreader, C. A. 1995. Design and evaluation of Bacteroides DNA probes for the specific detection of human fecal pollution. Appl. Environ. Microbiol. 61:1171-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leser, T. D., J. Z. Amenuvor, T. K. Jensen, R. H. Lindecrona, M. Boye, and K. Møller. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüβmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyons, S. R., A. L. Griffen, and E. J. Leys. 2000. Quantitative real-time PCR for Porphyromonas gingivalis and total bacteria. J. Clin. Microbiol. 38:2362-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mau, M., and K. N. Timmis. 1998. Use of subtractive hybridization to design habitat-based oligonucleotide probes for investigation of natural bacterial communities. Appl. Environ. Microbiol. 64:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meinkoth, J., and G. Wahl. 1984. Hybridization of nucleic acids immobilized on solid supports. Anal. Biochem. 138:267-284. [DOI] [PubMed] [Google Scholar]
- 24.Perrin, A., X. Nassif, and C. Tinsley. 1999. Identification of regions of the chromosome of Neisseria meningitidis and Neisseria gonorrhoeae which are specific to the pathogenic Neisseria species. Infect. Immun. 67:6119-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suau, A., R. Bonnet, M. Sutren, J.-J. Godon, G. R. Gibson, M. D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wassill, L., W. Ludwig, and K. H. Schleifer. 1998. Development of a modified subtraction hybridization technique and its application for the design of strain specific PCR systems for lactococci. FEMS Microbiol. Lett. 166:63-70. [Google Scholar]
- 27.Zwirglmaier, K., L. Wasill, W. Ludwig, and L. H. Schleifer. 2001. Subtraction hybridization in microplates: an improved method to generate strain-specific PCR primers. Syst. Appl. Microbiol. 24:108-115. [DOI] [PubMed] [Google Scholar]