Abstract
Genes encoding three putative endopeptidases were identified from a draft-quality genome sequence of Lactobacillus helveticus CNRZ32 and designated pepO3, pepF, and pepE2. The ability of cell extracts from Escherichia coli DH5α derivatives expressing CNRZ32 endopeptidases PepE, PepE2, PepF, PepO, PepO2, and PepO3 to hydrolyze the model bitter peptides, β-casein (β-CN) (f193-209) and αS1-casein (αS1-CN) (f1-9), under cheese-ripening conditions (pH 5.1, 4% NaCl, and 10°C) was examined. CNRZ32 PepO3 was determined to be a functional paralog of PepO2 and hydrolyzed both peptides, while PepE and PepF had unique specificities towards αS1-CN (f1-9) and β-CN (f193-209), respectively. CNRZ32 PepE2 and PepO did not hydrolyze either peptide under these conditions. To demonstrate the utility of these peptidases in cheese, PepE, PepO2, and PepO3 were expressed in Lactococcus lactis, a common cheese starter, using a high-copy vector pTRKH2 and under the control of the pepO3 promoter. Cell extracts of L. lactis derivatives expressing these peptidases were used to hydrolyze β-CN (f193-209) and αS1-CN (f1-9) under cheese-ripening conditions in single-peptide reactions, in a defined peptide mix, and in Cheddar cheese serum. Peptides αS1-CN (f1-9), αS1-CN (f1-13), and αS1-CN (f1-16) were identified from Cheddar cheese serum and included in the defined peptide mix. Our results demonstrate that in all systems examined, PepO2 and PepO3 had the highest activity with β-CN (f193-209) and αS1-CN (f1-9). Cheese-derived peptides were observed to affect the activity of some of the enzymes examined, underscoring the importance of incorporating such peptides in model systems. These data indicate that L. helveticus CNRZ32 endopeptidases PepO2 and PepO3 are likely to play a key role in this strain's ability to reduce bitterness in cheese.
The proteolytic enzyme system of lactic acid bacteria (LAB) includes diverse enzymes whose primary physiological functions involve housekeeping needs and acquisition of essential amino acids to support growth (9). Intracellular peptidases of LAB consist of both endopeptidases and aminopeptidases. Endopeptidases, due to their ability to hydrolyze peptide bonds within a peptide, are of particular interest, since they promote hydrolysis of peptides that are resistant to aminopeptidase activity. The proteolytic enzymes of LAB are also of practical interest because they play a major role in cheese maturation and flavor.
Bitterness, a common flavor defect in Cheddar and Gouda cheeses, results from the accumulation of hydrophobic bitter peptides to concentrations higher than their taste threshold. Formation of these peptides during cheese ripening is directly related to the activity and specificity of the cell envelope proteinase and chymosin (2, 14, 24). Degradation of these peptides is related to the activity of peptidases derived from the starter and nonstarter bacteria present (24). Bitter peptides typically contain relatively high levels of proline (24, 36), underscoring the importance of proline-specific endopeptidases in debittering cheese.
Proteolytic systems of lactobacilli have been studied extensively due to their ability to retain activity under the low-pH and high-salt conditions found in the cheese-ripening process (8). This is particularly true for the proteolytic system of Lactobacillus helveticus CNRZ32, a commercially used adjunct that can reduce bitterness in Cheddar and Gouda cheeses (8, 9, 26). While numerous enzymes of the proteolytic system of L. helveticus CNRZ32 have been identified (8), our understanding of the specific enzymes responsible for this strain's ability to reduce bitterness in cheese is incomplete. As part of this endeavor, previously characterized L. helveticus CNRZ32 aminopeptidase PepN and endopeptidase PepO2 were shown to hydrolyze model bitter peptides, β-casein (β-CN) (f193-209) and αS1-casein (αS1-CN) (f1-9) (5, 8). However, the hydrolysis specificities of previously characterized CNRZ32 endopeptidases PepE, PepO, and PepO2 toward these peptides under cheese-ripening conditions (pH 5.1, 4% NaCl, and 10°C) and in the presence of other cheese-derived peptides is largely unknown.
Recently, a draft-quality (4×) genome sequence for L. helveticus CNRZ32 was assembled and screened for genes encoding additional proteolytic enzymes (3). That effort identified coding sequences for three additional endopeptidases, designated PepE2, PepF, and PepO3, that appeared to be paralogs or orthologs of known endopeptidases. To utilize these enzymes to reduce bitterness in cheese, it would be advantageous to overexpress them in Lactococcus, a common cheese starter.
In this study, we independently cloned the genes encoding these three endopeptidases into Escherichia coli DH5α and examined the hydrolysis specificities of these peptidases and previously characterized CNRZ32 endopeptidases PepE (16), PepO (6), and PepO2 (5) against β-CN (f193-209) and αSI-CN (f1-9) under cheese-ripening conditions. Additionally, we cloned pepO2, pepO3, pepE, and pepF under the control of the pepO3 promoter and examined the expression of these peptidases in Lactococcus lactis LM0230 using the high-copy vector pTRKH2. We examined the rate of hydrolysis and specific activity of the individual peptidases toward αS1-CN (f1-9) and β-CN (f193-209) in single-peptide reactions, in a defined mixture of cheese-derived peptides, and in Cheddar cheese serum (CCS).
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strains and plasmids used in this study are presented in Table 1. E. coli DH5α (Gibco-BRL Life Technologies, Inc., Gaithersburg, MD) and derivatives were grown in Luria-Bertani (33) medium at 37°C with aeration. L. lactis was grown at 30°C without aeration in M17 (Difco, Detroit, MI) supplemented with 0.5% (wt/vol) glucose (G-M17) or lactose (L-M17). L. helveticus CNRZ32 (23) was grown in MRS broth (Difco) (12) at 37°C without aeration. Agar plates were prepared by adding 1.5% (wt/vol) granulated agar (Difco) to liquid media. To select for E. coli strains carrying pBluescript II SK (+) (Stratagene, La Jolla, CA) and its derivatives, ampicillin (Sigma, St. Louis, MO) was added to media to a final concentration of 100 μg ml−1. Erythromycin (Sigma) was added to liquid media or agar plates to select for pJDC9, pTRKH2, and their derivatives in E. coli and L. lactis at 500 μg ml−1 and 5 μg ml−1, respectively. Bacteria were maintained as frozen stocks at −80°C in liquid media containing 12% glycerol.
TABLE 1.
List of bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | Strain with high-efficiency cloning, enables α-complementation | Bethesda Research Laboratories |
| ABLE C | Cloning strain, reduces copy number of ColE1-based vectors by fourfold | Strategene, La Jolla, CA |
| L. helveticus CNRZ32 | Wild type | Steele Laboratory collection |
| L. lactis LM0230 | Plasmid-free laboratory strain | 13 |
| Plasmids | ||
| pTRKH2 | EmrlacZ; 6.9 kb | 30 |
| pJDC9 | EmrlacZ; 6.85 kb | 4 |
| pTRKH-N | 3.8-kb SmaI-SalI fragment containing L. helveticus CNRZ32 pepN ligated into pTRKH2; Emr | J. E. Christensen (Steele Laboratory collection) |
| pSUWL29 | 3.0-kb PstI fragment containing CNRZ32 pepO2 ligated into pJDC9 | Y.-S. Chen (Steele Laboratory collection) |
| pSUW650 | 2.6-kb BamHI-KpnI fragment containing pepO3 ligated into pJDC9 | This study |
| pSUW651 | 2.5-kb BamHI-KpnI fragment containing pepF ligated into pJDC9 | This study |
| pSUW652 | 2.1-kb BamHI-KpnI fragment containing pepE2 ligated into pJDC9 | This study |
| pSUW653 | 2.0-kb SmaI-XbaI fragment containing pepE ligated into pJDC9 | This study |
| pSUW660 | 0.2-kb KpnI-BamHI fragment containing CNRZ32 PpepO3 ligated into pBlueskript II SK (+) | This study |
| pSUW661 | 2.4-kb BamHI-XbaI ORF containing CNRZ32 pepO2 ligated into pSUW660 | This study |
| pSUW662 | 1.7-kb BamHI-XbaI ORF containing CNRZ32 pepE ligated into pSUW660 | This study |
| pSUW663 | 2.5-kb BamHI-SacI ORF containing CNRZ32 pepF ligated into pSUW660 | This study |
| pSUW664 | 2.6-kb PCR fragment containing CNRZ32 pepO3 from pSUW650 ligated into pTRKH2 | This study |
| pSUW665 | 2.6-kb PvuII-XbaI fragment containing CNRZ32 pepO2 from pSUW661 ligated into pTRKH2 | This study |
| pSUW666 | 1.9-kb PvuII-XbaI fragment containing CNRZ32 pepE from pSUW662 ligated into pTRKH2 | This study |
Molecular biology techniques.
DNA cloning and plasmid isolation techniques were performed according to the method of Sambrook et al. (33). Restriction and modifying enzymes were used as recommended by the manufacturer (Invitrogen, Carlsbad, CA). Transformation of E. coli was performed with a GenePulser following the manufacturer's recommended instructions (Bio-Rad Laboratories, Richmond, CA). For transformation of L. lactis, the procedure of Holo and Nes (21) was utilized. L. helveticus CNRZ32 chromosomal DNA was isolated as described by Marmur (27). Lactococcal plasmid DNA was isolated from a 50-ml culture using a modified alkaline lysis method (33) with an addition of lysozyme (30 mg ml−1) to the resuspension buffer. DNA used for cloning was further purified by passing it through minicolumns from a Qiaquick PCR purification kit (QIAGEN, Valencia, CA) and was resuspended in a final volume of 30 to 50 μl. For isolation of DNA from gels, the Qiaquick gel extraction kit (QIAGEN) was used.
DNA amplification via PCR.
The DNA primers listed in Table 2 were synthesized by Invitrogen. Amplification reactions were typically performed using Taq DNA polymerase; for high fidelity reactions, Platinum Pfx DNA polymerase (Invitrogen) was utilized. The PCR cycling conditions for amplification of DNA both from E. coli and L. lactis normally included 95°C for 5 min, followed by 25 to 30 cycles of 94°C for 30 s, 50 to 60°C for 30 s, and 72°C for 1 min per kb of the fragment amplified, followed by a single cycle of 72°C for 7 min. “Direct colony” PCR was used to screen transformants; the fragment of interest was amplified directly from the colonies without initial DNA template isolation. A sterile plastic pipette tip was used to pick colonies from plates, and cells were mixed with 20 μl of standard PCR mix containing Taq DNA polymerase (Invitrogen). To lyse the cells prior to standard cycling conditions, samples were heated to 98°C for 10 min.
TABLE 2.
List of sequence-specific primers
| Primer name | Description | Sequence (5′-3′)a |
|---|---|---|
| PepO3-For-BamHI | Forward, amplification of pepO3 | CGGGATCCTTTTGACTTTGGGTGAAT |
| PepO3-Rev-KpnI | Reverse, amplification of pepO3 | GGGGTACCACGAGAAGTGGTTAGTTGA |
| PepF-For-BamHI | Forward, amplification of pepF | CGGGATCCCTTAAGGGAGTTCGGAG |
| PepF-Rev-KpnI | Reverse, amplification of pepF | GGGGTACCTTGGAGGAATTCATCTTTAG |
| PepE2-For-BamHI | Forward, amplification of pepE2 | CGGGATCCTATAACAAGAACGCTAAGAA |
| PepE2-Rev-KpnI | Reverse, amplification of pepE2 | GGGGTACCCAGATAATGGCAAATGATA |
| PepE-For-SmaI | Forward, amplification of pepE | TCCCCCGGGATTAGATTAAGCAAG |
| PepE-Rev-XbaI | Reverse, amplification of pepE | GCTCTAGAGAAATTCGCCCTGGTC |
| KpnI-PpepO3-For | Forward, amplification of PpepO3 | GGGGTACCGACTTTGGGTGAATC |
| BamHI-PpepO3-Rev | Reverse, amplification of PpepO3 | CGGGATCCCATTTTATTATTCAAAGAGAA |
| BamHI-PepF-ORF-For | Forward, amplification of pepF ORF | CGGGATCCCCAACAAGAAGCGAAGTC |
| SacI-PepF-ORF-Rev | Reverse, amplification of pepF ORF | GCTGGAGCTCGTCAGCTTTTTGTATGG |
| BamHI-PepO2-ORF-For | Forward, amplification of pepO2 ORF | CGCGGATCCAATTTAGCAAAAATC |
| XbaI-PepO2-ORF-Rev | Reverse, amplification of pepO2 ORF | GCTCTAGATCAATTATATAACTGATAC |
| BamHI-PepE-ORF-For | Forward, amplification of pepE ORF | CGGGATCCGAATTAACTGTGCAGG |
| XbaI-PepE-ORF-Rev | Reverse, amplification of pepE ORF | GCTCTAGAGAAATTCGCCCTGGTC |
The restriction site included in each primer is underlined.
DNA sequencing and sequence analysis.
DNA sequencing was conducted with sequence-specific primers synthesized by Invitrogen (Table 2). Sequencing reactions were performed using the ABI Big Dye reaction mix (Applied Biosystems, Foster City, CA) and a Perkin-Elmer model 480 thermal cycler (Perkin-Elmer Corp., Norwalk, CT). Sequence analysis was done on an ABI 377XL DNA sequencer by the Nucleic Acid and Protein facility of the University of Wisconsin Biotechnology Center (Madison, WI). The sequences were assembled and analyzed using Lasergene (DNASTAR Inc., Madison, WI) sequence analysis software.
Cloning of L. helveticus CNRZ32 endopeptidases.
DNA and protein analyses of the draft sequence using online BLAST search engines (http://www.ncbi.nlm.nih.gov/BLAST/) detected several new putative endopeptidases (Table 3). Sequence-specific primers with added BamHI and KpnI linkers were designed for the CNRZ32 pepE2, pepF, and pepO3 genes (Table 2) to amplify fragments including the ribosome binding site, promoter regions, coding sequence, and inverted repeats located within 300 to 400 bp downstream of the coding sequence. The respective genes were amplified from the total genomic DNA of L. helveticus CNRZ32 using Platinum Pfx DNA polymerase (Invitrogen). The resulting amplicons were purified, digested with BamHI and KpnI, ligated to similarly double-digested pJDC9 (4), and then transformed into E. coli DH5α. L. helveticus pepE (16) was amplified with its native promoter from the total DNA template of L. helveticus CNRZ32 using Platinum Pfx DNA polymerase and gene-specific primers (Table 2) with SmaI and XbaI linkers. The resulting amplicon of ∼2.0 kb was digested with SmaI and XbaI, ligated to similarly digested pJDC9, and transformed into DH5α. Putative transformants for all constructs were screened using gene-specific primers by direct colony PCR. Restriction digest analysis and sequencing of the gene were performed to confirm the presence of cloned endopeptidase genes in transformants, and representative isolates containing cloned pepO3, pepF, pepE2, and pepE and were designated pSUW650, pSUW651, pSUW652, and pSUW653, respectively (Table 1). All cloned fragments were sequenced and found to be identical to the L. helveticus CNRZ32 genome sequence of the respective genes. The nucleotide sequences of pepO3, pepF, and pepE2 have been deposited in GenBank (see “Nucleotide sequence accession number” below).
TABLE 3.
L. helveticus CNRZ32 genes encoding known or putative endopeptidases
| Gene | GenBank accession no. | Known (reference) or predicted product |
|---|---|---|
| pepE | AAB52540 | Thiol-dependent endopeptidase (16) |
| pepE2 | AY365130 | PepE paralog, 52% identical to CNRZ32 endopeptidase PepE (16) |
| pepF | AY365129 | PepF ortholog, 46% identical to L. lactis endopeptidase PepF |
| pepO | AF019410 | Endopeptidase O ortholog (6) |
| pepO2 | AF321529 | Post-prolyl endopeptidase (5) |
| pepO3 | AY355128 | PepO/PepO2 paralog, 62% identical to CNRZ32 endopeptidases PepO and PepO2 |
| gcp | Gcp ortholog, 63% identical to predicted O-sialoglycoprotein endopeptidase Gcp from L. plantarum | |
| ydiC | Glycoprotein endopeptidase ortholog, 37% identical to predicted glycoprotein endopeptidase from L. plantarum |
Construction of PpepO3 transcriptional fusion plasmids.
The promoter region of pepO3 (GenBank accession number AF019410) was amplified from pSUW650 using Platinum Pfx DNA polymerase (Invitrogen) with primers KpnI-PpepO3-For and BamHI-PpepO3-Rev (Table 2). The amplified pepO3 promoter fragment was digested with BamHI and KpnI and then ligated to similarly digested pBluescript II SK(+) (Stratagene) to generate transcriptional fusion plasmid pSUW660. The ligation mixture was electroporated into E. coli DH5α, and blue-white screening by α-complementation using isopropyl-β-d-thiogalactopyranoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; Invitrogen) was employed to identify putative transformants. Direct colony PCR using the above primers, restriction digest analysis, and nucleotide sequencing confirmed the presence of the promoter fragment of pepO3 in pSUW660.
Cloning of L. helveticus CNRZ32 endopeptidase genes as PpepO3 transcriptional fusions.
The open reading frame (ORF) of L. helveticus CNRZ32 pepO2 starting with the second codon and with transcription terminators was amplified from pSUWL29 (Table 1) via PCR using Platinum Pfx polymerase and primers BamHI-PepO2-ORF-For and XbaI-PepO2-ORF-Rev (Table 2). A similar approach was used to prepare comparable fragments of the L. helveticus CNRZ32 pepE and pepF genes using PCR primer pairs BamHI-PepE-ORF-For-XbaI-PepE-ORF-Rev and BamHI-PepF-ORF-For-SacI-PepF-ORF-Rev, respectively (Table 2). The pepO2 and pepE fragments were digested with BamHI and XbaI and ligated to similarly digested pSUW660 to generate pSUW661 and pSUW662, respectively (Table 1). The pepF fragment was digested with BamHI and SacI and ligated to similarly digested pSUW660 to generate pSUW663. Ligation mixtures were transformed into E. coli ABLE C, and then representative transformants with each plasmid were identified via direct colony PCR using the respective gene-specific primers. Endopeptidase activity was confirmed with chromogenic substrates as described by Chen et al. (5).
Cloning of L. helveticus CNRZ32 endopeptidases into L. lactis using pTRKH2.
The L. helveticus CNRZ32 pepO3 gene with its promoter region was cleaved from pSUW650 by digestion with BamHI and SacI and ligated to similarly digested pTRKH2 to generate pSUW664 (Table 1). L. helveticus CNRZ32 PpepO3:pepO2 and PpepO3:pepE were cleaved from pSUW661 and pSUW662, respectively, using PvuII and XbaI sites and ligated to SmaI- and XbaI-digested pTRKH2 to generate pSUW665 and pSUW666, respectively. L. helveticus CNRZ32 PpepO3:pepF was removed from pSUW663 via digestion with PvuII and SacI and ligated to SmaI- and SacI-digested pTRKH2 to generate pSUW667. Ligation mixtures were electroporated into L. lactis LM0230, and transformants were isolated from L-M17 plates containing 5 μg ml−1 erythromycin after 48 h of anaerobic incubation at 30°C. The presence of the constructs in the transformants was confirmed using direct colony PCR with primers specific to the promoter of pepO3 and the pTRKH2-specific primer. However, several attempts to ligate PpepO3:pepF to pTRKH2 followed by direct transformation into L. lactis LM0230 were unsuccessful. Endopeptidase activity was confirmed with chromogenic substrates as described by Chen et al. (5).
Preparation of CCS.
CCS was prepared as described by Morris et al. (29) using custom-made molds designed by Hassan (20). Briefly, 850 g of a 3-week-old Cheddar cheese was grated and mixed with an equal quantity of sterile sea sand. The sea sand-cheese mixture was placed in the molds and squeezed using a Carver manual hydraulic press (model 3912; Fred S. Carver, Inc., Summit, NJ). Pressure was increased up to 10,000 lb/in2 over 6 h and then held at that pressure for ∼3 h. The CCS and cheese liquid fat were collected and kept at 4°C for 2 h. This storage temperature allowed the expressed fat to solidify as the upper layer, which was removed using a spatula. Residual fat was removed by centrifugation at 1,380 × g for 10 min using an induction drive centrifuge (model J2-21 M; Beckman Coulter, Fullerton, CA). Additionally, CCS was passed through a 1.6-μm glass microfiber filter (Whatman International Ltd., England), filter sterilized by sequential passages through 0.45-μm and 0.22-μm cellulose nitrate filters (Nalgene Filtration Products, Rochester, NY), and stored at −80°C until used. For peptide hydrolysis, CCS was prepared as described above, boiled for 5 min at 100°C, extracted with equal volumes of 100% ethyl ether, vacuum dried using a Savant Speed Vac (SC 210A; Global Medical Instrumentation, Inc., Albertville, MN), and then resuspended in a volume of 50 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer (pH 5.0) to yield an effective 2× concentrated CCS.
Synthesis of peptide substrates.
The peptides β-CN (f193-209), αS1-CN (f1-6), αS1-CN (f1-9), αS1-CN (f1-13), and αS1-CN (f1-16) were synthesized and then purified by preparatory reverse-phase high-performance liquid chromatography (RP-HPLC) by the University of Wisconsin Biotechnology Center. Peptide purity and identity was established by mass spectrometry (MS) using a Bruker Reflex II for matrix-assisted laser desorption ionization-time of flight. The peptides were lyophilized and stored at −80°C. Stock solutions were prepared in sterile, double-distilled water and stored at −80°C.
Peptide hydrolysis reactions.
Cultures were grown to late log phase (A600, ∼2.0), and cell extracts (CFEs) were prepared in 50 mM MES buffer (pH 5.1). Cells were broken by vortexing with 300 mg of glass beads for 3 min using a Turbomix attachment to a Vortex Genie 2 (Scientific Industries, NY). CFEs from E. coli expressing CNRZ32 pepE, pepE2, pepF, and pepO3 transcriptional fusion genes were assayed for endopeptidase activity against chromogenic substrates as described by Chen et al. (5). CFE protein concentrations were determined using the protein assay kit I from Bio-Rad with bovine serum albumin as the protein standard.
Peptide hydrolysis reactions with CFEs from E. coli or L. lactis were performed in a 50-μl total volume. Each sample contained 10 μl of CFE (2.0 to 2.5 mg of protein ml−1), 10 μl of peptide substrate solution, and 30 μl of buffer. The buffer used for single-peptide reactions and defined peptide mix reactions was 120 mM MES (pH 5.1)-0.68 M NaCl (4% NaCl). CCS concentrate with a final pH of 5.2 and 4% NaCl was used for the cheese model system. The reactions were initiated by substrate addition, and then samples were incubated at 10°C for 2, 4, 6, 12, 24, and 48 h. Initial substrate concentrations in individual peptide reactions were 1 mg ml−1 for β-CN (f193-209) and 10 mg ml−1 for αS1-CN (f1-9). In the defined peptide mix reactions, peptides αS1-CN (f1-9), αS1-CN (f1-13), αS1-CN (f1-16), αS1-CN (f1-6), and β-CN (f193-209) were present at 10, 5, 2.5, 1, and 1 mg ml−1 concentrations, respectively. αS1-CN (f1-9) and β-CN (f193-209) were added to the peptide mix and CCS at levels known to accumulate in bitter cheeses (2), while the other peptides were added at levels that would mimic our spiked CCS. Reactions were stopped by the addition of trifluoroacetic acid (TFA) to a final concentration of 5%, and samples were frozen at −20°C until analyzed by HPLC.
Peptide separation and identification of hydrolysis products.
Peptides were separated by RP-HPLC on an HP1100 series system (Agilent Technologies, Palo Alto, CA) using 0.1% TFA in HPLC-grade water (solvent A) and 0.085% TFA, 80% acetonitrile, 20% HPLC-grade water (solvent B). The samples were analyzed on an Ultima C18 column (250 by 2.1 mm, 5-μm particle size, 100-Å pore size; Alltech Associates, Inc., Deerfield, IL). The initial condition was 10% solvent B, and a linear gradient of solvent B from 10% to 60% was generated over the course of 35 min for the separation of β-CN (f193-209), the defined peptide mix, and CCS samples. A linear gradient from 10 to 20% was generated over the course of 15 min for the separation of αS1-CN (f1-9) at a flow rate of 0.25 ml/min at 25°C. The eluted peaks were detected by absorbance at 214 nm using a photodiode array detector spectrometer (HP1100 series). Before injection, samples inactivated with TFA were thawed at room temperature and centrifuged (14,000 × g for 5 min at 25°C); 20 μl of the supernatant was injected directly into the column using an HP1100 series autosampler equipped with a dilutor module that contained a water-acetonitrile (1:1) wash solution (Agilent Technologies). A standard curve for pure peptide was generated for every run of HPLC with an R2 of 0.99 when determining concentrations of the substrates by peak area. The substrate was hydrolyzed to ∼10 to 20%, and reaction rates were verified to be in the linear range (R2 = 0.99) when calculating specific activity. In the case of αS1-CN (f1-9), the reaction rates were calculated from a nonlinear range to include at least three different time points (R2 = 0.93 to 0.95). Average specific activities are reported for this peptide. Specific activity was calculated as nmoles of peptide hydrolyzed per h per mg of protein, and reported values were corrected by subtracting the mean values obtained in the control treatments.
Mass values for RP-HPLC-separated peptides were determined using a triple-quadrupole mass spectrometer (Quattro II; Micromass Ltd., Manchester, United Kingdom) with electrospray ionization sources at the University of Wisconsin Biotechnology Center. To identify hydrolysis products, individual mass values were compared to the calculated molecular masses of peptides and amino acids derived from β-CN (f193-209) and αS1-CN (f1-9). Sample preparation and identification of CCS peptides using tandem mass spectrometry were also performed at the University of Wisconsin Biotechnology Center. Data-dependent MS/MS switching on quadrupole-time of flight was done using MassLynx software. Raw data were analyzed using MASCOT (Matrix Science Ltd., London, United Kingdom) and Spectrum Mill (Agilent Technologies) software, allowing for oxidized methionine, phosphotyrosine, and phosphoserine modifications. The identities of individual peptides were determined by comparison against the SWISS PROT database.
Statistical analysis.
The rate of hydrolysis of the peptides and the specific activity were calculated in nmoles of peptide hydrolyzed per h per mg of protein. Results reported are mean values ± standard deviations of the results from three independent trials. Student's t test with a P value of ≤0.05 was used to report significant differences between values obtained from control samples and individual peptidases. A single-factor analysis of variance was performed, and the least significant differences were calculated (34) for separation of means of individual peptidases in single-peptide reactions, in the defined peptide mix, and in CCS.
Nucleotide sequence accession number.
The nucleotide sequences of pepO3, pepF, and pepE2 have been deposited in the GenBank database under accession numbers AY355128, AY365129, and AY365130, respectively.
RESULTS
Sequence analysis.
The ORF of pepO3 was 1,929 bp, encoding a polypeptide of 643 amino acid residues with a deduced mass of 71.4 kDa. PepO3 was 62% identical to PepO2 and PepO from L. helveticus CNRZ32 (GenBank accession number AF321529 and AF019410, respectively) and 78% identical to a predicted metalloendopeptidase from Lactobacillus gasseri (protein accession number ZP_00045894.1). The pepF ORF was 1,794 bp, encoding a polypeptide of 598 amino acid residues with a deduced mass of 66.4 kDa. PepF had 52%, and 46% identity to previously characterized PepF proteins from Lactobacillus plantarum (protein accession number NP_785715.1) and L. lactis (A55485) (28), respectively. It had 75% identity to oligopeptidase F from L. gasseri (protein accession number ZP_00046654.1). The pepE2 ORF was 1,311 bp, encoding a polypeptide of 437 amino acid residues with a deduced mass of 48.5 kDa. PepE2 had 52% identity to a previously characterized PepE protein from L. helveticus CNRZ32 and 80% identity to aminopeptidase C from L. gasseri (protein accession number ZP_00047232.1). The ORFs of pepO3 and pepF encode the sequences HEISH and HETGH, respectively, which are characteristic of the HEXXH motif present in zinc metallopeptidases (1). The amino acid residues (Q, H, N, and W at positions 64, 361, 382, and 384, respectively) important for substrate binding and catalysis by cysteine proteinases of prokaryotic and eukaryotic origin are conserved in PepE2 (16).
Peptide hydrolysis and specificity by E. coli CFE.
CFE of E. coli derivatives expressing PepO2 or PepO3 had significantly greater activity (P ≤ 0.05) with both β-CN (f193-209) and αS1-CN (f1-9) than the control, CFE from E. coli DH5α (pJDC9); the highest activity was observed with PepO2 (Table 4). PepF activity was detected with β-CN (f193-209) but not with αS1-CN (f1-9). PepE activity with αS1-CN (f1-9) was not higher than the control and, therefore, is reported as not detectable (Table 4). There was no PepE activity detected with β-CN (f193-209). PepO and PepE2 activities were not detected with either peptide.
TABLE 4.
Specific activities of CFEs of E. coli DH5α expressing L. helveticus CNRZ32 endopeptidases toward αS1-CN (f1-9) and β-CN (f193-209)1
| Peptidaseb | Mean (SD) sp act towarda:
|
|
|---|---|---|
| αS1-CN (f1-9) | β-CN (f193-209) | |
| Control | 46 (19)‡ | 0.3 (0.2)§ |
| PepE | ND | ND |
| PepE2 | ND | ND |
| PepF | ND | 14 (3.0)* |
| PepO | ND | ND |
| PepO2 | 3,700 (125)* | 290 (19)† |
| PepO3 | 85 (21)† | 88 (5.0)‡ |
Specific activity is measured in nmoles of substrate hydrolyzed per h per mg of protein. Values were corrected by subtracting the mean values obtained in the control treatments from CFE of E. coli DH5α (pJDC9). Means with different symbols are statistically different within a column at an α value of ≤0.05. Assays were performed under cheese-ripening conditions (pH 5.0 to 5.2, 4% NaCl, 10°C). ND, not detected.
CFEs were prepared from E. coli DH5α(pJDC9) (control), E. coli DH5α(pSUW653) (PepE), E. coli DH5α(pSUW652) (PepE2), E. coli DH5α(pSUW651) (PepF), E. coli DH5α(pSUW51) (PepO), E. coli DH5α(pSUW29) (PepO2), or E. coli DH5α(pSUW650) (PepO3).
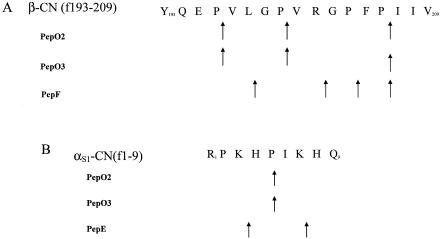
Hydrolysis specificities of PepO2, PepO3, PepE, and PepF with β-CN (f193-209) and αS1-CN (f1-9) were determined. Like PepO2 (5), PepO3 was a postproline endopeptidase that hydrolyzed the Pro206-Ile207, Pro196-Val197, and Pro200-Val201 bonds of β-CN(f193-209) and the Pro5-Ile6 bond of αS1-CN (f1-9) (Fig. 1). PepF also demonstrated postproline specificity at the Pro204-Phe205 and Pro206-Ile207 bonds but additionally hydrolyzed the X-Gly Lys198-Gly199 and Arg202-Gly203 bonds of β-CN(f193-209). PepE hydrolyzed αS1-CN (f1-9) at the Lys3-His4 and Lys7-His8 bonds.
FIG. 1.
Specificity of Lactobacillus helveticus CNRZ32 endopeptidases toward β-CN (f193-209) (A) and αS1-CN (f1-9) (B) under simulated cheese-ripening conditions (pH 5.0 to 5.2, 4% NaCl, 10°C).
Identification of peptides in CCS.
The peptide/protein summary of CCS generated by Spectrum Mill (Agilent Technologies) and MASCOT (Matrix Science Ltd.) identified peptides and phosphopeptides from αS1-CN and β-CN. The majority of the peptides identified from β-CN were from premature (uncleaved) β-CN, β-CN (f60-81) and β-CN (f107-118). β-CN (f193-209) was not identified. The peptides identified from αS1-CN were αS1-CN (f1-9), αS1-CN (f1-13), αS1-CN (f1-16), αS1-CN (f1-17), αS1-CN (f24-41), and αS1-CN (f24-39).
Peptide hydrolysis by L. lactis CFE.
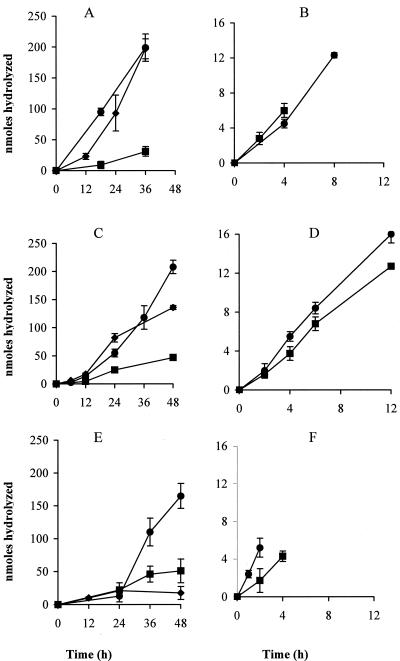
The time course of hydrolysis by CFE of LM0230 expressing PepO2, PepO3, and PepE of αS1-CN (f1-9) at a concentration of 10 mg ml−1 exhibited a slow initial rate (lag phase), which increased after 12 h of incubation with a single peptide and a defined peptide mix and after 24 h of incubation in CCS (Fig. 2). A lag phase (40) was not observed when lower concentrations (1 mg ml−1) of αS1-CN (f1-9) were used (data not shown), suggesting a possibility for substrate inhibition at higher concentrations (10 mg ml−1). LM0230 derivatives expressing PepO2 and PepO3 under the control of the pepO3 promoter hydrolyzed both β-CN (f193-209) and αS1-CN (f1-9) at a significantly greater rate (P ≤ 0.05) than the control, CFE from LM0230(pTRKH2), in all three systems (Fig. 2; Table 5). The peptide β-CN (f193-209) was almost completely hydrolyzed within 12 h when spiked CCS was incubated with CFE of strains expressing PepO2 or PepO3. The activity of PepO2 with β-CN (f193-209) was significantly higher (α ≤ 0.05) by twofold in spiked CCS than in single-peptide reactions or in the defined peptide mix (Table 5). The activity of PepO2 with αS1-CN (f1-9) was lower in CCS and the defined peptide reactions compared to the activity observed on αS1-CN (f1-9) in single-peptide reactions (Table 5).
FIG. 2.
Rate of β-CN (f193-209) (right panels) and αS1-CN (f1-9) (left panels) hydrolysis at pH 5.2, 4% NaCl at 10°C in a single-peptide system (A, B), in a defined peptide mix system (C, D), and in Cheddar cheese serum (E, F) by cell extracts from Lactococcus lactis LM0230 derivatives expressing Lactobacillus helveticus CNRZ32 endopeptidase PepO2 (circles), PepO3 (squares), or PepE (diamonds). Values were corrected by subtracting the values obtained from the control cell extract, prepared from L. lactis LM0230(pTRKH2). Error bars represent the standard errors of the means (n = 3). Initial substrate concentrations in individual peptide reactions were 1 mg ml−1 for β-CN (f193-209) and 10 mg ml−1 for αS1-CN (f1-9).
TABLE 5.
Specific activities of CFEs of L. lactis LM0230 expressing L. helveticus CNRZ32 endopeptidases towards β-CN (f193-209) and αS1-CN (f1-9) in a single-peptide reaction, in the defined peptide mix, and in Cheddar cheese serum
| Peptide and peptidaseb | Mean (SD) spact ina:
|
||
|---|---|---|---|
| Single-peptide reaction | Defined peptide mix | Cheddar cheese serum | |
| β-CN (f193-209) | |||
| Control | 16 (2.0)†,* | 9.0 (2.0)†,† | 27 (3.0)‡,* |
| PepE | ND | ND | ND |
| PepO2 | 64 (1.5)*,† | 42 (10)*,* | 120 (32)*,* |
| PepO3 | 81 (7.0)*,* | 40 (6.0)*,* | 61 (23)†,* |
| αS1-CN (f1-9)c | |||
| Control | 4.0 (4.0)‡,* | 4.0 (0.2)§,* | 23 (3.0)‡,† |
| PepE | 190 (14)*,* | 120 (7)*,† | 31 (17)‡,‡ |
| PepO2 | 240 (10)*,* | 84 (20)†,‡ | 150 (40)*,† |
| PepO3 | 41 (19)†,* | 38 (9.0)‡,* | 63 (25)†,* |
Specific activity is measured in nmoles of substrate hydrolyzed per h per mg of protein. Values were corrected by subtracting the mean values obtained in the control treatments from CFE of L. lactis LM0230(pTRKH2). Means with different symbols are statistically different at an α value of ≤0.05. Lightface symbols compare values within a column, and boldface symbols compare values within a row. Assays were performed under cheese-ripening conditions (pH 5.0 to 5.2, 4% NaCl, 10°C). ND, not detected.
CFEs were prepared from L. lactis LM0230(pTRKH2) (control), L. lactis LM0230(pSUW666) (PepE), L. lactis LM0230(pSUW665) (PepO2), or L. lactis LM0230(pSUW664) (PepO3).
In the case of αS1-CN (f1-9) hydrolysis, average specific activities are reported.
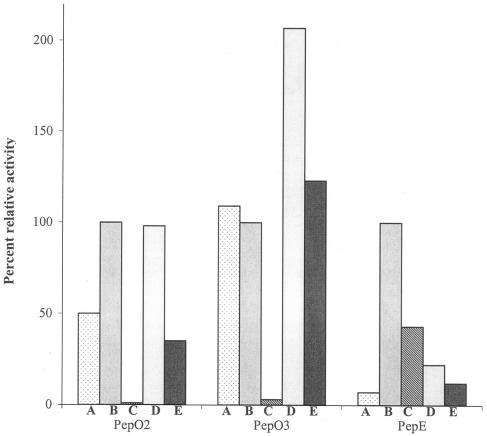
The CFEs from LM0230 derivatives expressing PepE hydrolyzed αS1-CN (f1-9) at a significantly greater rate (P ≤ 0.05) than the control CFE in single-peptide and defined peptide mix reactions (Fig. 2; Table 5). In CCS, PepE activity was significantly inhibited (α ≤ 0.05) (Table 5). There was higher background activity detected in the control CFE, L. lactis LM0230 (pTRKH2), for peptides αS1-CN (f1-9) and β-CN (f193-209) in CCS than in single-peptide reactions or in defined peptide mix reactions (data not shown); however, the peptide profiles of the control reactions were similar to those observed at time zero. The relative activity toward the other peptides in the defined peptide mix was also calculated, with activity toward αS1-CN (f1-9) taken as 100% (Fig. 3). Each peptidase hydrolyzed the different peptides at different rates. Among αS1-CN-derived peptides, for example, PepO2 had the highest activity toward peptides αS1-CN (f1-9) and αS1-CN (f1-13), PepO3 had the highest activity toward αS1-CN (f1-13), and PepE had the highest activity toward αS1-CN (f1-9) and αS1-CN (f1-6) (Fig. 3).
FIG. 3.
Percent relative activity of Lactococcus lactis LM0230 derivatives expressing Lactobacillus helveticus CNRZ32 PepO2, PepO3, or PepE toward peptides in the defined peptide mix at pH 5.2, 4% NaCl, and 10°C. (A) β-CN (f193-209); (B) αS1-CN (f1-9); (C) αS1-CN (f1-6); (D) αS1-CN (f1-13); (E) αS1-CN (f1-16). Values were corrected by subtracting the values obtained in the control treatments. Activity with αS1-CN (f1-9) was arbitrarily set to 100%.
DISCUSSION
Bitterness is a major flavor defect in Cheddar and Gouda cheeses (37). This defect is primarily caused by the accumulation of hydrophobic peptides, such as β-CN (f193-209) and αS1-CN (f1-9), to concentrations greater than their taste thresholds (2, 14, 24). The reduction of bitterness in cheese is believed to be the result of preferential hydrolysis of bitter peptides to nonbitter hydrolysis products by specific peptidases from starter or nonstarter bacteria (2, 19, 24).
In this investigation, we conducted functional studies on three putative endopeptidase genes, pepE2, pepF, and pepO3, which were identified from a draft-quality genome sequence of L. helveticus CNRZ32. Although two additional glycoprotein endopeptidases were identified (Table 3), genes encoding the latter enzymes were not included in this study, since glycoproteins are not thought to influence bitterness. We also included three previously characterized endopeptidases, PepE, PepO, and PepO2, in this investigation, since their hydrolysis specificities had not been determined under cheese-ripening conditions (pH of 5.0 to 5.2, 4% NaCl, 10°C). Additionally, we attempted to express these CNRZ32 peptidases in L. lactis under the control of the L. helveticus CNRZ32 pepO3 promoter on a high-copy vector. Peptide hydrolysis studies in single-peptide reactions, in a defined peptide mix, and in CCS were then conducted to evaluate the debittering potential of the genetically modified strains.
We successfully cloned and expressed the newly identified endopeptidases PepF and PepO3 from L. helveticus CNRZ32 in E. coli and determined their hydrolysis specificities toward two bitter peptides. In contrast, no measurable PepE2 activity was detected against chromogenic substrates or bitter peptides; this result may be due to either enzyme specificity or insufficient expression. L. helveticus CNRZ32 PepO3 appeared to be a functional paralog to the postprolyl endopeptidase PepO2 (5) and hydrolyzes β-CN (f193-209) and αS1-CN (f1-9) in a manner that was indistinguishable from that observed with the latter enzyme. PepF was also found to be a postproline endopeptidase, but this enzyme did not hydrolyze αS1-CN (f1-9). In contrast, PepE hydrolyzed αS1-CN (f1-9), but not β-CN (f193-209), and does not have postproline endopeptidase activity. Proline constitutes 16.7% of β-CN and 8.7% of αS1-CN and is present in the majority of the known bitter peptides (24, 38). The postproline hydrolysis of PepF, PepO2, and PepO3 toward β-CN (f193-209) and αS1-CN (f1-9) under cheese-ripening conditions suggests that these enzymes may contribute to the debittering activity of L. helveticus CNRZ32.
Genes from lactobacilli have been previously expressed in L. lactis (10, 39) using the inducible nisA promoter, the L. helveticus pepX promoter, or their own promoters (22, 25). A previous study of L. helveticus CNRZ32 promoter strength, which used β-glucuronidase as the reporter gene, found that pepO and pepO2 promoters were weakly expressed in L. lactis (7). In this study, we cloned pepO3 on pTRKH2 and observed relatively high PepO3 activity in L. lactis. Therefore, the ORFs of pepO2 and pepE were subsequently cloned under the control of the pepO3 promoter, and significant PepO2 and PepE activities were observed in the respective constructs, higher than when expressed using their native promoters. These results suggest that the pepO3 promoter may have utility for the expression of heterologous genes in lactococci.
Peptidase specificities for bitter peptides may be different in cheeses than in in vitro model systems. However, assessing the performance of the different strains expressing these peptidases during cheese ripening would be time-consuming and expensive; therefore, the development of a cheese-like system was desirable. Previously, researchers have used various types of cheese model systems that included cheese slurries, cheese pastes, and miniature cheeses (11, 15, 32, 35, 41). In this study, we used a cheese-like buffer system based on cheese serum that had been previously used for studying inorganic constituents and proteolysis in Cheddar and Emmental cheeses, respectively (18, 20, 29).
Peptides αS1-CN (f1-9), (f1-13) (f1-16), and (f1-17) were identified in CCS and are known to accumulate in cheese due to the activity of the lactococcal cell envelope proteinase and a lactococcal endopeptidase on αS1-CN (f1-23) (2, 17). We did not identify β-CN (f193-209) in our CCS; this was expected because accumulation of this peptide typically occurs after 1 month of ripening (2). A previous study demonstrated that peptides αS1-CN (f1-6) and αS1-CN (f1-16) were derived from αS1-CN (f1-23) by L. helveticus CNRZ32 PrtH (31); therefore, we believed that CNRZ32 would likely contain peptidases capable of hydrolyzing these peptides.
Recombinant lactococcal strains expressing specific lactobacillus peptidases have been used in numerous studies to study cheese flavor development and reduce bitterness (10, 11, 22). In this study, we demonstrated that three CNRZ32 endopeptidases (PepO2, PepO3, and PepE) had relatively high specific activities toward the peptides αS1-CN (f1-9) and β-CN (f193-209) when present alone, in the defined peptide mix, or in CCS under cheese-ripening conditions (pH 5.2, 4% NaCl, 10°C). Interestingly, the activities of PepO2 and PepO3 for both αS1-CN (f1-9) and β-CN (f193-209) were greater in CCS than in the defined peptide mix. One reason could be less inhibition by competing peptides, αS1-CN (f1-13) and αS1-CN (f1-16), that were present at a higher concentration in the defined peptide mix than in CCS. Alternatively, the ions present in CCS, particularly Ca2+, may activate PepO2 and PepO3, as they are metallopeptidases. In contrast, we noted significant inhibition of PepE activity in CCS. One possible explanation is competitive inhibition of PepE by peptides present in CCS.
In conclusion, we successfully expressed L. helveticus CNRZ32 PepO2, PepO3, and PepE under the control of PpepO3 in L. lactis LM0230 using a high-copy vector. The results indicated that PepO2 and PepO3 had equally high activities toward β-CN (f193-209), while PepO2 had greater activity for αS1-CN (f1-9) and αS1-CN (f1-13) in CCS at pH 5.2, 4% NaCl, and 10°C. We also determined that other cheese peptides can affect the activity of CNRZ32 endopeptidases, especially PepE, underscoring the importance of using simple but cheese-like model systems such as CCS for studying peptide hydrolysis. Moreover, we have identified two key enzymes from L. helveticus CNRZ32, PepO2 and PepO3, that have the potential to hydrolyze bitter peptides in cheese when expressed in a lactococcal strain. Future studies will include examining food-grade strains of lactococci that express CNRZ32 peptidases PepO2 and PepO3 in combination with aminopeptidase PepN to reduce bitterness in bacterial ripened cheeses, such as Cheddar and Gouda.
Acknowledgments
We thank Gary Case, Amy Harms, and James Brown of the University of Wisconsin—Madison Biotechnology Center for synthesis of β-CN (f193-209) and αS1-CN (f1-9) and for providing mass spectrometry expertise. We also thank Leslie Plhak for technical assistance with RP-HPLC and Kurt Fenster for critical evaluation of the manuscript.
Support for this research was provided by Dairy Management, Inc., Chr. Hansen, Inc., Wisconsin Center for Dairy Research, University of Wisconsin—Madison, and Utah State University.
REFERENCES
- 1.Barret, A. J., N. D. Rawlings, and J. F. Woessner. 1998. Introduction: metallopeptidases and their clans, p. 989-991. In A. J. Barret, N. D. Rawlings, and J. F. Woessner (ed.), Handbook of proteolytic enzymes. Academic Press, London, United Kingdom.
- 2.Broadbent, J. R., M. Barnes, C. Brennand, M. Strickland, K. Houck, M. E. Johnson, and J. L. Steele. 2002. Contribution of Lactococcus lactis cell envelope proteinase specificity to peptide accumulation and bitterness in reduced-fat cheddar cheese. Appl. Environ. Microbiol. 68:1778-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broadbent, J. R., J. L. Steele, D. L. Welker, and J. E. Hughes. 2002. Global analysis of the Lactobacillus helveticus CNRZ32 proteolytic enzyme system, abstr. K13. 7th International Symposium of Lactic Acid Bacteria Genetics, Metabolism, and Applications.
- 4.Chen, J., and D. A. Morrison. 1987. Cloning of Streptococcus pneumoniae DNA fragments in Escherichia coli requires vectors protected by strong transcriptional terminators. Gene 55:179-187. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Y.-S., J. E. Christensen, M. Strickland, and J. L. Steele. 2003. Identification and characterization of Lactobacillus helveticus PepO2, an endopeptidase with post-proline specificity. Appl. Environ. Microbiol. 69:1276-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, Y.-S., and J. L. Steele. 1998. Genetic characterization and physiological role of endopeptidase O from Lactobacillus helveticus CNRZ32. Appl. Environ. Microbiol. 64:3411-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Y.-S., and J. L. Steele. 2005. Analysis of promoter sequences from Lactobacillus helveticus CNRZ32 and their activity in other lactic acid bacteria. J. Appl. Microbiol. 98:64-72. [DOI] [PubMed] [Google Scholar]
- 8.Christensen, J. E., J. R. Broadbent, and J. L. Steele. 2003. Hydrolysis of casein-derived peptides αS1-casein(f1-9) and β-casein(f193-209) by Lactobacillus helveticus peptidase deletion mutants indicates the presence of a previously undetected endopeptidase. Appl. Environ. Microbiol. 69:1283-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen, J. E., E. G. Dudley, J. A. Pederson, and J. L. Steele. 1999. Peptidases and amino acid catabolism in lactic acid bacteria. Antonie Leeuwenhoek 76:217-246. [PubMed] [Google Scholar]
- 10.Christensson, C., H. Bratt, L. J. Collins, T. Coolbear, R. Holland, M. W. Lubbers, P. W. O'Toole, and J. R. Reid. 2002. Cloning and expression of an oligoendopeptidase, PepO, with novel specificity from Lactobacillus rhamnosus HN001 (DR20). Appl. Environ. Microbiol. 68:254-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Courtin, P., M. Nardi, U. Wegmann, V. Joutsjoki, J. C. Ogier, J. C. Gripon, A. Palva, B. Henrich, and V. Monnet. 2002. Accelerating cheese proteolysis by altering Lactococcus lactis proteolytic system with lactobacilli peptidases. Int. Dairy J. 12:447-454. [Google Scholar]
- 12.DeMan, J., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 13.Efstathiou, J. D., and L. L. Mckay. 1976. Inorganic salts resistance associated with a lactose-fermenting plasmid in Streptococcus lactis. J. Bacteriol. 130:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Exterkate, F. A. 1995. The lactococcal cell envelope proteinases: differences, calcium-binding effects and role in cheese ripening. Int. Dairy J. 5:995-1018. [Google Scholar]
- 15.Farkye, N. Y., S. A. Madkor, and H. G. Atkins. 1995. Proteolytic abilities of some lactic acid bacteria in a model cheese system. Int. Dairy J. 5:715-725. [Google Scholar]
- 16.Fenster, K. M., K. L. Parkin, and J. L. Steele. 1997. Characterization of a thiol-dependent endopeptidase from Lactobacillus helveticus CNRZ32. J. Bacteriol. 179:2529-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox, P. F., J. Law, and P. L. H. Sweeney. 1994. Proteolysis in cheese during ripening, p. 1-31. In A. T. Andrews and J. Varley (ed.), Biochemistry of milk products. Royal Society of Chemistry, Cambridge, United Kingdom.
- 18.Gagnaire, V., S. Lortal, and J. Léonil. 1998. Free active peptidases are detected in Emmental juice extracted before ripening in the warm room. J. Dairy Res. 65:119-128. [Google Scholar]
- 19.Gomez, M. J., P. Gaya, M. Nunez, and M. Medina. 1996. Effect of Lactobacillus plantarum as adjunct starter on the flavor and texture of a semi-hard cheese made from pasteurized cows' milk. Lait 76:461-472. [Google Scholar]
- 20.Hassan, A. 2001. Development of analytical methods to quantify the insoluble and soluble calcium content in Cheddar cheese and a study of its influence on cheese functionality. M.S. thesis. University of Wisconsin, Madison.
- 21.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joutsjoki, V., S. Luoma, M. Tamminen, M. Kilpi, E. Johansen, and A. Palva. 2002. Recombinant Lactococcus starters as a potential source of additional peptidolytic activity in cheese ripening. J. Appl. Microbiol. 92:1159-1166. [DOI] [PubMed] [Google Scholar]
- 23.Khalid, N. M., and E. H. Marth. 1990. Purification and partial characterization of a prolyldipeptidyl aminopeptidase from Lactobacillus helveticus CNRZ32. Appl. Environ. Microbiol. 56:381-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemieux, L., and R. E. Simard. 1992. Bitter flavor in dairy products. II. A review of bitter peptides from CNs: their formation, isolation and identification, structure masking and inhibition. Lait 72:335-382. [Google Scholar]
- 25.Luoma, S., K. Peltoniemi, V. Joutsjoki, T. Rantanen, M. Tamminen, I. Heikkinen, and A. Palva. 2001. Expression of six peptidases from Lactobacillus helveticus in Lactococcus lactis. Appl. Environ. Microbiol. 67:1232-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madkor, S. A., P. S. Tong, and M. El-Soda. 2000. Ripening of Cheddar cheese with added attenuated adjunct cultures of lactobacilli. J. Dairy Sci. 83:1684-1691. [DOI] [PubMed] [Google Scholar]
- 27.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 28.Monnet, V., M. Nardi, A. Chopin, M.-C. Chopin, and J.-C. Gripon. 1994. Biochemical and genetic characterization of PepF, an oligopeptidase from Lactococcus lactis. J. Biol. Chem. 269:32070-32073. [PubMed] [Google Scholar]
- 29.Morris, H. A., C. Holt, B. E. Brooker, J. M. Banks, and W. Manson. 1988. Inorganic constituents of cheese: analysis of juice from a one-month-old Cheddar cheese and the use of light and electron microscopy to characterize the crystalline phases. J. Dairy Res. 55:255-268. [Google Scholar]
- 30.O'Sullivan, D. J., and T. R. Klaenhammer. 1993. High and low copy number Lactococcus shuttle cloning vectors with features for clone screening. Gene 137:227-231. [DOI] [PubMed] [Google Scholar]
- 31.Pederson, J. A., G. J. Mileski, B. C. Weimer, and J. L. Steele. 1999. Genetic characterization of a cell envelope-associated proteinase from Lactobacillus helveticus CNRZ32. J. Bacteriol. 181:4592-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salles, C., S. Dalmas, C. Septier, S. Issanchou, Y. Noël, P. Etiévant, and J. L. Le Quéré. 1995. Production of a cheese model for sensory evaluation of flavour compounds. Lait 75:535-549. [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 34.SAS. 1985. SAS user's guide: statistics, version 5 ed. SAS Institute, Inc., Cary, N.C.
- 35.Shakeel-Ur-Rehman, A., H. Pripp, P. L. H. McSweeney, and P. F. Fox. 1999. Assessing the proteolytic and cheese ripening properties of single strains of Lactococcus in miniature cheeses. Lait 79:361-383. [Google Scholar]
- 36.Smid, E. J., B. Poolman, and W. N. Konings. 1991. Casein utilization by lactococci. Appl. Environ. Microbiol. 57:2447-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smukowski, M., W. L. Wendorff, Y. Ping, and R. D. Rao. 2003. Impact of cheese defects on U.S. graded cheeses. J. Dairy Sci. 86(Suppl. 1):364. [Google Scholar]
- 38.Swaisgood, H. E. 1982. The chemistry of milk protein, p. 1-59. In P. F. Fox (ed.), Developments in dairy chemistry, vol. 1. Elsevier, London, United Kingdom.
- 39.Wegmann, U., J. R. Klein, I. Drumm, O. P. Kuipers, and B. Henrich. 1999. Introduction of peptidase genes from Lactobacillus delbrueckii subsp. lactis into Lactococcus lactis and controlled expression. Appl. Environ. Microbiol. 65:4729-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitaker, J. R. 1994. Principles of enzymology for the food sciences, 2nd ed. Marcel Dekker, Inc., New York, N.Y.
- 41.Wijesundera, C., M. Roberts, and G. K. Y. Limsowtin. 1997. Flavour development in aseptic cheese curd slurries prepared with single-strain starter bacteria in the presence and absence of adjuncts. Lait 77:121-131. [Google Scholar]