Abstract
A two-phase cultivation system was developed which will enable studies of streptomycete differentiation by molecular biological and global techniques such as transcriptomics and proteomics. The system is based on a solid phase formed by glass beads corresponding to particles in soil, clay, or sand natural habitats of streptomycetes. The beads are immersed in a liquid medium that allows easy modification or replacement of nutrients and growth factors as well as radioactive labeling of proteins. Scanning electron microscopy was used to analyze morphological differentiation of streptomycetes on glass beads and two-dimensional protein electrophoresis to demonstrate the potential of the system for analyses of protein synthesis profiles during the developmental program. This system facilitates studies of differentiation including expression and posttranslation modifications of streptomycetes proteins, secondary metabolite biosynthesis, and morphological development.
Streptomycetes are saprophytic filamentous gram-positive bacteria inhabiting particulate soil ecosystems and marine sediments throughout the world. They are a particularly interesting group of bacteria, having a complex life cycle, diverse metabolic capabilities, and one of the largest of bacterial genomes (2). They excrete extracellular enzymes to hydrolyze polymers such as starch and cellulose and utilize the soluble breakdown products. Their ability to produce many important antibiotics and other useful metabolites has been of interest to academics and exploited by many pharmaceutical companies. The life cycle of streptomycetes is initiated by spore germination followed by the development of a branched hyphal network covering the surface of soil particles or organic debris. In response to complex but still poorly defined signals, the substrate mycelium produces aerial hyphae that eventually undergo septation to yield chains of unigenomic spores. This has proven to be a complex process linked to primary metabolism (17) and production of secondary metabolites, including many antibiotics (5, 6, 9, 18). Therefore, it is important to study streptomycete morphological differentiation, ideally under conditions that mimic its natural environment as closely as possible. To achieve this, streptomycetes have been cultivated on agar plates, sometimes covered with sheets of cellophane (1, 7) to facilitate harvesting of cells. However, there are several disadvantages. Agar gels do not simulate the particulate nature of soil ecosystems. Furthermore, it is not well adapted to allowing adjustment of the medium composition during growth, to providing radiolabeled nutrients or growth factors in a controllable manner, or to analyzing secondary metabolite production during cell differentiation. The problem of obtaining large homogenous samples from differentiating streptomycetes in a reproducible manner is encountered in all the transcriptomic and proteomic studies.
Here, we present a novel system for cultivation of the mycelial bacteria in a two-phase system composed of glass beads immersed in a liquid medium. The size of the beads can be selected to mimic as nearly as possible the particles of soil or sand found in natural habitats of streptomycetes. The defined liquid medium, filling the space between the beads, can be easily modified by the addition or replacement of nutrients, growth factors, or radiolabeling of proteins and nucleic acids. The use of glass beads thus allowed us to develop simple and inexpensive techniques for sequential monitoring of antibiotic production, labeling of differentiating cells, and disintegration of mycelia. Differentiation of mycelia cultivated on the beads was studied by scanning electron microscopy (SEM). Preparation and analysis of samples for proteomic studies was demonstrated using high-resolution two-dimensional (2D) polyacrylamide gel electrophoresis (PAGE) of cell-free protein homogenates from Streptomyces mycelia grown on glass beads in the presence of [35S]methionine or [32P]phosphate labels. The potential use of this cultivation system in studies of functional genomics and metabolomics of streptomycetes will be discussed.
MATERIALS AND METHODS
Materials.
The glass beads “Balotina” no. 10 from ORNELA a.s. (Desna, in Jizerské mountains, Czech Republic) were in the size range of 265 to 325 μm. Originally used as a reflective additive in street paints, we adopted them for growing and breaking Streptomyces cells. The glass beads were boiled in 0.1 M HCl for 30 min, rinsed many times with distilled water until the washing water remained neutral, and dried at 120°C before use. l-[35S]methionine (SJ 1015, cell labeling grade) was purchased from Amersham Biosciences; [32P]orthophosphate was from ICN Biomedicals. All other chemicals were of reagent or analytical grade.
Strains and cultivation conditions.
Streptomyces coelicolor A3(2) J1501 (John Innes Center, Norwich, United Kingdom), Streptomyces granaticolor (ETH 7437), Streptomyces collinus (ETH 24318), and Streptomyces aureofaciens CCM3239 (Czechoslovak Collection of Microorganisms, Brno, Czech Republic) were used to test the cultivation system. The mycelia used to inoculate the glass beads were grown in a minimal NMP medium (14) containing 1% glucose and buffered by TES [N-tris(hydroxylmethyl)methyl-2-aminoethanesulfonic acid]-NaOH. About 106 Streptomyces spores/ml were inoculated into 50 ml NMP medium in 500-ml flasks and cultivated until the cultures reached late exponential phase [48 h for S. coelicolor A3(2) and 20 h for S. aureofaciens CCM3239, the difference being due to their different growth rates]. The same cultivation conditions were used for the other tested strains (not shown). For morphological studies, plastic 5-ml petri dishes were used. Each dish contained 19 g of the glass beads, which were autoclaved separately in glass test tubes at 121°C for 30 min. Beads were evenly spread in the dish, and thereafter, a 4-ml aliquot of the liquid culture was carefully deposited dropwise and spread onto the glass bead surface, and the plates were incubated at 28°C. To prevent cultures from drying out, they were kept in large plastic boxes where high humidity was provided by water-soaked cotton wool. Alternatively, glass beads were inoculated with a suspension of 108 dormant spores in 4 ml of NMP medium.
SEM.
For sample preparation, we developed a technique (13) which involves postharvesting replacement of liquid cultivation medium surrounding glass beads by low-melting agarose. This fills the free space between beads and fixes them together. Osmium-fixed samples were examined in an AQUASEM scanning electron microscope (TESCAN) at 15 kV. Samples of cultures collected at different times during differentiation were subjected to SEM analysis.
Pulse-labeling of proteins by l-[35S]methionine and [32P]orthophosphate and protein extraction for two-dimensional gel electrophoresis.
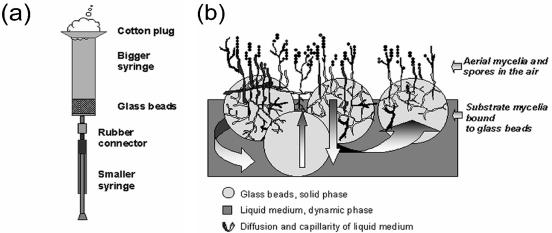
For protein pulse-labeling with l-[35S]methionine, cells were grown either in 5-ml syringes containing 4.8-g glass beads or on plates as described above. The culture was evenly deposited dropwise onto the surface of glass beads, and the volume of liquid medium was calculated so that it filled the space between the glass beads. To initiate labeling, a sterile 1-ml syringe containing 3.7 × 106 Bq of l-[35S]methionine in 100 μl of sterile medium was connected to the syringe containing the glass bead culture through sterile soft rubber tubing (Fig. 1). The liquid medium in the bigger syringe was first sucked into the smaller syringe and mixed with the radioactive component and then pushed back gently to the larger syringe. This allowed the cultivation medium to be replaced or supplemented with any soluble additives. The culture was then kept in a 28°C incubator for 2 h before the cell mass was harvested. When petri dishes were used, the label was similarly deposited dropwise onto the surface of the glass bead substrate.
FIG. 1.
Cartoon representing the two-phase culture system for in vivo labeling of Streptomyces strains by radioactive compounds (a) and a side view of a Streptomyces culture growing on glass beads (b). The bigger syringe in panel a containing the glass bead layer serves as an artificial habitat for growth of streptomycetes. To initiate labeling, the smaller syringe functions as a pump to withdraw spent medium and inject it back after supplementing radioactivity. The spent medium can similarly be modified or supplemented with signal molecules. During development, the submerged hyphae shown on panel b probably undergo lysis through an orderly process and serve as a source of nutrients for emerging aerial mycelium (15).
For in vivo phosphate labeling, cultures were pulse-labeled with 3.7 × 107 Bq ml−1 of carrier-free [32P]orthophosphate (specific activity > 3.14 TBq mmol−1) for 1 h.
Labeling was stopped by aspirating away the radioactive medium in cultures growing in syringes or by removing the surface layer of glass beads supporting growth on plates and washing the mixture of cell mass and beads with cold buffer containing 10 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, and a protease inhibitor cocktail (Boehringer). The washed mixture was then transferred to vials and disrupted in a Mini-BeadBeater (two 30-s cycles at 4,200 rpm; Biospec Products Inc.); the samples were kept on ice throughout the procedure. Cell debris and glass beads were removed by centrifugation at 12,000 rpm in a Beckman Minifuge at 4°C. Protein concentration was measured by the Bradford method (3). The total proteins were then precipitated with cold acetone and kept for 2 h at −70°C. After centrifugation (12,000 rpm for 15 min), the precipitate was solubilized in 2D gel electrophoresis sample buffer containing 9.5 M urea, 2% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 1.6% ampholyte 5-7, 0.4% ampholyte 3-10, and 2% 2-mercaptoethanol. Samples were kept for 2 h at room temperature before being loaded onto isoelectric focusing (IEF) gels. The experimental set up for pulse-labeling is shown in Fig. 1a.
High-resolution 2D PAGE.
Radioactivity incorporated into proteins was measured in an LKB Wallac scintillation counter. For [35S]methionine labeled proteins, about 106 dpm (disintegrations per minute) were loaded onto each IEF tube gel.
Total protein cell extracts prepared as described above were analyzed by a large-format two-dimensional (IEF/sodium dodecyl sulfate-PAGE) gel electrophoresis (16) system (Investigator; Oxford Glycosystem). IEF was carried out in cylindrical gels within 26-cm tubes of 1-mm inside diameter. IEF gels containing carrier ampholytes (Sigma), pH 3 to 10 and pH 5 to 7, in a 1:4 ratio were run for 17,000 Vh. After equilibration in sodium dodecyl sulfate buffer, tube gels were laid on top of 12.5% polyacrylamide slab gels and run at 16 W per gel at 20°C. Gels were air dried between two cellophane sheets. Radioactive protein spots were visualized in the Fuji phosphorimaging plate scanner BAS5000.
Analysis of antibiotic production.
The procedures for detection and analysis of pigmented antibiotics of S. coelicolor A3(2) followed the methods described previously by Bystrykh et al. (4), with modifications. To analyze intracellular blue pigment (actinorhodin), glass bead cultures were washed twice with 0.1 M HCl and resuspended in 1 M KOH, thoroughly mixed for 1 h, and centrifuged at 5,000 rpm for 5 min. To extract the antibiotics for thin-layer chromatography (TLC), samples in KOH were acidified by 4 M HCl to pH 1 to 2 and allowed to stand on ice for 15 min. Precipitates were redissolved in dioxane and loaded onto silica-coated glass plates (Kieselgel 60) containing a UV 254 fluorescent indicator (Merck, Germany). The solvent used for TLC was benzene-acetic acid (9:1), and the spots were visualized under normal or UV light.
RESULTS AND DISCUSSION
Growth and morphological differentiation of streptomycetes on glass beads.
Many Streptomyces species, including S. coelicolor A3(2), S. aureofaciens CCM 3239 (both analyzed as described below), S. collinus, and S. granaticolor underwent morphological differentiation and produced pigments when cultured on our biphasic cultivation system. Before the cells were transferred onto the surface of glass beads to facilitate morphological differentiation, the substrate mycelium was cultivated until late exponential phase. A slowdown in growth was reflected by a decreased rate of incorporation of radiolabeled methionine into proteins (see Fig. 3 and 5a). Alternatively, the cultures were started from dormant spores, which were able to germinate and form primary vegetative hyphae on the surface of glass beads (Fig. 2B).
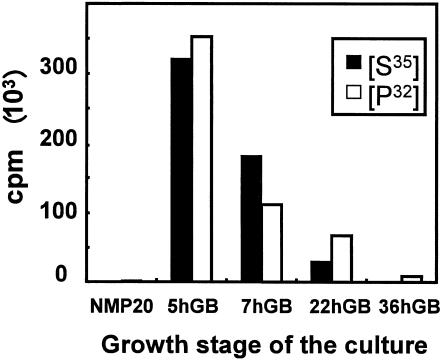
FIG. 3.
[35S]methionine and [32P]phosphate incorporation into total proteins of S. aureofaciens growing in liquid culture before transfer onto glass beads (NMP20) and during the course of morphological differentiation on glass beads (GB) (5 to 36 h). Ten microliters of 10-fold-diluted radioactive samples used for 2D electrophoresis was spotted onto a nitrocellulose filter and measured in a scintillation counter.
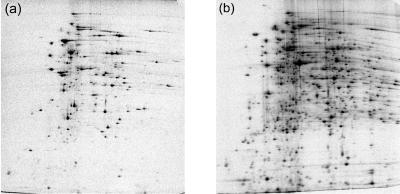
FIG. 5.
Representative two-dimensional electrophoresis autoradiographs of newly synthesized proteins of S. coelicolor A3(2) pulse-labeled with [35S]methionine. Mycelia grown on glass beads (b) are compared with those of the liquid culture inoculum just before being transferred onto glass beads (a).
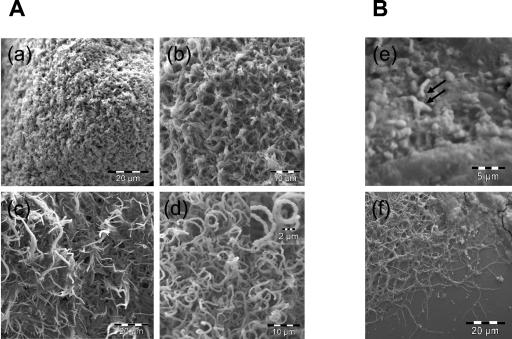
FIG. 2.
Morphological differentiation of S. coelicolor A3(2) grown on glass beads. (A) Image a shows a young colony composed of only substrate mycelia. Later on, aerial mycelia started to stand up into the air (b) and elongate (c), coincident with the appearance of pigments. Later, coiled aerial mycelia started to form septa and spores (d). Images a, b, c, and d were taken at 6, 16, 19, and 36 h after transfer to glass beads, respectively. The magnified image in d shows an aerial mycelium septated to form spores. Panel B shows spores germinating on glass beads after 8 h (e) and primary network of mycelium formed after 24 h (f). One or two germ tubes coming from the spore were observed (arrows).
The appearance of aerial mycelium on the surface of glass beads was observable using a binocular magnifier after 7 h in S. aureofaciens CCM 3239 cultures and after 17 h in S. coelicolor A3(2) cultures (Table 1). S. coelicolor A3(2) first grew on the glass bead surface as a dense network of branched and interlaced substrate hyphae (Fig. 2a), which later (16 h) differentiated into aerial hyphae (Fig. 2b and c). At this time, blue pigment appeared, later identified as actinorhodin. The transition from aerial mycelium to spores is shown in Fig. 2c and d (19 and 36 h). While Fig. 2c represents the time when aerial hyphae were elongated and erect, Fig. 2d shows the moment when aerial mycelia were septating to form coiled spore chains.
TABLE 1.
Transition points representing differentiation of S. coelicolor A3(2) and S. aureofaciens CCM 3239 on glass beads
| Life cycle stage | Transition point (h)
|
|
|---|---|---|
| S. coelicolor A3(2) | S. aureofaciens 3239 | |
| Cultivation in liquid medium | 48 | 20 |
| Aerial mycelium formationa | 17 (± 1) | 7 (± 1) |
| Pigment appearancea | 13 (± 3) | NDb |
| Spores observeda | 30 (± 1) | 36 (± 4) |
Measured from the time when the substrate mycelium was transferred onto the glass bead surface.
ND, not determined.
The fine structure of the aerial mycelium in streptomycetes was first documented by classical studies by Wildermuth (19) and more recently related to bacterial apoptosis (15). In those studies, streptomycetes were grown on agar plates with or without cellophane. The cultivation system described here offers an alternative for studies of morphological and biochemical differentiation under conditions more similar to those in nature. The system not only mimics surface dehydration of soil but also provides mycelial growth and developmental compartments that include deeper regions where mycelia are submerged in liquid medium. This creates environmental-like conditions for signaling in the population. Methionine incorporation into protein increased dramatically after transfer of the culture to glass beads (Fig. 3), probably reflecting rearrangement of metabolism before initiating differentiation. Times at which aerial mycelium, pigments, and spore were formed by S. coelicolor A3(2) and S. aureofaciens CCM 3239 are presented in Table 1. We could also demonstrate Streptomyces spore germination on glass beads (Fig. 2B), which allowed the use of the cultivation system to monitor transition periods during the complete cell cycle. The ability to start the life cycle either from spores or from liquid-grown culture and easy sampling of cultures at any point of differentiation make the system very flexible and useful for studies of gene expression and regulation.
Detection and analysis of pigmented antibiotics.
The erection of aerial mycelium, the most obvious visual manifestation of the developmental program, is linked to the biosynthesis of antibiotics. This step in streptomycete differentiation may involve a cascade of diffusible signaling molecules, which leads to the formation of aerial mycelium and production of secondary metabolites (for a review, see reference 12).
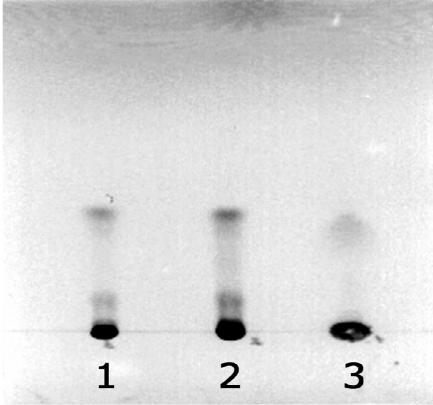
We observed production of pigments by a number of Streptomyces strains when they were grown on glass beads, including pigmented antibiotics made by S. granaticolor and S. coelicolor A3(2). Analysis of antibiotics produced by streptomycetes when grown on agar plates requires filtering/extraction procedures (17). With the glass bead cultures, this task is simplified by either removing the liquid phase containing the metabolites or using the beads to disintegrate the cells and then extracting intracellular metabolites from the homogenate. S. coelicolor A3(2) produced more visible blue pigment on glass beads than it did in liquid cultures based on the same medium. Extracts from glass bead and liquid cultures were analyzed on TLC plates (Fig. 4) and had the characteristic absorption spectra of actinorhodin (results not shown).
FIG. 4.
Thin-layer chromatography of actinorhodins produced by S. coelicolor A3(2) grown on glass beads (lane 1 and 2) compared with the same strain grown in the same volume of identical liquid medium (lane 3). The antibiotic in the lower bands, most probably γ-actinorhodin, is present in much higher amounts in the glass bead-grown samples than in those from the liquid culture.
Labeling and two-dimensional electrophoresis analysis of newly synthesized proteins.
The availability of the complete genome sequences of S. coelicolor A3(2) and S. avermitilis has stimulated proteomic studies of these and other Streptomyces strains (2, 10). Proteomic analyses of microorganisms begin with the labeling of their newly synthesized proteins under defined physiological conditions, followed by the separation and visualization of the protein mixture by 2D PAGE. Newly synthesized proteins are commonly labeled with radiolabeled amino acids, i.e., l-[35S]methionine.
For many Streptomyces species, morphological differentiation occurs only on a solid surface. However, labeling of cells differentiating on standard agar plates is complicated, and collecting mycelia which penetrate agar is difficult and never quantitative. These problems can be minimized by growing the cultures on cellophane sheets covering the agar (8). Labeling of colonies growing on the surface of the cellophane is done by transferring the cellophane sheet to another agar plate containing the radiolabel. However, moving the culture to a fresh plate inevitably disturbs their metabolism, and the agar or cellophane might be used as a carbon source by some strains. Furthermore, mycelia of some species can grow through the cellophane. The same problems accompany any analyses of extracellular or intracellular secondary metabolites on agar plates. Our glass bead-based cultivation conditions (see Materials and Methods) allowed studies of both morphological and biochemical differentiation. We were able to identify the newly synthesized proteins by pulse-labeling with l-[35S]methionine (Fig. 5) while the cells were undergoing differentiation. Although the gels were not rigorously compared, it was clear that the liquid-grown culture spot profiles were different from those undergoing differentiation on glass beads. Similarly, in vivo-phosphorylated proteins can be analyzed at the same time points during the course of morphological differentiation (results not shown). These above-described procedures, coupled with protein identification by mass spectrometry, could make this cultivation and labeling system a powerful tool for studies of functional proteomes in differentiating streptomycetes.
Conclusion.
The cultivation system we have developed permits easy manipulation of medium composition (nutrient supplies or additives) during differentiation of Streptomyces. It should also be possible to exchange conditioned media between cultures representing different stages of development and thus analyze the nature and effect of different signal molecules coordinating morphological and biochemical differentiation. In this way, proteomes induced by different stimuli (cyclic AMP, pH, starvation, etc.) or extracellular signaling molecules excreted by certain mutants of the bald cascade (20) could be analyzed. Comprehensive analysis of these proteomes could help elucidate signaling networks controlling streptomycete differentiation.
By taking advantage of the use of glass beads for breaking the cells by the “bead beater” (11), cultures differentiating on glass beads might also be used to extract RNA for cDNA microarrays, Northern blots, reverse transcription-PCR, etc. Using high-performance liquid chromatography and mass spectrometry of extracts from the glass bead cultures, the production of secondary metabolites during streptomycete development could be studied. We believe that the technique could be further developed to study not only proteomes but also transcriptomes and metabolomes of streptomycetes in order to gain insight into the physiology and differentiation of these interesting bacteria.
Acknowledgments
We thank Silvia Bezoušková for technical assistance in preparing 2D gels.
This work was supported by the Grant Agency of the Czech Republic (grants 204/00/1252 and 204/03/1014) and by Institutional Research Concept no. AV0Z502090. L.D.N. was supported by a UNESCO fellowship.
REFERENCES
- 1.Allan, E. J., and J. I. Prosser. 1983. Mycelial growth and branching of Streptomyces coelicolor A3(2) on solid medium. J. Gen. Microbiol. 129:2029-2036. [Google Scholar]
- 2.Bentley, S. D., K. F. Chater, A. M. Cerdeño-Tárraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Bystrykh, L. V., M. A. Fernández-Moreno, J. K. Herrema, F. Malpartida, D. A. Hopwood, and L. Dijkhuizen. 1996. Production of actinorhodin-related “blue pigments” by Streptomyces coelicolor A3(2). J. Bacteriol. 178:2238-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chater, K. F. 2001. Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr. Opin. Microbiol. 4:667-673. [DOI] [PubMed] [Google Scholar]
- 6.Chater, K. F. 1989. Multilevel regulation of Streptomyces differentiation. Trends Genet. 5:372-377. [DOI] [PubMed] [Google Scholar]
- 7.Erikson, D. 1949. The morphology, cytology, and taxonomy of the actinomycetes. Annu. Rev. Microbiol. 3:23-54. [Google Scholar]
- 8.Granozzi, C., R. Billetta, R. Passantino, M. Sollazo, and A. M. Puglia. 1990. A breakdown in macromolecular synthesis preceding differentiation in Streptomyces coelicolor A3(2). J. Gen. Microbiol. 136:713-716. [DOI] [PubMed] [Google Scholar]
- 9.Hodgson, D. A. 2000. Primary metabolism and its control in streptomycetes: a most unusual group of bacteria. Adv. Microb. Physiol. 42:47-238. [DOI] [PubMed] [Google Scholar]
- 10.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Ómura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, B. 1998. Breaking up isn't hard to do: a cacophony of sonicators, cell bombs, and grinders. Scientist 12:23. [Google Scholar]
- 12.Kelemen, G. H., and M. J. Buttner. 1998. Initiation of aerial mycelium formation in Streptomyces. Curr. Opin. Microbiol. 1:656-662. [DOI] [PubMed] [Google Scholar]
- 13.Kofroňová, O., L. D. Nguyen, J. Weiser, and O. Benada. 2002. Streptomycetes cultured on glass beads—sample preparation for SEM. Microsc. Res. Tech. 58:111-113. [DOI] [PubMed] [Google Scholar]
- 14.Kormanec, J., A. Lempelová, R. Nováková, B. Řežuchová, and D. Homérová. 1997. Expression of the Streptomyces aureofaciens glyceraldehyde-3-phosphate dehydrogenase gene (gap) is developmentally regulated and induced by glucose. Microbiology 143:3555-3561. [DOI] [PubMed] [Google Scholar]
- 15.Miguelez, E. M., C. Hardisson, and M. B. Manzanal. 1999. Hyphal death during colony development in Streptomyces antibioticus: morphological evidence for the existence of a process of cell deletion in a multicellular prokaryote. J. Cell Biol. 145:515-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 17.Süsstrunk, U., J. Pidoux, S. Taubert, A. Ullmann, and C. J. Thompson. 1998. Pleiotropic effects of cAMP on germination, antibiotic biosynthesis and morphological development in Streptomyces coelicolor. Mol. Microbiol. 30:33-46. [DOI] [PubMed] [Google Scholar]
- 18.Vaněk, Z., and J. Janeček. 1997. The physiology and biosynthesis of secondary metabolites. Acta Biol. Hung. 48:339-358. [PubMed] [Google Scholar]
- 19.Wildermuth, H. 1970. Development and organization of the aerial mycelium in Streptomyces coelicolor. J. Gen. Microbiol. 60:43-50. [DOI] [PubMed] [Google Scholar]
- 20.Willey, J., J. Schwedock, and R. Losick. 1993. Multiple extracellular signals govern the production of a morphogenetic protein involved in aerial mycelium formation by Streptomyces coelicolor. Genes Dev. 7:895-903. [DOI] [PubMed] [Google Scholar]